Abstract
Subgenomic selectable RNAs of the hepatitis C virus (HCV) have recently been shown to self-replicate to high levels in the human hepatoma cell line Huh-7 (V. Lohmann, F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager, Science 285:110–113, 1999). Taking advantage of this cell culture system that allows analyses of the interplay between HCV replication and the host cell, in this study we characterized two replicon-harboring cell lines that have been cultivated for more than 1 year. During this time, we observed no signs of cytopathogenicity such as reduction of growth rates or ultrastructural changes. High levels of HCV RNAs were preserved in cells passaged under continuous selection. When selective pressure was omitted replicon levels dropped, but depending on culture conditions the RNAs persisted for more than 10 months. A tight coupling of the amounts of HCV RNA and proteins to host cell growth was observed. Highest levels were found in exponentially growing cells, followed by a sharp decline in resting cells, suggesting that cellular factors required for RNA replication and/or translation vary in abundance and become limiting in resting cells. Studies of polyprotein processing revealed rapid cleavages at the NS3/4A and NS5A/B sites resulting in a rather stable NS4AB5A precursor that was processed slowly into individual products. Half-lives (t1/2s) of mature proteins ranged from 10 to 16 h, with the exception of the hyperphosphorylated form of NS5A, which was less stable (t1/2, ∼7 h). Results of immunoelectron microscopy revealed an association of the majority of viral proteins with membranes of the endoplasmic reticulum, suggesting that this is the site of RNA replication. In summary, replicon-bearing cells are a good model for viral persistence, and they allow the study of various aspects of the HCV life cycle.
The hepatitis C virus (HCV) is a major leading cause of chronic liver disease (reviewed in reference 45). Infections with HCV are usually subclinical, and most patients do not develop acute hepatitis or have only mild symptoms. However, most infected individuals are unable to eliminate the virus, resulting in a persistent infection in ∼80% of all cases, and these patients are at high risk to develop liver fibrosis, liver cirrhosis, or hepatocellular carcinoma.
HCV was classified as the distinct genus Hepacivirus in the family Flaviviridae (38). Other members of this family are the pestiviruses, to which the Classical swine fever virus (CSFV) belongs, and the flaviviruses, with the prototype member Yellow fever virus. These viruses have in common a plus-strand RNA genome carrying a single long open reading frame (ORF) that is flanked at both termini by nontranslated regions (NTRs). In case of HCV, the 5′ NTR has a length of 341 nucleotides and carries an internal ribosome entry site (IRES) permitting the binding of ribosomes in close proximity of the start codon of the ORF (57, 58). The 3′ NTR has a tripartite structure composed of a variable region following the stop codon of the ORF, a polyuridine tract of variable length, and a 98-nucleotide sequence, designated the X-tail, that is highly conserved among all HCV genotypes and that is essential for replication in vivo (31, 32, 52, 53, 61). The ORF encodes a polyprotein of ∼3,000 amino acid residues and it is cleaved into at least 10 different products: core (C), envelope proteins E1 and E2, p7, and nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B (reviewed in references 9 and 47). C, E1, and E2 are the structural proteins that are processed by host cell signal peptidases (22). The function of the small hydrophobic polypeptide p7 so far is not known. NS2 and the amino-terminal domain of NS3 constitute the NS2-3 proteinase responsible for cleavage between NS2 and NS3 (20, 23). NS3 harbors three different enzymatic activities. The amino-terminal ∼180 residues constitute a chymotrypsin-like serine proteinase responsible for cleavage of the NS3-5B region (7, 21). The carboxy-terminal remainder possesses nucleoside triphosphatase and helicase activities (29, 51). NS4A is an NS3 proteinase cofactor forming a stable complex with this enzyme and enhancing its proteolytic activity (10, 16, 33, 54). The function of NS4B is currently unknown. NS5A is a highly phosphorylated protein that at least with some genotypes is produced in two phosphorylation states: a basal and a hyperphosphorylated form that can be separated because of their different apparent molecular weights (27, 48, 55). The requirements for hyperphosphorylation appear to differ with respect to HCV genotypes. For instance, in case of the genotype 1b HCV-J isolate NS5A hyperphosphorylation is increased upon coexpression of NS4A, and this increase depends on complex formation between both proteins (1). Recently, two groups have reported independently for two other genotype 1b isolates that a continuous NS3-5A region is required for hyperphosphorylation (30, 40). Whether NS5A is directly involved in RNA replication is not known. A more indirect role could be the inhibition of the effector proteins of the antiviral state induced in the cell after stimulation with alpha interferon. It was shown that NS5A of at least some HCV isolates can interact with the double-stranded RNA activated protein kinase R (18, 19). This enzyme normally induces a reduction of translation via the phosphorylation of translation initiation factor eIF-2α. Upon interaction of NS5A with protein kinase R, kinase activity is blocked, allowing continued translation in cells in the presence of alpha interferon. However, the in vivo relevance of this observation is not known (17). The most carboxy-terminal domain of the polyprotein is the RNA-dependent RNA polymerase NS5B (11, 34, 60).
Despite great progress in understanding the genomic organization of the virus and the functions of viral proteins, fundamental aspects of HCV replication, pathogenesis, and persistence remain unknown. A major barrier in gaining experimental access to these issues is the lack of an efficient cell culture system allowing production of infectious virus particles. Although infection of primary cell cultures and certain human cell lines has been reported, the amounts of virus produced in these systems and the levels of HCV replication have been too low to permit detailed studies (for review, see reference 9). As a first step towards establishing a more productive system, we have recently described the construction of selectable subgenomic HCV RNAs that replicate to high levels in the human hepatoma cell line Huh-7 (35). These replicons were derived from a cloned full-length HCV consensus genome of genotype 1b by removing the C-p7 or C-NS2 region and insertion of the neomycin phosphotransferase gene (neo) downstream of the HCV IRES. Translation of the HCV NS2-5B or NS3-5B region was directed by the IRES of the encephalomyocarditis virus (EMCV) inserted downstream of neo. After transfection of Huh-7 cells only those supporting HCV RNA replication amplified neo and developed resistance against the drug G418. Cell lines derived from such G418-resistant colonies contained high levels of replicon RNAs and viral proteins (35).
Since the availability of such a system for the first time allowed an analysis of the interplay between an autonomously replicating subgenomic HCV RNA and the host cell, we performed a detailed characterization of two of these cell lines carrying an NS3-5B replicon (cell lines 9-13 and 5-15 [35]). We analyzed the stabilities of HCV RNAs under different conditions of cell passage, polyprotein processing kinetics, the half-lives of the cleavage products, and their subcellular localization. A strong dependence of HCV RNA replication on cell growth was found, suggesting that cellular factors are limiting in resting cells. Finally, no ultrastructural changes or alterations of growth properties were found in cells with a replicon, suggesting that these HCV RNAs and the viral NS3-5B proteins are not cytopathogenic.
MATERIALS AND METHODS
Cell culture.
Cell monolayers of the human hepatoma cell line Huh-7 (39) were routinely grown in Dulbecco's modified mininal essential medium (Life Technologies GmbH, Karisruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin, 100 μg of streptomycin, and 10% fetal calf serum (complete DMEM). In case of cell lines carrying HCV replicons, various concentrations of G418 (Geneticin; Life Technologies GmbH) were added to the medium as given in the Results section. These concentrations refer to the amount of total substance and are not corrected for the percentage of active substance as given by the manufacturer. Unless otherwise stated cells were passaged three times a week at a dilution of 1:3 to 1:4, depending on confluency.
Northern blot analysis.
Total RNA was prepared by a single-step isolation method (13) from cell pellets that had been washed once with phosphate-buffered saline (PBS), and RNA was quantified by measuring the optical density at 260 nm. Total RNA (2 to 15 μg) was denatured by treatment with 5.9% glyoxal in 50% dimethyl sulfoxide and 10 mM sodium phosphate buffer (pH 7.0) and separated by denaturing agarose gel electrophoresis. RNA was transferred to positively charged nylon membranes (Hybond-N+; Amersham Pharmacia Biotech, Freiburg, Germany) with 50 mM NaOH using a vacuum manifold and, after drying, cross-linked to the membrane by UV irradiation. Hybridization was done using standard procedures (2). Prior to hybridization, RNA bound to the membrane was stained with 0.03% methylene blue in 0.3 M sodium acetate for 5 min and briefly destained with water, and the membrane was cut 1 cm below the 28S rRNA band. The upper strip, which contained the HCV replicon RNA was hybridized with a 32P-labeled negative sense riboprobe complementary to the HCV IRES and neo. The lower strip was hybridized with a β-actin-specific antisense riboprobe. HCV- and β-actin-specific bands were quantified by phosphoimaging using a BAS 2500 scanner (Fuji), and the number of replicon molecules was determined by comparison with a serial dilution of in vitro transcripts loaded in parallel onto the gel. β-Actin was used to correct for different amounts of total RNA loaded in each lane of the gel.
Western blot and immunofluorescence analysis.
Cells were washed twice with PBS and detached from the plate by treatment with 0.05% trypsin–0.02% EDTA. Cells contained in a small aliquot of the suspension were counted, whereas the remaining cells were lysed by a 1-min sonification in denaturing protein sample buffer (200 mM Tris-HCl [pH 8.8], 5 mM EDTA, 0.1% bromophenol blue, 10% sucrose, 3.3% sodium dodecyl sulfate [SDS], and 2% 2-mercaptoethanol). Aliquots of cell lysates corresponding to 106 cells were loaded onto SDS–10% polyacrylamide gels and after electrophoresis transferred to a polyvinylidene difluoride membrane (PolyScreen; NEN Life Science Products, Zaventem, Belgium) using a semidry blotter (Bio-Rad, Munich, Germany) according to the instruction of the manufacturer. Membranes were incubated overnight in blocking buffer (PBS containing 0.5% Tween 20 and 2% milk powder [wt/vol]), and an NS5B-specific monoclonal antibody (kindly provided by Darius Moradpour) was added thereafter at a dilution of 1:1,000 for 1 h. After being washed three times with 0.5% Tween 20 in PBS, the membrane was incubated with a mouse-specific antibody conjugated with peroxidase (Sigma, Deisenhofen, Germany) in blocking buffer for 1 h and washed three times as described above, and bound antibodies were detected by chemiluminescence using luminol and a specific enhancer (SuperSignal West Dura Extended Duration Substrate; KMF Laborchemie, St. Augustin, Germany). For immunofluorescence, cells were grown on glass coverslips for various times and fixed after being washed three times with PBS in an ice cold mixture of acetone and methanol (90 and 10%, respectively). After 10 min of incubation at −20°C, cells were washed three times with PBS and incubated for 1 h in IF buffer (PBS, 3% bovine serum albumin, 0.1% Triton X-100) at 4°C. An NS5B-specific mouse monoclonal antibody was added at a dilution of 1:100 in IF buffer, and after 1 h cells were washed three times with PBS followed by incubation with a mouse-specific antibody conjugated with fluorescein isothiocyanate (Sigma) in IF buffer. Coverslips were washed three times with PBS and mounted on glass slides with Permafluor (Immunotech, Marseille, France), and cells were examined under a fluorescence microscope (Zeiss, Jena, Germany).
Metabolic radiolabeling of proteins and immunoprecipitation.
A total of 2 × 105 cells of cell line 9-13 or parental Huh-7 cells were seeded in a 3-cm-diameter culture dish in complete DMEM supplemented with G418 (1 mg/ml). About 60 h later, cells were washed three times with PBS and incubated in methionine-free DMEM for 60 min. Medium was replaced by methionine-free DMEM supplemented with Express labeling mix (150 to 250 μCi/ml; NEN Life Science Products) followed by incubation of cells for various times. Cells were lysed either directly or, for pulse-chase analysis, washed several times and incubated in complete DMEM for time periods given in the Results section. Cells were harvested by lysis in NPB (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS) supplemented with 1 mM phenylmethylsulfonyl fluoride, 40 μg of leupeptin per ml and 0.001 Units of aprotinin per ml (all proteinase inhibitors from Sigma), and cell lysates were cleared by centrifugation at 13,800 × g for 15 min at 4°C. The cleared lysate was used for immunoprecipitation using rabbit polyclonal antisera monospecific for NS3, NS4B, NS5A, or NS5B (8). The NS4B-specific antiserum was obtained by immunizing rabbits with a purified recombinant full-length NS4B protein carrying a carboxy-terminal hexahistidine affinity tag. This protein was expressed in insect cells by using a recombinant baculovirus and purified from supernatant 2 as described recently for the NS5B protein (34). Details of immunoprecipitation are given in reference 8. Immunocomplexes were analyzed by SDS–10% polyacrylamide gelelectrophoresis (SDS–10% PAGE), and separated proteins were detected by autoradiography using a low-energy intensifying screen (TransScreen LE; Kodak).
Electron microscopy.
Cells were seeded as described above and after 3 to 4 days prepared for electron microscopy. After being washed two times with ice-cold PIPES buffer [100 mM piperazine-N,N′-bis(2-ethane sulfonic acid), pH 6.9] cells were fixed in petri dishes in 2.5% glutaraldehyde for 30 min. Fixed cells were washed three times with PIPES buffer, scraped off the plates, and collected by low-speed centrifugation. Cell pellets were embedded in melted agarose (low-gelling temperature), cut into small blocks, and fixed in 1% osmium tetroxid in PIPES buffer for 30 min at 4°C. After extensive washing in PIPES buffer cells were treated with 1% tannic acid in water, washed again, dehydrated in ethanol, and embedded in ERL resin in gelatin capsules (50). Silver-gray sections were stained with lead citrate and uranyl acetate and examined under a Philips CM120 electron microscope at 60 kV. For immunoelectron microscopy, cells were washed twice with ice cold PIPES buffer and fixed in petri dishes for 30 min in a mixture of 4% formaldehyde, 0.25% glutaraldehyde, and 0.2% picric acid in PIPES buffer. Cells were embedded in agarose as described above, and small blocks were stained in 1% uranyl acetate dissolved in water for 30 min at 4°C. The material was further dehydrated in ethanol and embedded in London resin white (41) in gelatin capsules. Polymerization was initiated by the addition of 0.5% benzoinmethylether, and the reaction was incubated under UV for 2 days at 4°C. Specimen blocks were cut with an ultratome, and silver sections were mounted on nickel grids without supporting film. All labelings were conducted on grids according to the method of Sparkman and White (49). After 15 min of etching the grids with 1% sodium periodate in water, they were washed three times with water and blocked for 30 min in TCG (150 mM NaCl, 20 mM Tris-HCl [pH 7.5], 0.5% casein, and 0.2% gelatin). Grids were incubated overnight at 4°C with a mixture of rabbit polyclonal antisera directed against NS3, NS4B, NS5A, and NS5B (8) diluted 1:1,000 in TCG. After being washed five times in TCG, sections were treated for 1 h at room temperature with goat anti-rabbit antibodies conjugated with colloidal gold (12-nm diameter). Grids were rinsed first with TCG and then with distilled water and finally were stained with 4% aqueous uranyl acetate and Reynold's lead citrate. After air drying, samples were examined as described above.
RESULTS
Absence of cytopathic effects in HCV replicon-harboring cells.
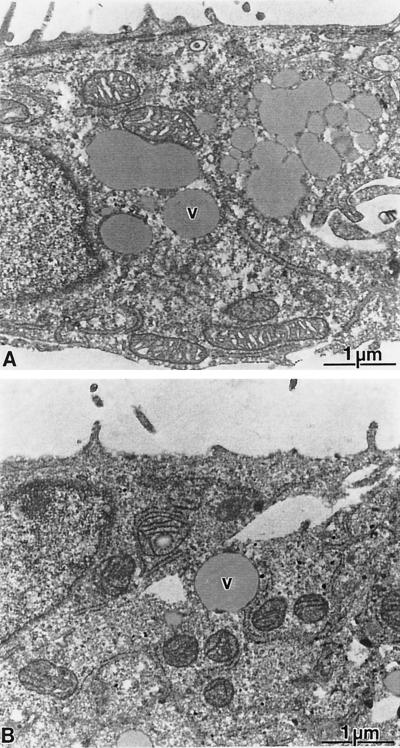
It has been suggested that liver cell damage is caused by the immune response targeted against HCV-infected cells rather than by virus replication itself (discussed in reference 43). However, owing to the lack of an efficient cell culture system this aspect could not be studied in detail thus far. To further clarify this important question, we took advantage of several cell lines which carry subgenomic self-replicating HCV RNAs (35). Two of these cell lines (designated 9-13 and 5-15) harbor NS3-5B replicons, whereas the cell line 8-1 carries only the neo gene and was therefore used as a negative control in addition to the parental Huh-7 cells. A comparison of the growth rates of these cell lines revealed no significant differences (Table 1). When cells were examined by light microscopy, no obvious differences in morphology were found (not shown). Cells with the replicon occasionally had some vacuoles, but this was also the case with control cells (see below). To identify potential ultrastructural alterations in replicon-harboring cells, they were further analyzed by electron microscopy. Cultures that had been seeded 5 days earlier were fixed with glutaraldehyde and osmium tetroxide and embedded in ERL resin as described in Materials and Methods. Under these conditions a strong cytoplasmic vesiculation of all cultures (9-13, 5-15, 8-1, and parental Huh-7 cells) was found (Fig. 1). The vesicles were heterogenous in size and showed a gray staining. Since the frequency of these cytoplasmic vesicles and their distribution did not depend on the presence of the replicon but rather on the time the cells had been kept in culture since the last passage, we concluded that this was a normal structural feature of Huh-7 cells. Further examination of replicon-harboring cell lines did not reveal morphological differences to control cells. These data show that the HCV replicons did neither influence growth properties of the cells nor induce a cytopathogenic effect.
TABLE 1.
Growth rates of Huh-7 cell lines with or without HCV repliconsa
| Cell line | Mean doubling time ± SD (h) |
|---|---|
| Huh-7 (parental) | 22 ± 1 |
| 8-1 | 22 ± 1.5 |
| 5-15 | 25.5 ± 2.5 |
| 9-13 | 21.5 ± 1 |
Growth rates were determined by daily counting of regularly passaged cells during a 3-week observation period.
FIG. 1.
Lack of ultrastructural changes in cells with HCV replicons. Analysis of ultrathin sections obtained from cultures of Huh-7 control cells (A) or replicon cell line 5-15 (B). Both cultures display a similar cellular architecture with a variable number of vesicles (v) of various sizes. The vesiculation was dependent on the cell density of the cultures and not on the expression of HCV proteins. No specific morphological alterations could be detected in the replicon-bearing cells.
Stability of HCV replicons in the presence or absence of G418.
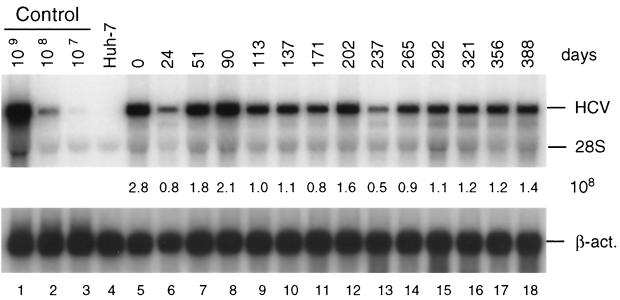
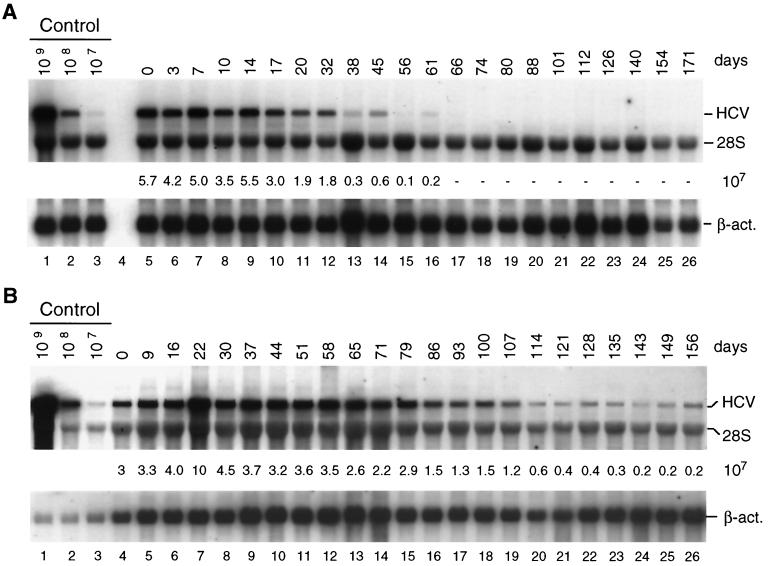
To assess the stability of HCV RNAs in cells kept under continuous selection, cells were passaged at regular intervals in the presence of G418, and the amount of replicon RNA was quantified by Northern blot analysis. As shown in Fig. 2, no significant reduction of replicon RNA was found even after 1 year of culturing. A different picture was seen with cells passaged in the absence of selective pressure. In this case we found a gradual reduction in the amount of replicon RNA over time, with the kinetics of the drop being determined by the frequency of cell passage and the density of cells prior to splitting. When cells were kept in a confluent state for a rather long time prior to passage a rapid reduction of replicon RNA was found. As exemplified in Fig. 3A when cells were passaged three times a week at a dilution of 3:5, RNA levels dropped by more than 90% within the first ∼50 days of culture and after 70 days were at the limit of detection by Northern blotting. In contrast, when cells were passaged at a much higher dilution (1:5) at the same interval, replicon RNA levels were much more stable (Fig. 3B). In this case no significant reduction of HCV RNA was found within the first ∼80 days of culture, with the levels declining slowly thereafter down to 10% of the original one after ∼135 days.
FIG. 2.
Stability of HCV replicon in cell line 9-13 passaged under continuous G418 selection. Cells were regularly passaged three times a week in the presence of G418 (1 mg/ml) and total RNA was isolated from cells at given time points. After denaturing agarosegel electrophoresis, HCV- and β-actin-specific RNAs were detected by Northern bloting using 32P-labeled riboprobes complementary to the HCV IRES and neo or a β-actin-specific antisense RNA, respectively, and quantified by phosphorimaging. The number of replicon RNA molecules was determined by comparison with the serial dilution of in vitro transcripts (lanes 1 to 3). β-Actin RNA served as a control to correct for the amount of total RNA loaded in each lane of the gel (∼2 μg). The result obtained with total RNA from the parental Huh-7 cells is shown in lane 4. Numbers between both panels refer to the number of replicon RNA molecules (108) contained in 1 μg of total RNA of the respective sample. Note that the control RNA is a mixture of given numbers of in vitro transcripts and 2 μg of total RNA from the parental Huh-7 cells that was used as carrier. The positions of HCV RNA, 28S rRNA, and β-actin mRNA (β-act.) are given to the right. Analogous results were obtained with cell line 5-15 (not shown).
FIG. 3.
Stability of HCV replicons in cells passaged in the absence of selection. (A) Cells were passaged three times a week in a way that two-fifths of the cells were harvested for RNA preparation and three-fifths of the cells were seeded in a new culture flask. (B) Cells were passaged three times a week at a dilution of 1:5; i.e., four-fifths were harvested and one-fifth was seeded in a new culture flask. Total RNA was prepared from harvested cells and 4 μg of each preparation was analyzed for the amount of HCV RNA by Northern blotting as described in the legend to Fig. 2.
To substantiate this observation cells were passaged for 10 months in the absence of G418 at a dilution of 1:3 to 1:5, depending on confluency. After this time RNA could no longer be detected by Northern blotting (not shown). To analyze whether these cells still contained the replicon, they were subjected to a reselection with G418. Owing to the low levels of HCV RNA, selection was performed with only a 100-μg/ml concentration of the drug which is 1/10 of the concentration regularly used for selection. Under these conditions ∼10% of selected cells formed colonies (Table 2), demonstrating that even after such a long time in the absence of selective pressure, replicon RNAs were present in a significant percentage of cells. From this result we concluded that replicon-bearing cells may be a good reflection of viral persistence in vivo.
TABLE 2.
Percentage of replicon-harboring 9-13 cells after 10 months of passage in the absence of selective pressurea
| No. of seeded cells | No. of G418-resistant cells | % Resistant cells |
|---|---|---|
| 1,000 | 114 | 11.4 |
| 1,250 | 144 | 11.5 |
| 1,500 | 160 | 10.6 |
| 1,750 | 208 | 11.9 |
| 2,000 | 259 | 13 |
| 2,250 | 272 | 12 |
Given numbers of 9-13 cells passaged in the absence of G418 for 10 months were seeded in 3-cm-diameter cell culture dishes and subjected to selection with G418 (100 μg/ml). After ∼3 weeks drug-resistant colonies were stained and counted.
Influence of cell growth on HCV RNA replication.
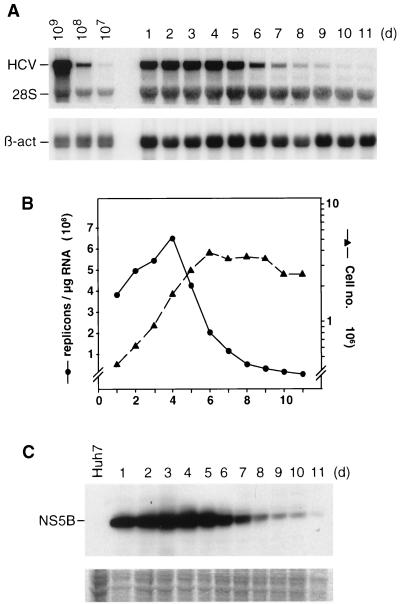
During the course of the analysis of cell lines harboring the replicon, we observed significant fluctuations in the amounts of HCV RNAs that appeared to depend on the conditions of cell growth. To analyze this observation in detail, a time course experiment was performed. Cells that had been passaged three times a week in the presence of G418 were seeded in multiple cell culture plates, and cells were harvested regularly to determine the number of viable cells and the amounts of HCV RNA and proteins. In addition, cells seeded in parallel on glass coverslips were processed for immunofluorescence. As shown in Fig. 4, the level of replicon RNA increased approximately twofold during the first 4 days of culture but dropped sharply thereafter to ∼1/40 of the original amount (6.6 × 108 replicon molecules per μg of total RNA on day 4 versus 0.17 × 108 replicon molecules per μg of total RNA on day 10). The same kinetic was found in the NS5B-specific Western blot (Fig. 4C). A correlation was observed between the level of replicon RNA and cell growth (Fig. 4B). While cells in the early logarithmic phase of growth carried the highest amounts of HCV RNA, the level dropped drastically with cells in the late logarithmic and the stationary phase. This fluctuation of HCV RNA replication and protein expression, in particular the sharp decline in resting cells, suggested that the amounts or activities of host cell factors required for RNA translation and replication varied during cell growth and became limiting in resting cells.
FIG. 4.
Fluctuations of HCV RNA replication levels during cell passage. Cells of cell line 9-13 that had been regularly passaged under continuous selection were seeded in multiple culture dishes and harvested at regular intervals between 1 and 11 days postseeding. Harvested cells were counted, and each half of these cells were used to prepare total RNA or a total lysate for protein analysis. (A) Total RNA was analyzed by Northern blotting as described in the legend to Fig. 2. (B) Quantification of HCV RNA and the number of cells at the time of harvest. The amounts of replicon RNA (left y axis) were determined by phosphorimaging using the Northern blot shown in panel A. The numbers of cells are given in a log scale (right y axis). (C) Proteins contained in 8 × 105 cells were loaded in each lane of the gel, and NS5B was detected by Western bloting To demonstrate comparable amounts of proteins in each lane, a gel was loaded in parallel and stained after electrophoresis with Coomassie brilliant blue. A portion of this gel is shown in the lower panel. The result obtained with the parental Huh-7 cells is shown in the left lane. d, day.
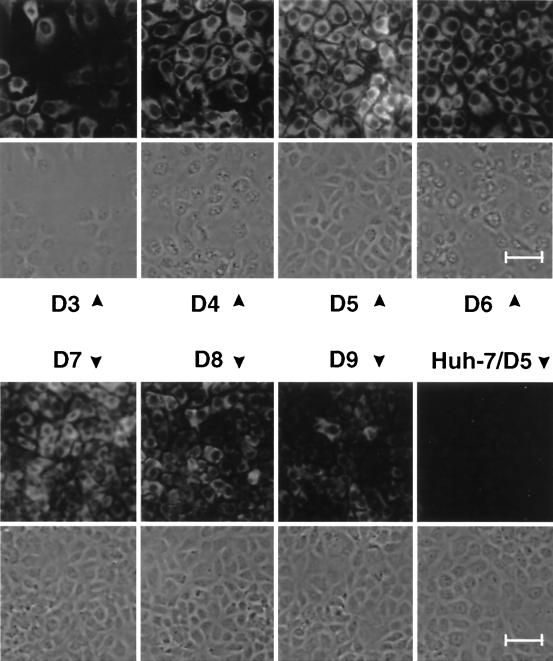
To analyze whether the fluctuations in RNA replication and protein levels occurred in all cells of a culture or only a fraction thereof, cells from the same experiment were analyzed by immunofluorescence. In agreement with the previous results, NS5B was clearly detected at day three postseeding (Fig. 5). The amounts of the HCV protein increased thereafter up to day 5 and then gradually declined. At the time of the peak nearly all cells were positive for immunofluorescence, but the expression levels differed between the cells. As exemplified with the sample stained on day 5 postseeding, a nest of strongly positive cells was found surrounded by cells containing less antigen. This difference in the expression levels of individual cells was found throughout the experiment. For instance, in the case of the sample stained on day 3 postseeding, strongly and weakly positive cells were found simultaneously, and the same was true with resting cells (e.g., day 9). Since HCV RNA replication and protein expression most likely are dependent on host cell factors that may vary in abundance or activity during cell growth, the heterogeneity observed in the immunofluorescence with individual cells probably reflected differences between the cells in the availability of these factors. Based on these observations, all subsequent analyses were performed with regularly passaged cells that had been seeded 2 to 3 days prior to the experiment.
FIG. 5.
Fluctuations of NS5B expression in a growing culture of cell line 9-13. Cells were seeded on coverslips and harvested at given times postseeding (days [D]). NS5B was detected by indirect immunofluorescence. A representative section of each sample is shown both by phase-contrast microscopy and immunofluorescence. All pictures were taken at a magnification of × 180. The smaller size of the older cells is due to confluency (compare, e.g., day 4 with day 7). Bar, 50 μm.
Kinetics of polyprotein processing and NS5A hyperphosphorylation.
In the past few years numerous studies have been performed to analyze processing of the HCV polyprotein as well as stabilities and modification of cleavage products. However, these studies had to rely on recombinant expression systems, most often the Vaccinia virus/T7 hybrid system. Therefore, it is still unclear whether polyprotein processing and modification of cleavage products, in particular hyperphosphorylation of NS5A, follows the same kinetics in vivo. Since the cells carrying a self-replicating HCV RNA to some extent mimic an infected cell, we performed a series of experiments to analyze the kinetics of polyprotein cleavage and NS5A phosphorylation.
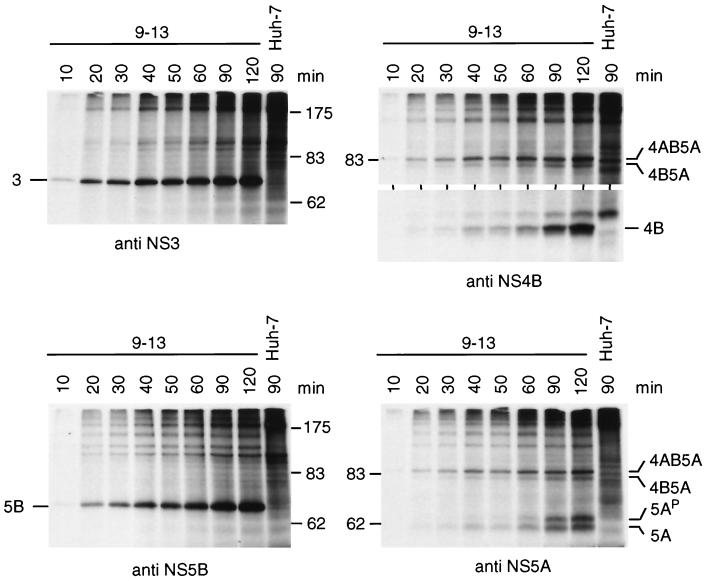
Processing kinetics were determined by pulse labeling. Cells were incubated for various times with [35S]methionine-cysteine, and HCV proteins were analyzed after immunoprecipitation by SDS-PAGE (Fig. 6). The first cleavage products detectable after a 10-min labeling period were NS3, NS5B, and an NS4AB5A precursor. Generation of NS4B and the basal phosphorylated form of NS5A was delayed, and they were detectable simultaneously after 20 min of labeling (10 min with films exposed longer). The hyperphosphorylated NS5A (5AP) was generated last and became detectable after the basal phosphorylated form (Fig. 6, lower right panel). Thus, we concluded that the NS3-5B polyprotein was rapidly cleaved at the NS3/4A and NS5A/B sites, whereas processing of the NS4AB5A precursor was delayed.
FIG. 6.
Processing kinetics of the NS3-5B polyprotein as determined by pulse labeling. 9-13 cells were seeded 60 h prior to metabolic radiolabeling of proteins with 100 μCi of labeling mix per ml for the time periods indicated above the lanes. Cells were lysed, and HCV-specific proteins were isolated by immunoprecipitation using antisera monospecific for NS3, NS4B, NS5A, or NS5B. HCV proteins and the positions of protein molecular weight standards (in kilodaltons) are specified in each panel. Results obtained with the parental Huh-7 cells radiolabeled for 90 min are shown in the right lane of each panel. Only the upper and lower portion of the gel is shown in case of the immunoprecipitations with the NS4B-specific antiserum.
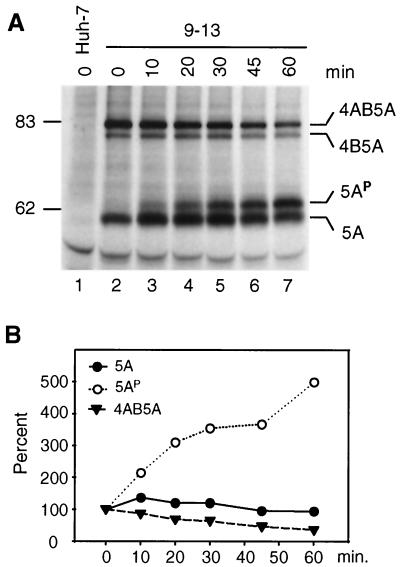
To clarify the kinetics of NS5A hyperphosphorylation, a pulse-chase experiment was performed (Fig. 7). In agreement with the previous results, both the NS4AB5A precursor and the basal phosphorylated NS5A were detected first (Fig. 7A, lane 2). During the chase period, the amounts of the precursor decreased in parallel with an initial increase in the amount of basal phosphorylated NS5A (up to 30 min of chase). The hyperphosphorylated form of this protein was barely detectable after the 10 min of labeling, but its level increased constantly during the 60-min chase period, concomitant with a decrease in the amounts of basal phosphorylated NS5A after 45 and 60 min of chase (Fig. 7). These results suggest that the basal phosphorylated form of NS5A was generated from the NS4AB5A and perhaps also from the NS4B5A precursors and that 5AP was derived from the basal phosphorylated form with rather slow kinetics. However, the possibility that a certain fraction of 5AP was directly produced from one of the precursors cannot be excluded. We noted an additional shift towards higher molecular weights for both NS5A phosphoproteins after the 60-min chase, indicating additional phosphorylation events late after polyprotein processing (Fig. 7A, lane 7, and Fig. 8A; see below).
FIG. 7.
Kinetics of generation of NS5A proteins. (A) 9-13 cells were seeded 60 h prior to radiolabeling with [35S]methionine-cysteine (200 μCi/ml) for 10 min followed by incubation with nonradioactive medium for the times given above the lanes. Cells were lysed, and NS5A proteins were analyzed by immunoprecipitation, SDS-PAGE, and autoradiography. (B) Quantification of NS5A-specific bands by phosphorimaging. Values obtained at time point zero were set at 100%.
FIG. 8.
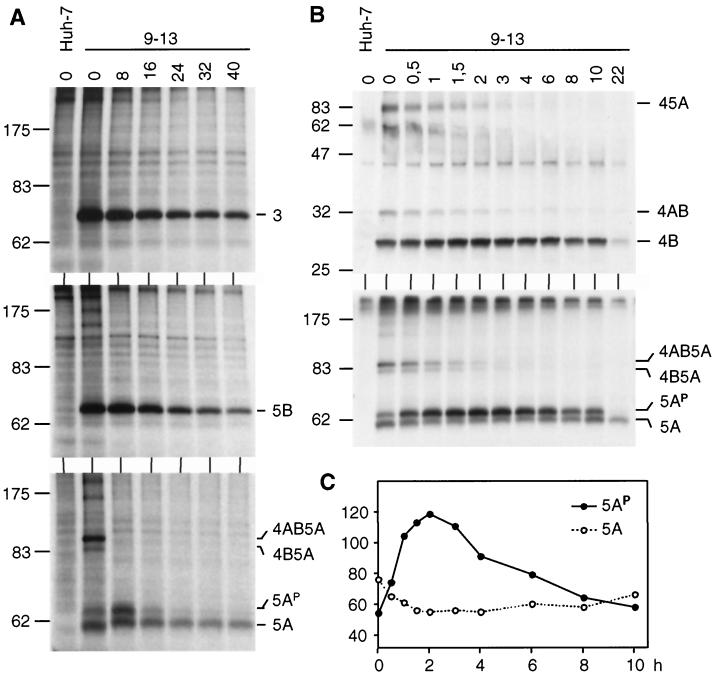
Half-lives of HCV proteins. (A) Cells of cell line 9-13 were pulse-labeled for 30 min with [35S]methionine-cysteine, followed by incubation with nonradioactive medium for various times as given above each lane (hours). Cells were lysed, and HCV proteins were analyzed by immunoprecipitation, SDS-PAGE, and autoradiography. (B) Pulse-chase experiments were performed as in panel A, but cells were lysed at shorter intervals during the chase period. HCV-specific bands are identified to the right of each panel; the positions of molecular weight standards (in kilodaltons) are given to the left. (C) The fully processed NS5A and its hyperphosphorylated form shown in panel B were quantified by phosphorimaging, and the values (given in photo-stimulated light units) are plotted against the chase time.
Half-lives and subcellular localization of HCV proteins.
Using pulse-chase experiments we also determined the half-lives of the polyprotein cleavage products and of the differentially phosphorylated NS5A forms. As shown in Fig. 8 and summarized in Table 3, all but one of the HCV proteins had similar half-lives ranging from ∼10 to 16 h. The exception was 5AP, with a half-life of only ∼7 h, which was much shorter than that of the basal phosphorylated NS5A form (half-life, ∼16 h [Fig. 8A]). Although not directly tested, the fact that NS4A exists intracellularly as a very stable NS3/4A proteinase complex 10, 16, 33, 54) suggests that the half-lives of both proteins are similar.
TABLE 3.
Half-lives of HCV nonstructural proteins as determined by pulse-chase analysis
| Proteina | Mean half-life ± SD (h) |
|---|---|
| NS3 | 14.5 ± 2.5 |
| NS4B | 10.8 ± 0.8 |
| NS5A | 16 ± 1.6 |
| NS5AP | 7 ± 1.6 |
| NS5B | 12 ± 1.6 |
NS5SP, hyperphosphorylated form of NS5A.
In order to determine the subcellular localization of HCV proteins, cells of cell line 5-15 were fixed in a paraformaldehyde-glutaraldehyde solution as described in Materials and Methods. A postfixation step with osmium tetroxid was avoided to preserve the antigenicity of the proteins. In a first set of experiments, antisera monospecific for NS3, NS4B, NS5A, or NS5B were used, but under these conditions the staining was generally weak and difficult to interpret (G. Rutter, unpublished results). However, when we applied a mixture of all four antisera, a clear and specific signal was found, revealing an exclusive cytoplasmic distribution of HCV proteins (Fig. 9B). Within a cell, several small intensively stained areas could be found distributed between vesicles. Within these areas the gold label was usually bound to cytoplasmic elongated structures representing cisternae of the endoplasmic reticulum (ER) (Fig. 9B and C). In addition, a more compact staining pattern was found in a few areas that were probably derived from smooth ER. In cells embedded for immunostaining, vesicular structures were devoid of contrast, suggesting that part of their contents were lipids. Most of the immunogold label detected was associated with the ER and not with the surface of these vesicles as described for the HCV core protein (3). The labeling of the nonstructural proteins was specific, because sections of control cells prepared in parallel were negative (not shown). Furthermore, only a few gold particles were found associated with the vesicles or mitochondria or outside the labeled areas.
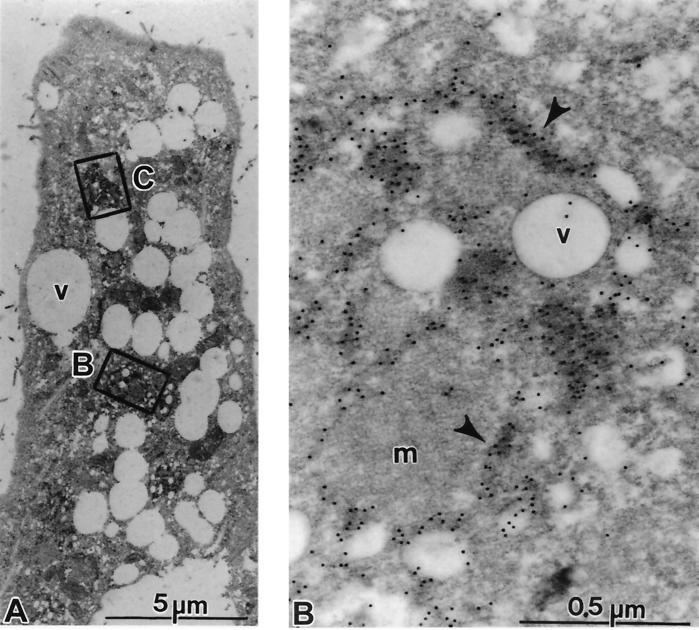
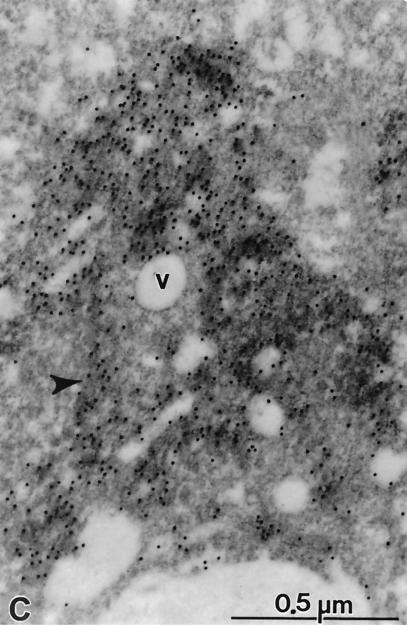
FIG. 9.
Subcellular localization of HCV proteins. Cells of cell line 5-15 were processed 4 days postseeding for immunoelectron microscopy without osmium fixation as described in Materials and Methods. The sections were probed with a mixture of polycional antisera directed against NS3, NS4B, NS5A, and NS5B, followed by 12-nm-diameter colloidal gold particles conjugated to anti-rabbit antibodies. (A) The cell in low magnification overview displays a strong vesiculation and a couple of gold-labeled areas (boxed). (B and C) Enlargements of the areas indicated by rectangles in panel A. (B) Accumulations of gold label in elongated structures representing cisternae of the ER (arrowheads). Note the scarcity of gold particles on vesicles (v), on mitochondria (m), or outside the labeled cisternae. (C) Gold-labeled cisternae are easily identified (arrowhead). Additional antibody binding could be seen on numerous submembranous structures around small vesicles (v).
DISCUSSION
In this study we have characterized two HCV replicon-harboring cell lines that have been cultured for more than 1 year. During this time, we did not observe signs of cytopathogenicity such as a reduced growth rate or structural alterations. Although this lack of cytopathic effects is consistent with the majority of evidence suggesting that liver cell damage is primarily due to the immune reaction targeted against infected cells rather than to a direct cytotoxicity of HCV (43), it does not necessarily exclude this possibility. Since the cell lines can only survive when HCV RNA replication is not cytotoxic, we might have selected for mutations in the replicon leading to a reduction or loss of cytopathogenicity. It must also be noted that the replicons used in this study lack the coding region for the structural proteins that appear to be cytotoxic when expressed at high levels (38). An experimental examination of these questions requires the development of a transient-replication assay that does not depend on selection of stable cell lines and that allows expression of all viral proteins.
The replicons were found to be very stable when cells were passaged under continuous selection pressure. Interestingly, even when selection was omitted the RNAs persisted for several months, albeit at low levels. This reduction of HCV RNA strongly depended on the way cells were passaged. When cells were kept confluent for extended periods, replicon levels dropped rapidly, whereas a rather constant amount of HCV RNA was found in cells passaged regularly. Based on the result that replicon RNA levels dropped in confluent cells (Fig. 4), the most likely explanation is that replication in senescent cells stops and, therefore, RNA degradation is no longer compensated by de novo synthesis. Consequently, the drop in the amount of viral RNA is highest in cells that are left in a confluent state for prolonged times. When these cells are passaged, the replicon amplifies again in growing cells but the RNA level in these cells is lower than it was in cells of the previous passage. Upon successive repetition of this passage scheme, HCV RNA is rapidly lost. In contrast, when cells are passaged regularly around the time of confluency and are not left for prolonged times in a confluent or senescent state, RNA amounts drop much less and amplify again to high levels in passaged cells at the time of exponential growth. Under these conditions, the replicon can be preserved for prolonged times even in the absence of selective pressure, demonstrating that HCV replicons have an intrinsically high propensity for persistence. This property is not unique to HCV. In fact, we have generated an analogous selectable replicon of the pestivirus CSFV (V. Lohmann, R. Devos, J.-D. Tratschin, and R. Bartenschlager, unpublished observation). When cells harboring this replicon RNA were passaged for 7 months in the absence of selective pressure and then cultured again in G418-containing medium, 5 to 10% of the cells formed colonies, demonstrating that comparable to what we show for HCV, the CSFV replicon persists in a significant fraction of the cells, too (V. Lohmann et al., unpublished observation).
In our view the most interesting finding was the dependence of HCV RNA replication and/or translation on host cell growth. This observation could be explained by the availability of host cell factors being high in exponentially growing cell cultures but low in resting cells. One potential candidate is PTB, which has been found to specifically interact with sequences at the 3′ NTR (14, 26, 56). Another host cell factor may be glyceraldehyde-3-phosphate dehydrogenase binding to the poly(U) sequence (44). Finally, cellular proteins tentatively designated p87 and p130 were identified by UV–cross-linking experiments with the X-tail sequence, but the nature of these proteins remains to be determined (25). It is unclear whether these factors are required for RNA replication, but if they are, a reduction of their expression levels in resting cells would lead to an analogous reduction of HCV RNA replication.
The variations in the amounts of replicon RNA could also be explained by fluctuations in the efficiency of RNA translation. In this respect it should be kept in mind that the replicons we used to generate the cell lines carry two functional IRES elements: the HCV IRES, which directs translation of neo, and the EMCV IRES, which is required for translation of the HCV nonstructural region. Owing to this bicistronic design, fluctuations in the amounts of host cell factors required for translation from the EMCV IRES rather than the HCV IRES might lead to variations in the amounts of replicase proteins. However, at least some of them are required for activity of both IRES elements. These are conventional translation factors such as eIF-2 and -3 as well as noncanonical translation factors like PTB (reviewed in reference 42). Therefore, the availability of these factors might affect production of both neomycin phosphotransferase and the replicase proteins.
Studies of processing kinetics of the nonstructural polyprotein expressed from the replicon RNA revealed results very similar to the ones obtained with heterologous expression systems (reviewed in reference 6). Rapid cleavages take place at the NS3/4A and the NS5A/B sites, resulting in a rather stable NS4AB5A precursor that is processed slowly by alternative pathways. Most remarkable were the slow generation of 5AP and the very different t1/2s of the two phosphoprotein variants, suggesting that they serve different functions. The role NS5A may play in RNA replication and the importance of its phosphorylation state for this process so far are not known. However, the conservation of this biochemical property among members of the Flaviviridae indicates an important function (46). Furthermore, for several RNA viruses phosphoproteins have been shown to be important regulators of replication. For instance, in the case of dengue virus, NS5 is found in two phosphorylated forms: a nuclear hyperphosphorylated form and a cytoplasmic hypophosphorylated NS5 that was found to interact with NS3 (28). Based on these observations, a model was suggested in which hyperphosphorylation of NS5 at a late stage of replication leads to disruption of the NS3/5 interaction, allowing NS5 to be transported to the nucleus. Regulation of replication by phosphorylation of viral proteins has also been described for several other viruses, such as vesicular stomatitis virus, parainfluenza virus, or human respiratory syncytial virus (4, 5, 15, 36). Based on HCV's analogies to these viruses one might speculate that HCV RNA replication is linked to the phosphorylation state of NS5A. Since phosphorylation is mediated by a cellular kinase(s) (24, 48) that may vary in abundance or activity during the cell cycle, NS5A might couple HCV RNA replication to host cell growth.
Using immunoelectron microscopy we observed a clear association of HCV proteins with membranes derived from the ER, similar to what was described by other groups when using recombinant expression systems (9, 47). This observation is in keeping with the notion that RNA replication occurs in membrane-associated replicase complexes, as is the case with several other plus-strand RNA viruses, such as poliovirus or flaviviruses (12, 59). The most obvious benefits of this property are the possibility of coupling functions residing in different polypeptide chains and the sequestration of viral proteins and nucleic acids in a distinct cytoplasmic compartment with high local concentrations of viral components.
In summary, we provide the first characterization of cell lines carrying autonomously replicating HCV RNAs. Replication of these RNAs was stable over an extended period of time even without selective pressure, suggesting that replicon-harboring cells are a good model system for persistence. Most notably, we found that HCV replication and/or RNA translation depended on host cell growth. It will certainly be an important and challenging task to identify the cellular factors and to unravel the mechanisms responsible for the tight coupling of HCV replication to the host cell.
ACKNOWLEDGMENTS
We are grateful to Hans-Georg Kräusslich for helpful discussion and support with electron microscopy studies, Ulrike Herian for excellent technical assistance, Ingrid Ellhof for help with the preparation of Fig. 1 and 9, and Jan-Oliver Koch for initial help with the characterization of the cell lines. We also thank Darius Moradpour for his gift of the NS5B-specific monoclonal antibody, Rene Devos and Jon-Duri Tratschin for gift of the CSFV replicon, Nicole Krieger for technical support, and Michael Frese for a critical reading of the manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB490, Teilprojekt A2).
REFERENCES
- 1.Asabe S I, Tanji Y, Satoh S, Kaneko T, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J Virol. 1997;71:790–796. doi: 10.1128/jvi.71.1.790-796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 3.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman M J, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barik S, Banerjee A K. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc Natl Acad Sci USA. 1992;89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barik S, Banerjee A K. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J Virol. 1992;66:1109–1118. doi: 10.1128/jvi.66.2.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartenschlager R. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepat. 1999;6:165–181. doi: 10.1046/j.1365-2893.1999.00152.x. [DOI] [PubMed] [Google Scholar]
- 7.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 10.Bartenschlager R, Lohmann V, Wilkinson T, Koch J O. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens S E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 12.Bolten R, Egger D, Gosert R, Schaub G, Landmann L, Bienz K. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent In situ hybridization. J Virol. 1998;72:8578–8585. doi: 10.1128/jvi.72.11.8578-8585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Chung R T, Kaplan L M. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem Biophys Res Commun. 1999;254:351–362. doi: 10.1006/bbrc.1998.9949. [DOI] [PubMed] [Google Scholar]
- 15.De B P, Gupta S, Banerjee A K. Cellular protein kinase C isoform zeta regulates human parainfluenza virus type 3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Failla C, Tomei L, De Francesco R. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J Virol. 1995;69:1769–1777. doi: 10.1128/jvi.69.3.1769-1777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francois C, Duverlie G, Rebouillat D, Khorsi H, Castelain S, Blum H E, Gatignol A, Wychowski C, Moradpour D, Meurs E F. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J Virol. 2000;74:5587–5596. doi: 10.1128/jvi.74.12.5587-5596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale M J, Blakely S M, Kwieciszewski B, Tan S-L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanism of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale M J, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 20.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-alpha catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 25.Inoue Y, Miyazaki M, Ohashi R, Tsuji T, Fukaya K, Kouchi H, Uemura T, Mihara K, Namba M. Ubiquitous presence of cellular proteins that specifically bind to the 3′ terminal region of hepatitis C virus. Biochem Biophys Res Commun. 1998;245:198–203. doi: 10.1006/bbrc.1998.8315. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 28.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 29.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 30.Koch J O, Bartenschlager R. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J Virol. 1999;73:7138–7146. doi: 10.1128/jvi.73.9.7138-7146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolykhalov A A, Mihalik K, Feinstone S M, Rice C M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin C, Thomson J A, Rice C M. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J Virol. 1995;69:4373–4380. doi: 10.1128/jvi.69.7.4373-4380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohmann V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohmann V, Körner F, Koch J O, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 36.Mazumder B, Barik S. Requirement of casein kinase II-mediated phosphorylation for the transcriptional activity of human respiratory syncytial viral phosphoprotein P: transdominant negative phenotype of phosphorylation-defective P mutants. Virology. 1994;205:104–111. doi: 10.1006/viro.1994.1624. [DOI] [PubMed] [Google Scholar]
- 37.Moradpour D, Wakita T, Wands J R, Blum H E. Tightly regulated expression of the entire hepatitis C virus structural region in continuous human cell lines. Biochem Biophys Res Commun. 1998;246:920–924. doi: 10.1006/bbrc.1998.8727. [DOI] [PubMed] [Google Scholar]
- 38.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Classification and nomenclature of viruses: sixth report of the international committee on taxonomy of viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 424–426. [Google Scholar]
- 39.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 40.Neddermann P, Clementi A, De Francesco R. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J Virol. 1999;73:9984–9991. doi: 10.1128/jvi.73.12.9984-9991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman G R, Jasani B, Williams E D. A simple post-embedding system for the rapid demonstration of tissue antigens under the electron microscope. Histochem J. 1983;15:543–555. doi: 10.1007/BF01954145. [DOI] [PubMed] [Google Scholar]
- 42.Niepmann N. Internal initiation of translation of picornaviruses, hepatitis C virus and pestiviruses. Recent Res Dev Virol. 1999;1:229–250. [Google Scholar]
- 43.Pawlotsky J M. Hepatitis C virus infection: virus/host interactions. J Viral Hepat. 1998;5(Suppl. 1):3–8. doi: 10.1046/j.1365-2893.1998.0050s1003.x. [DOI] [PubMed] [Google Scholar]
- 44.Petrik J, Parker H, Alexander G M. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J Gen Virol. 1999;80:3109–3113. doi: 10.1099/0022-1317-80-12-3109. [DOI] [PubMed] [Google Scholar]
- 45.Poynard T, Ratziu V, Benhamou Y, Opolon P, Cacoub P, Bedossa P. Natural history of HCV infection. Best Pract Res Clin Gastroenterol. 2000;14:211–228. doi: 10.1053/bega.1999.0071. [DOI] [PubMed] [Google Scholar]
- 46.Reed K E, Gorbalenya A E, Rice C M. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol. 1998;72:6199–6206. doi: 10.1128/jvi.72.7.6199-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed K E, Rice C M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 48.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sparkman D R, White C L. A simple apparatus for processing large numbers of specimens for colloidal gold immunoelectron microscopy: application to paired helical filaments of Alzheimer's disease. J Electron Microsc Tech. 1989;13:152–153. doi: 10.1002/jemt.1060130207. [DOI] [PubMed] [Google Scholar]
- 50.Spurr A R. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 51.Suzich J A, Tamura J K, Palmer H F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka T, Kato N, Cho M J, Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka T, Kato N, Cho M J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 61.Yanagi M, St. Claire M, Emerson S U, Purcell R H, Bukh J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci USA. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]