Abstract
Introduction:
Our aim was to investigate the expression of miRNAs, C-reactive protein as a blood inflammation marker, and alanine aminotransferase as a tissue inflammation marker, in recovered and not-recovered COVID-19 patients.
Methods:
This cross-sectional project was conducted at three medical centers in Iran from December to March 2021. In total, 20 confirmed cases of COVID-19 with grade III severity and 20 healthy subjects were enrolled in the study. Subsequently, the neuroinflammatory expression of miRNAs (miR-199, miR-203, and miR-181), C-reactive protein, and alanine aminotransferase was investigated during hospitalization from week 0 to week 2.
Results:
Among COVID-19 subjects who did not recover, the expression levels of miR-199, miR-203, and miR-181 were decreased, while the levels of C-reactive protein and alanine aminotransferase increased during hospitalization. Conversely, in recovered COVID-19 subjects, the relative expression of miR-199, miR-203, and miR-181 increased and the levels of C-reactive protein and alanine aminotransferase decreased during hospitalization.
Conclusion:
The expression pattern of neuroinflammatory miRNAs depends on whether the COVID-19 patient is recovering or deteriorating. Their expression is downregulated in COVID-19 patients who do not recover and upregulated in those who do recover.
Keywords: miRNAs, COVID-19, Neuroinflammatory, Hospitalization
Highlights
The expression pattern of neuroinflammatory miRNAs depends on whether the COVID-19 patient is recovering or deteriorating.
Their expression is downregulated in COVID-19 patients who do not recover and upregulated in those who do recover.
Plain Language Summary
In this cross-sectional project, which done in three medical centers in Iran from December to March 2021, 20 confirmed cases of COVID-19 with grade III severity and 20 healthy subjects were enrolled and the neuroinflammatory expression of miRNAs (miR-199, miR-203, and miR-181), C-reactive protein, and alanine aminotransferase was investigated during hospitalization from week 0 to week 2. We found that among COVID-19 subjects who did not recover, the expression levels of miR-199, miR-203, and miR-181 were decreased, while the levels of C-reactive protein and alanine aminotransferase increased during hospitalization. Conversely, in recovered COVID-19 subjects, the relative expression of miR-199, miR-203, and miR-181 increased and the levels of C-reactive protein and alanine aminotransferase decreased during hospitalization.
1. Introduction
Acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the Coronaviridae family and shares similarities with the 2003 SARS outbreak (Wang et al., 2020). RNA viruses undergo rapid evolution aided by mutations (Girardi et al., 2018). On the other hand, the human host employs various mechanisms to combat viral infections, including innate immunity. Viral components also lead to produce different cytokines to limit the virus. In the case of SARS-CoV-2, different pathways associated with stress, apoptosis, autophagy, and innate immunity can be activated (Fung & Liu, 2019).
It has been found that the activation and regulation of immune response are modulated by miRNAs, which are non-coding RNAs consisting of 19–24 nucleotides (Nejad et al., 2018). They regulate gene expression through binding to mRNA and this process can regulate cell differentiation, cell growth, apoptosis, and disease progression (Lou et al., 2019). During viral infections, miRNAs play a crucial role in various signaling pathways, interfering with virus transmission and pathogenicity by regulating host genes and activating antiviral immune responses (Li et al., 2012). Previously, the role of miRNAs in other viral diseases, such as HIV, herpes, and Ebola has been determined (Bernier & Sagan, 2018). Recently, some miRNAs have been identified in COVID-19 using both experimental and bioinformatics tools (Ahmed et al., 2020; Khan et al., 2020). However, these reports are limited and there is insufficient information on neuroinflammatory miRNAs in COVID-19 patients during hospitalization.
We investigated the expression of miRNAs, C-reactive protein (CRP) as a blood inflammation marker, and alanine aminotransferase (ALT) as a tissue inflammation marker in recovered and not-recovered COVID-19 patients during hospitalization.
2. Materials and Methods
Blood sample collection
This study was done in three medical centers, in Yazd, Zahedan, and Tehran provinces of Iran from December to March 2021. Twenty COVID-19-confirmed patients with grade III were enrolled, according to World Health Organization (WHO) guidance and diagnostic criteria (Sohrabi et al., 2020). Also, 20 healthy subjects were enrolled in the study. We collected 5 mL of whole blood from each patient at week 0, week 1, and week 2 and kept the samples at −80°C.
The expression of miRNAs
Total RNAs were first isolated by mirPremier isolation kit (Sigma-Aldrich, USA) and confirmed by a Nano-Drop ND-1000 UV-VIS spectrophotometer. In the next step, Mir-X™ miRNA First Strand Synthesis Kit (Takara Bio Inc, USA) was used to synthesize cDNA. Mir-X miRNA qPCR SYBR (Invitrogen, UK) was applied for real-time- polymerase chain reaction (PCR). In this study, RNU 48 was considered as the internal reference.
Quantification of CRP and ALT
Quantification of CRP and ALT in serum was done according to Aceti et al. 2020.
Severity of disease
According to WHO guidelines (Aceti et al., 2020), COVID-19 patients are divided into five grades, according to the severity of the disease, including grade I or asymptomatic infection, grade II or mild illness, grade III or moderate illness, grade IV or severe illness, and grade V or critical illness.
Statistical analysis
SPSS software, version 29 was applied for the statistical analysis. Mean±SD was used to express data and one-way ANOVA was used to indicate significant differences at a P<0.05.
3. Results
The analysis of microRNA expression in recovered and not-recovered COVID-19 subjects
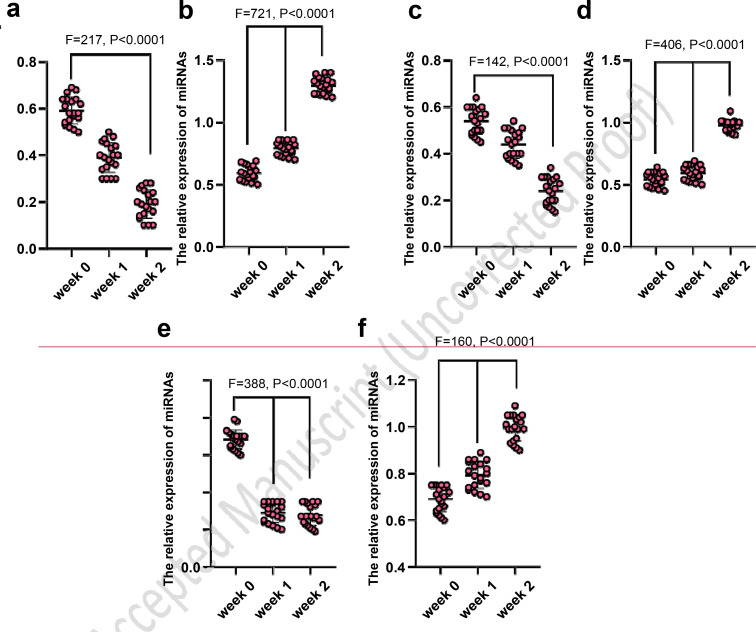
In not-recovered COVID-19 subjects, the expression levels of miR-199 (Figure 1a), miR-203 (Figure 1c), and miR-181 (Figure 1e) significantly decreased (P<0.0001). Conversely, in recovered COVID-19 subjects, the expression levels of miR-199 (Figure 1b), miR-203 (Figure 1d), and miR-181 (Figure 1f) significantly increased (P<0.0001).
Figure 1.
The analysis of microRNA expression in recovered and not-recovered COVID-19 subjects
The expression in not-recovered COVID-19 subjects: a) miR-199, c) miR-203 and e) miR-181 (significantly decreased); The expression in recovered COVID-19 subjects: b) miR-199, d) miR-203 and f) miR-181 (significantly increased)
The analysis of inflammation markers in recovered and not-recovered COVID-19 subjects
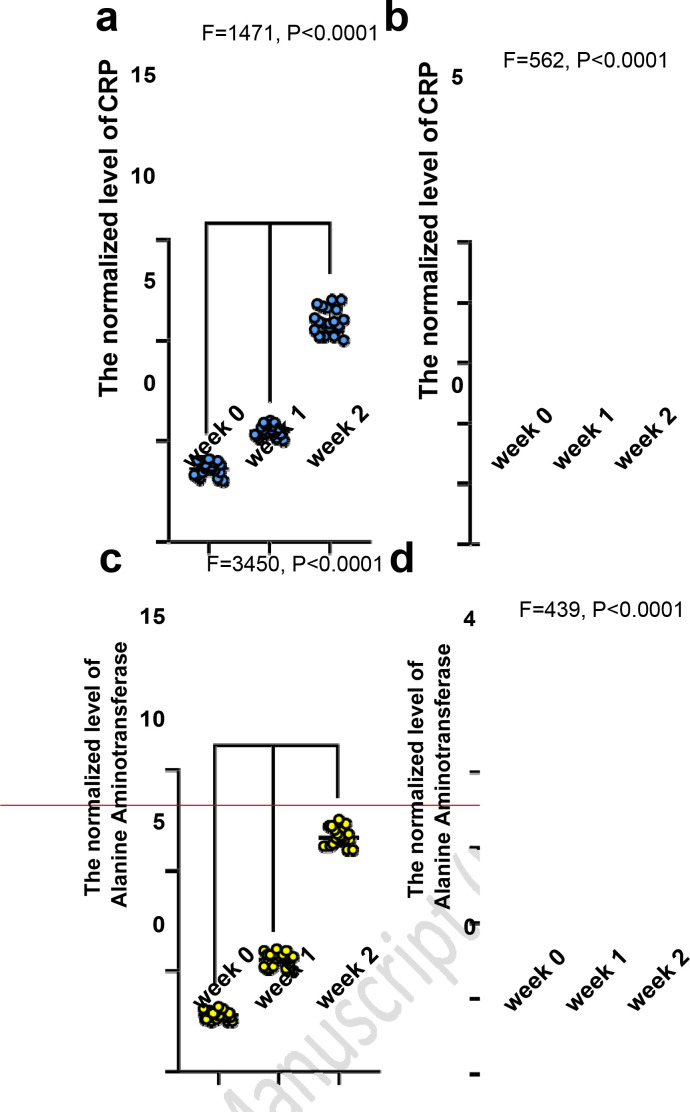
In not-recovered COVID-19 subjects, the normalized levels of CRP (Figure 2a) and ALT (Figure 2c) significantly increased (P<0.0001). Conversely, in recovered COVID-19 subjects, the normalized levels of CRP (Figure 2b) and ALT (Figure 2d) significantly decreased (P<0.0001).
Figure 2.
The analysis of inflammation markers in recovered and not-recovered COVID-19 subjects
The normalized levels in not-recovered COVID-19 subjects: a) C-reactive protein, c) Alanine aminotransferase (significantly increased from week 0 to week; The normalized levels in recovered COVID-19 patients: b) C-reactive protein, d) Alanine aminotransferase (significantly decreased)
The analysis of disease grade in recovered and not-recovered COVID-19 subjects
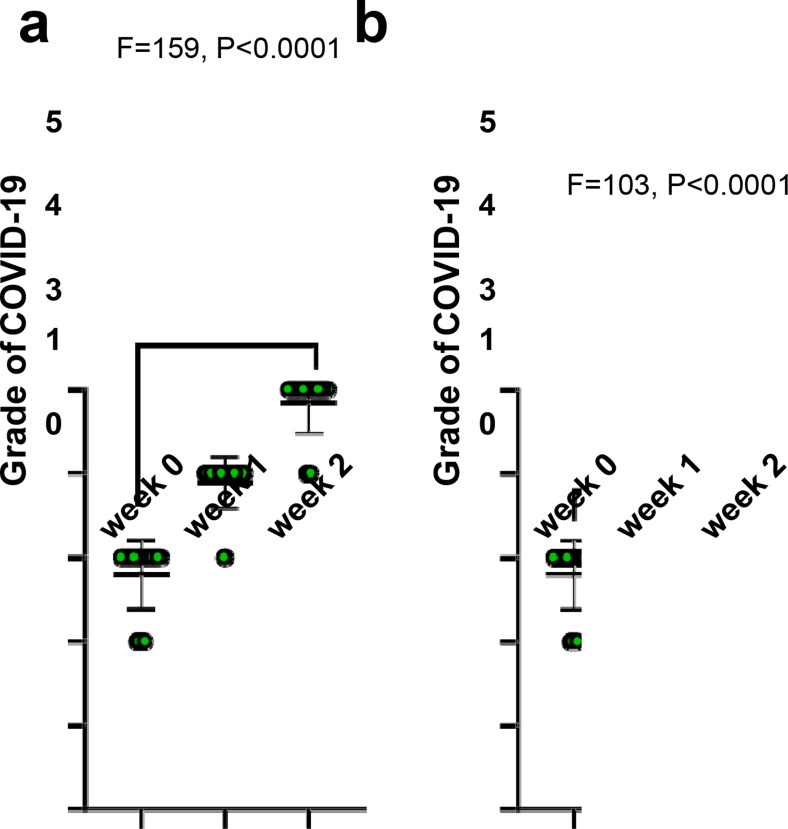
In not-recovered COVID-19 subjects, the grade of disease significantly increased (P<0.0001) (Figure 3a) whereas, in recovered COVID-19 subjects (Figure 3b), this parameter significantly decreased (P<0.0001).
Figure 3.
The analysis of grade of disease in recovered and not-recovered COVID-19 subjects
Not-recovered COVID-19 subjects: a) The grade of disease significantly increased (P<0.0001) and conversely; Recovered COVID-19 subjects: b) The grade of disease significantly decreased (P<0.0001)
4. Discussion
Although SARS-CoV-2 attacks the lungs, the virus can spread to other tissues, such as the brain, and cause neurological complications, especially in the elderly (Huang et al., 2020; Wolfel et al., 2019; Wu & McGoogan, 2020). Up to 20–30% of COVID-19 patients have brain complications (Chen et al., 2020; Varatharaj et al., 2020). An important question is how SARS-CoV-2 damages the central nervous system (CNS). Although SARSCoV-2 enters the brain by several routes, the virus can directly infect nerve cells (Song et al., 2021). The binding of the virus to ACE2, which is present on the surface of cells (Zeisel et al., 2015), neurons, and neuroglial cells (Nemoto et al., 2020) is critical. Moreover, endosomal proton pump and NAADP-sensitive intracellular two-pore channel 2 are important parts of endocytosis (Petersen et al., 2020). Unfortunately, the virus can enter the brainstem through the olfactory bulb after initial entry through the nose and eyes, subsequently spreading to various brain regions (Li et al., 2016). The situation worsens when the virus breaches the blood-brain barrier, spreading throughout the brain or body, and potentially triggering a cytokine storm that can lead to death (Coperchini et al., 2020). As the virus infiltrates the brain, peripheral immune cells enter, followed by microglial cells and astrocytes, an occurrence not typical under normal circumstances (Engelhardt et al., 2017). One study has demonstrated that the astrocytic calcium-binding protein S100b, as well as tissue and blood inflammatory markers, are elevated in COVID-19 patients (Aceti et al., 2020). In response to viral infections, microglia and astrocytes become activated and secrete proinflammatory cytokines (Shabab et al., 2017). These events are commonly referred to as neuroinflammation and serve to protect the brain against pathogens (Kempuraj et al., 2016).
Although neuroinflammation is observed in various disorders, the underlying molecular processes are often unclear (DiSabato et al., 2016). Microglia, which are static macrophages originating from myeloid progenitor cells in early embryonic life (Wieghofer & Prinz, 2016), are the primary cells responsible for neuroinflammation. Under normal conditions, microglia remain in a resting state, continuously surveying the environment, phagocytosing debris, and releasing neurotrophic factors (Kabba et al., 2018). Equipped with molecular recognition receptors, cytokine receptors, and various neural receptors, microglia can respond to diverse and unfamiliar molecules (Harry, 2013). Activation of microglia induces morphological changes, adopting an amoeboid shape (Kabba et al., 2018). Microglia can assume two functional states: M1 (pro-inflammatory) and M2 (anti-inflammatory). M1 microglia release inflammatory mediators such as TNFα, IL-6, and IL-1β (Tang & Le, 2016), while M2 microglia release anti-inflammatory cytokines including IL-10, TGF-β, IL-4, and IL-13 [9]. Astrocytes, another type of glial cell in the brain, also contribute to the inflammatory response within the CNS (Cekanaviciute & Buckwalter, 2016). In a healthy brain, astrocytes promote synaptogenesis, clear neurotransmitters, and regulate nervous signaling to maintain neural homeostasis (Vasile et al., 2017). Astrocytes also play an important role in responding to pathogens (Dossi et al., 2018).
In this study, we found that in not-recovered COVID-19 subjects, the expression of miR-199, miR-203, and miR-181 significantly decreased from week 0 to week 2. Conversely, in recovered COVID-19 patients, their relative expression was significantly increased from week 0 to week 2. Also, we found that in not-recovered COVID-19 patients, the normalized levels of C- reactive protein, ALT, and COVID-19 grade significantly increased and conversely, in recovered COVID-19 patients, these parameters significantly decreased.
Host-directed therapies, such as miRNAs, represent a novel therapeutic approach targeting factors influencing host cells. In the case of SARS-CoV, researchers are identifying specific molecules to block crucial pathways. Certain cytokines can activate antiviral defense mechanisms during viral infections. Interestingly, various miRNAs play a role in regulating antiviral genes. For instance, Sardar et al. identified 2197 human miRNAs capable of targeting host genes (Sardar et al., 2020). Satyam et al. identified six putative miRNAs in SARSCoV-2, noting that hsa-miR-214-3p levels could be elevated by SARS-CoV-2 infection (Satyam et al., 2020). Brogaard et al. demonstrated late regulation of hsa-miR-223-5p in circulating leukocytes of a pig model of influenza A (Brogaard et al., 2016). Moffett et al. illustrated that miR-31 could inhibit CD8+ T cell function and is an important regulator in chronic infections (Moffett et al., 2017). Nersisyan et al. introduced six miRNAs, including miR-21-3p, miR-195-5p, miR-16-5p, miR-3065-5p, miR-424- 5p and miR-421, in the coronavirus-host interaction (Nersisyan et al., 2020). Jafarinejad et al showed that the miR-29 family had a high affinity for the SARSCoV-2 genome (Jafarinejad-Farsangi et al., 2020).
5. Conclusion
In summary, this study revealed that in COVID-19 patients who did not recover, the expression of miR-199, miR-203, and miR-181 significantly decreased from week 0 to week 2. Conversely, in recovered COVID-19 patients, their relative expression significantly increased during the same period. Additionally, it was observed that in non-recovered COVID-19 patients, the normalized levels of CRP, ALT, and COVID-19 severity grade significantly increased, whereas in recovered COVID-19 patients, these parameters significantly decreased.
Acknowledgments
The authors thank the Reference Laboratory of Zahedan University of Medical Sciences.
Ethical Considerations
Compliance with ethical guidelines
This study was approved by the Ethics Committee of Zahedan University of Medical Sciences (Code: IR.ZAUMS.REC.1399.316).
Funding
This article was financially supported by the Zahedan University of Medical Sciences (Grant No.: 9936).
Authors’ contribution
Conceptualization, study design, writing and final approval: All authors; Data collection, data interception and data analysis: Reza Keikha and Ali Jebali.
Conflict of interest
The authors declared no conflict of interest.
References
- Aceti A., Margarucci L. M., Scaramucci E., Orsini M., Salerno G., Di Sante G., et al. (2020). Serum S100B protein as a marker of severity in Covid-19 patients. Scientific Reports, 10(1), 18665. [DOI: 10.1038/s41598-020-75618-0] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S. S. S. J., Paramasivam P., Raj K., Kumar V., Murugesan R., Ramakrishnan V. (2020). Regulatory cross talk between SARS-CoV-2 receptor binding and replication machinery in the human host. Frontiers in Physiology, 11, 802. [DOI: 10.3389/fphys.2020.00802] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier A., Sagan S. M. (2018). The diverse roles of microRNAs at the host-virus interface. Viruses, 10(8), 440. [DOI: 10.3390/v10080440] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard L., Heegaard P. M., Larsen L. E., Mortensen S., Schlegel M., Dürrwald R., et al. (2016). Late regulation of immune genes and microRNAs in circulating leukocytes in a pig model of influenza A (H1N2) infection. Scientific Reports, 6, 21812. [DOI: 10.1038/srep21812] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekanaviciute E., Buckwalter M. S. (2016). Astrocytes: Integrative regulators of neuroinflammation in stroke and other neurological diseases. Neurotherapeutics, 13(4), 685–701. [DOI: 10.1007/s13311-016-0477-8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., et al. (2020). Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ (Clinical research ed.), 368, m1091. [DOI: 10.1136/bmj.m1091] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. (2020). The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine & Growth Factor Reviews, 53, 25–32. [DOI: 10.1016/j.cytogfr.2020.05.003] [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSabato D. J., Quan N., Godbout J. P. (2016). Neuroinflammation: The devil is in the details. Journal of Neurochemistry, 139(Suppl 2), 136–153. [DOI: 10.1111/jnc.13607] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossi E., Vasile F., Rouach N. (2018). Human astrocytes in the diseased brain. Brain Research Bulletin, 136, 139–156. [DOI: 10.1016/j.brainresbull.2017.02.001] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt B., Vajkoczy P., Weller R. O. (2017). The movers and shapers in immune privilege of the CNS. Nature Immunology, 18(2), 123–131. [DOI: 10.1038/ni.3666] [DOI] [PubMed] [Google Scholar]
- Fung T. S., Liu D. X. (2019). Human Coronavirus: Host-pathogen interaction. Annual Review of Microbiology, 73, 529–557. [DOI: 10.1146/annurev-micro-020518-115759] [DOI] [PubMed] [Google Scholar]
- Girardi E., López P., Pfeffer S. (2018). On the importance of host MicroRNAs during viral infection. Frontiers in Genetics, 9, 439. [DOI: 10.3389/fgene.2018.00439] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry G. J. (2013). Microglia during development and aging. Pharmacology & Therapeutics, 139(3), 313–326. [DOI: 10.1016/j.pharmthera.2013.04.013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. [DOI: 10.1016/S0140-6736(20)30183-5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarinejad-Farsangi S., Jazi M. M., Rostamzadeh F., Hadizadeh M. (2020). High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis. Non-Coding RNA Research, 5(4), 222–231. [DOI: 10.1016/j.ncrna.2020.11.005] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabba J. A., Xu Y., Christian H., Ruan W., Chenai K., Xiang Y., et al. (2018). Microglia: Housekeeper of the central nervous system. Cellular and Molecular Neurobiology, 38(1), 53–71. [DOI: 10.1007/s10571-017-0504-2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempuraj D., Thangavel R., Natteru P. A., Selvakumar G. P., Saeed D., Zahoor H., et al. (2016). Neuroinflammation induces neurodegeneration. Journal of Neurology, Neurosurgery and Spine, 1(1), 1003. [PMC free article] [PubMed] [Google Scholar]
- Khan M. A., Sany M. R. U., Islam M. S., Islam A. B. M. M. K. (2020). Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Frontiers in Genetics, 11, 765. [DOI: 10.3389/fgene.2020.00765] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wohlford-Lenane C., Perlman S., Zhao J., Jewell A. K., Reznikov L. R., et al. (2016). Middle East respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. The Journal of Infectious Diseases, 213(5), 712–722. [DOI: 10.1093/infdis/jiv499] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan X., He X., Sun H., Zou Z., Yuan H., et al. (2012). MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-alpha. Cellular & Molecular Immunology, 9(6), 497–502. [DOI: 10.1038/cmi.2012.35] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou W., Liu J., Ding B., Chen D., Xu L., Ding J., et al. (2019). Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC. Journal of Translational Medicine, 17(1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett H. F., Cartwright A. N. R., Kim H. J., Godec J., Pyrdol J., Äijö T., et al. (2017). The microRNA miR-31 inhibits CD8+ T cell function in chronic viral infection. Nature Immunology, 18(7), 791–799. [DOI: 10.1038/ni.3755] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejad C., Stunden H. J., Gantier M. P. (2018). A guide to miRNAs in inflammation and innate immune responses. The FEBS Journal, 285(20), 3695–3716. [DOI: 10.1111/febs.14482] [DOI] [PubMed] [Google Scholar]
- Nemoto W., Yamagata R., Nakagawasai O., Nakagawa K., Hung W. Y., Fujita M., et al. (2020). Effect of spinal angiotensin-converting enzyme 2 activation on the formalin-induced nociceptive response in mice. European Journal of Pharmacology, 872, 172950. [DOI: 10.1016/j.ejphar.2020.172950] [DOI] [PubMed] [Google Scholar]
- Nersisyan S., Engibaryan N., Gorbonos A., Kirdey K., Makhonin A., Tonevitsky A. (2020). Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ, 8, e9994. [DOI: 10.7717/peerj.9994] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen O. H., Gerasimenko O. V., Gerasimenko J. V. (2020). Endocytic uptake of SARS-CoV-2: The critical roles of pH, Ca2+, and NAADP. Function, 1(1), zqaa003. [DOI: 10.1093/function/zqaa003] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar R., Satish D., Gupta D. (2020). Identification of novel SARS-CoV-2 drug targets by host microRNAs and transcription factors co-regulatory interaction network analysis. Frontiers in Genetics, 11, 571274. [DOI: 10.3389/fgene.2020.571274] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyam R., Bhardwaj T., Goel S., Jha N. K., Jha S. K., Nand P., et al. (2020). miRNAs in SARS-CoV 2: A Spoke in the wheel of pathogenesis. Current Pharmaceutical Design, 27(13), 1628–1641. [DOI: 10.2174/1381612826999201001200529] [DOI] [PubMed] [Google Scholar]
- Shabab T., Khanabdali R., Moghadamtousi S. Z., Kadir H. A., Mohan G. (2017). Neuroinflammation pathways: A general review. International Journal of Neuroscience, 127(7), 624–633. [DOI: 10.1080/00207454.2016.1212854] [DOI] [PubMed] [Google Scholar]
- Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., et al. (2020). World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). International Journal of Surgery, 76, 71–76. [DOI: 10.1016/j.ijsu.2020.02.034] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A. V., Skriabine S., et al. (2021). Neuroinvasion of SARS-CoV-2 in human and mouse brain. Journal of Experimental Medicine, 218(3), e20202135. [DOI: 10.1084/jem.20202135] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Le W. (2016). Differential roles of M1 and M2 microglia in neurodegenerative diseases. Molecular Neurobiology, 53(2), 1181–1194. [DOI: 10.1007/s12035-014-9070-5] [DOI] [PubMed] [Google Scholar]
- Varatharaj A., Thomas N., Ellul M. A., Davies N. W. S., Pollak T. A., Tenorio E. L., et al. (2020). Neurological and neuropsychiatric complications of COVID-19 in 153 patients: A UK-wide surveillance study. The Lancet. Psychiatry, 7(10), 875–882. [DOI: 10.1016/S2215-0366(20)30287-X] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile F., Dossi E., Rouach N. (2017). Human astrocytes: Structure and functions in the healthy brain. Brain Structure and Function, 222(5), 2017–2029. [DOI: 10.1007/s00429-017-1383-5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li X., Li T., Zhang S., Wang L., Wu X., et al. (2020). The genetic sequence, origin, and diagnosis of SARSCoV-2. European Journal of Clinical Microbiology & Infectious Diseases, 39(9), 1629–1635. [DOI: 10.1007/s10096-020-03899-4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieghofer P., Prinz M. (2016). Genetic manipulation of microglia during brain development and disease. Biochimica et Biophysica Acta, 1862(3), 299–309. [DOI: 10.1016/j.bbadis.2015.09.019] [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V. M., Guggemos W., Seilmaier M., Zange S., Müller M. A., et al. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature, 581(7809), 465–469. [DOI] [PubMed] [Google Scholar]
- Wu Z., McGoogan J. M. (2020). Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA, 323(13), 1239–1242. [DOI: 10.1001/jama.2020.2648] [DOI] [PubMed] [Google Scholar]
- Zeisel A., Muñoz-Manchado A. B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., et al. (2015). Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science, 347(6226), 1138–1142. [DOI: 10.1126/science.aaa1934] [DOI] [PubMed] [Google Scholar]