Abstract
Quantitative evaluation of tissue images is crucial for computational pathology (CPath) tasks, requiring the objective characterization of histopathological entities from whole-slide images (WSIs). The high resolution of WSIs and the variability of morphological features present significant challenges, complicating the large-scale annotation of data for high-performance applications. To address this challenge, current efforts have proposed the use of pretrained image encoders through transfer learning from natural image datasets or self-supervised learning on publicly available histopathology datasets, but have not been extensively developed and evaluated across diverse tissue types at scale. We introduce UNI, a general-purpose self-supervised model for pathology, pretrained using more than 100 million images from over 100,000 diagnostic H&E-stained WSIs (>77 TB of data) across 20 major tissue types. The model was evaluated on 34 representative CPath tasks of varying diagnostic difficulty. In addition to outperforming previous state-of-the-art models, we demonstrate new modeling capabilities in CPath such as resolution-agnostic tissue classification, slide classification using few-shot class prototypes, and disease subtyping generalization in classifying up to 108 cancer types in the OncoTree classification system. UNI advances unsupervised representation learning at scale in CPath in terms of both pretraining data and downstream evaluation, enabling data-efficient artificial intelligence models that can generalize and transfer to a wide range of diagnostically challenging tasks and clinical workflows in anatomic pathology.
Introduction
The clinical practice of pathology involves performing a large range of tasks: from tumor detection and subtyping to grading and staging, and, given the thousands of possible diagnoses, a pathologist must be adept at solving an incredibly diverse group of problems, often simultaneously1–4. Contemporary computational pathology (CPath) has expanded this array even further by enabling prediction of molecular alterations5,6, prognostication7–9, and therapeutic response prediction10, among other applications11–14. With a vast array of tasks, training models from scratch has practical limitations due to challenges in gathering pathologist annotations, building large histology collections.
The transfer learning, generalization and scaling capabilities of self-supervised (or pretrained) models are dependent on the size and diversity of the training data21–23. In general computer vision, the development and evaluation of many fundamental self-supervised models24–27 are based on the ImageNet Large Scale Visual Recognition Challenge28,29 and other large datasets30–32. Such models have also been described as ‘foundation models’ due to their ability to adapt to a wide range of downstream tasks when pretrained on massive amounts of data33,34. In CPath, The Cancer Genome Atlas (TCGA; ~29,000 formalin-fixed paraffin-embedded and frozen H&E whole-slide images (WSIs), 32 cancer types)35 similarly serves as the basis for many self-supervised models36–46 along with other histology datasets47–53, with a number of prior works demonstrating great progress in learning meaningful representations of histology tissue for clinical pathology tasks37,38,54–66. However, current pretrained models for CPath remain constrained by the limited size and diversity of pretraining data, given that the TCGA comprises mostly primary cancer histology slides, and by the limited evaluation of generalization performance across diverse tissue types, and many pan-cancer analyses and popular clinical tasks in CPath are also based on annotated histology regions of interest (ROIs) and slides from TCGA6,9,16,17,61,67–74. Addressing these limitations is critical in the broader development of foundation models in CPath that can generalize and transfer to real-world clinical settings with widespread applications.
In this work we build upon these prior efforts by introducing a general-purpose, self-supervised vision encoder for pathology, UNI, a large vision transformer (ViT-Large or ViT-L)75 pretrained on one of the largest histology slide collections created for self-supervised learning, termed ‘Mass-100K’. Mass-100K is a pretraining dataset that consists of more than 100 million tissue patches from 100,426 diagnostic H&E WSIs across 20 major tissue types collected from Massachusetts General Hospital (MGH) and Brigham and Women’s Hospital (BWH), as well as the Genotype–Tissue Expression (GTEx) consortium76, and provides a rich source of information for learning objective characterizations of histopathologic biomarkers (Fig. 1a and Supplementary Tables 1–3). In the pretraining stage, we use a self-supervised learning approach called DINOv2 (ref. 22), which has been shown to yield strong, off-the-shelf representations for downstream tasks without the need for further fine-tuning with labeled data (Fig. 1b). We demonstrate the versatility of UNI on diverse machine learning settings in CPath, including ROI-level classification, segmentation and image retrieval, and slide-level weakly supervised learning (Fig. 1c). In total, we assess UNI on 34 clinical tasks across anatomic pathology and a range of diagnostic difficulty, such as nuclear segmentation, primary and metastatic cancer detection, cancer grading and subtyping, biomarker screening and molecular subtyping, organ transplant assessment, and several pan-cancer classification tasks that include subtyping to 108 cancer types in the OncoTree cancer classification system77 (Figs. 1d and 2a). In addition to outperforming previous state-of-the-art models such as CTransPath37 and REMEDIS38, we also demonstrate capabilities such as resolution-agnostic tissue classification and few-shot class prototypes for prompt-based slide classification (Fig. 2d), highlighting the potential of UNI as a foundation model for the further development of artificial intelligence (AI) models in anatomic pathology.
Fig. 1: Overview of UNI.

UNI is a general-purpose, self-supervised vision encoder for anatomic pathology based on the vision transformer architecture, achieving state-of-the-art performance across 34 clinical tasks in anatomic pathology. a, Slide distribution of Mass-100K, a large-scale and diverse pretraining dataset of 100 million tissue patches sampled from over 100,000 diagnostic WSIs across 20 major organ types. b, UNI is pretrained on Mass-100K using the DINOv2 self-supervised training algorithm22, which consists of a mask image modeling objective118 and a self-distillation objective25. c, UNI generally outperforms other pretrained encoders across 34 clinical tasks in anatomical pathology (average performance of the 8 SegPath tasks reported). d, The evaluation tasks consist of ROI-level classification, segmentation, retrieval and prototyping, and slide-level classification tasks. Further details are given in Methods. class., classification; seg., segmentation; det., detection; assess., assessment.
Fig. 2: Slide-level tasks for OT-43 and OT-108, and slide-level task performance.
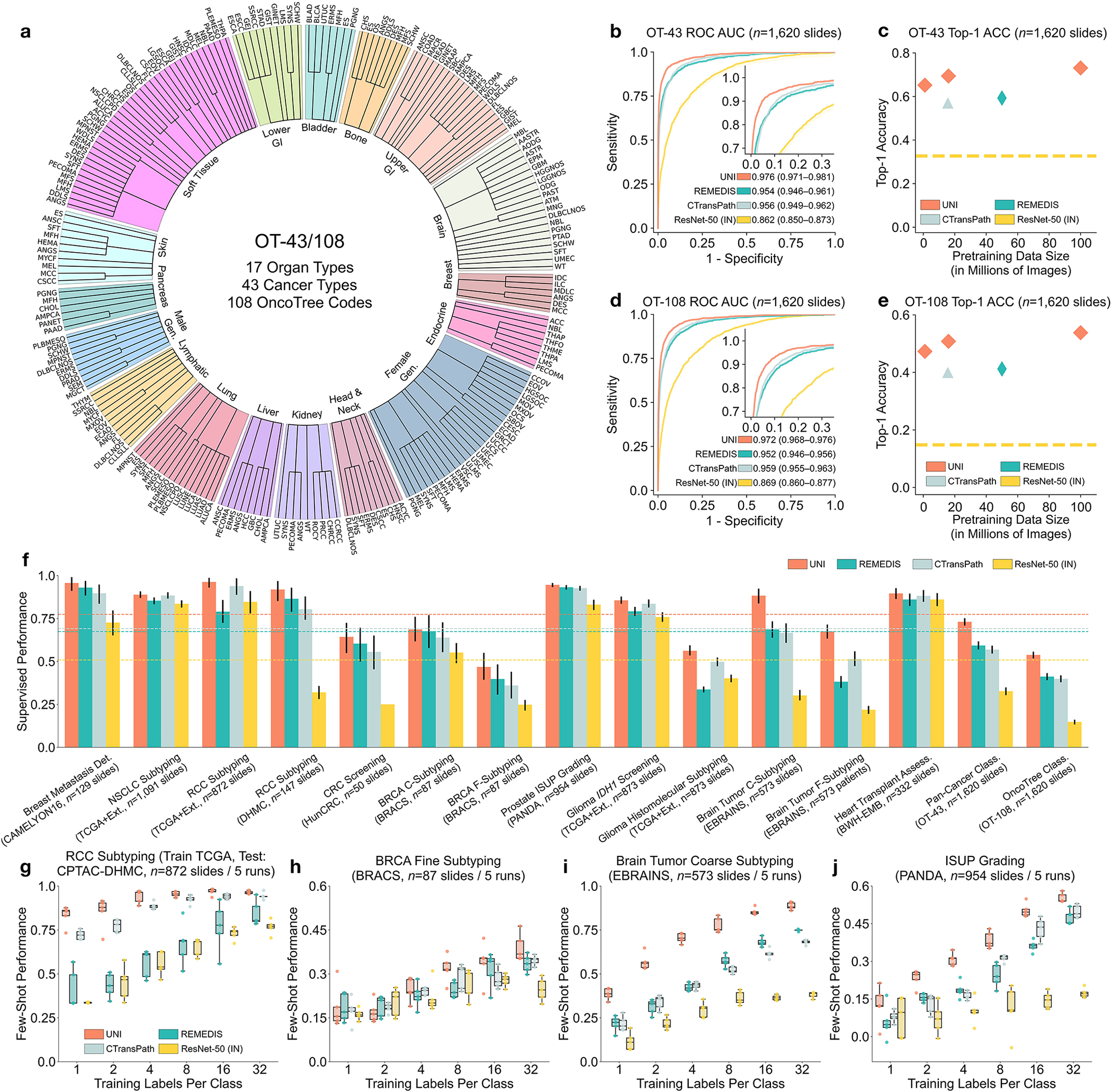
a, Organ and OncoTree code distribution for the slide-level OT-43 and OT-108 classification tasks. All comparisons with UNI are evaluated on 43-way cancer type classification and 108-way OncoTree code classification tasks with OT-43 and OT-108, respectively. Further details regarding data distribution are provided in Supplementary Table 4. Gen., genitalia; GI, gastrointestinal. b,d, Comparison of macro-averaged AUROC of UNI and other pretrained encoders for OT-43 (b) and OT-108 (d) (n = 1,620 slides each). c,e, Top-1 accuracy of UNI across different pretraining data scales (Mass-1K, Mass-22K, Mass-100K) for OT-43 (c) and OT-108 (e) (n = 1,620 slides each). f, Supervised performance of UNI and its comparisons across 15 weakly supervised slide-level classification tasks. Dashed lines represent the average performance of each model across all tasks. All data are given as balanced accuracy, except for ISUP grading, which is given as quadratic weighted Cohen’s κ. Error bars represent 95% confidence intervals and the centers correspond to computed values of each metric as specified above. Detailed results for all tasks are provided in Supplementary Tables 12–35. Ext., external test set. g–j, Few-shot slide-level performance with K ∈ {1, 2, 4, 8, 16, 32} slides per class reported for four tasks. g, RCC subtyping (train, TCGA; test, CPTAC-DHMC; n = 872 slides). h, BRCA fine-grained subtyping (BRACS, n = 87 slides). i, Brain tumor coarse-grained subtyping (EBRAINS, n = 573 slides). j, ISUP grading (PANDA, n = 954 slides). Boxes indicate quartile values of model performance (n = 5 runs), and whiskers extend to data points within 1.5-fold the interquartile range. Few-shot results for all tasks are given in Extended Data Fig. 1.
Results
Pretraining Scaling Laws in CPath
A pivotal characteristic of foundation models lies in their capability to deliver improved downstream performance on various tasks when trained on larger datasets. Although datasets such as CAMELYON16 (Cancer Metastases in Lymph Nodes Challenge 2016 (ref. 78) and the TCGA nonsmall cell lung carcinoma subset (TCGA-NSCLC)79 are commonly used to benchmark pretrained encoders using weakly supervised multiple instance learning (MIL) algorithms15,37,40,80, they source tissue slides only from a single organ and are often used for predicting binary disease states81, which is not reflective of the broader array of disease entities seen in real-world anatomic pathology practice. Instead, we assess the generalization capabilities of UNI across diverse tissue types and disease categories by constructing a large-scale, hierarchical, and rare cancer classification task for CPath that follows the OncoTree cancer classification system77. Using in-house BWH slides, we defined a dataset that comprises 5,564 WSIs from 43 cancer types further subdivided into 108 OncoTree codes, with at least 20 WSIs per OncoTree code. A total of 90 out of the 108 cancer types are designated as rare cancers as defined by the RARECARE project82 and the National Cancer Institute’s Surveillance, Epidemiology, and End Results (NCI-SEER) Program. The dataset forms the basis of two tasks that vary in diagnostic difficulty: 43-class OncoTree cancer type classification (OT-43), and 108-class OncoTree code classification (OT-108) (Fig. 2a and Supplementary Table 4). The goal of these large multi-class classification task is not necessarily clinical utility but to assess the capabilities of the foundation model and richness of the feature representations in comparison with other models. To assess scaling trends, we also pretrain UNI across varying data scales, with Mass-100K subsetted to create Mass-22K (16 million images, 21,444 WSIs) and Mass-1K (1 million images, 1,404 WSIs). We also assess model scale by ablating UNI using two different ViT architecture sizes: ViT-Base (or ViT-B) and ViT-Large (or ViT-L). Last, we also assess the impact of self-supervised learning algorithm choice, compared also against MoCoV3 (ref. 24). For weakly supervised slide classification, we follow the conventional paradigm of first pre-extracting patch-level features from tissue-containing patches in the WSI using a pretrained encoder, followed by training an attention-based MIL (ABMIL) algorithm83. To reflect the label complexity challenges of these tasks, we report top-K accuracy (K = 1, 3, 5) as well as weighted F1 score and area under the receiver operating characteristic curve (AUROC) performance. Additional details regarding the OT-43 and OT-108 tasks, experimental setup, implementation details and performance are provided in Methods, Supplementary Tables 1–11 and Supplementary Tables 12–18, respectively.
Overall, we demonstrate model and data scaling capabilities of self-supervised models in UNI, with the scaling trend for UNI on OT-43 and OT-108 shown in Fig. 2c,e. On OT-43 and OT-108, we observe a +4.2% performance increase (P < 0.001, two-sided paired permutation test) in top-1 accuracy when scaling UNI using VIT-L from Mass-1K to Mass-22K, and a similar +3.5% performance increase (P < 0.001) on OT-108. From Mass-22K to Mass-100K, performance increases further: +3.7% and +3.0% on OT-43 and OT-108, respectively (P < 0.001). Similar trends are observed using VIT-B, with performance plateauing from Mass-22K to Mass-100K (Supplementary Tables 13 and 16). Supplementary Tables 14 and 17 show the impact of data diversity and pretraining length, with monotonic improvement from 50,000 to 125,000 training iterations on both tasks. Overall, these scaling trends align with findings observed in many ViT models applied to natural images21,31,75, in which the performance of larger ViT variants improves as the pretraining dataset grows. Exploring other self-supervised learning algorithms, we also trained MoCoV3 (ref. 24) (using ViT-L and ResNet-50 backbones) on Mass-1K, which performed worse against DINOv2 (Supplementary Table 18). To scale performance with increasing model and data size, the choice of algorithms and their hyper-parameters is also important in developing CPath foundation models.
We compare UNI using ViT-L pretrained on Mass-100K to publicly available pretrained encoders used in CPath, on OT-43 and OT-108 tasks: ResNet-50 (ref. 84) pretrained on ImageNet-1K; CTransPath37 pretrained on TCGA and PAIP (Pathology AI Platform)85; and REMEDIS38 pretrained on TCGA. We observe that UNI outperforms all baselines by a wide margin. On OT-43, UNI achieves a top-5 accuracy of 93.8% and an AUROC of 0.976, outperforming the next best-performing model (REMEDIS) by +6.3% and +0.022 on these respective metrics (both P < 0.001) (Fig. 2b and Supplementary Table 12). On OT-108 we observe a similar margin of performance increase, +10.8% and +0.020 (P < 0.001), respectively, over REMEDIS (Fig. 2c and Supplementary Table 15). Overall, we find that UNI is able to classify rare cancers in OT-43 and OT-108 with wide margins of performance improvement.
Weakly supervised slide classification
Furthermore, we investigate UNI’s capabilities across a diverse over all pretrained encoders. range of 15 slide-level classification tasks, which include breast cancer metastasis detection (CAMELYON16)78, International Society of Urological Pathology (ISUP) grading in prostate cancer (Prostate Cancer Grade Assessment, PANDA)18, cardiac transplant assessment (in-house BWH slides)86, and brain tumor subtyping (EBRAINS; representing 30 rare cancers defined by the RARECARE project), among others. Similar to OT-43 and OT-108 evaluation, we compare the pre-extracted features from UNI with that of other pretrained encoders using ABMIL83. Given that CTransPath and REMEDIS were trained using almost all TCGA slides, the reported performance of these models on TCGA tasks may be contaminated with data leakage and thus unfairly inflated. Additional details regarding slide tasks, experimental setup, and performance are provided in Methods, Supplementary Tables 19–21 and Supplementary Tables 22–35, respectively.
Across all 15 slide-level tasks, UNI consistently outperforms other pretrained encoders (average performance increases of +26.4% over ResNet-50, +8.3% over CTransPath, and +10.0% over REMEDIS), with greater improvements observed on tasks classifying rare cancer types or characterized by higher diagnostic complexity (Fig. 2f). On prostate ISUP grading (PANDA), UNI achieves a quadratic weighted Cohen’s κ of 0.946, outperforming the next best-performing model (REMEDIS) by +0.014 (P < 0.05) (Supplementary Table 29). On hierarchical classification tasks (which also involve rare disease categories) such as glioma biomarker prediction (2-class IDH1 mutation prediction and 5-class histomolecular subtyping using TCGA87,88 and EBRAINS89) and brain tumor subtyping (12-class coarse-grained and 30-class fine-grained brain tumor subtyping using EBRAINS), UNI outperforms the next best-performing model (either CTransPath or REMEDIS), by +2.0% (P = 0.076), +6.4% (P = 0.001), +19.6% (P < 0.001) and +16.1% (P < 0.001) (Supplementary Tables 31–34). Similar to OT-43 and OT-108, we find that UNI has the largest impact on the evaluation of brain tumor subtyping tasks, which involve only rare cancer types.
On comparison of existing leaderboards, we find that ABMIL with UNI features outperforms many sophisticated MIL architectures. On breast cancer metastasis detection (CAMELYON16), ABMIL with UNI outperforms all state-of-the-art MIL methods on this task (Supplementary Table 36), and is one of the few MIL results that outperforms the human pathologist performance (AUROC of 0.966) without time constraints in the original challenge78. On tasks with detailed comparisons such as prostate ISUP grading (PANDA) and cellular-mediated allograft rejection (BWH-EMB), ABMIL with UNI outperforms methods such as WholeSIGHT90 and CRANE86 (Supplementary Tables 37 and 38). Although many of these comparisons are not equivalent due to the use of ResNet-50 with ImageNet transfer (ResNet-50IN) features, we note that their proposed MIL architectures are often motivated and developed specifically for solving these challenging tasks. Our comparisons highlight the strength of having a better-pretrained encoder versus MIL architecture.
Data contamination is a concern in foundation models trained on large collections of public datasets91–95. Although labels may not be explicitly leaked into the model during self-supervised training, models pretrained on the evaluated test set may exhibit optimistically biased performance, observed in other CPath studies96. We additionally compare UNI against CTransPath and REMEDIS on TCGA test sets from the nonsmall cell lung cancer (NSCLC) subtyping, renal cell carcinoma (RCC) subtyping, glioma IDH1 mutation prediction and glioma histomolecular subtyping tasks, observing performance decreases when comparing the in-domain versus out-of-domain performance. On NSCLC subtyping, REMEDIS outperforms UNI on TCGA evaluation (97.3% versus 94.7%), but underperforms on CPTAC (Clinical Proteomic Tumor Analysis Consortium) evaluation (79.0% versus 96.3%) (Supplementary Table 23). On glioma IDH1 mutation prediction, CTransPath and REMEDIS outperform UNI on TCGA evaluation (89.1% and 81.9% versus 80.8%), but underperform on EBRAINS evaluation (83.6% and 79.2% versus 85.6%) (Supplementary Tables 31 and 32). We emphasize that data contamination exists only in how the models are used, not in the models themselves, which have been shown to transfer well in settings independent of TCGA38,59,97. Given that many CPath studies use the TCGA for studying diverse cancer types, UNI is more flexible than CTransPath and REMEDIS in developing pathology AI models on public histology datasets and benchmarks.
Label efficiency of few-shot slide classification
We additionally evaluate UNI in few-shot MIL across all slide-level tasks. Few-shot learning is an evaluation scheme that studies the generalization capabilities of models on new tasks (C classes) given a limited number of examples (K training samples per class, also called supports or shots). For all pretrained encoders, we trained an ABMIL model with K ∈ {1, 2, 4, 8, 16, 32} training examples per class, where K is limited to 32 due to small support sizes in rare disease categories. Given that the performance can fluctuate depending on which K examples are chosen for each class, we repeat experiments over five runs with C × K training examples randomly sampled each time. Additional details regarding few-shot MIL experimentation and performance are provided in Methods and Extended Data Fig. 1.
UNI generally outperforms other pretrained encoders and with superior label efficiency across all tasks, especially in classifying rare diseases (Fig. 2g–j and Extended Data Fig. 1). When comparing the 4-shot performance of UNI with that of other encoders (using the median performance), the next best-performing encoder needs up to eightfold as many training examples per class to reach the same 4-shot performance of UNI. On prostate ISUP grading (PANDA), UNI is consistently twice as label efficient across all few-shot settings (Fig. 2j). On challenging rare cancer subtyping tasks such as fine-grained brain tumor subtyping (EBRAINS), the 4-shot performance of UNI outperforms other encoders by a large margin, matched only by the 32-shot performance of REMEDIS (Fig. 2i). Overall, our comprehensive evaluation of slide classification tasks demonstrates UNI’s potential as a foundational model that can be used in histopathology workflows that screen for rare and underrepresented diseases.
Supervised ROI classification in linear classifiers
In addition to slide-level tasks, we also assess UNI on a diverse range of 11 ROI-level tasks, which include colorectal tissue and polyp classification (CRC-100K-NONORM98, HunCRC99, UniToPatho100), prostate adenocarcinoma (PRAD) tissue classification (Automated Gleason Grading Challenge 2022 (AGGC)101), pan-cancer tumor-immune lymphocyte detection (TCGA-TILS67), 32-class pan-cancer tissue classification (TCGA Uniform Tumor68), and others. For evaluation and comparisons, we perform logistic regression and K-nearest neighbors (KNN) on top of the pre-extracted features of each encoder, a common practice referred to as linear probing and KNN probing, which measure discriminative performance and the representation quality of pre-extracted features, respectively23. We evaluate all tasks using balanced accuracy, with PRAD tissue classification evaluated using weighted F1 score101. Additional details regarding ROI tasks, experimental setup and performance are provided in Methods and Supplementary Tables 39–60.
Across all 11 ROI-level tasks, UNI outperforms nearly all baselines on all tasks, with average performance increases of +18.8%, +7.58% and +5.75% on linear probing for ResNet-50, CTransPath and REMEDIS, respectively (Fig. 3a). On KNN probing, UNI similarly outperforms ResNet-50, CTransPath and REMEDIS with average performance increases of +15.6%, +8.6% and +9.4%. We find larger gains on challenging tasks such as PRAD tissue classification (in weighted F1 score, +0.131, P < 0.001; +0.020, P < 0.001; +0.027, P < 0.001) and esophageal carcinoma subtyping (+25.3%, P < 0.001; +10.1%, P < 0.001; +5.5%, P < 0.001) compared with the other three pretrained encoders, respectively. Figure 3b shows the UNI predictions on prostate cancer grading, in which a simple linear classifier trained with pre-extracted UNI features can achieve high agreement with pathologist annotations (Extended Data Fig. 2). On 32-class pan-cancer tissue classification (19 out of 32 of which are rare cancers), UNI achieves the highest overall balanced accuracy and AUROC of 65.7% and 0.975, respectively, outperforming the next best-performing model (REMEDIS) by +4.7% and +0.017 (both P < 0.001).
Fig. 3: ROI-level tasks.
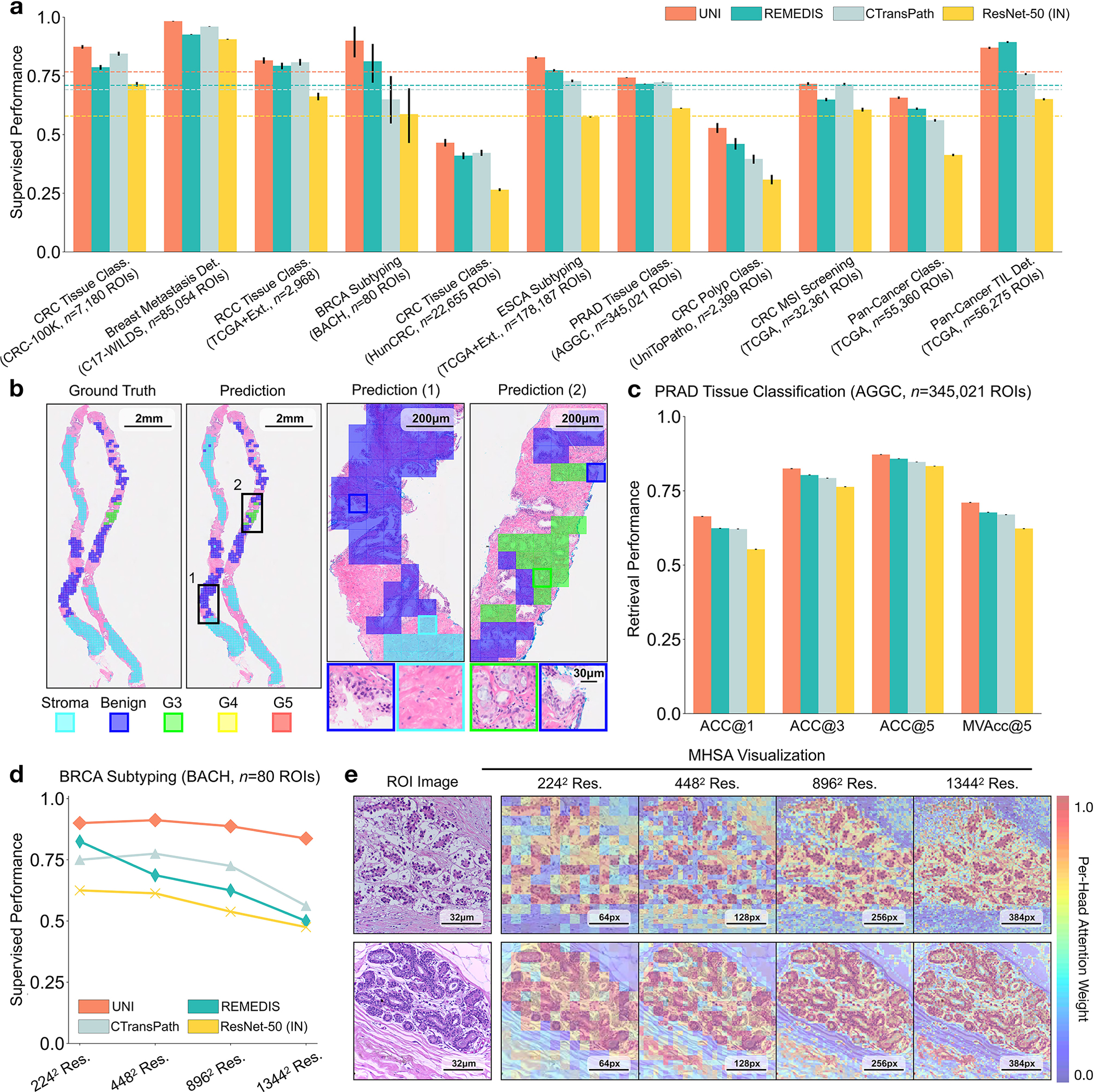
a, Supervised linear probe performance of UNI and its comparisons across 11 ROI-level classification tasks. All results are given as balanced accuracy except for PRAD tissue classification, which is given as weighted F1 score. Dashed lines represent the average performance of each model across all tasks. Error bars represent 95% confidence intervals and the centers correspond to computed values of each metric as specified above. Detailed results for all tasks are provided in Supplementary Tables 39–60. b, Examples of UNI on ROI classification for PRAD tissue classification in AGGC. Left: ground-truth ROI- level labels overlaid on the WSI. Right: predicted patch labels. ROIs are enlarged for better visualization, with further comparisons shown in Extended Data Fig. 2. c, ROI retrieval performance of UNI on PRAD tissue classification (AGGC, n = 345,021 ROIs). We report Recall@K for K ∈ {1, 3, 5} and the mean recall, with error bars representing 95% confidence intervals and the centers corresponding to computed values of each metric. d, Supervised KNN probe performance of UNI across various image resolutions (res., in pixels) in BRCA subtyping in BACH (n = 80 ROIs). Retrieval performance for all tasks is provided in Extended Data Fig. 3 and Supplementary Tables 63–68. e, Multi-head self-attention (MHSA) heatmap visualization of UNI across different image resolutions (in pixels) in BACH. Each colored square represents a 16 × 16 pixel patch token encoded by UNI, with heatmap color corresponding to the attention weight of that patch token to the global [CLS] (that is, classification) token of the penultimate layer in UNI. Top and bottom, respectively: visualizations for the invasive- and normal-labeled images, with further visualizations and interpretations provided in Extended Data Figs. 4–6. Scale bars: b, ground truth and prediction, 2 mm; prediction(1) and prediction(2), 200 μm; insets, 30 μm; e, ROI image, 32 μm; 2242, 64 pixels; 4482, 128 pixels; 8962, 256 pixels; 1,3442, 384 pixels.
We also compare UNI’s performance against that on the official leaderboards. For tumor-immune lymphocyte detection, compared with the best model in the ChampKit benchmark, which reports an AUROC of 0.974 and a false-negative rate (FNR) of 0.246, UNI has an AUROC of 0.978 and an FNR of 0.193 (without stain normalization) (Supplementary Table 61). For breast cancer metastasis detection (CAMELYON17-WILDS leaderboard), compared with the best model to date, which has accuracies of 95.2% and 96.5% on the out-of-domain validation and test sets, UNI reaches 97.4% and 98.3%, respectively (Supplementary Table 62). We note that many of these comparisons are end-to-end fine-tuned with transfer learning from natural images (and not from pathology). Although not equivalent in experimentation to UNI, these comparisons highlight the versatility of UNI given that out-of-the-box evaluation using linear classifiers is competitive with state-of-the-art techniques using end-to-end fine-tuning.
ROI retrieval
In addition to using representations in UNI to build task-specific classifiers, representations can also be used for image retrieval. Retrieval is similar to KNN in that we evaluate how well a query image can retrieve other images of the same class, given that visually similar images should be closer in representation space than visually distinct images. Different to KNN evaluation, we consider the accuracy of retrieval, that is, Acc@K for K ∈ {1, 3, 5}, in which the retrieval is successful if a correctly labeled image is within the top-K retrieved images, and MVAcc@5, which uses the majority vote of the top-5 retrieved images. We evaluate histology image retrieval on six ROI-level tasks (tasks with at least 5 classes). Additional details regarding ROI retrieval experimentation and performance are provided in Methods, Extended Data Fig. 3 and Supplementary Tables 63–68.
UNI outperforms other encoders on all tasks, demonstrating superior retrieval performance across diverse settings. On PRAD tissue classification (AGGC), UNI outperforms the next best-performing encoders (REMEDIS) by +4% and +3.3% on Acc@1 and MVAcc@5, respectively (both P < 0.001) (Fig. 2c). On colorectal cancer (CRC) tissue classification (CRC-100K), the gap between the top performing encoders is relatively smaller (by +3.1%, P < 0.001 and +0.01%, P = 0.188, respectively, compared with REMEDIS), presumably because the different tissue types have very distinct morphology, as shown by the relatively high classification performance in linear probing. On the more challenging 32-class pan-cancer tissue classification task, which contains many rare cancer types, UNI outperforms the second-best performing encoder (REMEDIS) by a large margin of +4.6% for Acc@1 and +4.1% for MVAcc@5 (both P < 0.001).
Robustness to high image resolution
Although visual recognition models are commonly evaluated on resized 224 × 224 pixel (2242 pixel) images, image resizing changes the microns per pixel (mpp) and may alter the interpretation of morphological features such as cellular atypia. We study how feature quality in UNI is affected at varying resolutions in breast invasive carcinoma (BRCA) subtyping (Grand Challenge on Breast Cancer Histology images, BACH) (2242 pixels at 2.88 mpp to 1,3442 pixels at 0.48 mpp) and CRC polyp classification (UniToPatho) (2242 pixels at 3.60 mpp to 1,7922 pixels at 0.45 mpp) with linear and KNN probing. Additional details regarding multiple resolution experimentation and performance are provided in Methods, Extended Data Fig. 4 and Supplementary Tables 45, 46, 51 and 52.
On both tasks we demonstrate the robustness of UNI to different image resolutions, as well as biases introduced into image resizing for high-resolution ROI tasks. When scaling the image resolutions used for evaluation, we observe that other encoders have worse performance degradation, with KNN performance decreases of −18.8% in CTransPath and −32.5% in REMEDIS on BRCA subtyping (2242 pixels versus 1,3442 pixels), compared with −6.3% in UNI. In CRC polyp classification, although other encoders do not have significant performance decreases (2242 pixels versus 1,7922 pixels), UNI increases by +5.1% via KNN probe. Figure 2e and Extended Data Figs. 5 and 6 show how UNI highlights finer-grained visual features when evaluating high-resolution images. In CRC polyp classification, resizing to 2242 pixels obscures important fine-grained details localizing the crypts that are otherwise detected at high resolution by UNI. These observations suggest that UNI can encode semantically meaningful representations agnostic to most image resolutions, which can be valuable in CPath tasks known to be optimal at different image magnifications.
ROI cell type segmentation
We assess UNI on the largest, public ROI-level segmentation dataset, SegPath102, a dataset for segmenting eight major cell types in tumor tissue: epithelial cells, smooth muscle cells, red blood cells, endothelial cells, leukocytes, lymphocytes, plasma cells, and myeloid cells. All pretrained encoders are fine-tuned end-to-end using Mask2Former103, a flexible framework commonly used for evaluating the off-the-shelf performance of pretrained encoders22,104. Given that the SegPath dataset divides the cell types into separate dense prediction tasks (eight tasks in total), each encoder is individually fine-tuned per cell type, with the dice score used as the primary evaluation metric. Additional details regarding segmentation tasks and performance are provided in Methods and Supplementary Table 69.
Although hierarchical vision backbones such as Swin transformers (CTransPath) and convolutional neural networks (CNNs; ResNet-50 and REMEDIS) have well-known advantages over vision transformers (UNI) for segmentation, we observe that UNI still outperforms all comparisons on a majority of cell types in SegPath. On individual segmentation tasks for the epithelial, smooth muscle and red blood cell types, UNI achieves dice scores of 0.827, 0.690 and 0.803, respectively, outperforming the next best-performing encoder (REMEDIS) by +0.003 (P = 0.164), +0.016 (P < 0.001) and +0.008 (P = 0.001), respectively. Across all eight cell types in SegPath, UNI achieves the overall performance with an average dice score of 0.721, outperforming ResNet-50 (0.696), CTransPath (0.695) and REMEDIS (0.716). Extended Data Fig. 7 shows segmentation visualizations for all cell types by UNI and other encoders, with all comparisons performing well in matching the ground truth segmentation. Overall, we find that UNI can outperform state-of-the-art CNNs and hierarchical vision models on segmentation tasks, extending its versatility in less conventional settings.
Few-shot ROI classification with class prototypes
Similar to slide-level classification, we also assess the label efficiency of UNI on ROI-level tasks. We evaluate all pretrained encoders using the nonparametric SimpleShot framework105, a strong baseline in the few-shot classification literature that proposes averaging extracted feature vectors of each class as the support examples in K = 1 nearest neighbors (or nearest centroid) classification106. These averaged feature vectors can also be viewed as ‘class prototypes’, a set of one-shot exemplars that are unique in representing semantic information such as class labels (for example, lung adenocarcinoma (LUAD) versus lung squamous cell carcinoma (LUSC) morphologies). At test time, unseen test examples are assigned the label of the nearest class prototype via Euclidean distance (Fig. 4a). For all pretrained encoders, we evaluate their pre-extracted features using SimpleShot with K ∈ {1, 2, 4, 8, ..., 256} training examples per class for a majority of tasks, with experiments repeated over 1,000 runs where C × K training examples are randomly sampled for each run. Additional details regarding few-shot ROI experimentation and performances are provided in Methods and Extended Data Fig. 8.
Fig. 4: Few-shot ROI- and slide-level prototyping.
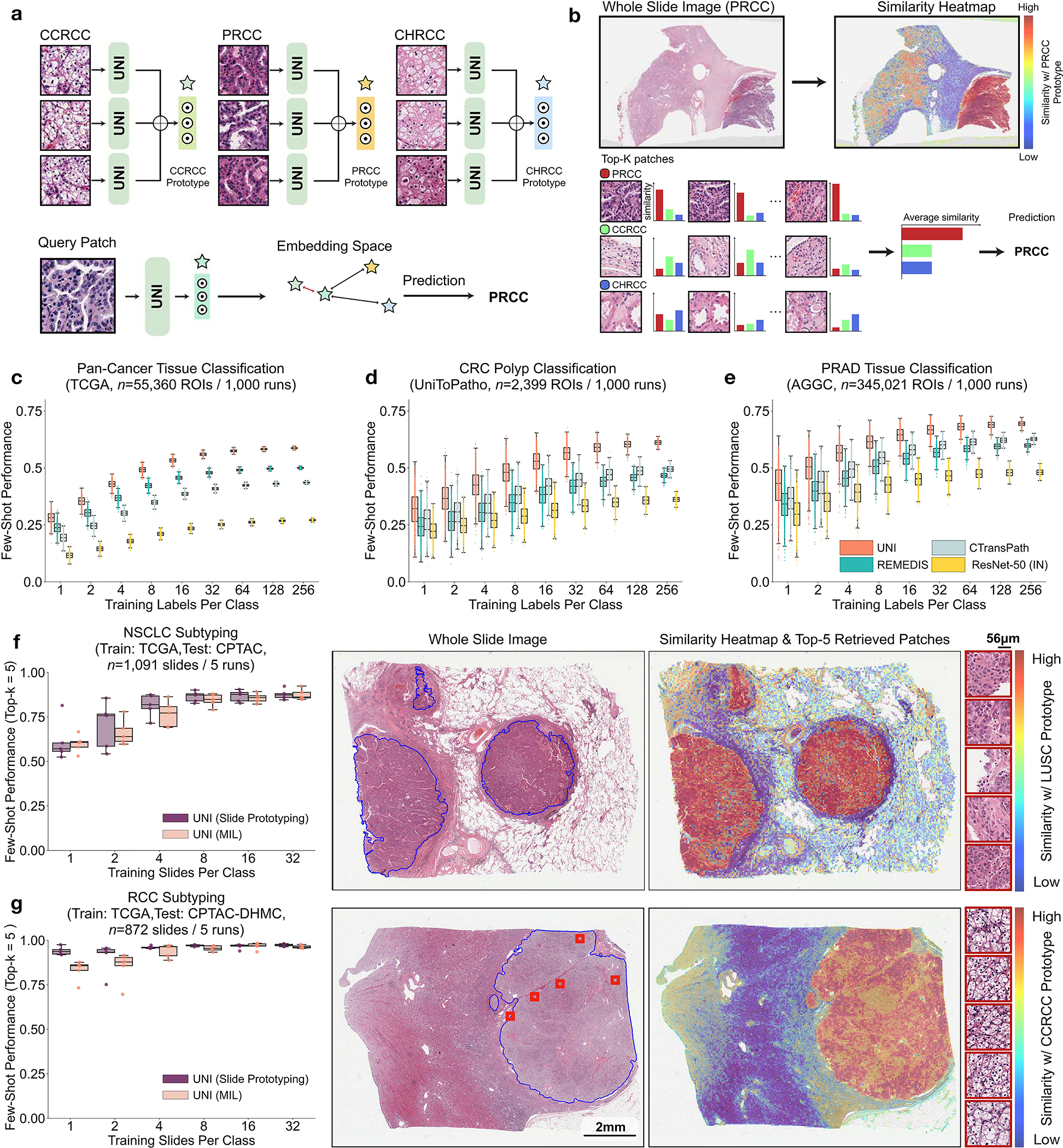
a, Prototypical few-shot ROI classification via SimpleShot. A class prototype is constructed by averaging the extracted features from ROIs of the same class. For a test ROI, SimpleShot assigns the class of the most similar class prototype (smallest Euclidean distance) as the predicted ROI label. b, Prototypical few-shot slide classification via MI- SimpleShot. Using a pre-computed set of ROI-level class prototypes (sharing the same class labels as the slide), MI-SimpleShot predicts the slide label using the class prototype with the highest average similarity of top-K patches queried from the WSI. The similarity heatmap visualizes the similarity between the ground- truth class prototype and each patch in the WSI. c–e, Few-shot ROI classification performance via SimpleShot on three tasks, with boxes indicating quartiles of model performance (n = 1,000 runs) and whiskers extending to data points within 1.5-fold the interquartile range. c, Pan-cancer tissue classification (TCGA, n = 55,360 ROIs). d, CRC polyp classification (UniToPatho, n = 2,399 ROIs). e, PRAD tissue classification (AGGC, n = 345,021 ROIs). Few-shot ROI performances for all tasks are provided in Extended Data Fig. 8. f,g, Few-shot slide classification performance and similarity heatmaps via MI-SimpleShot for NSCLC subtyping (train, TCGA; test, CPTAC; n = 1,091 slides) (f) and RCC subtyping (train, TCGA; test, CPTAC-DHMC; n = 872 slides) (g). In both tasks, using pre-extracted features from UNI, we compare MI-SimpleShot in the same few-shot settings as ABMIL (boxes indicate quartile values of model performance with n = 5 runs and whiskers extend to data points within 1.5-fold the interquartile range), and visualize similarity heatmaps and the top-5 similar patches (indicated in red bounding boxes) for a LUSC (f) and CCRCC (g) slide. Scale bars: WSI, 2 mm; top-5 retrieved patches, 56 μm. Further details, comparisons and visualizations are provided in Methods and Extended Data Figs. 8–10.
Across various tasks and evaluation settings, we find that UNI is a strong few-shot learner and is much more label efficient than other pretrained encoders. When comparing the median 8-shot performance of UNI with that of other encoders, UNI consistently exceeds the 128-shot and 256-shot performance of the next best-performing encoder on many tasks (Fig. 4c–e and Extended Data Fig. 8). We note that the variance in 1and 2-shot performances for all encoders can be high due to the choice of ROIs randomly selected as prototypes, potentially affected by H&E stain variability. However, given that the number of support examples increases in forming the class prototypes, we observe a monotonic decrease in variance of few-shot performance runs (0.32–1.59% standard deviation across tasks in UNI’s 256-shot performance), which demonstrates performance stability in permuting training examples to average as class prototypes in SimpleShot. Still, we observe that the lowest few-shot performance of UNI can sometimes exceed the maximum few-shot performance reported across 1,000 runs of other encoders. In pan-cancer tissue classification, the lowest-performing run for UNI in 2-shot, 8-shot and 32-shot evaluation outperforms the best possible run for ResNet-50, CTransPath and REMEDIS, respectively. These findings demonstrate the superior label efficiency and representation quality of UNI, given that averaging the extracted features from only a few ROIs can create effective class prototypes.
Prompt-based slide classification using class prototypes
Although weakly supervised learning via MIL has shifted slide-level classification such that ROI annotations are no longer required81, accessing and curating histology slide collections may still exist as barriers for clinical tasks that address rare and underrepresented diseases. From observing the strong retrieval performance and few-shot capabilities in UNI, we re-visit the problem of few-shot slide classification using class prototypes. Similar to textual prompting55, we used the class prototypes from SimpleShot also as ‘prompts’ for majority voting on the top-K retrieved patches (top-K pooling), which we call multiple instance SimpleShot (MI-SimpleShot) (Fig. 4b). We evaluate MI-SimpleShot on the same folds as trained ABMIL models in few-shot slide classification, with prototypes created using annotated ROIs (from training slides) from the pan-cancer tissue classification task68. We also compare MI-SimpleShot using other pretrained encoders, as well as the MIL baseline for UNI. We also develop similarity heatmaps that show the normalized Euclidean distances of all patches in a slide with respect to the class prototype of the ground-truth label, with pathologist annotations of tissue regions that match the slide label outlined in blue. Additional details regarding MI-SimpleShot experimentation and performance are provided in Methods, Extended Data Figs. 9 and 10 and Supplementary Tables 70 and 71.
Using only a few annotated ROI examples per class as prototypes, we demonstrate the potential of applying UNI with MI-SimpleShot as a simple but highly efficient system for slide-level disease subtyping and detection. On NSCLC and RCC subtyping (trained on TCGA and tested on external cohorts), MI-SimpleShot with top-5 pooling achieves better performance than ABMIL when using 1, 2 and 4 training slides per class for creating prototypes, and achieves similar performance to ABMIL when using more slides (Fig. 4f,g). Using similarity heatmaps, we also observe that retrieved patches of UNI (corresponding to the slide label) have strong agreement with pathologist annotations, as observed in the right-hand side of Fig. 4f,g for LUSC and clear cell renal cell carcinoma (CCRCC) slides. We believe that the effectiveness of MI-SimpleShot can be attributed to not requiring trainable parameters (ABMIL models may still over- and under-fit in few-shot settings) and the strong representation quality of UNI features for ROI retrieval. Although other pretrained encoders can be used for learning prototypes in MI-SimpleShot, UNI is potentially less sensitive to H&E staining variability. This is seen in the high standard deviation of one-shot performances for RCC subtyping (both in ABMIL in Extended Data Fig. 1 and MI-SimpleShot in Extended Data Fig. 9), with only one site used for learning a class prototype in MI-SimpleShot. This is also underscored in SimpleShot evaluation of breast metastasis detection (CAMELYON17-WILDS), given that CTransPath and REMEDIS have larger performance disparities than UNI between the two out-of-domain hospital test cohorts (accuracy differences of 12.3% and 12.8% versus 5.1%, respectively), alluding to the potential effects of H&E stain intensity skewing retrieval performance (Supplementary Table 42). In Extended Data Fig. 10 we observe instances of incorrect retrieval performance with respect to the predicted label and the pathologist annotations. Overall, our evaluation of UNI via MI-SimpleShot showcases how visual-centric foundation models with strong retrieval capabilities may enable applications in anatomic pathology.
Discussion
In this study, we demonstrate the versatility of UNI, a general-purpose, self-supervised model pretrained on one of the largest histology slide collections (for self-supervised learning) to date in CPath. We curated Mass-100K, a pretraining dataset containing more than 100 million tissue patches from 100,426 WSIs across 20 major organ types, including normal tissue, cancerous tissue and other pathologies. Using the DINOv2 self-supervised learning approach (demonstrated to scale to large datasets)22, we developed and validated a ViT-L (pretrained on Mass-100K) that consistently outperforms other histopathology image encoders. Depending on the task, although CTransPath and REMEDIS may achieve similar performances, our findings suggest that these encoders have limitations with regard to retrieval capabilities, label efficiency and potential biases to H&E staining intensity in out-of-domain evaluation.
As a visual-centric foundation model that may enable versatile clinical applications in CPath, several challenges emerged in developing UNI with regard to how factors such as model and data scaling would affect transfer performance. Although many empirical studies explore these components to achieve good generalization of natural images, many solutions may not be translatable due to differences between pathology and natural images. For example, although MoCoV3 has a lower but still competitive performance against DINOv2 on ImageNet, the same training configurations mirrored for developing a ViT-L on Mass-1K demonstrate large gaps in performance on OT-108. Following our study, we note several other studies that have recently emerged in training on larger histology slide datasets and collections107–109. Distinct from prior and recent works, our study is unique in providing unique insights into scaling laws and transfer learning capabilities of self-supervised models in CPath. Although model and data scale are important components for building visual-centric self-supervised learning, we find that the self-supervised learning (SSL) algorithm choice is the most impactful, with MoCoV3 (ViT-L on Mass-1K) under-performing not only against its DINOv2 counterpart, but also against CTransPath and REMEDIS. Increasing model scale (ViT-B to ViT-L) and data scale (Mass-1K and Mass-100K) does reflect performance increase, but note that performances of UNI ablations on OT-43 and OT-108 are relatively close and have consistent improvement over CTransPath and REMEDIS, which suggests that competitive pretrained encoders can still be developed with smaller models and less data. In tandem with the many clinical applications demonstrated by UNI, we believe that our testing of the aforementioned factors would guide CPath practitioners in developing their own foundation models using private in-house slide collections.
With regard to the wide range of clinical tasks to which UNI can be applied, compared with other encoders, we find that UNI excels in classifying rare and underrepresented diseases, such as the 90 out of 108 rare cancer types in the OT-108 benchmark, the 30 rare brain tumor diagnoses in the EBRAINS Digital Tumor Atlas, and the 19 out of 32 cancer subtypes in pan-cancer tissue classification sourced from TCGA. On these tasks and others, UNI demonstrates consistent and significant performance increases over the next best-performing encoder (REMEDIS or CTransPath). We hypothesize that UNI’s performance is attributed to the strong representation quality of the pre-extracted features, as seen in few-shot ROI and slide classification using class prototypes. In weakly supervised paradigms in which rare cancer types are infrequent and underrepresented in current slide datasets, MI-SimpleShot using UNI shows that annotating four slides per class can outperform task-specific MIL algorithms. Overall, we believe that UNI and other visual-centric foundation models that are being developed can be transformative in enabling creative clinical applications that would ordinarily require orders of magnitude more data.
On comparison with public leaderboards, we believe that UNI also presents an important shift from task-specific model development to generalist AI models110 in CPath. Beyond the 34 clinical tasks evaluated in this study, UNI evaluated out of the box is competitive when compared with published results of other works, outperforming leading models that are often trained end-to-end or that use carefully designed training recipes implemented for solving these specific public challenges. Altogether, our findings highlight the strength of having a better-pretrained encoder versus developing task-specific models that target narrow clinical problems, which we hope would shift research directions in CPath toward the development of generalist AI models that would have greater performance and flexibility in targeting diverse clinical applications in pathology. Following the conventional nomenclature of self-supervised models in computer vision22,75, labels such as ‘foundation model’ may create misleading expectations.
Our study has several limitations. Based on the ViT-L architecture, UNI lacks vision-specific biases for solving dense prediction tasks in CPath, and we note that performance increases for cell type segmentation in SegPath are not as drastic as observed in other tasks. We envision further improvement as better recipes emerge for adapting ViT architectures for segmentation tasks111. Our study also does not evaluate the best-performing ViT-Giant architecture in DINOv2, an even larger model that would likely translate well in CPath but demands more computational resources for pretraining. Although our study organizes the largest collection of clinical tasks for evaluating pretrained models in CPath (to our knowledge), other clinical tasks, such as those in cytopathology or hematopathology, are not represented in our analyses. Due to the breadth of our evaluation and small (or missing) validation sets for certain tasks, hyper-parameters were fixed, which follows other works in CPath25,37,40,112,113. Further hyper-parameter tuning and other training recipes may be likely to improve results further; however, our evaluation protocol was implemented for ranking the representation quality of pretrained encoder backbones. In developing UNI, although Mass-100K was developed intentionally to not overlap significantly with most public histology collections, biases such as data contamination and image acquisition shift should be further studied if the same model is re-used across many applications, especially if it were to have a disparate impact on diverse populations114. UNI is a unimodal model for CPath, meaning that multimodal capabilities such as cross-modal retrieval and visual question answering remain out of scope, which we explore in concurrent work115,116. Last, UNI is also only a ROI-level model for CPath, with the majority of clinical tasks in pathology performed at the slide or patient level. Future work will focus on using UNI as the building block for slide-level self-supervised models117 and general slide-level pathology AI development in anatomic pathology.
Methods
Large-scale visual pretraining
Mass General Brigham institutional review board approved the retrospective analysis of internal pathology images and corresponding reports used in this study. All internal digital data, including WSIs, pathology reports and electronic medical records were de-identified before computational analysis and model development. Patients were not directly involved or recruited for the study. Informed consent was waived for the retrospective analysis of archival pathology slides. In developing and evaluating self-supervised models in CPath, an important and relatively under-discussed challenge is the difficulty in developing large-scale models that can also be used for evaluation on public histology datasets. For natural images, ImageNet-1K is an integral dataset for the model development and evaluation lifecycle of self-supervised learning methods. Specifically, models are first pretrained on the training set of ImageNet-1K and then evaluated with fine-tuning and linear probe performance on the validation set (treated as the test set), reported as a community-accepted ‘goodness of fit’113,119, with further evaluation of generalization performance via other downstream tasks such as fine-grained classification and activity video recognition. Although such off-the-shelf self-supervised learning methods can readily be adapted to CPath, we note that there is considerably less public data for pretraining in CPath than natural images and that pretraining on large, public collections of histology slides also restricts their adaptability to public CPath benchmarks. Specifically, the development of many self-supervised pathology models has been limited to pretraining on TCGA35, one of the largest and most diverse public histology datasets for CPath, with many models opting to use the entire TCGA collection to realize data scaling benefits in self-supervised learning37,38,117. However, their applicability to public CPath benchmarks may be restricted to transductive inference37,40,41,44,46,57,117, given that many popular clinical tasks in CPath are also derived from TCGA (for example, pan-cancer analyses6,9,16,17,61,67–74) and thus extensive evaluation of out-of-domain, generalization performance is limited. Although datasets such as CAMELYON78,120 and PANDA18 can be used to evaluate TCGA-pretrained models, we note that these datasets are limited to single tissue types with limited disease categories.
Dataset curation for Mass-100K.
To overcome this limitation, we developed Mass-100K, a large-scale and diverse pretraining dataset consisting of in-house histology slides from MGH and BWH, and external histology slides from the GTEx consortium. Following natural image datasets, we also created three partitions of Mass-100K that vary in size to evaluate the data scaling laws, an empirical observation found in natural language and image foundation models that scaling of dataset size would also increase model performance21–23,75. Analogous to ImageNet-22K and ImageNet-1K, we developed the Mass-22K dataset, which contains 16,059,454 histology image patches sampled from 21,444 diagnostic formalin-fixed paraffin-embedded (FFPE) H&E WSIs across 20 major tissue types consisting mostly of cancer tissue, as well as its subset, Mass-1K (1,064,615 images, 1,404 WSIs). All histology slides in Mass-22K and Mass-1K were collected from BWH, and scanned using an Aperio GT450 scanner or a Hamamatsu S210 scanner. To make the image dataset sizes approximately equivalent to that of ImageNet-22K and ImageNet-1K, we sample approximately 800 image patches from histology tissue regions of each WSI, with image resolutions of 256 × 256 pixels at ×20 magnification. For slide preprocessing, we adapted the WSI preprocessing in the CLAM (clustering-constrained attention-based multiple-instance learning) toolbox15, which performs tissue segmentation at a low resolution via binary thresholding of the saturation channel in RGB → HSV color space; median blurring, morphological closing and filtering of contours below a minimum area to smooth tissue contours and remove artifacts; and patch coordinate extraction of non-overlapping 256 × 256 tissue patches in the segmented tissue regions of each WSI at ×20 magnification. The distribution of major tissue types in Mass-22K and Mass-1K are given in Supplementary Tables 2 and 3, respectively.
Inspired by even larger natural image datasets such as LVD-142M22 and JFT-300M30, we developed Mass-100K, which combines Mass-22K with further in-house FFPE H&E histology slide collections (including renal and cardiac transplant tissue) and GTEx76, which consists of 24,782 noncancerous, human autopsy WSIs. Additional in-house slides were collected from both BWH and MGH, and scanned using an Aperio GT450 scanner or a Hamamatsu S210 scanner. We purposefully excluded other public histology slide collections such as TCGA, CPTAC and PAIP for the external evaluation of UNI. Altogether, Mass-100K includes 100,426 histology slides, with the distribution of major tissue types given in Supplementary Table 1. Following the slide preprocessing protocol reported above, sampling approximately 800 histology tissue patches per WSI in Mass-100K yielded 75,832,905 images at 256 × 256 pixels at ×20 magnification. For high-resolution fine-tuning in DINOv2, we sampled an additional 24,297,995 images at 512 × 512 pixels at ×20 magnificatin, which altogether yielded 100,130,900 images for pretraining in Mass-100K.
Network architecture and pretraining protocol.
For large-scale visual pretraining on Mass-100K we used DINOv222, a state-of-the-art self-supervised learning method based on student–teacher knowledge distillation for pretraining large ViT architectures. DINOv2 is an extension of two previous methods, DINO25 and iBOT118, and uses two main loss objectives: self-distillation loss (that is, alignment loss in Fig. 1b) and masked image modeling loss (that is, reconstruction loss in Fig. 1b), to achieve state-of-the-art results in linear probe accuracy. DINOv2 also demonstrates capabilities in understanding the semantic layout of histopathology images when pretrained using knowledge distillation117. Self-distillation, introduced in BYOL27 for CNN pretraining and DINO25 for ViT pretraining, minimizes the predictive categorical distributions from the teacher (UNI Teacher in Fig. 1b) and student network (UNI in Fig. 1b) obtained from two augmented views of the same image by minimizing their cross-entropy loss. The teacher is updated as an exponential moving average of previous iterations of the student. Masked image modeling using an online tokenizer, introduced in iBOT118, involves strategically masking specific regions in an input image and training the model to predict the masked regions based on the remaining contextual information. This approach captures high-level visual features and context, inspired by masked language modeling in BERT121. Specifically, we denote two augmented views of an input image x as u and v, which are subsequently randomly masked. The masked images of u and v are represented as and , respectively. While u and v are propagated through the teacher network, the student network receives and as inputs. For the self-distillation objective, we compute cross-entropy loss between the [CLS] (that is, classification) token from the teacher network and the [CLS] token from the student network. For the masked image modeling objective, DINOv2 uses the output of the masked tokens from the student network to predict the patch tokens from the teacher network, where the teacher network can be regarded as an online tokenizer. We used DINOv2 because an important property for pretrained vision models in histopathology is linear probe performance, given that these models are often used as frozen feature extractors for pre-extracting patch features in weakly supervised slide-level tasks. Although other ViT-based self-supervised methods have demonstrated superior fine-tuning performance21,122, their linear probe performance is not comparable, and note that full fine-tuning in ROI-level and slide-level tasks is not always feasible due to cost in collecting annotations.
For smaller-scale visual pretraining on Mass-1K and Mass-22K we used iBOT, which has the same loss objectives introduced above for DINOv2. We note that iBOT and DINOv2 are overlapping methods that exist in the same family of ViT pretraining techniques, given that both methods extend the original DINO method (which introduced student–teacher knowledge distillation for ViTs), with iBOT extending DINO via the introduction of an online tokenizer component for masked image modeling, and DINOv2 extending iBOT via the introduction of additional modifications, thereby improving training stability and efficiency for larger ViT architectures. These six modifications can be summarized as follows: untying of the head weights between the above loss objectives instead of tying these objectives as performed in iBOT118; Sinkhorn–Knopp centering instead of teacher softmax-centering performed in iBOT118; KoLeo regularization to improve token diversity123; high-resolution fine-tuning toward the end of pretraining124; an improved code implementation that implements FlashAttention125, fully sharded data-parallel training and an efficient stochastic depth; and an improved pretraining recipe of the ViT-Large architecture on large-scale datasets. Last, although iBOT and DINOv2 use the same two loss objectives, the training recipes of these methods were developed for different data scales: iBOT was developed for ViT-Base and ViT-Large models on ImageNet-1K and ImageNet-22K, while DINOv2 was developed for ViT-Large and ViT-Giant models on LVD-142M, which is a dataset of 142 million curated natural images. To leverage the improved training recipe for ViT-Large on large-scale datasets in DINOv2 while also making comparisons fair to iBOT-trained ViT-Base models, we excluded the first two modifications of DINOv2 that modified the iBOT loss objective (untying of head weights and use of Sinkhorn–Knopp centering), as outlined in Supplementary Table 5. High-resolution fine-tuning was also conducted on the last 12,500 iterations of pretraining (out of 125,000 iterations in total).
Evaluation setting
Comparisons and baselines.
For slide- and ROI-level evaluation, we compare UNI against three pretrained encoders commonly used in the CPath community. For comparison to models with ImageNet Transfer, we compare against a ResNet-5084 pretrained on ImageNet28 (truncated after the third residual block, 8,543,296 parameters), which is a commonly used baseline in many slide-level tasks15,20. For comparison to the current state-of-the-art encoders, we compare against CTransPath37, which is a Swin transformer126 using the ‘tiny’ configuration with a window size of 14 (Swin-T/14, 28,289,038 parameters) pretrained mostly on the TCGA via MoCoV3 (ref. 24), and REMEDIS38, a ResNet-152 × 2 (232,230,016 parameters) initialized with the ‘Big Transfer’-medium protocol127 on ImageNet-22K and then pretrained with SimCLR26. Regarding data distributions, CTransPath was pretrained using 29,753 WSIs across 25 anatomic sites in TCGA (including both FFPE and frozen tissue slides) and 2,457 WSIs from PAIP85 across six anatomic sites, with 15,580,262 tissue patches and 32,120 WSIs used for pretraining altogether. REMEDIS was pretrained with a random sample of ~50 million patches from 29,018 WSIs also across 25 anatomic sites in TCGA. For self-supervised learning, CTransPath was trained using the MoCoV3 (ref. 24) algorithm for 100 epochs, with ~1.56 × 109 (or 1.56 billion) images seen during pretraining, and REMEDIS was trained using the SimCLR algorithm for a maximum of 1,000 epochs, with upwards of ~50 × 109 (or 50 billion) images seen during pretraining. In our implementation of these pretrained encoders, we use the truncated ResNet-50 implementation provided by CLAM15, and use the official model checkpoints for CTransPath and REMEDIS. The image embeddings outputted by these models are 1,024, 768 and 4,096, respectively. Similar to ResNet-50 and other ResNet models in which the penultimate feature layer before the classification head is a grid-like feature map of [1 × 7 × 7 × 4,096]-dimensions, we apply a two-dimensional (2D) adaptive average pooling layer to output a single [1 × 4,096]-dimensional image embedding. For all images used in ROI tasks and extracted patches for MIL in slide tasks, across all models, all feature extraction operations are performed on resized 224 × 224 images at ×20 magnification. We note that the Swin-T/14 architecture used by CTransPath has constraints in which it can take only image dimensions in which the length is divisible by 224. We also note that although CTransPath was pretrained on ×10 magnification, it demonstrates state-of-the-art performance at ×20 magnification55,59,128. All pretrained encoders use ImageNet mean and standard deviation parameters for image normalization (including UNI). To compare against transfer learning from a general pathology task, we also trained a ViT-L/16 architecture (initialized with ImageNet-22K transfer) end-to-end on the 32-class pan-cancer tissue classification task in TCGA. In several benchmarking tasks, we note that this ablation study performed worse than UNI, even on in-domain tasks such as pan-cancer tumor-immune lymphocyte detection in TCGA (Supplementary Table 72).
Last, we note that although many slide and ROI tasks are created using annotated data from the TCGA, CTransPath and REMEDIS were also trained using almost all slides in the TCGA, which can result in information leakage that inflates the performance of these models on TCGA benchmarks. When possible, we report evaluation on external cohorts outside of TCGA for all tasks. This may not be possible for all tasks, given that the official train–validation–test folds may all be developed using TCGA.
Weakly supervised slide classification.
Training and evaluation for weakly supervised slide classification tasks follow the conventional two-stage MIL paradigm consisting of pre-extraction of ROI-level features as instances from non-overlapping tissue patches of segmented tissue regions of the WSI, and the learning of a trainable permutation-invariant pooling operator that aggregates patch-level (or instance) features into a single slide-level (or bag) feature. For slide preprocessing, we use the same WSI preprocessing pipeline as described in the dataset curation section, which uses the CLAM toolbox15, with additional patch feature extraction using a pretrained encoder performed on the patched coordinates. Images are resized down to 224 × 224 pixels and normalized using ImageNet mean and standard deviation parameters. As a quality control, we performed the additional following steps: first, for slides with under- or over-segmented tissue masks, we adjusted the segmentation parameters in CLAM (threshold value and downsample level) to segment only tissue regions; second, we removed slides that were non-H&E and nonFFPE; and third, for slides that did not have a downsample level equivalent to ×20 magnification in their WSI pyramidal format, we patched the tissue into non-overlapping 512 × 512 pixel tissue patches at ×40 magnification and then later resized these images to 224 × 224 pixels during feature extraction. Pre-extracted features for all pretrained encoders used the same set of patch coordinates for feature extraction of each WSI.
For comparison of pre-extracted features of pretrained encoders in weakly supervised learning, we used the ABMIL algorithm83 across all tasks, which is a canonical weakly supervised baseline in slide classification tasks. We use the two-layer gated variant of the ABMIL architecture with all input embeddings mapped to an embedding dimension of 512 in the first fully connected layer, followed by hidden dimensions of 384 in the following intermediate layers. For regularization, we use dropout with P = 0.10 applied to the input embeddings and P = 0.25 after each intermediate layer in the network. Aside from the first fully connected layer, which is dependent on the embedding dimension of the pre-extracted features, all comparisons used the same ABMIL model configuration. We trained all ABMIL models using the AdamW optimizer129 with a cosine learning rate scheduler, a learning rate of 1 × 10−4, cross-entropy loss, and a maximum of 20 epochs. We additionally performed early stopping on the validation loss if a validation fold was available. For all slide classification tasks, we case-stratified and label-stratified the slide dataset into train–validation–test folds, or used official folds if available. Given that CTransPath and REMEDIS were pretrained using all slides in TCGA, we considered TCGA slide tasks in which additional external evaluation was possible (for example, NSCLC subtyping was included due to availability of LUAD and LUSC slides in CPTAC, whereas BRCA subtyping was excluded). For glioma IDH1 mutation prediction and histomolecular subtyping, train–validation–test folds were additionally site-stratified to mitigate potential batch effects.
Linear and K-nearest neighbors probe evaluation in ROI classification.
For ROI-level classification tasks, we follow previous works that use logistic regression (linear) probing and KNN probing130 to evaluate, respectively, discriminative transfer performance and the representation quality of pre-extracted feature embeddings on downstream tasks23. For linear probing, following the practice recommended by the self-supervised learning community, we fix the l2 regularization coefficient λ to 100/MC, where M is the embedding dimension and C is the number of classes, and use the L-BFGS solver131 with a maximum of 1,000 iterations. KNN probing is an additional evaluation technique advocated by the self-supervision community for measuring representation quality of pre-extracted features25,132,133. In comparison with linear probing, KNN probing is nonparametric (aside from the choice of K), given that it classifies unseen test examples based on only their feature similarity to labeled training examples (for example, similar examples in representation space should also be visually similar and share the same class label). We use the KNN implementation from Scikit-Learn134, trained using K = 20 and Euclidean distance as the distance metric, following observed stability of this evaluation setup of other self-supervision works25. For all ROI tasks, we approximately case-stratified and label-stratified datasets into train–test folds or used official folds if available.
For all tasks, we resize images to 224 × 224 pixels (or 448 × 448 pixels if available) and normalize using ImageNet mean and standard deviation parameters. Additionally, we note that many ROI datasets consist of images with high image resolutions, with image resizing to a fixed 224 × 224 pixels or 448 × 448 pixels resolution also changing the image magnification and mpp. For example, resizing ROIs in the CRC polyp classification task in UniToPatho (ROIs having an original image resolution of 1,812 × 1,812 pixels at 0.45 mpp) to 224 × 224 pixels would change the magnification to 3.6 mpp. For CRC polyp classification as well as BRCA subtyping (BACH), we carry out evaluations using resized image resolutions of {2242 pixels, 4482 pixels, 8962 pixels, 1,7922 pixels} and {2242 pixels, 4482 pixels, 8962 pixels, 1,3442 pixels}, with multiples of 224 chosen due to constraints with CTransPath. To pre-extract features from high-resolution images, for ViTs such as the plain ViT-large architecture in UNI and the hierarchical Swin transformer-T architecture in CTransPath, the forward passes of these architectures are not modified, and interpolation of positional embeddings is performed to have the same sequence length as patch tokens in the ROI. To illustrate, in the patch embedding layer of our ViT-Large architecture in UNI that has a patch token size of 16 × 16, a 224 × 224 pixel image would be converted into a [14 × 14 × D]-dimension 2D grid of patch embeddings using a 2D convolutional layer (kernel and stride size of 16, three incoming channels from RGB-input image inputs and D-dimension outgoing channels set as a hyper-parameter for feature embedding length), followed by flattening and transposing (now a [196 × D]-dimension sequence of patch embeddings), which can now be used in transformer attention (called ‘patchifying’). For a 1,792 × 1,792 pixel image in CRC polyp classification, patchifying this image using the same patch embedding layer would result in a [112 × 112 × D] → [12,544 × D]-dimension sequence of patch embeddings. Feeding this sequence into the forward pass of transformer attention, although computationally expensive, is still tractable via memory-efficient implementations such as FlashAttention or MemEffAttention. For positional embedding interpolation, we used the implementation provided in DINO25. For multi-head self-attention (MHSA) visualization, we visualize the weights from the last attention layer using the notebook implementation provided by the HIPT codebase117, which we note is applicable only for plain VIT architectures.
ROI retrieval.
To assess the quality of embeddings produced by different encoders for content-based image retrieval of histopathology images, we use ROI-level classification datasets, in which the goal is to retrieve similar images (that is, images with the same class label) to a given query image. For each benchmark, we first embed all images into a low-dimensional feature representation using the pretrained encoders. We treat each image in the test set as a query. Each query image is compared with each image from the ROI-level classification training set, which serves as a database of candidates (keys). Note that no supervised learning takes place in these experiments and the class labels are used only for evaluation purposes (that is, to assess whether retrieved images share the same class label as the query). We first center the database of keys by subtracting their Euclidean centroid from each embedding followed by l2 normalization of each key to unit length. For each new query, we apply the same shift and normalization steps and then measure it against each key in the database via the l2 distance metric, where lower distance is interpreted as higher similarity. The retrieved images are sorted by their similarity scores and their corresponding class labels are used to evaluate the success of a given retrieval using Acc@K for K ∈ 1, 3, 5 and MVAcc@5, which are described in Evaluation metrics.
ROI-level cell type segmentation.
For training and evaluation of ROI-level cell type segmentation tasks, we follow previous works in using Mask2Former, which is a flexible framework commonly used for evaluating off-the-shelf performance of pretrained vision encoders103. In the case of the ViT architecture, which is nonhierarchical, we additionally use the ViT-Adapter framework alongside the Mask2Former head111. For both ViT-Adapter and Mask2Former, we use the same hyper-parameters used for ADE20k semantic segmentation. Specifically, we use the AdamW129 optimizer along with a step learning rate schedule. The initial learning rate was set to 0.0001 and a weight decay of 0.05 was applied. To adjust the learning rate specifically for the backbone, we apply a learning rate multiplier of 0.1. Additionally, we decay the learning rate by a factor of 10 at 0.9 and 0.95 fractions of the total number of training steps. For all backbones, we fine-tune the full model for 50 epochs with a batch size of 16. The model’s performance on the validation set is evaluated every 5 epochs, and the optimal model based on validation performance is saved for testing. To augment the data, we use the large-scale jittering (LSJ) augmentation135, with a random scale sampled from a range of 0.5–2.0, followed by a fixed size crop to 896 × 896 pixels to accommodate the size constraints of CTransPath. At inference time, we resize the image dimensions to their nearest multiples of 224.
Few-shot ROI classification and prototype learning.
For few-shot classification, we follow previous works using the SimpleShot framework to evaluate the few-shot learning performance of prototypical representations of self-supervised models105,136. Prototypical (or prototype) learning is a longstanding task in the few-shot learning community106,137,138,and it has also been posed (in many related forms) in CPath as well43,139–142. In contrast with traditional few-shot learners based on meta-learning, SimpleShot and related works demonstrate that strong feature representations combined with specific transformations and simple classifiers can reach state-of-the-art performance on few-shot tasks105,136,143. SimpleShot is similar to nearest neighbors classification, in which the training set (called ‘supports’ in few-shot learning literature) is drawn from C classes (‘ways’) with K examples per class (‘shots’) for predicting unseen images in the test set (‘queries’). Instead of nearest neighbors, SimpleShot uses a nearest-centroid approach based on ProtoNet106, in which the average feature vector (centroid) for each class is used as a prototypical ‘one-shot’ example for labeling the query set via distance similarity. As noted, these averaged feature vectors can also be viewed as ‘class prototypes’, a set of one-shot representative examples that are unique in representing semantic information such as class labels (for example, LUAD versus LUSC morphologies). Given that SimpleShot is a simple and surprisingly strong baseline in the few-shot learning community and popularized in evaluating self-supervised models136, we adopt this baseline in evaluating UNI and its comparisons in few-shot ROI classification tasks. We follow the recommendations in SimpleShot that suggest centering (subtracting the mean computed on the support set) and l2 normalizing the support set before computing the class prototypes, with the query set also transformed (also centered using the mean of the support set) before nearest centroids classification.
Conventional few-shot learners on natural image classification tasks are evaluated by drawing 10,000 C-way, K-shot episodes from the training set with 15 query images per class as the test set. For equivalent comparison with metrics in linear and KNN probing, we instead draw 1,000 C-way, K-shot episodes but use all images in the test set per episode. Due to the relatively larger number of training examples available in ROI tasks than that of slide tasks, we vary the number of labeled examples per class from K ∈ {1, 2, 4, 8, 16, 32, ...256} or the maximum number of labeled examples available for a given class. To compare with linear and KNN probing that use all training examples, we also evaluate SimpleShot by averaging all training examples per class, which we denote as ‘1-NN’ in Supplementary Tables 40–60.
Prompt-based slide classification using multiple instance SimpleShot.
To evaluate the quality of extracted representations serving as the class prototype for slide classification tasks, we adapt class prototypes from SimpleShot (described above) as ‘prompts’ (similar to the use of textual prompts in zero-shot classification55), which we describe as MI-SimpleShot. As described in the main text, we use two slide-level datasets (NSCLC and RCC subtyping datasets), which have matching ROI training examples from datasets that can be used as the support set. In brief, we use the annotated LUAD and LUSC ROIs from the TCGA Uniform Tumor dataset for NSCLC subtyping, and annotated CCRCC, papillary renal cell carcinoma (PRCC) and chromophobe renal cell carcinoma (CHRCC) ROIs from the TCGA Uniform Tumor dataset for RCC subtyping. The TCGA Uniform Tumor dataset (described further in Methods) consists of 271,170 256 × 256 pixel ROIs at around 0.5 mpp of 32 cancer types annotated and extracted from 8,736 H&E FFPE diagnostic histopathology WSIs. We note that the number of annotated ROIs per slide ranges from 10 to 70 examples in the TCGA-LUAD, -LUSC, -CCRCC, -PRCC and -CHRCC cohorts. For each class, we first embed ROIs in the support set into a low-dimensional feature representation using the pretrained encoders, followed by average pooling of all ROI features in the class. The average-pooled feature representations are considered as the class prototypes, which are used as prompts for labeling the top-K ROIs for each slide in the query set via normalized Euclidean distance similarity. The slide-level prediction is then made by majority voting of the top-K ROI predictions. For each benchmark, we evaluate MI-SimpleShot with both top-5 average pooling and top50 average pooling and on {1, 2, 4, 8, 16, 32} training slides per class, similar to our evaluation in few-shot slide classification using the same five folds as the trained ABMIL models, with prototypes created from the annotated ROIs in the same training slides. We note little performance change in considering the average scores of the top-5 and top50 patches per class prototype. To compare with the performance that uses all training slides with ROI annotations, we also evaluate MI-SimpleShot by averaging all training ROI feature representations per class, with results detailed in Supplementary Tables 70 and 71. To create similarity heatmaps, we visualize the normalized Euclidean distances of all patches in a slide with respect to the ground-truth class prototype.
Evaluation metrics.
We report balanced accuracy, weighted F1 score, and AUROC for classification tasks. Balanced accuracy is computed by taking the unweighted average of the recall of each class, which takes into account class imbalance in the evaluation set. Weighted F1 score is computed by averaging the F1 score (the harmonic mean of precision and recall) of each class, weighted by the size of its respective support set. AUROC is the area under the receiver operating characteristic curve plotting true-positive rate against the false-positive rate as the classification threshold is varied. Additionally, we compute quadratic weighted Cohen’s κ (inter-annotator agreement between two sets of labels, for example, ground truth and predictions) which we perform for ISUP grading (PANDA), and top-K accuracy for K ∈ {1, 3, 5} (for a given test sample, a sample is scored correctly if the ground-truth label is among the top-K labels predicted) for OT-43 and OT-108. For retrieval, we consider Acc@K for K ∈ {1, 3, 5}, which represent the standard top-K accuracy scores in retrieving images with the same class label as the query. Specifically, a retrieval is considered successful if at least one image among the top-K retrieved images has the same class label as the query. We also report MVAcc@5, which, compared with Acc@5, more strictly requires that the majority vote of the top-5 retrieved images be in the same class as the query for retrieval to be considered successful. For segmentation, we report the Dice score (same definition as the F1 score), the precision and recall, macro averaged across all images and classes.
Statistical analysis.
For all semi- and fully supervised experiments, we estimate 95% confidence intervals for the model performance with nonparametric bootstrapping using 1,000 bootstrap replicates. For statistical significance, we use a two-sided paired permutation test with 1,000 permutations to assess observed differences in the performance of the two models. For all few-shot settings, we report results using box plots that indicate quartile values of model performance (n = 5 runs) with whiskers extending to data points within 1.5-fold the interquartile range. For ROI-level few-shot classification, for each C-way, K-shot setting, we randomly sample K training examples per C classes with 1,000 repeated experiments (called ‘episodes’ or ‘runs’) evaluated on the entire test set. For slide-level few-shot classification, we follow the same setting as above but with the number of runs limited to 5 due to small support sizes in rare disease categories.
Tasks, datasets and comparisons to leaderboard
In this section we outline the data preprocessing, number of samples per class, train–validation–test folds and other details per dataset (which may also span multiple tasks). We also add context and comparisons of our results to existing leaderboards and baselines of other studies when possible, and note that comparisons may not always be equivalent due to differences in hyper-parameters, splits and pre-extracted features (many existing baselines may not use histopathology-specific pretrained encoders). In comparing against leaderboards and in comparisons, we adopt the metrics used in public evaluation, elaborated further in the table captions.
OncoTree cancer classification based on in-house BWH data (43 cancer types, 108 OncoTree codes).
As described in the main text, OncoTree cancer classification is a large-scale hierarchical classification task for CPath that follows the OncoTree (OT) cancer classification system77. This task was devised to assess the generalization capabilities of pretrained models in classifying diverse disease categories and tissue types. Using in-house BWH slides, we defined a dataset consisting of 5,564 WSIs from 43 cancer types further subdivided into 108 OncoTree codes, with at least 20 WSIs per OncoTree code. The dataset forms the basis of two tasks that vary in diagnostic difficulty: 43-class cancer type classification (OT-43) and 108-class OncoTree code classification (OT-108). Due to the small support sizes for several OncoTree codes in OT-108, all ABMIL models were trained using train–test folds and without early stopping. For training and evaluation, we approximately label-stratified the dataset into 71:29 train–test folds (a ratio of 3,944:1,620 slides) using the same folds for OT-43 and OT-108, with 15 slides used per OncoTree code in the test set and a minimum of 5 slides used per OncoTree code in the training set. The hierarchical classification of the coarseand fine-grained task is reported in Supplementary Table 4. Except for bladder urothelial carcinoma (BLCA), invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC), colon adenocarcinoma (COAD), rectum adenocarcinoma (READ), uterine endometrioid carcinoma (UEC), stomach adenocarcinoma (STAD), head and neck squamous cell carcinoma (HNSC), diffuse large B cell lymphoma not otherwise specified (DLBCLNOS), melanoma (MEL), LUAD, LUSC, pancreatic adenocarcinoma (PAAD), PRAD, cutaneous squamous cell carcinoma (CSCC), small-cell lung cancer (SCLC), adenocarcinoma of the gastroesophageal junction (GEJ) and chronic lymphocytic leukemia/small lymphocytic lymphoma (CLLSLL), cancer types in this task are rare cancers designated by the RARECARE project82 and the National Cancer Institute’s Surveillance, Epidemiology and End Results (NCI-SEER) Program. We note that slides in the training fold of OT-43 and OT-108 were included in OP-1K and OP-22K pretraining, with the test set held out from these pretraining sources (following practices in ImageNet).
Due to storage limitations in repeatedly extracting features for all non-overlapping tissue patches per WSI for all pretrained models (including intermediate checkpoints), we sampled 200 representative patches per WSI for feature extraction. To select these patches, we first extracted ResNet-50IN features, followed by clustering144, used previously in other works such as WSISA145, DeepAttnMISL146,147, and others148. We note that these works are inspired by visual bag-of-words (vBOW)149,150, which has been adapted to pathology for formulating high-resolution ROIs and WSIs as smaller but representative collections of tissue patches via clustering applied to deep features151,152, with downstream applications such as MIL145–148 and retrieval69,153. For all pretrained encoders, we extract features from the same sampled collection of patches. Although additional computational steps were taken to derive these sampled patches, we note that this does not fall under transductive inference, given that the entire test set (all WSI samples) is never made visible to any learning component (clustering is fitted per WSI, with ‘samples’ defined at the slide level instead of the patch level). To validate this approach as having comparable performance using features for all tissue patches per WSI, we compare the performance of sampled versus full features of UNI, CTransPath, REMEDIS and ResNet-50IN, which we also report in Supplementary Tables 12 and 15. We observe not only marginal performance decrease when using sampled features (maximum decrease of −0.9% in top-1 accuracy, −0.007 in AUROC), but also performance increases for many models. For REMEDIS we observe that the performance of ABMIL models collapses when using full features, with top-1 accuracy performances of 4.0% and 11.8%, respectively, on OT-43 and OT-108 (compared with 59.3% and 41.2%, respectively, with sampled features). We hypothesize that these performance increases are due to the difficult nature of OT-43 and OT-108, with patch sampling reducing the input data complexity for ABMIL (for example, instead of finding diagnostically relevant features in a bag of 10,000+ patches, only 200 representative patches are considered).
Breast metastasis detection based on CAMELYON16 (2 classes).
The breast metastasis detection task from the Cancer Metastases in Lymph Nodes Challenge 2016 (CAMELYON16) consists of 400 H&E FFPE histopathology WSIs of sentinel lymph node from Radboud University Medical Center and the University Medical Center Utrecht for metastasis detection78. We removed one mislabeled slide from the test set, resulting in 399 slides (239 normal, 160 metastasis). For training and evaluation we used the official train–test folds and label-stratified the training set into 90:10 train–validation, resulting in 61:7:32 train–validation–test folds (243:27:129 slides). In addition to internal comparisons, we also compare our results with the leaderboard taken at the time of the challenge, provide a chronological timeline of best-performing models reported in recent peer-reviewed literature, and add context to the comparison of state-of-the-art methods in Supplementary Table 36. We note that comparisons with UNI may not be equivalent, with many proposed methods using ResNet-50IN features and also more sophisticated MIL architectures.
NSCLC subtyping based on TCGA and CPTAC (LUAD versus LUSC, 2 classes).
The NSCLC subtyping task consists of NSCLC H&E FFPE diagnostic histopathology WSIs sourced from TCGA and CPTAC for classifying two subtypes: primary LUAD and LUSC cases79,154,155. For quality control, in TCGA we excluded slides with missing or incorrect metadata, which resulted in 1,041 slides (529 LUAD and 512 LUSC). In CPTAC we excluded slides that were frozen tissue, nontumor tissue or were not labeled as having acceptable tumor segments, which resulted in 1,091 slides (578 LUAD and 513 LUSC). For training and evaluation, we label-stratified the TCGA-NSCLC cohort into 80:10:10 train–validation–test folds (848:97:98 slides), with external evaluation using the held-out CPTAC cohort.
RCC subtyping based on DHMC (CCRCC versus PRCC versus CHRCC versus ROCY versus Benign, 5 classes).
The RCC subtyping task consists of 563 RCC H&E FFPE diagnostic histopathology WSIs (485 resections and 78 biopsies) from the Dartmouth-Hitchcock Medical Center (DHMC) for classifying five subtypes: primary CCRCC, 344 slides), PRCC (101 slides) and CHRCC (23 slides), renal oncocytomas (ROCY, 66 slides) and benign cases (29 slides)156. For training and evaluation of both tasks, we used a modified configuration of the train–validation–test folds with a 70:4:26 ratio (393:23:147 slides), with eight CHRCC cases moved from the test to the train fold due to CHRCC being absent in the train fold.
RCC subtyping based on TCGA, DHMC and CPTAC (CCRCC versus PRCC versus CHRCC, 3 classes).
The RCC subtyping task consists of 1,794 RCC H&E FFPE diagnostic histopathology WSIs from TCGA, DHMC and CPTAC for classifying three subtypes: primary CCRCC, PRCC and CHRCC156–160. For quality control, in TCGA we excluded slides with missing low-resolution downsamples, which resulted in 922 slides (519 CCRCC, 294 PRCC and 109 CHRCC). In the DHMC set we filtered out oncocytomas in the previously described DHMC-Kidney cohort, which resulted in 468 slides (344 CCRCC, 101 PRCC and 23 CHRCC). In CPTAC we excluded slides that were frozen tissue, nontumor tissue or were not labeled as having acceptable tumor segments, which resulted in 404 slides (404 CCRCC). For training and evaluation, we label-stratified the TCGA-NSCLC cohort into 80:10:10 train–validation–test folds (736:89:97 slides), with external evaluation on the held-out DHMC and CPTAC cohorts. Given that CPTAC includes only CCRCC cases, we combined DHMC and CPTAC into a single evaluation cohort.
CRC screening based on HunCRC (4 classes).
The CRC screening task consists of 200 H&E FFPE diagnostic histopathology WSIs of colorectal biopsies from the Hungarian Colorectal Cancer Screening (HunCRC) dataset from Semmelweis University99. In this dataset we defined a 4-way coarse-grained subtyping task using the categories of negative (10 slides), non-neoplastic lesion (38 slides), CRC (46 slides), and adenoma (106 slides), in which the ground-truth label was set by the study’s pathologist. For training and evaluation we label-stratified the HunCRC slide dataset into 50:25:25 train–validation–test folds (158:21:21 slides).
BRCA coarseand fine-grained subtyping based on BRACS (3 and 7 classes).
The BRCA coarseand fine-grained subtyping tasks consist of 547 breast carcinoma H&E slides from 187 patients sourced from the Breast Carcinoma Subtyping (BRCA) task sourced from IRCCS Fondazione Pascale, The Institute for High-Performance Computing and Networking (ICAR) of the National Research Council (CNR), and IBM Research-Zurich161. In this dataset we defined a 3-way coarse-grained subtyping task using the ‘benign tumor’, ‘atypical tumor’ and ‘malignant tumor’ labels. Furthermore, we define a 7-way fine-grained subtyping task that subtypes benign tumors as ‘normal’, ‘pathological benign’, ‘usual ductal hyperplasia’, atypical tumors as ‘flat epithelial atypia’ and ‘atypical ductal hyperplasia’, and malignant tumors as ‘ductal carcinoma in situ’ and ‘invasive carcinoma’. The hierarchical classification of the coarseand fine-grained tasks is reported in Supplementary Table 19. For training and evaluation of both tasks, we used the official train–validation–test folds with a 72:12:16 ratio (395:65:87 slides), using the same folds for both coarseand fine-grained tasks.
Glioma IDH1 mutation prediction and histomolecular subtyping based on TCGA and EBRAINS (2 and 5 classes).
The glioma IDH1 mutation prediction and histomolecular subtyping task consists of 1,996 H&E FFPE diagnostic histopathology WSIs from cases of glioblastoma, astrocytoma and oligodendroglioma with molecular status from the TCGA and the EBRAINS Digital Tumor Atlas87–89. We first defined a 5-way glioma histomolecular subtyping task with the following labels: IDH1-mutant astrocytomas (257 slides), IDH1-mutant glioblastomas (93 slides), IDH1-mutant and 1p/19q codeleted oligodendrogliomas (408 slides), IDH1-wild-type glioblastomas (1,094 slides), and IDH1-wild-type astrocytomas (144 slides). Additionally, we defined a simpler 2-way task that predicts only IDH1 status: IDH1-wild-type (1,238 slides) and IDH1-mutant (756 slides). All brain tumors in these tasks are designated as rare cancers by the RARECARE project and the NCI-SEER program. The hierarchical classification of the coarseand fine-grained tasks is reported in Supplementary Table 21. For training and evaluation of both tasks, we approximately label-stratified the TCGA-GBMLGG (TCGA Glioblastoma lower-grade glioma) dataset into a train–validation–test fold with a 47:22:31 ratio (525:243:355 slides), with external evaluation using the held-out EBRAINS cohort (873 slides), using the same folds for both coarseand fine-grained tasks.
Brain tumor coarseand fine-grained subtyping based on EBRAINS (12 and 30 classes).
The brain tumor coarseand fine-grained subtyping tasks consists of 2,319 H&E FFPE diagnostic histopathology WSIs from the EBRAINS Digital Tumor Atlas sourced from the University of Vienna89. With an original dataset size of 3,114 slides, we defined a 30-way fine-grained brain tumor subtyping task limited to diagnostic labels that have at least 30 slides: IDH1-wildtype glioblastoma (474 slides), pilocytic astrocytoma (173 slides), meningothelial meningioma (104 slides), pituitary adenoma (99 slides), IDH1-mutant and 1p/19q codeleted anaplastic oligodendroglioma (91 slides), ganglioglioma (88 slides), hemangioblastoma (88 slides), adamantinomatous craniopharyngioma (85 slides), IDH1-mutant and 1p/19q codeleted oligodendroglioma (85 slides), atypical meningioma (83 slides), schwannoma (81 slides), IDH1-mutant diffuse astrocytoma (70 slides), transitional meningioma (68 slides), diffuse large B cell lymphoma of the central nervous system (CNS) (59 slides), gliosarcoma (59 slides), fibrous meningioma (57 slides), anaplastic ependymoma (50 slides), IDH1-wild-type anaplastic astrocytoma (47 slides), metastatic tumors (47 slides), DH1-mutant anaplastic astrocytoma (47 slides), ependymoma (46 slides), anaplastic meningioma (46 slides), secretory meningioma (41 slides), lipoma (38 slides), hemangiopericytoma (34 slides), IDH1-mutant glioblastoma (34 slides), non-WNT/ Non-SHH medulloblastoma (32 slides), Langerhans cell histiocytosis (32 slides), angiomatous meningioma (31 slides), and hemangioma (30 slides). From the same 2,319 slide dataset in the fine-grained task, we also defined a 12-way coarse-grained brain tumor subtyping task that groups the above labels into the following categories: adult-type diffuse gliomas (837 slides), meningiomas (430 slides), mesenchymal, non-meningothelial tumors involving the CNS (190 slides), tumors of the sellar region (184 slides), circumscribed astrocytic gliomas (173 slides), ependymal tumors (96 slides), hematolymphoid tumors involving the CNS (91 slides), glioneuronal and neuronal tumors (88 slides), cranial and paraspinal nerve tumors (81 slides), pediatric-type diffuse low-grade gliomas (70 slides), metastatic tumors (47 slides), and embryonal tumors (32 slides). All brain tumors in these tasks are designated as rare cancers by the RARECARE project and the NCI-SEER program. The hierarchical classification of the coarseand fine-grained tasks is reported in Supplementary Table 20. For training and evaluation of both tasks, we approximately label-stratified the dataset into a train–validation–test fold with a 50:25:25 ratio (1,151:595:573 slides), using the same folds for both coarseand fine-grained tasks.
Prostate ISUP grading based on PANDA (6 classes).
The ISUP grading task is derived from the PANDA challenge, which consists of 10,616 prostate cancer core needle biopsies of prostate cancer sourced from the Radboud University Medical Center and the Karolinska Institute18,162. Each slide is assigned an ISUP score that defines prostate cancer grade (6-class grading task). For quality control, we follow prior work90 in excluding slides that were erroneously annotated (https://www.kaggle.com/competitions/prostate-cancer-grade-assessment/discussion/169230) or had noisy labels (https://www.kaggle.com/competitions/prostate-cancer-grade-assessment/discussion/169230), which resulted in 9,555 slides (2,603 G0, 2,399 G1, 1,209 G2, 1,118 G3, 1,124 G4, 1,102 G5). For training and evaluation, we label-stratified PANDA into 80:10:10 train–validation–test folds (7,647:954:954 slides). In addition to internal comparisons, we also re-evaluate our results using the same splits of public MIL baselines of recent work90. In evaluation with public baselines, we adopt the evaluation strategy in WholeSIGHT90 of also evaluating the Karolinska and Radboud cohorts separately. Supplementary Table 30 reports the performance of UNI and its internal comparisons with the public splits, with Supplementary Table 37 reporting our results against the public MIL baselines. We also note the same caveat from the CAMELYON description in this task, given that comparisons with public MIL performances may not be equivalent due to using ResNet-50IN features, but note that these baselines also adopt more sophisticated MIL architectures.
Endomyocardial assessment based on in-house BWH data (2 classes).
The BWH-EMB dataset consists of 5,021 H&E FFPE histopathology WSIs from 1,688 in-house endomyocardial biopsies (EMBs) collected from BWH for cellular-mediated allograft rejection (ACR) (2,444 ACR, 2,577 others)86. For training and evaluation, we caseand label-stratified the dataset into train–validation–test folds (3,547:484:900 slides, 1,192:164:332 patients), with evaluation performed at the patient level. In addition to internal comparisons, we also compare our results with the CRANE86 results (which shares the same splits) (Extended Data Fig. 8). We also note the same caveat from the CAMELYON description in this task, given that comparison with UNI may not be equivalent due to CRANE using ResNet-50IN features, but note that this baseline also uses multi-task learning with other clinical endpoints for EMB assessment.
CRC tissue classification based on CRC-100K (9 classes).
The CRC tissue classification task is based on the CRC-100K dataset, which consists of 107,180 224 × 224 pixel ROIs at 0.5 mpp annotated and extracted from H&E FFPE diagnostic histopathology WSIs of 136 colorectal adenocarcinoma samples from the National Center for Tumor Diseases (NCT) biobank and the University Medical Center Mannheim (UMM) pathology archive98. ROIs were labeled with the following 9 classes: adipose (11,745 ROIs), background (11,413 ROIs), debris (11,851 ROIs), lymphocyte (12,191 ROIs), mucus (9,931 ROIs), smooth muscle (14,128 ROIs), normal colon mucosa (9,504 ROIs), cancer-associated stroma (10,867 ROIs) and colorectal adenocarcinoma epithelium (15,550 ROIs). For training and evaluation we used the official case-stratified train–test folds (100,000:7,180 ROIs), with the training fold constructed from 100,000 ROIs (86 WSIs) from the NCT biobank and UMM pathology archive (referred to as ‘NCT-CRC-HE-100K’), and the test fold constructed from 7,180 ROIs (50 WSIs) from the NCT biobank (referred to as ‘CRC-VAL-HE-7K’). Additionally, we use the version of NCT-CRC-HE-100K without stain normalization. We use the same folds for linear probe, KNN and SimpleShot evaluation. We evaluate this dataset on ROIs of 224 × 224 pixels at 0.5 mpp.
Breast metastasis detection based on CAMELYON17-WILDS (2 classes).
The breast metastasis detection task is based on the patch-based variant of the CAMELYON17 dataset120 (called PatchCAMELYON or ‘PCAM’)163, with folds created by WILDS164 for testing the models’ robustness under distribution shift. The dataset consists of 417,894 96 × 96 pixel histopathology ROIs at ~0.92–1.00 mpp extracted from WSIs of breast cancer metastases in lymph nodes sections, obtained from the CAMELYON17 challenge120. The ROI label refers to whether the patch contains tumor. For training and evaluation, we used the official train–validation–test folds provided by WILDS. The training set contains 302,436 patches from three hospitals, and the model is evaluated on two out-of-distribution (OD) datasets containing 34,904 patches (ValOD) and 80,554 patches (TestOD) collected from two other hospitals, respectively. We bilinearly upsampled all images to 224 × 224 pixels for equivalent comparisons with CTransPath. In addition to internal comparisons, we also compare our results with the public leaderboard on the WILDS benchmark (https://wilds.stanford.edu/leaderboard/), which we report in Supplementary Table 62. The in-domain validation fold was not combined with the training set or used for hyper-parameter tuning. We note that comparisons with public results may not be equivalent to our evaluation, because many methods are end-to-end fine-tuned with transfer learning from natural images (and not from pathology).
CRC tissue classification based on HunCRC (9 classes).
The CRC tissue classification task is based on the HunCRC dataset, which consists of 101,398 512 × 512 pixel ROIs at 0.48 mpp, annotated and extracted from the same 200 H&E FFPE diagnostic histopathology WSIs of colorectal biopsies also described in the slide-level task99. ROIs were labeled with the following nine classes: adenocarcinoma (4,315 ROIs), high-grade dysplasia (2,281 ROIs), low-grade dysplasia (55,787 ROIs), inflammation (763 ROIs), tumor necrosis (365 ROIs), suspicious for invasion (570 ROIs), resection edge (534 ROIs), technical artifacts (3,470 ROIs), and normal (31,323 ROIs). For training and evaluation we case-stratified and approximately label-stratified the dataset into train–test folds (151:49 cases, 76,753:22,655 ROIs) for use in linear probe, KNN and SimpleShot evaluation. We evaluate this dataset on resized ROIs of 448 × 448 pixels at 0.55 mpp.
BRCA subtyping based on BACH (4 classes).
The BRCA subtyping task is based on the Breast Carcinoma Subtyping (BACH) dataset, which consists of 400 2,048 × 1,536 pixel ROIs at 0.42 mpp, annotated and extracted from H&E FFPE diagnostic histopathology WSIs of breast carcinoma samples from the International Conference on Image Analysis and Recognition (ICIAR) 2018 grand challenge on breast cancer histology images (BACH)165. ROIs were labeled with the following four classes: normal (100 ROIs), benign (100 ROIs), in situ carcinoma (100 ROIs) and invasive carcinoma (100 ROIs). For training and evaluation we label-stratified the dataset into train–test folds (320:80 ROIs) for use in linear probe, KNN and SimpleShot evaluation. Additionally, we evaluate this dataset across the following center-cropped and resized image resolutions: 224 × 224 pixels at 2.88 mpp, 448 × 448 pixels at 1.44 mpp, 896 × 896 pixels at 0.72 mpp and 1,344 × 1,344 pixels at 0.48 mpp
CCRCC tissue classification based on TCGA and HEL (3 classes).
The CCRCC tissue classification task consists of 52,713 256 × 256 pixel and 300 × 300 pixel ROIs at approximately 0.25 mpp, annotated and extracted from H&E FFPE diagnostic histopathology WSIs of CCRCC samples from TCGA (502 samples) and Helsinki University Hospital (HEL) (64 samples)166. ROIs were labeled with the following six classes: cancer (13,057 ROIs), normal (8,652 ROIs), stroma (5,460 ROIs), red blood cells (996 ROIs), empty background (16,026 ROIs), and other textures (8,522 ROIs). For this task we considered only the cancer, normal and stroma labels due to label imbalance when stratifying by data source and ambiguities in the ‘other’ category. We used ROIs from TCGA (21,095 ROIs) and HEL (6,074 ROIs) as the train and test cohorts, respectively (train–test fold with a ratio of 21,095:6,074), which we used for linear probe, KNN and SimpleShot evaluation. We evaluate this dataset on resized ROIs of 224 × 224 pixels at approximately 0.29 mpp.
PRAD tissue classification based on AGGC (5 classes).
The PRAD tissue classification task is based on the Automated Gleason Grading Challenge 2022 (AGGC) from the National University Hospital and Agency of Science, Technology and Research (A*STAR) in Singapore101. It consists of 203 WSIs obtained from prostatectomies (105 training, 45 testing) and biopsies (37 training, 16 testing) digitized using an Akoya Biosciences scanner at ×20 magnification at 0.5 mpp. Each slide includes partial pixel-level annotations delineating different Gleason patterns and stromal regions. From the original WSIs and annotations we built a ROI dataset consisting of 1,125,640 nonoverlapping 256 × 256 pixel ROIs (train–test fold with a ratio of 780,619:345,021), which we used for linear probe, KNN and SimpleShot evaluation. ROIs with more than one Gleason pattern were assigned the most aggressive grade. We evaluate this dataset on resized ROIs of 224 × 224 pixels at approximately 0.57 mpp.
ESCA tissue classification based on UKK, WNS, TCGA and CHA (11 classes).
The ESCA (esophageal carcinoma) tissue classification task consists of 367,229 256 × 256 pixel ROIs at 0.78 mpp, annotated and extracted from 320 H&E FFPE diagnostic histopathology WSIs of esophageal adenocarcinoma and adenocarcinoma of the esophagogastric junction from four sources: University Hospital Cologne (UKK, 22 slides), Landesklinikum Wiener Neustadt (WNS, 62 slides), TCGA (22 slides) and the University Hospital Berlin–Charité (CHA, 214 slides)167. ROIs were labeled with the following 11 classes: adventitia (71,131 ROIs), lamina propria mucosae (2,173 ROIs), muscularis mucosae (2,951 ROIs), muscularis propria (83,358 ROIs), regression tissue (56,490 ROIs), mucosa gastric (44,416 ROIs), muscosa esophagus (18,561 ROIs), submucosa (22,117 ROIs), submucosal glands (1,516 ROIs), tumor (63,863 ROIs) and ulceration (753 ROIs). For training and evaluation we combined UKK, WNS and TCGA into one training cohort (189,142 ROIs) and used CHA as a test cohort (178,187 ROIs), with a train–test fold ratio of 51:49, which we then used for linear probe, KNN and SimpleShot evaluation. We evaluate this dataset on resized ROIs of 224 × 224 pixels at approximately 0.89 mpp.
CRC polyp classification based on UniToPatho (6 classes).
The CRC polyp classification task is based on the UniToPatho dataset, which consists of 9,536 1,812 × 1,812 pixel ROIs at 0.44 mpp, annotated and extracted from 292 H&E FFPE diagnostic histopathology WSIs of colorectal polyp samples from the University of Turin100. ROIs were labeled with the following six classes: normal (950 ROIs), hyperplastic polyp (545 ROIs), tubular adenoma with high-grade dysplasia (454 ROIs), tubular adenoma with low-grade dysplasia (3,618 ROIs), tubulo-villous adenoma with high-grade dysplasia (916 ROIs), and tubulo-villous adenoma with low-grade dysplasia (2,186 ROIs). For training and evaluation we used the official train–test folds (6,270:2,399 ROIs). We evaluate this dataset across the following resized image resolutions: 224 × 224 pixels at 3.60 mpp, 448 × 448 pixels at 1.80 mpp, 896 × 896 pixels at 0.90 mpp, and 1,792 × 1,792 pixels at 0.45 mpp.
CRC MSI screening based on TCGA CRC-MSI (2 classes).
The CRC microsatellite instability (MSI) prediction task is based on the TCGA CRC-MSI dataset, which consists of 51,918 512 × 512 pixel ROIs at approximately 0.5 mpp, extracted from H&E FFPE diagnostic histopathology WSIs of colorectal adenocarcinoma samples annotated and extracted from TCGA and also pre-normalized using Macenko normalization6. ROIs were labeled with the following two classes according to the patient-level label of the sample: microsatellite instable (15,002 ROIs) and microsatellite stable (36,916 ROIs). For training and evaluation, we used the official train–test folds (19,557:32,361 ROIs) in linear probe, KNN, and SimpleShot evaluation. We evaluate this dataset on resized ROIs of 448 × 448 pixels at 0.57 mpp.
Pan-cancer tissue classification based on TCGA Uniform Tumor (32 classes).
The pan-cancer tissue classification task is based on the TCGA Uniform Tumor dataset, which consists of 271,170 256 × 256 pixel ROIs at around 0.5 mpp of 32 cancer types annotated and extracted from 8,736 H&E FFPE diagnostic histopathology WSIs in TCGA68. Images were labeled with the following 32 classes: adrenocortical carcinoma (ACC) (4,980 ROIs), bladder urothelial carcinoma (BLCA) (9,990 ROIs), brain lower-grade glioma (LGG) (23,530 ROIs), BRCA (23,690 ROIs), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (6,270 ROIs), cholangiocarcinoma (CHOL) (900 ROIs), COAD (8,150 ROIs), ESCA (3,380 ROIs), glioblastoma multiforme (GBM) (23,740 ROIs), HNSC (11,790 ROIs), kidney chromophobe (KICH) (2,460 ROIs), kidney renal clear cell carcinoma (KIRC) (11,650 ROIs), kidney renal papillary cell carcinoma (KIRP) (6,790 ROIs), liver hepatocellular carcinoma (LIHC) (8,370 ROIs), LUAD (16,460 ROIs), LUSC (16,560 ROIs), lymphoid neoplasm diffuse large B cell lymphoma (DLBC) (840 ROIs), mesothelioma (MESO) (2,090 ROIs), ovarian serous cystadenocarcinoma (OV) (2,520 ROIs), PAAD (4,090 ROIs), pheochromocytoma and paraganglioma (PCPG) (1,350 ROIs), PRAD (9,810 ROIs), 23) READ (1,880 ROIs), sarcoma (SARC) (13,480 ROIs), skin cutaneous melanoma (SKCM) (10,060 ROIs), STAD (9,670 ROIs), testicular germ cell tumor (TGCT) (6,010 ROIs), thymoma (THYM) (3,600 ROIs), thyroid carcinoma (THCA) (11,360 ROIs), uterine carcinosarcoma (UCS) (2,120 ROIs), uterine corpus endometrial carcinoma (UCEC) (12,480 ROIs), and uveal melanoma (UVM) (1,640 ROIs). Except for BLCA, BRCA, COAD, HNSC, LUAD, LUSC, PAAD, PRAD, READ, SKCM, STAD, THCA and UCEC, all other cancer types in this task are designated as rare cancers by the RARECARE project and the NCI-SEER program. For training and evaluation we case-stratified and approximately label-stratified the dataset into train–test folds (216,350:55,360 ROIs), for use in linear probe, KNN and SimpleShot evaluation. We evaluate this dataset on resized ROIs of 224 × 224 pixels at approximately 0.57 mpp. To mitigate potential biases from site-specific H&E staining variability in TCGA168, we used Macenko normalization169 to normalize all ROIs.
Pan-cancer TIL detection based on TCGA-TILS (2 classes).
The tumor-immune lymphocyte (TIL) detection task is based on the TCGA-TILs dataset, which consists of 304,097 100 × 100 pixel histopathology ROIs at approximately 0.5 mpp, annotated and extracted from H&E FFPE diagnostic histopathology WSIs in TCGA61,67,170. ROIs were labeled with the following two classes: TIL-positive (if there are at least two TILs present in the image, 54,910 ROIs) and TIL-negative (249,187 ROIs). For training and evaluation we used the official train–validation–test folds (209,221:38,601:56,275 ROIs) and combine the train and validation folds into a single training fold. We bilinearly upsampled all images to 224 × 224 pixels at 0.20 mpp for equivalent comparisons with CTransPath. To mitigate potential biases from site-specific H&E staining variability in TCGA, we used Macenko normalization169 to normalize all ROIs. In addition to internal comparisons, we also compare our results with the ChampKit leaderboard, which we report in Supplementary Table 61. We note that comparisons with public results may not be equivalent to our evaluation, given that many methods are end-to-end fine-tuned with transfer learning from natural images (and not from pathology).
Pan-cancer cell type segmentation based on SegPath (8 cell types treated as individual tasks).
The cell type segmentation tasks are derived from the SegPath dataset, which consists of 158,687 984 × 984 pixel ROIs at 0.22 mpp, annotated and extracted from H&E FFPE diagnostic histopathology WSIs of eight major cell types in cancer tissue from University of Tokyo Hospital102. Immunofluorescence and DAPI nuclear staining were performed on ROIs and used as image masks for the following classes: endothelium (10,647 ROIs), epithelium (26,509 ROIs), leukocyte (24,805 ROIs), lymphocyte (12,273 ROIs), myeloid cell (14,135 ROIs), plasma cell (13,231 ROIs), red blood cell (25,909 ROIs), and smooth muscle (31,178 ROIs). Each cell type in the dataset forms an independent tissue segmentation task with two classes, tissue/cell region and non-tissue/cell region. For training and evaluation we used the official train–validation–test split with an approximate 80:10:10 ratio. Furthermore, we compare our results using the public evaluation of this dataset, which we also report in Supplementary Table 69. We note that individual model performances are not made public in the official dataset, and thus we interpolated the performance bound of the best-performing model for each cell type.
Computing hardware and software
We used Python (v3.8.13) and PyTorch171 (v2.0.0, CUDA 11.7) (https://pytorch.org) for all experiments and analyses in the study (unless specified), which can be replicated using open-source libraries as outlined below. To train UNI via DINOv2, we modify the vision transformer implementation maintained by the open-source timm library (v0.9.2) from Hugging Face (https://huggingface.co) for the encoder backbone and use the original DINOv2 self-supervised learning algorithm (https://github.com/facebookresearch/dinov2) for pretraining, which used 4 × 8 80 GB NVIDIA A100 GPU (graphics processing unit) nodes configured for multi-GPU, multi-node training using distributed data-parallel (DDP). All other computations for downstream experiments were conducted on single 24 GB NVIDIA 3090 GPUs. All WSI processing was supported by OpenSlide (v4.3.1), openslide-python (v1.2.0), and CLAM (https://github.com/mahmoodlab/CLAM). We use Scikit-learn134 (v1.2.1) for its implementation of K-nearest neighbors, and the logistic regression implementation and SimpleShot implementation provided by the LGSSL codebase (https://github.com/mbanani/lgssl). Implementations of other visual pretrained encoders benchmarked in the study are found at the following links: ResNet-50 with ImageNet Transfer (https://github.com/mahmoodlab/CLAM), CTransPath (https://github.com/Xiyue-Wang/TransPath), and REMEDIS (https://github.com/google-research/medical-ai-research-foundations). We note that REMEDIS requires fulfillment of a data use agreement, which can be accessed and submitted at the PhysioNet website (https://physionet.org/content/medical-ai-research-foundation)172,173. For multi-head attention visualization, we used the visualization tools provided by the HIPT codebase (https://github.com/mahmoodlab/HIPT). For training weakly supervised ABMIL models, we adapted the training scaffold code from the CLAM codebase (https://github.com/mahmoodlab/CLAM). For training semantic segmentation, we use the original Mask2Former implementation (https://github.com/facebookresearch/Mask2Former), which is based on detectron2 (ref. 174) (v0.6), and required the following older packages for compatibility: Python (v3.8) and PyTorch (v1.9.0, CUDA 11.1). For adding ViT-Adapter to UNI, we adapt its original implementation (https://github.com/czczup/ViT-Adapter) in detectron2 to train it using Mask2Former. Pillow (v9.3.0) and OpenCV-python were used to perform basic image processing tasks. Matplotlib (v3.7.1) and Seaborn (v0.12.2) were used to create plots and figures. Use of other miscellaneous Python libraries is detailed in the Reporting Summary.
Extended Data
Extended Data Fig. 1: Few-shot slide classification.
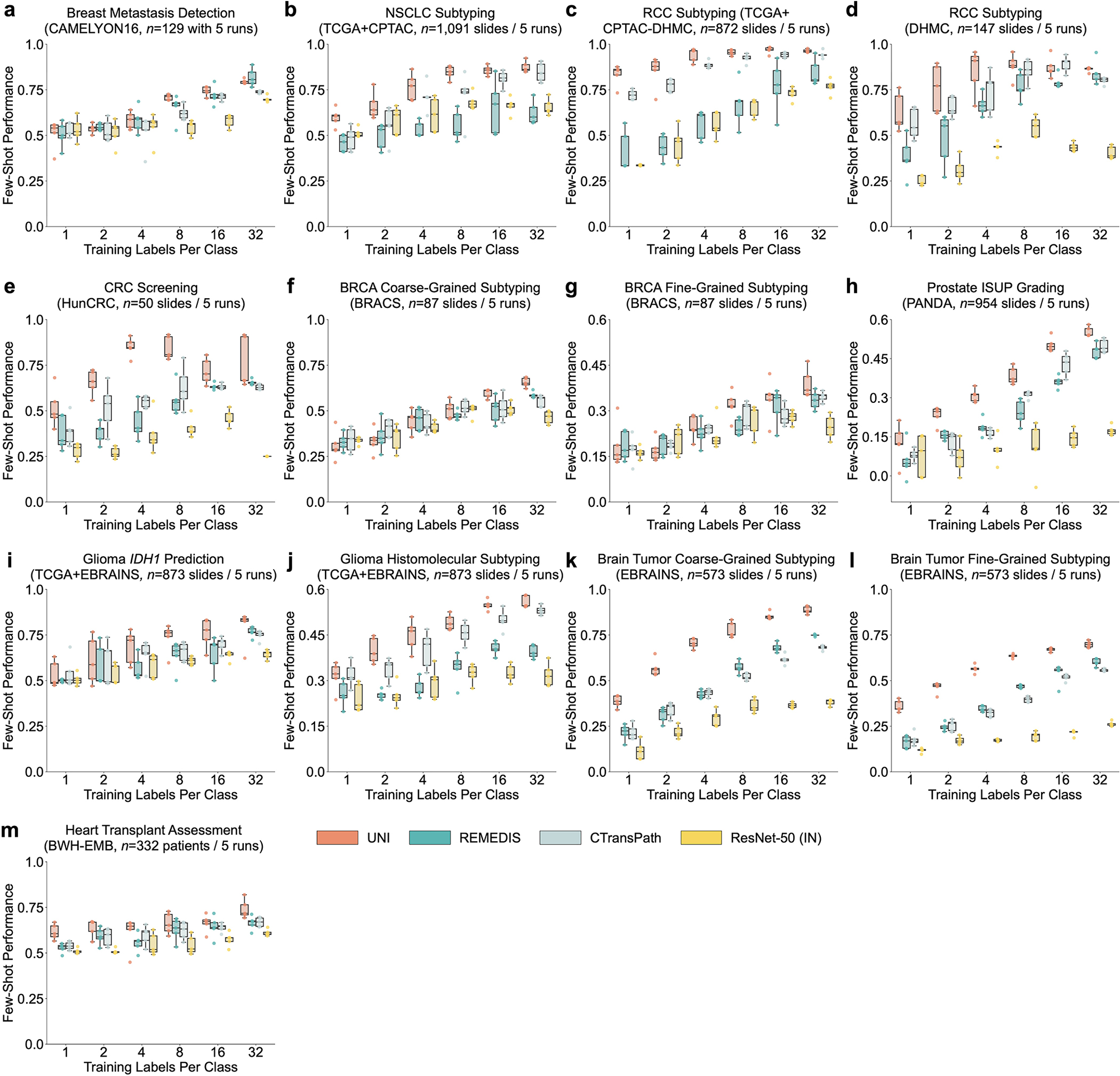
To study the label efficiency of UNI in slide classification, we compare UNI with other pretrained encoders on: a. breast metastasis detection in CAMELYON16, b. NSCLC subtyping in CPTAC (trained on TCGA) c. RCC subtyping in CPTAC-DHMC (trained on TCGA), d. RCC subtyping in DHMC, e. BRCA coarse-grained subtyping in BRACS, f. BRCA fine-grained subtyping in BRACS, g. CRC screening in HunCRC, h. Prostate ISUP Grading in PANDA, i. glioma IDH1 prediction in EBRAINS (trained on TCGA), j. glioma histomolecular subtyping in EBRAINS (trained on TCGA), k. brain tumor coarse-grained subtyping in EBRAINS, l. brain tumor fine-grained subtyping in EBRAINS, and m. heart transplant assessment in BWH-EMB. The performance is measured across different few-shot settings with K ∈ 1, 2, 4, 8, 16, 32 training examples used per class. Boxes indicate quartile values of model performance (n = 5 runs) and whiskers extend to data points within 1.5 × the interquartile range. Overall, we observe that UNI consistently demonstrates superior label efficiency over other baselines.
Extended Data Fig. 2: Comparing supervised performance on PRAD tissue classification in AGCC.
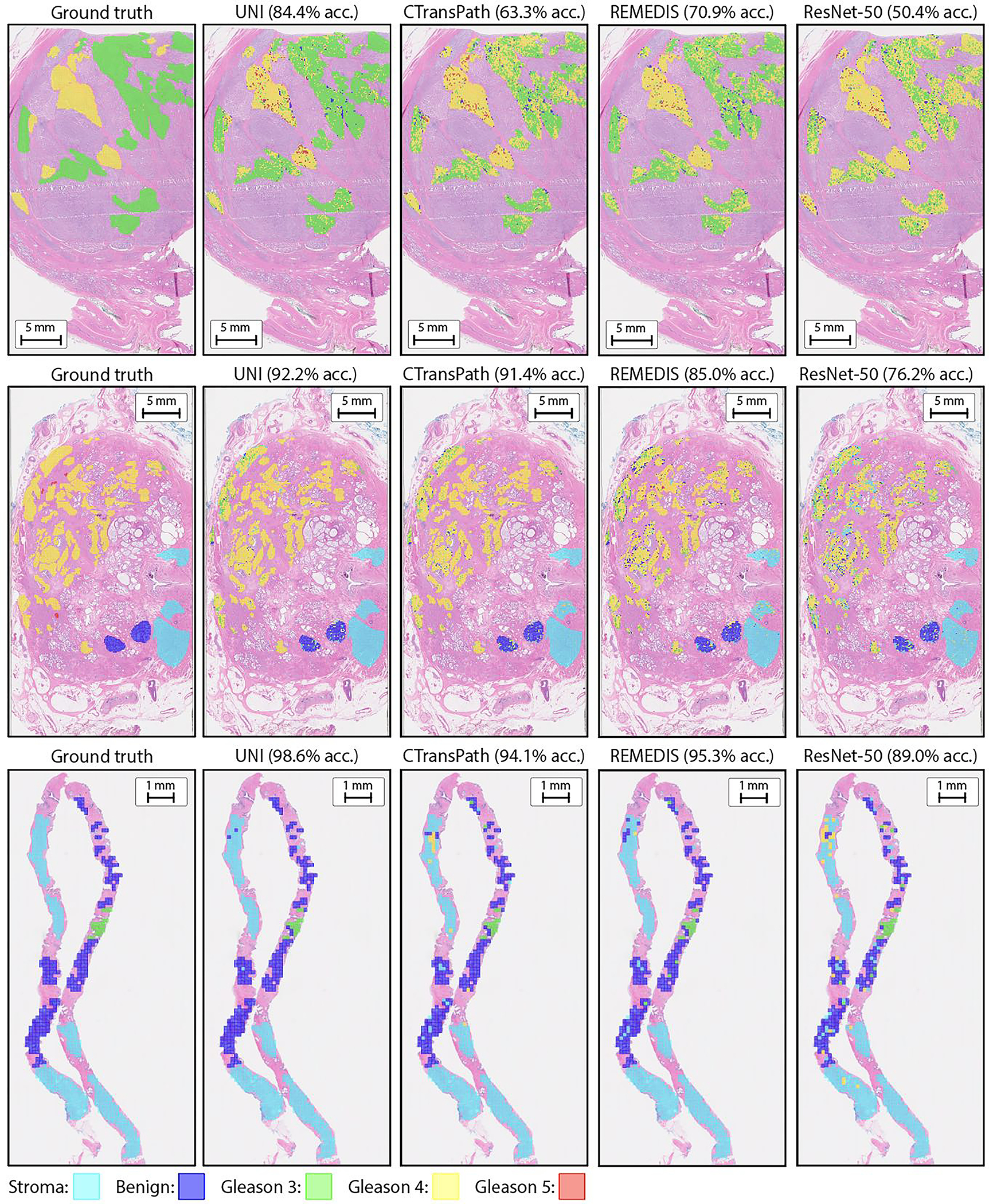
Qualitative illustrations comparing UNI to CTransPath, REMEDIS, and ResNet-50 (IN) via KNN probing on PRAD tissue classification in AGCC. UNI achieves better accuracy (acc.) on all three examples. The reported results are based on partial annotations (left-most panel) provided by pathologists.
Extended Data Fig. 3: ROI retrieval.
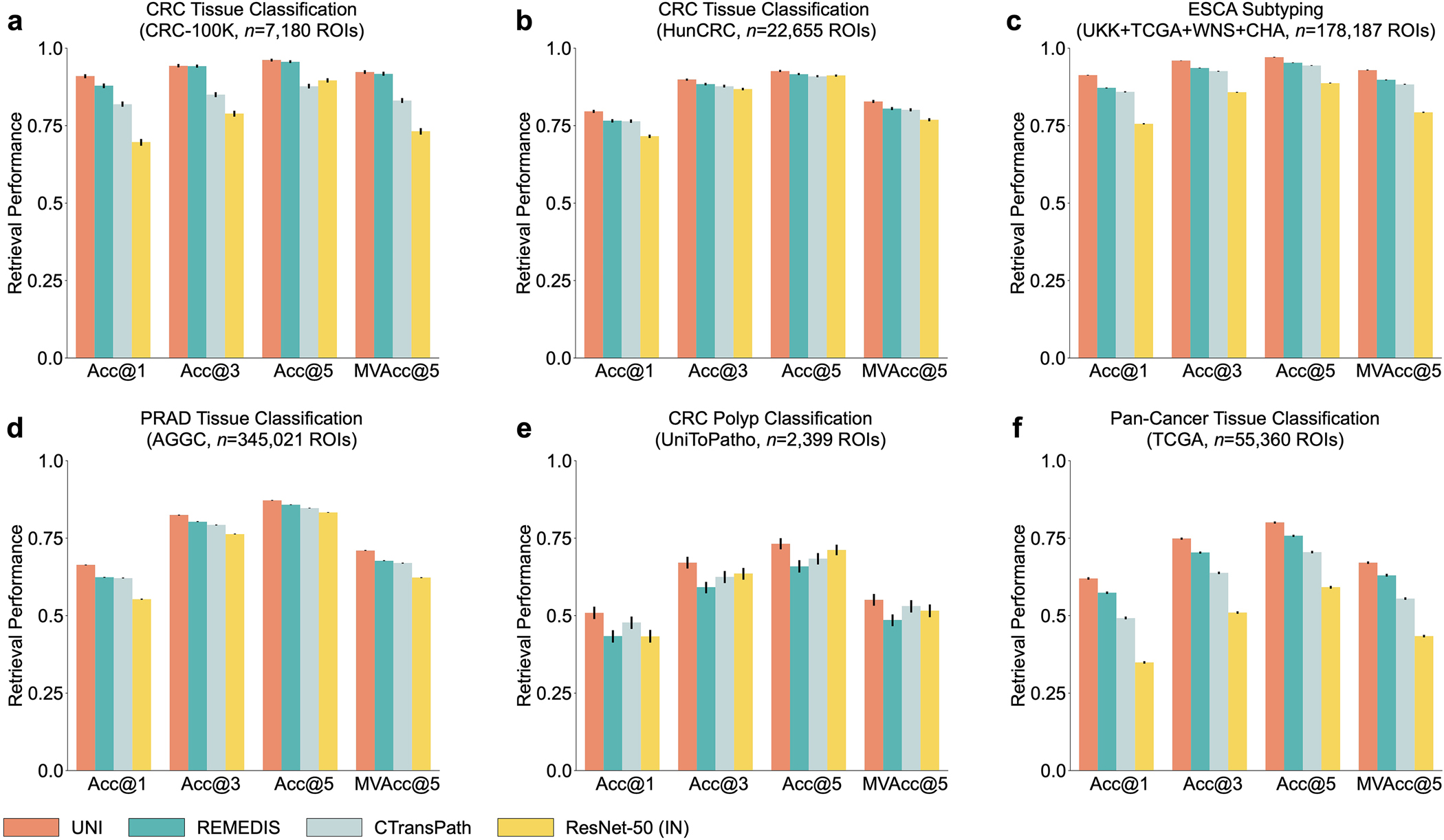
We evaluate content-based image retrieval for ROI-level classes with at least 5 classes, for a. CRC tissue classification in CRC-100K, b. CRC tissue classification in HunCRC, c. ESCA subtyping on CHA (trained on UKK, WNS and TCGA), d. PRAD tissue classification in AGGC, e. CRC polyp classification in UniToPatho, and f. pan-cancer tissue classification in TCGA, and. UNI consistently outperforming all pretrained encoders. Error bars represent 95% confidence intervals and the center is the computed value of the corresponding retrieval metric. Detailed performance metrics are further provided in Supplementary Tables 63–68.
Extended Data Fig. 4: ROI classification across different image resolutions.
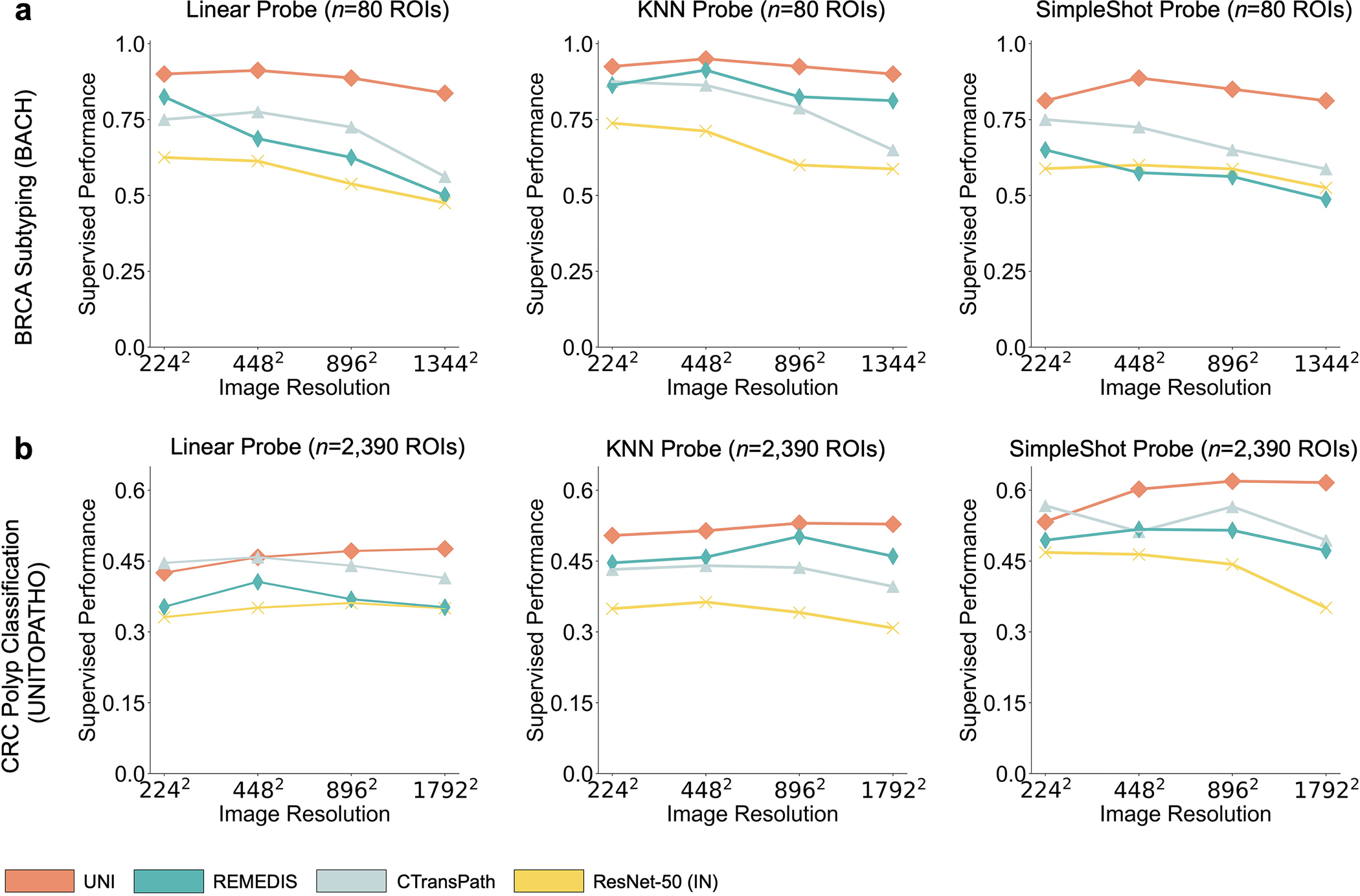
To assess how image resolution affects performance, we compare UNI and other baselines on various resized and center-cropped ROIs for a. BRCA subtyping and b. CRC polyp classification tasks. The original image sizes are 2048 × 1536 and 1812 × 1812 pixels, respectively. All models are evaluated on linear, SimpleShot (1-NN), and KNN (20-NN) probe settings. UNI consistently outperforms all baselines across all resolutions. The performance metrics are further provided in Supplementary Tables 45, 46, 51, 52.
Extended Data Fig. 5: Multi-head self-attention (MHSA) heatmap visualization of UNI across different image resolutions in BRCA Subtyping in BACH.
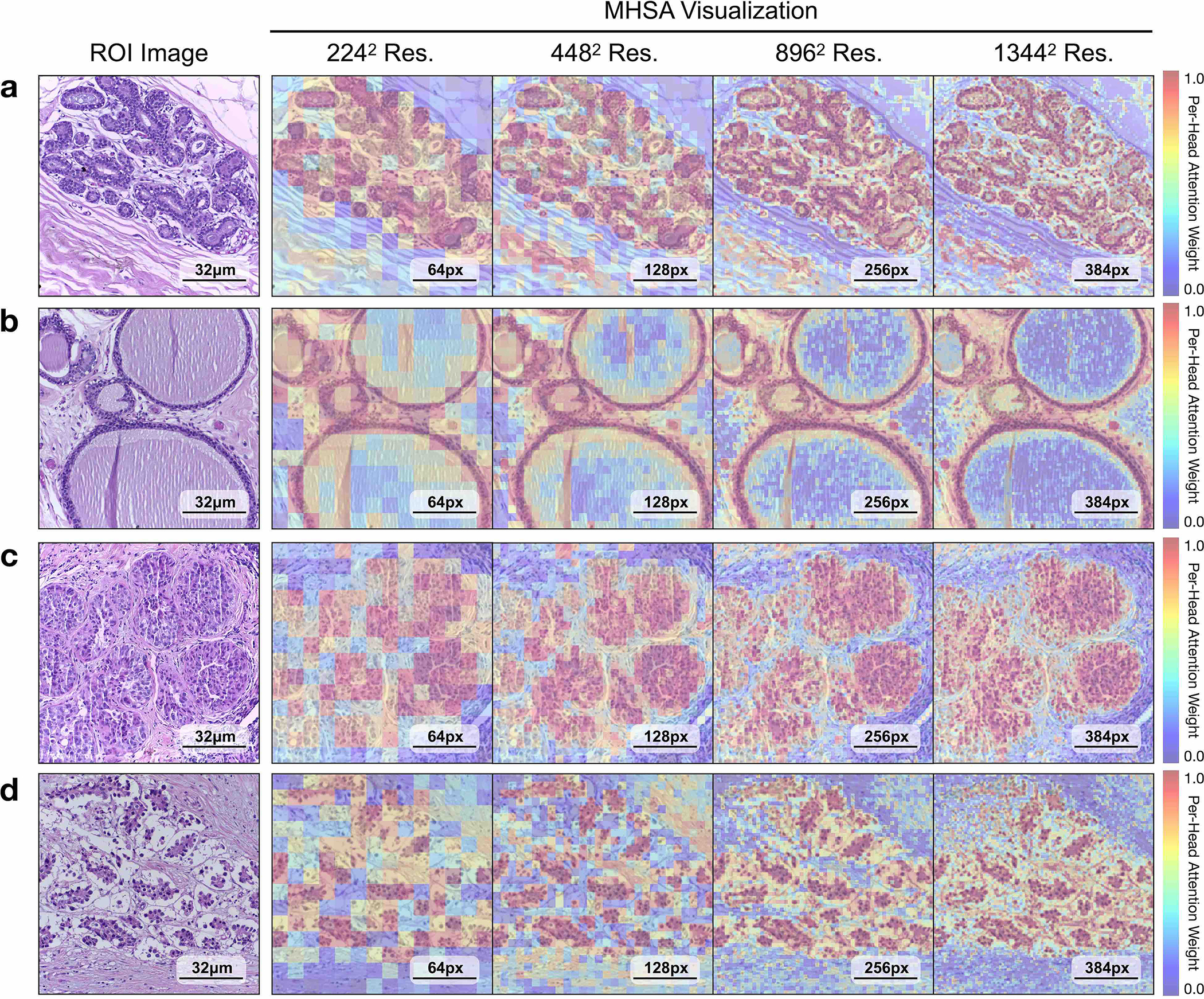
Each colored square represents a 16 × 16 patch token encoded by UNI, with heatmap color corresponding to the attention weight of that patch token to the global [CLS] token of the penultimate layer in UNI. We show MHSA visualizations for resized and center-cropped ROIs at 2242, 4482, 8962, 1,3442 resolutions for the a. normal, b. benign, c. in situ, and d. invasive classes in BACH. In each, the left-most image is the original H&E ROI and the right four images are the MHSA visualizations. For comparative purposes, we resize all images within the figure to have the same dimension, but note that at higher resolutions, each colored square has an original image resolution of 16 × 16 pixels at 0.42 mpp. As the resolution increases, the heatmaps demonstrate increasing and increasingly fine-grained attention focused on epithelial structures, with relatively lower attention on stroma or other background, neither of which are contributory to the diagnoses in these ROIs.
Extended Data Fig. 6: Multi-head self-attention (MHSA) heatmap visualization of UNI across different image resolutions for CRC polyp classification in UniToPatho.
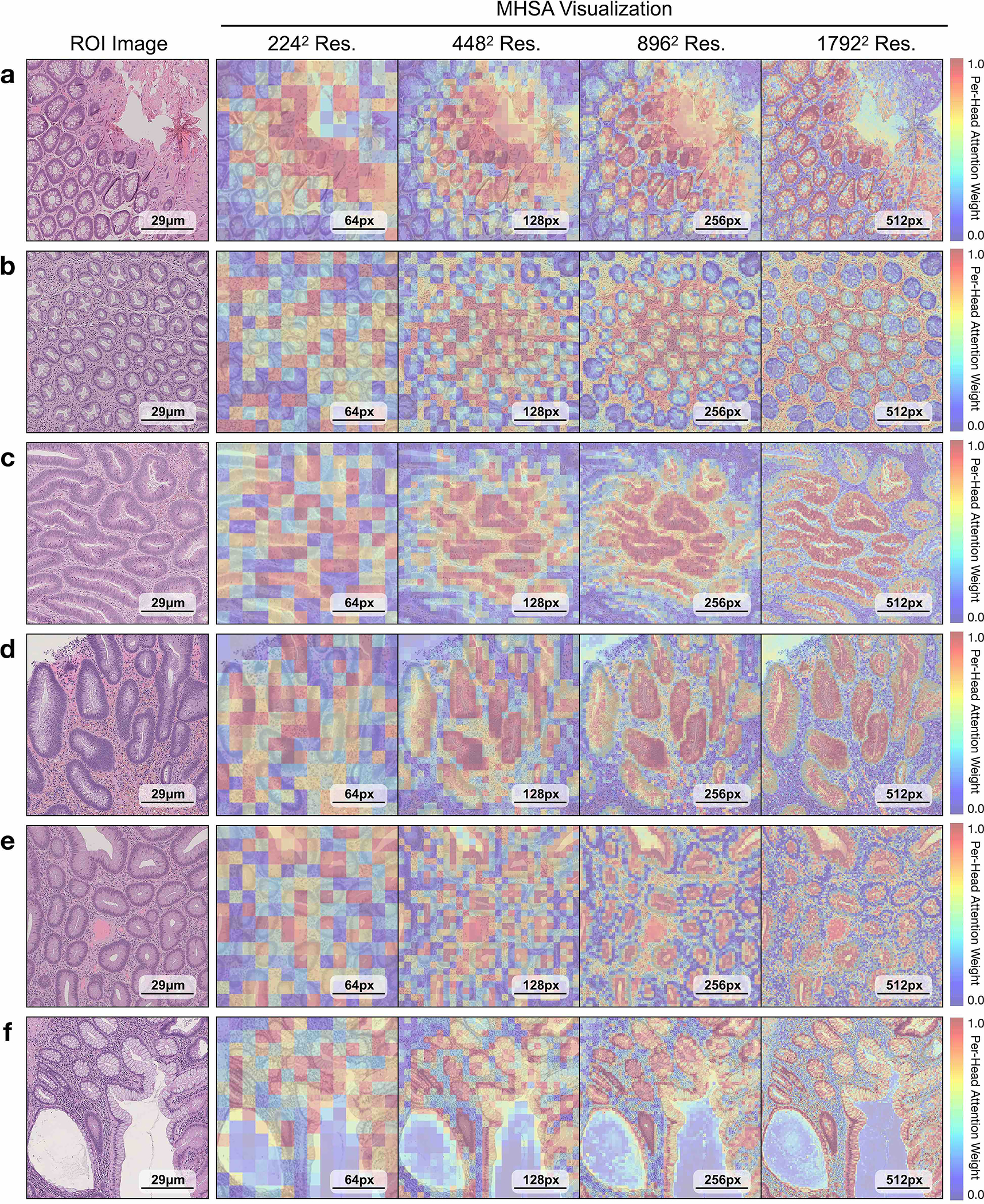
Each colored square represents a 16 × 16 patch token encoded by UNI, with heatmap color corresponding to the attention weight of that patch token to the global [CLS] token of the penultimate layer in UNI. We show MHSA visualizations for resized and center-cropped ROIs at 2242, 4482, 8962, 17922 resolutions for a. normal tissue, b. hyperplastic polyp, c. tubular adenoma with low-grade dysplasia, d. tubular adenoma with high- grade dysplasia, e. tubulo-villous adenoma with high-grade dysplasia, and f. tubulo-villous adenoma with low-grade dysplasia. In each, the left-most image is the original H&E ROI and the right four images are the MHSA visualizations. For comparative purposes, we resize all images within the figure to have the same dimension, but note that at higher resolutions, each colored square has an original image resolution of 16 × 16 pixels at 0.48 mpp. As resolution increases, the heatmaps demonstrate increasing and increasingly fine-grained attention focused on the crypts, in all cases except the hyperplastic polyp in b, focusing on areas a pathologist would use to make the diagnosis.
Extended Data Fig. 7: Visualizing segmentation results in SegPath.

Using the Mask2Former head, we visualize the tissue segmentation of each class in SegPath created by all pretrained encoders. Overall, we find that UNI is competitive with convolutional and hierarchical models like CTransPath and REMEDIS in matching the segmentation masks obtained via immunofluorescence and DAPI nuclear staining.
Extended Data Fig. 8: Few-shot ROI classification using class prototypes.
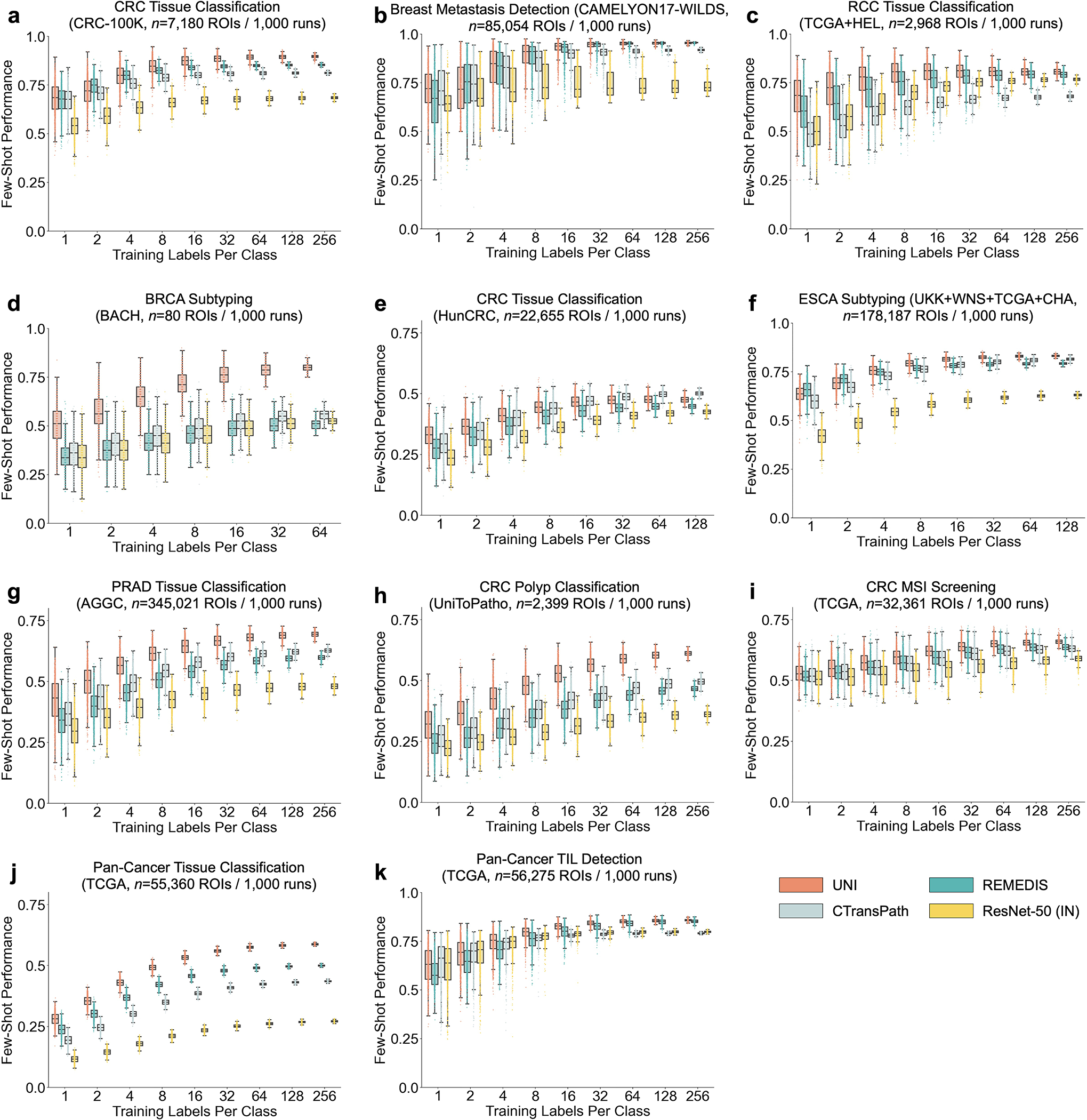
Similar to slide-level classification, we also assess the label efficiency of UNI on ROI-level tasks, and observe superior label efficiency of UNI on most tasks except CRC tissue classification on HunCRC. We evaluate all pretrained encoders using the nonparametric SimpleShot framework for a. CRC tissue classification in CRC-100K, b. Breast metastasis detection in CAMELYON17-WILDS, c. RCC tissue classification on HEL (trained on TCGA), d. BRCA subtyping in BACH, e. CRC tissue classification in HunCRC, f. ESCA subtyping on CHA (UKK+WNS+TCGA), g. PRAD tissue classification in AGGC, h. CRC polyp classification in UniToPatho, i. CRC MSI screening in TCGA, j. pan-cancer tissue classification in TCGA, and k. pan-cancer TIL detection in TCGA. The performance is measured across different few-shot settings with K ∈ 1, 2, 4, 8, 16, 32, 64, 128, 256 training examples used per class (or support set size). Boxes indicate quartile values of model performance (n = 1000 runs) and whiskers extend to data points within 1.5 × the interquartile range. We adapt the SimpleShot framework for slide-level classification, called ‘MI- SimpleShot’. ROI class prototypes are constructed by averaging the pre-extracted ROI features for each class using the ‘TCGA Uniform Tumor’ dataset, which we use as ‘prompts’ for assigning the slide-level label. We assess and compare the few-shot performance of all pretrained encoders on NSCLC subtyping (a) and RCC subtyping task (b), using the same runs (n = 5) in the few-shot setting for ABMIL for K ∈ 1, 2, 4, 8, 16, 32 training examples used per class. We compare performance of top-5 and top-50 pooling of nearest patches in the test set, as well as show performance on both the internal test fold in TCGA and external cohort. Boxes indicate quartile values of model performance (n = 5 runs) and whiskers extend to data points within 1.5 × the interquartile range. Overall, we observe that the formed prototypes by UNI can be used to classify slides based on the MI-SimpleShot frame- work. a. On NSCLC subtyping, we observe that 2-shot and 4-shot performance from UNI outperforms the 32-shot performance of all other models. b. On RCC subtyping, 1-shot performance of UNI also outperforms the 32-shot performance of other models. We also observe that MI-SimpleShot can be combined with other pretrained encoders as well, but generally require more annotated ROIs for creating prototypes.
Extended Data Fig. 9: Few-shot slide classification using class prototypes.
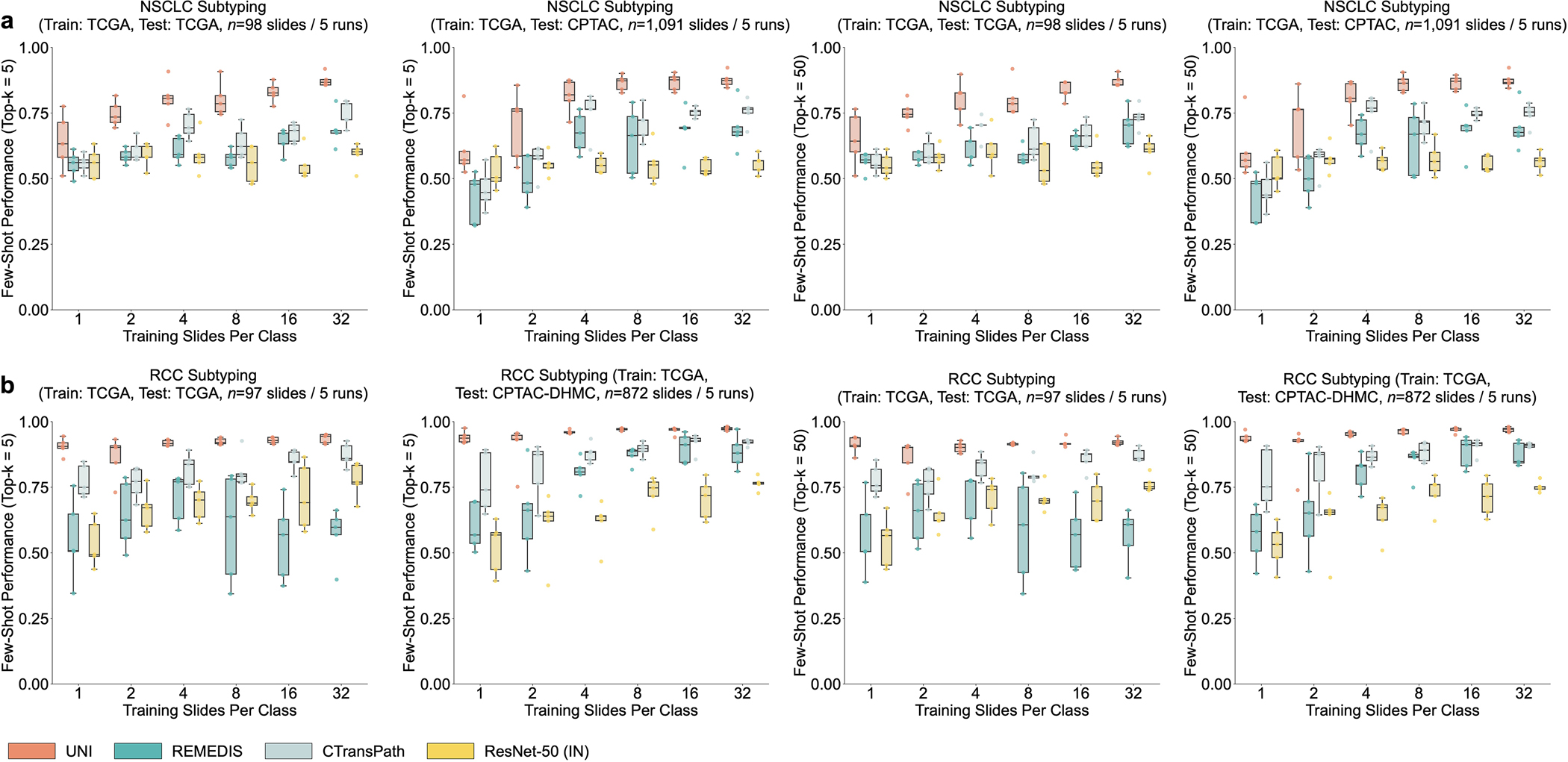
We adapt the SimpleShot framework for slide-level classification, called ‘MI- SimpleShot’. ROI class prototypes are constructed by averaging the pre-extracted ROI features for each class using the ‘TCGA Uniform Tumor’ dataset, which we use as ‘prompts’ for assigning the slide-level label. We assess and compare the few-shot performance of all pretrained encoders on NSCLC subtyping (a) and RCC subtyping task (b), using the same runs (n = 5) in the few-shot setting for ABMIL for K ∈ 1, 2, 4, 8, 16, 32 training examples used per class. We compare performance of top-5 and top-50 pooling of nearest patches in the test set, as well as show performance on both the internal test fold in TCGA and external cohort. Boxes indicate quartile values of model performance (n = 5 runs) and whiskers extend to data points within 1.5 × the interquartile range. Overall, we observe that the formed prototypes by UNI can be used to classify slides based on the MI-SimpleShot frame- work. a. On NSCLC subtyping, we observe that 2-shot and 4-shot performance from UNI outperforms the 32-shot performance of all other models. b. On RCC subtyping, 1-shot performance of UNI also outperforms the 32-shot performance of other models. We also observe that MI-SimpleShot can be combined with other pretrained encoders as well, but generally require more annotated ROIs for creating prototypes.
Extended Data Fig. 10: Comparing 1-shot similarity heatmaps of pretrained encoders with class prototype.
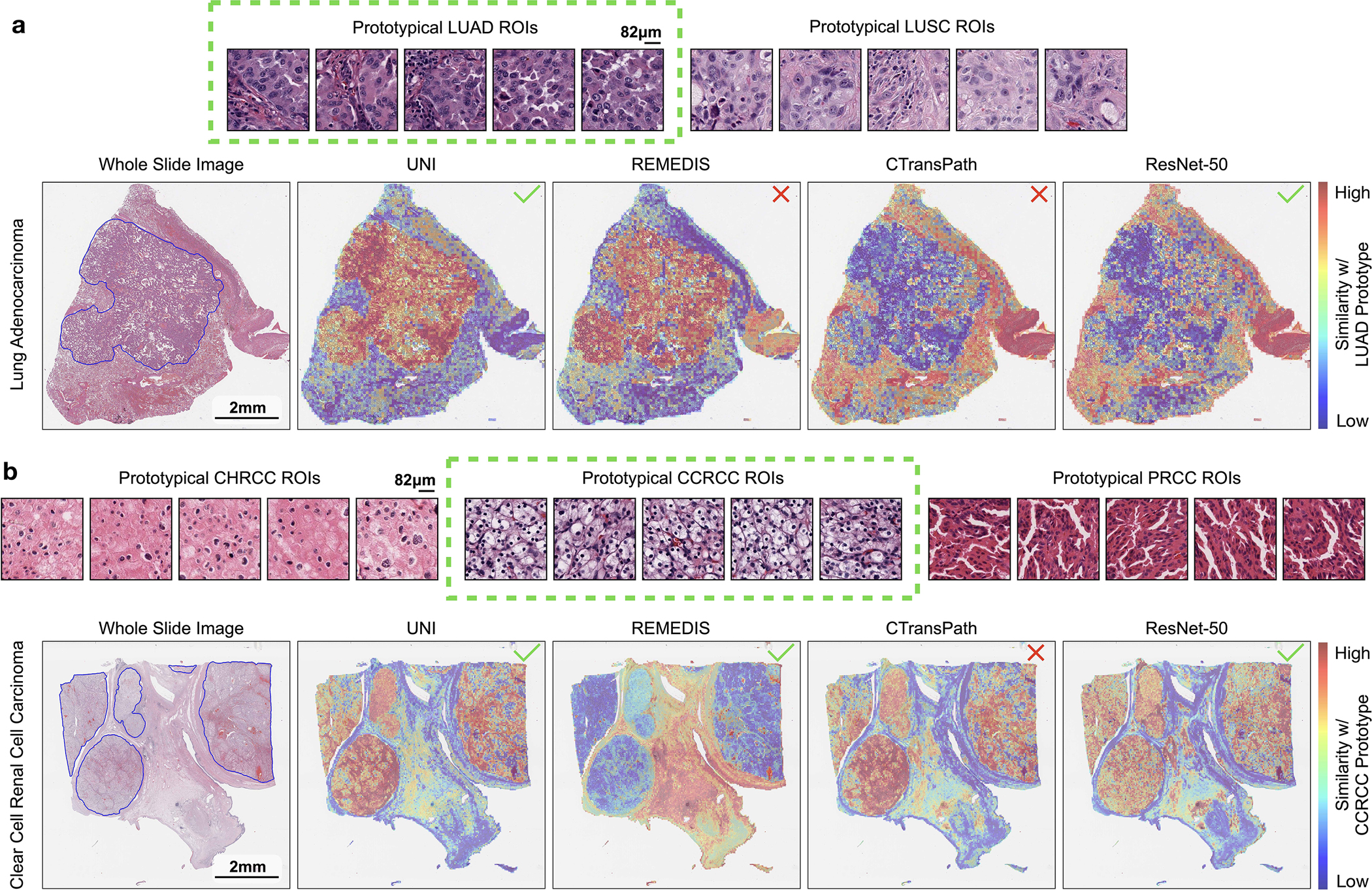
We compare the similarity heatmaps of all pretrained encoders using annotated ROIs from a single slide per class for forming class prototypes in MI-SimpleShot (with top-5 pooling) on NSCLC subtyping (a) and RCC subtyping task (b), with top visualizing example ROIs used for each class, and bottom showing similarity heatmaps. Outlined in blue are pathologist annotations of ROIs that match the label of the histology slide. Similarity heatmaps are created with respect to the class prototype of the correct slide label (indicated in green), with a ✓ indicating a correct prediction and ✗ indicating incorrect prediction. Note that since the visualizations are created with respect to the ground truth label, the model may retrieve correct patches that match pathologist annotations but still misclassify the slide. a. On a LUAD slide, we observe strong agreement of the pathologist’s annotations with retrieved LUAD patches by UNI. Although retrieved patches by REMEDIS also have strong agreement with the pathologist’s annotations, we note that slide was misclassified as LUSC, indicating that the top-5 retrieved patches of the LUSC prototype was higher than that of the LUAD prototype. Vice versa, ResNet-50IN classifies the slide correctly but incorrectly retrieves the correct patches that agree with the pathologist’s annotations, indicating that non-LUAD patches in the slide were more LUAD-like than the pathologist-annotated LUAD patches with respect to the LUAD prototype. The similarity heatmap for CTransPath both misclassified the slide and retried incorrect patches. b. On an CCRCC slide, we observe strong agreement of the pathologist’s annotations with retrieved CCRCC patches by UNI. We observe similar mismatch in predicted class label and retrieved patches, in which REMEDIS classifies the slide correctly but retrieves the incorrect patches, and CTransPath misclassifies the slide but retrieves the correct patches.
Supplementary Material
Footnotes
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Code and model weights for UNI can be accessed for academic research purposes at https://github.com/mahmoodlab/UNI. We have documented all technical deep learning methods and software libraries used in the study while ensuring that the paper is accessible to the broader clinical and scientific audience.
Data availability
TCGA and CPTAC data consisting of whole-slide images and labels can be accessed through the NIH genomic data commons (https://portal.gdc.cancer.gov) and proteomics data commons (https://proteomic.datacommons.cancer.gov), respectively. GTEx data added to the pretrainingdatasetcanbeaccessedthroughtheGTExportal(https://www.gtexportal.org/home/). CPTAC data consisting of all publicly available datasets analyzed in this work can be can accessed in their respective data portals: CRC-100K (https://zenodo.org/record/1214456), HunCRC ROIs (10.6084/m9.figshare.c.5927795.v1), HunCRC slides (10.7937/ tcia.9cjf-0127), BACH (https://iciar2018-challenge.grand-challenge.org/Dataset/), TCGA CRC-MSI (https://zenodo.org/record/3832231), CCRCC tissue classification (https://zenodo.org/record/7898308), TCGA-TILs (https://zenodo.org/record/6604094), TCGA Uniform (https://zenodo.org/record/5889558), UniToPatho (https://zenodo.org/record/4643645), ESCA(https://zenodo.org/record/7548828), CAMELYON17-WILDS (https://wilds.stanford.edu/datasets), EBRAINS (10.25493/WQ48-ZGX), DHMC (https://bmirds.github.io/KidneyCancer), BRACS (https://bracs.icar.cnr.it), PANDA (https://panda.grand-challenge.org), SegPath (https://zenodo.org/record/7412731) and AGGC (https://zenodo.org/record/6460100). TCGA, CPTAC, HunCRC and TCGA-TILS can also be accessed using The Cancer Imaging Archive175. Links for all datasets are also listed in Supplementary Table 73. We note that data from AGGC were obtained from a public grand challenge (of the same name (https://aggc22.grand-challenge.org)) with a pending publication101, with permission granted by the challenge organizers to present results from this dataset. No internal patient data were specifically collected for this study. This study relies on retrospective analysis of anonymized whole-slide images. Following institution policies, all requests for data collected or curated in-house will be evaluated on a case-by-case basis to determine whether the data requested and the use case comply with intellectual property or patient privacy obligations.
References
- 1.Song AH et al. Artificial intelligence for digital and computational pathology. Nat. Rev. Bioeng. 1, 930–949 (2023). [Google Scholar]
- 2.Bera K, Schalper KA, Rimm DL, Velcheti V & Madabhushi A Artificial intelligence in digital pathology: new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipkova J et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 40, 1095–1110 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinz CN, Echle A, Foersch S, Bychkov A & Kather JN The future of artificial intelligence in digital pathology: results of a survey across stakeholder groups. Histopathology 80, 1121–1127 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Coudray N et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kather JN et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mobadersany P et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl Acad. Sci. USA 115, E2970–E2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amgad M et al. A population-level digital histologic biomarker for enhanced prognosis of invasive breast cancer. Nat. Med. 30, 85–97 (2024). [DOI] [PubMed] [Google Scholar]
- 9.Chen RJ et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell 40, 865–878 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanguri RS et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat. Cancer 3, 1151–1164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper M, Ji Z & Krishnan RG Machine learning in computational histopathology: challenges and opportunities. Genes Chromosomes Cancer 62, 540–556 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Graham S et al. Screening of normal endoscopic large bowel biopsies with interpretable graph learning: a retrospective study. Gut 72, 1709–1721 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozyoruk KB et al. A deep-learning model for transforming the style of tissue images from cryosectioned to formalin-fixed and paraffin-embedded. Nat. Biomed. Eng. 6, 1407–1419 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Lu MY et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 594, 106–110 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Lu MY et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kather JN et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Bulten W et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat. Med. 28, 154–163 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foersch S et al. Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat. Med. 29, 430–439 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Chen RJ et al. Multimodal co-attention transformer for survival prediction in gigapixel whole slide images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 4015–4025 (2021). [Google Scholar]
- 21.He K et al. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 16000–16009 (2022). [Google Scholar]
- 22.Oquab M et al. DINOv2: learning robust visual features without supervision. Preprint at 10.48550/arxiv.2304.07193 (2023). [DOI]
- 23.Balestriero R et al. A cookbook of self-supervised learning. Preprint at 10.48550/arxiv.2304.12210 (2023). [DOI]
- 24.Chen X, Xie S & He K An empirical study of training self-supervised vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision (2021). [Google Scholar]
- 25.Caron M et al. Emerging properties in self-supervised vision transformers. In Proceedings of the IEEE/CVF international Conference on Computer Vision, 9650–9660 (2021). [Google Scholar]
- 26.Chen T, Kornblith S, Norouzi M & Hinton G A simple framework for contrastive learning of visual representations. In International Conference on Machine Learning, 1597–1607 (PMLR, 2020). [Google Scholar]
- 27.Grill J-B et al. Bootstrap your own latent: a new approach to self-supervised learning. Adv. Neural Inf. Process. Syst. 33, 21271–21284 (2020). [Google Scholar]
- 28.Deng J et al. ImageNet: a large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition, 248–255 (IEEE, 2009). [Google Scholar]
- 29.Russakovsky O et al. ImageNet large scale visual recognition challenge. International Journal of Computer Vision 115, 211–252 (2015). [Google Scholar]
- 30.Sun C, Shrivastava A, Singh S & Gupta A Revisiting unreasonable effectiveness of data in deep learning era. In Proceedings of the IEEE International Conference on Computer Vision, 843–852 (2017). [Google Scholar]
- 31.Zhai X, Kolesnikov A, Houlsby N & Beyer L Scaling vision transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 12104–12113 (2022). [Google Scholar]
- 32.Goyal P, Mahajan D, Gupta A & Misra I Scaling and benchmarking self-supervised visual representation learning. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 6391–6400 (2019). [Google Scholar]
- 33.Bommasani R et al. On the opportunities and risks of foundation models. Preprint at 10.48550/arxiv.2108.07258 (2021). [DOI]
- 34.Yuan L et al. Florence: A new foundation model for computer vision. Preprint at 10.48550/arxiv.2111.11432 (2021). [DOI]
- 35.Weinstein JN et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen RJ & Krishnan RG Self-supervised vision transformers learn visual concepts in histopathology. In Learning Meaningful Representations of Life, NeurIPS 2021; (2022). [Google Scholar]
- 37.Wang X et al. Transformer-based unsupervised contrastive learning for histopathological image classification. Med. Image Anal. 81, 102559 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Azizi S et al. Robust and data-efficient generalization of self-supervised machine learning for diagnostic imaging. Nat. Biomed. Eng. 7, 756–779 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Kang M, Song H, Park S, Yoo D & Pereira S Benchmarking self-supervised learning on diverse pathology datasets. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 3344–3354 (2023). [Google Scholar]
- 40.Li B, Li Y & Eliceiri KW Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 14318–14328 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazard T, Lerousseau M, Decencière E & Walter T Giga-SSL: self-supervised learning for gigapixel images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 4304–4313 (2023). [Google Scholar]
- 42.Schirris Y, Gavves E, Nederlof I, Horlings HM & Teuwen J DeepSMILE: contrastive self-supervised pre-training benefits MSI and HRD classification directly from H&E whole-slide images in colorectal and breast cancer. Med. Image Anal. 79, 102464 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Vu QD, Rajpoot K, Raza SEA & Rajpoot N Handcrafted Histological Transformer (H2T): unsupervised representation of whole slide images. Med. Image Anal. 85, 102743 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y et al. Predicting lymph node metastasis using histopathological images based on multiple instance learning with deep graph convolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 4837–4846 (2020). [Google Scholar]
- 45.Wang X et al. RetCCL: clustering-guided contrastive learning for whole-slide image retrieval. Med. Image Anal. 83, 102645 (2023). [DOI] [PubMed] [Google Scholar]
- 46.Filiot A et al. Scaling self-supervised learning for histopathology with masked image modeling. Preprint at 10.1101/2023.07.21.23292757 (2023). [DOI]
- 47.Srinidhi CL, Kim SW, Chen F-D & Martel AL Self-supervised driven consistency training for annotation efficient histopathology image analysis. Med. Image Anal. 75, 102256 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Koohbanani NA, Unnikrishnan B, Khurram SA, Krishnaswamy P & Rajpoot N Self-Path: self-supervision for classification of pathology images with limited annotations. IEEE Trans. Med. Imaging 40, 2845–2856 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Ciga O, Xu T & Martel AL Self supervised contrastive learning for digital histopathology. Machine Learning with Applications 7, 100198 (2022). [Google Scholar]
- 50.Lin T et al. SGCL: spatial guided contrastive learning on whole-slide pathological images. Med. Image Anal. 89, 102845 (2023). [DOI] [PubMed] [Google Scholar]
- 51.Tellez D, Litjens G, van der Laak J & Ciompi F Neural image compression for gigapixel histopathology image analysis. IEEE Trans. Pattern Anal. Mach. Intell. 43, 567–578 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Huang Z, Bianchi F, Yuksekgonul M, Montine T & Zou J A visual–language foundation model for pathology image analysis using medical Twitter. Nat. Med. 29, 2307–2316 (2023). [DOI] [PubMed] [Google Scholar]
- 53.Jiang C et al. Hierarchical discriminative learning improves visual representations of biomedical microscopy. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 19798–19808 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saldanha OL et al. Self-supervised attention-based deep learning for pan-cancer mutation prediction from histopathology. NPJ Precis. Oncol. 7, 35 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu MY et al. Visual language pretrained multiple instance zero-shot transfer for histopathology images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 19764–19775 (2023). [Google Scholar]
- 56.Mokhtari R et al. Interpretable histopathology-based prediction of disease relevant features in inflammatory bowel disease biopsies using weakly-supervised deep learning. In Medical Imaging with Deep Learning; 479–495 (PMLR, 2023). [Google Scholar]
- 57.Jaume G et al. Modeling dense multimodal interactions between biological pathways and histology for survival prediction. Preprint at 10.48550/arxiv.2304.06819 (2023). [DOI]
- 58.Hörst F et al. Histology-based prediction of therapy response to neoadjuvant chemotherapy for esophageal and esophagogastric junction adenocarcinomas using deep learning. JCO Clin. Cancer Inform. 7, e2300038 (2023). [DOI] [PubMed] [Google Scholar]
- 59.Wagner SJ et al. Transformer-based biomarker prediction from colorectal cancer histology: a large-scale multicentric study. Cancer Cell 41, 1650–1661 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hörst F et al. CellViT: vision transformers for precise cell segmentation and classification. Preprint at 10.48550/arxiv.2306.15350 (2023). [DOI] [PubMed] [Google Scholar]
- 61.Kaczmarzyk JR et al. ChampKit: a framework for rapid evaluation of deep neural networks for patch-based histopathology classification. Computer Methods and Programs in Biomedicine 239, 107631 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J et al. Gigapixel whole-slide images classification using locally supervised learning. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 192–201 (Springer, 2022). [Google Scholar]
- 63.Nasrallah MP et al. Machine learning for cryosection pathology predicts the 2021 WHO classification of glioma. Med. 4, 526–540 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H et al. Task-specific fine-tuning via variational information bottleneck for weakly-supervised pathology whole slide image classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 7454–7463 (2023). [Google Scholar]
- 65.Ikezogwo WO et al. Quilt-1M: One million image-text pairs for histopathology. In Advances in Neural Information Processing Systems (2023). [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang D et al. Inferring super-resolution tissue architecture by integrating spatial transcriptomics with histology. Nat. Biotechnol, 10.1038/s41587-023-02019-9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saltz J et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 23, 181–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Komura D et al. Universal encoding of pan-cancer histology by deep texture representations. Cell Rep. 38, 110424 (2022). [DOI] [PubMed] [Google Scholar]
- 69.Kalra S et al. Yottixel: an image search engine for large archives of histopathology whole slide images. Med. Image Anal. 65, 101757 (2020) [DOI] [PubMed] [Google Scholar]
- 70.Schmauch B et al. A deep learning model to predict RNA-seq expression of tumours from whole slide images. Nat. Commun. 11, 3877 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Graham S et al. One model is all you need: multi-task learning enables simultaneous histology image segmentation and classification. Med. Image Anal. 83, 102685 (2023). [DOI] [PubMed] [Google Scholar]
- 72.Diao JA et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun. 12, 1613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wulczyn E et al. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS One 15, e0233678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riasatian A et al. Fine-tuning and training of DenseNet for histopathology image representation using TCGA diagnostic slides. Med. Image Anal. 70, 102032 (2021). [DOI] [PubMed] [Google Scholar]
- 75.Dosovitskiy A et al. An image is worth 16×16 words: transformers for image recognition at scale. In International Conference on Learning Representations (2021). [Google Scholar]
- 76.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kundra R et al. OncoTree: a cancer classification system for precision oncology. JCO Clin. Cancer Inform. 5, 221–230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bejnordi BE et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campbell JD et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48, 607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shao Z et al. TransMIL: transformer based correlated multiple instance learning for whole slide image classification. In 35th Conference on Neural Information Processing Systems (2021). [Google Scholar]
- 81.Campanella G et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gatta G et al. Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet – a population-based study. Lancet Oncol. 18, 1022–1039 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Ilse M, Tomczak J & Welling M Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning, 2132–2141 (2018). [Google Scholar]
- 84.He K, Zhang X, Ren S & Sun J Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (2016). [Google Scholar]
- 85.Kim YJ et al. PAIP 2019: liver cancer segmentation challenge. Med. Image Anal. 67, 101854 (2021). [DOI] [PubMed] [Google Scholar]
- 86.Lipkova J et al. Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies. Nat. Med. 28, 575–582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brennan CW et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 372, 2481–2498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roetzer-Pejrimovsky T et al. The Digital Brain Tumour Atlas, an open histopathology resource. Sci. Data 9, 55 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pati P et al. Weakly supervised joint whole-slide segmentation and classification in prostate cancer. Preprint at 10.48550/arxiv.2301.02933 (2023). [DOI] [PubMed]
- 91.Jacovi A, Caciularu A, Goldman O & Goldberg Y Stop uploading test data in plain text: practical strategies for mitigating data contamination by evaluation benchmarks. Preprint at 10.48550/arxiv.2305.10160 (2023). [DOI]
- 92.Magar I & Schwartz R Data contamination: from memorization to exploitation. In Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), 157–165 (2022). [Google Scholar]
- 93.Brown T et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020). [Google Scholar]
- 94.Dodge J et al. Documenting large webtext corpora: a case study on the colossal clean crawled corpus. In Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, 1286–1305 (2021). [Google Scholar]
- 95.Kapoor S & Narayanan A Leakage and the reproducibility crisis in machine-learning-based science. Patterns 4(9), 100804 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiang J & Zhang J Exploring low-rank property in multiple instance learning for whole slide image classification. In The Eleventh International Conference on Learning Representations (2022). [Google Scholar]
- 97.Niehues JM et al. Generalizable biomarker prediction from cancer pathology slides with self-supervised deep learning: a retrospective multi-centric study. Cell Rep. Med. 4, 100980 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kather JN et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 16, e1002730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pataki BÁ et al. HunCRC: annotated pathological slides to enhance deep learning applications in colorectal cancer screening. Sci. Data 9, 370 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barbano CA et al. UniToPatho, a labeled histopathological dataset for colorectal polyps classification and adenoma dysplasia grading. In 2021 IEEE International Conference on Image Processing (ICIP), 76–80 (IEEE, 2021). [Google Scholar]
- 101.Huo X et al. Comprehensive AI model development for Gleason grading: from scanning, cloud-based annotation to pathologist–AI interaction. Preprint at 10.2139/ssrn.4172090 (2022). [DOI]
- 102.Komura D et al. Restaining-based annotation for cancer histology segmentation to overcome annotation-related limitations among pathologists. Patterns 4, 100688 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng B, Misra I, Schwing AG, Kirillov A & Girdhar R Masked-attention mask transformer for universal image segmentation. In 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2022). [Google Scholar]
- 104.Fang Y et al. EVA: exploring the limits of masked visual representation learning at scale. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 19358–19369 (2023). [Google Scholar]
- 105.Wang Y, Chao W-L, Weinberger KQ & van der Maaten L SimpleShot: revisiting nearest-neighbor classification for few-shot learning. Preprint at 10.48550/arxiv.1911.04623 (2019). [DOI]
- 106.Snell J, Swersky K & Zemel R Prototypical networks for few-shot learning. In Advances in Neural Information Processing Systems 30 (2017). [Google Scholar]
- 107.Vorontsov E et al. Virchow: a million-slide digital pathology foundation model. Preprint at 10.48550/arxiv.2309.07778 (2023). [DOI]
- 108.Campanella G et al. Computational pathology at health system scale: self-supervised foundation models from three billion images. Preprint at 10.48550/arxiv.2310.07033 (2023). [DOI]
- 109.Lai J et al. Domain-specific optimization and diverse evaluation of self-supervised models for histopathology. Preprint at 10.48550/arxiv.2310.13259 (2023). [DOI]
- 110.Moor M et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023). [DOI] [PubMed] [Google Scholar]
- 111.Chen Z et al. Vision transformer adapter for dense predictions. In The Eleventh International Conference on Learning Representations (2023). [Google Scholar]
- 112.Wang X et al. SCL-WC: cross-slide contrastive learning for weakly-supervised whole-slide image classification. Advances in Neural Information Processing Systems 35, 18009–18021 (2022). [Google Scholar]
- 113.Kolesnikov A, Zhai X & Beyer L Revisiting self-supervised visual representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 1920–1929 (2019). [Google Scholar]
- 114.Chen RJ et al. Algorithmic fairness in artificial intelligence for medicine and healthcare. Nat. Biomed. Eng. 7, 719–742 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lu MY et al. A foundational multimodal vision language AI assistant for human pathology. Preprint at 10.48550/arxiv.2312.07814 (2023). [DOI] [Google Scholar]
- 116.Chen RJ et al. Scaling vision transformers to gigapixel images via hierarchical self-supervised learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2022). [Google Scholar]
- 117.Zhou J et al. iBOT: image BERT pre-training with online tokenizer. In International Conference on Learning Representations (2022). [Google Scholar]
- 118.Zhai X, Oliver A, Kolesnikov A & Beyer L S4L: self-supervised semi-supervised learning. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 1476–1485 (2019). [Google Scholar]
- 119.Bandi P et al. From detection of individual metastases to classification of lymph node status at the patient level: the CAMELYON17 challenge. IEEE Trans. Med. Imaging 38, 550–560 (2019). [DOI] [PubMed] [Google Scholar]
- 120.Devlin J, Chang M-W, Lee K & Toutanova K BERT: pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers; ) (2018). [Google Scholar]
- 121.Tian K et al. Designing BERT for convolutional networks: sparse and hierarchical masked modeling. In The Eleventh International Conference on Learning Representations (2023). [Google Scholar]
- 122.Sablayrolles A, Douze M, Schmid C & Jégou H Spreading vectors for similarity search. In International Conference on Learning Representations (2019). [Google Scholar]
- 123.Touvron H, Vedaldi A, Douze M & Jegou H Fixing the train–test resolution discrepancy. In Advances in Neural Information Processing Systems, Vol. 32 (eds Wallach H et al. ) (Curran Associates, 2019). [Google Scholar]
- 124.Dao T, Fu DY, Ermon S, Rudra A & Ré C FlashAttention: fast and memory-efficient exact attention with IO-awareness. In Advances in Neural Information Processing Systems (2022). [Google Scholar]
- 125.Liu Z et al. Swin transformer: hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 10012–10022 (2021). [Google Scholar]
- 126.Kolesnikov A et al. Big Transfer (BiT): general visual representation learning. In Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part V 16, 491–507 (Springer, 2020). [Google Scholar]
- 127.Lin T, Yu Z, Hu H, Xu Y & Chen C-W Interventional bag multi-instance learning on whole-slide pathological images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 19830–19839 (2023). [Google Scholar]
- 128.Loshchilov I & Hutter F Decoupled weight decay regularization. In International Conference on Learning Representations (2019). [Google Scholar]
- 129.Bentley JL Multidimensional binary search trees used for associative searching. Communications of the ACM 18, 509–517 (1975). [Google Scholar]
- 130.Zhu C, Byrd RH, Lu P & Nocedal J Algorithm 778: L-BFGS-B: Fortran subroutines for large-scale bound-constrained optimization. ACM Transactions on Mathematical Software 23, 550–560 (1997). [Google Scholar]
- 131.Sarıyıldız MB, Kalantidis Y, Alahari K & Larlus D No reason for no supervision: improved generalization in supervised models. In The Eleventh International Conference on Learning Representations (2023). [Google Scholar]
- 132.Fang Z et al. SEED: self-supervised distillation for visual representation. In International Conference on Learning Representations (2020). [Google Scholar]
- 133.Pedregosa F et al. Scikit-learn: machine learning in python. Journal of Machine Learning Research 12, 2825–2830 (2011). [Google Scholar]
- 134.Ghiasi G et al. Simple copy-paste is a strong data augmentation method for instance segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2918–2928 (2021). [Google Scholar]
- 135.El Banani M, Desai K & Johnson J Learning visual representations via language-guided sampling. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 19208–19220 (2023). [Google Scholar]
- 136.Koch G, Zemel R & Salakhutdinov R Siamese neural networks for one-shot image recognition. In Proceedings of the 32nd International Conference on Machine Learning (2015). [Google Scholar]
- 137.Vinyals O, Blundell C, Lillicrap T, Kavukcuoglu K & Wierstra D Matching networks for one shot learning. In Advances in Neural Information Processing Systems 29 (2016). [Google Scholar]
- 138.Yu J-G et al. Prototypical multiple instance learning for predicting lymph node metastasis of breast cancer from whole-slide pathological images. Med. Image Anal. 85, 102748 (2023). [DOI] [PubMed] [Google Scholar]
- 139.Yu Z, Lin T & Xu Y SLPD: slide-level prototypical distillation for WSIs. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 259–269 (Springer, 2023). [Google Scholar]
- 140.Quiros AC et al. Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unlabeled, unannotated pathology slides. Preprint at 10.48550/arxiv.2205.01931 (2022). [DOI] [PMC free article] [PubMed]
- 141.Yang J, Chen H, Yan J, Chen X & Yao J Towards better understanding and better generalization of low-shot classification in histology images with contrastive learning. In International Conference on Learning Representations (2021). [Google Scholar]
- 142.Tian Y, Wang Y, Krishnan D, Tenenbaum JB & Isola P Rethinking few-shot image classification: a good embedding is all you need? In Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, August 23–28, 2020, Proceedings, Part XIV 16, 266–282 (Springer, 2020). [Google Scholar]
- 143.Lloyd S Least squares quantization in PCM. IEEE Transactions on Information Theory 28, 129–137 (1982). [Google Scholar]
- 144.Zhu X, Yao J, Zhu F & Huang J WSISA: making survival prediction from whole slide histopathological images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 7234–7242 (2017). [Google Scholar]
- 145.Yao J, Zhu X & Huang J Deep multi-instance learning for survival prediction from whole slide images. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 496–504 (Springer, 2019). [Google Scholar]
- 146.Yao J, Zhu X, Jonnagaddala J, Hawkins N & Huang J Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med. Image Anal. 65, 101789 (2020). [DOI] [PubMed] [Google Scholar]
- 147.Li R, Yao J, Zhu X, Li Y & Huang J Graph CNN for survival analysis on whole slide pathological images. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 174–182 (Springer, 2018). [Google Scholar]
- 148.Sivic J & Zisserman A Video Google: A text retrieval approach to object matching in videos. In Proceedings of the Ninth IEEE International Conference on Computer Vision, 1470–1477 (IEEE, 2003). [Google Scholar]
- 149.Fei-Fei L & Perona P A Bayesian hierarchical model for learning natural scene categories. In 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05) Vol. 2, 524–531 (IEEE, 2005). [Google Scholar]
- 150.Cruz-Roa A, Caicedo JC & González FA Visual pattern mining in histology image collections using bag of features. Artif. Intell. Med. 52, 91–106 (2011). [DOI] [PubMed] [Google Scholar]
- 151.Xu Y et al. Weakly supervised histopathology cancer image segmentation and classification. Med. Image Anal. 18, 591–604 (2014). [DOI] [PubMed] [Google Scholar]
- 152.Chen C et al. Fast and scalable search of whole-slide images via self-supervised deep learning. Nat. Biomed. Eng. 6, 1420–1434 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gillette MA et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 182, 200–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Satpathy S et al. A proteogenomic portrait of lung squamous cell carcinoma. Cell 184, 4348–4371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhu M et al. Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Sci. Rep. 11, 7080 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 374, 135–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Davis CF et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li Y et al. Histopathologic and proteogenomic heterogeneity reveals features of clear cell renal cell carcinoma aggressiveness. Cancer Cell 41, 139–163 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Brancati N et al. BRACS: a dataset for breast carcinoma subtyping in H&E histology images. Database 2022, baac093 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bulten W et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 21, 233–241 (2020). [DOI] [PubMed] [Google Scholar]
- 162.Veeling BS, Linmans J, Winkens J, Cohen T & Welling M Rotation equivariant CNNs for digital pathology. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16–20, 2018, Proceedings, Part II 11, 210–218 (Springer, 2018). [Google Scholar]
- 163.Koh PW et al. WILDS: a benchmark of in-the-wild distribution shifts. In International Conference on Machine Learning, 5637–5664 (PMLR, 2021). [Google Scholar]
- 164.Aresta G et al. BACH: grand challenge on breast cancer histology images. Med. Image Anal. 56, 122–139 (2019). [DOI] [PubMed] [Google Scholar]
- 165.Brummer O, Pölönen P, Mustjoki S & Brück O Computational textural mapping harmonises sampling variation and reveals multidimensional histopathological fingerprints. British Journal of Cancer 129, 683–695 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tolkach Y et al. Artificial intelligence for tumour tissue detection and histological regression grading in oesophageal adenocarcinomas: a retrospective algorithm development and validation study. Lancet Digit. Health 5, e265–e275 (2023). [DOI] [PubMed] [Google Scholar]
- 167.Howard FM et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat. Commun. 12, 4423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Macenko M et al. A method for normalizing histology slides for quantitative analysis. In 2009 IEEE international Symposium on Biomedical Imaging: From Nano to Macro, 1107–1110 (IEEE, 2009). [Google Scholar]
- 169.Abousamra S et al. Deep learning-based mapping of tumor infiltrating lymphocytes in whole slide images of 23 types of cancer. Front. Oncol. 11, 806603 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Paszke A et al. PyTorch: an imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems 32 (2019). [Google Scholar]
- 171.Goldberger AL et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101, e215–e220 (2000). [DOI] [PubMed] [Google Scholar]
- 172.Azizi S et al. Medical AI research foundations: a repository of medical foundation models (version 1.0.0). PhysioNet; 10.13026/grp0-z205 (2023). [DOI] [Google Scholar]
- 173.Wu Y, Kirillov A, Massa F, Lo W-Y & Girshick R Detectron2. GitHub; https://github.com/facebookresearch/detectron2 (2019). [Google Scholar]
- 174.Clark K et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCGA and CPTAC data consisting of whole-slide images and labels can be accessed through the NIH genomic data commons (https://portal.gdc.cancer.gov) and proteomics data commons (https://proteomic.datacommons.cancer.gov), respectively. GTEx data added to the pretrainingdatasetcanbeaccessedthroughtheGTExportal(https://www.gtexportal.org/home/). CPTAC data consisting of all publicly available datasets analyzed in this work can be can accessed in their respective data portals: CRC-100K (https://zenodo.org/record/1214456), HunCRC ROIs (10.6084/m9.figshare.c.5927795.v1), HunCRC slides (10.7937/ tcia.9cjf-0127), BACH (https://iciar2018-challenge.grand-challenge.org/Dataset/), TCGA CRC-MSI (https://zenodo.org/record/3832231), CCRCC tissue classification (https://zenodo.org/record/7898308), TCGA-TILs (https://zenodo.org/record/6604094), TCGA Uniform (https://zenodo.org/record/5889558), UniToPatho (https://zenodo.org/record/4643645), ESCA(https://zenodo.org/record/7548828), CAMELYON17-WILDS (https://wilds.stanford.edu/datasets), EBRAINS (10.25493/WQ48-ZGX), DHMC (https://bmirds.github.io/KidneyCancer), BRACS (https://bracs.icar.cnr.it), PANDA (https://panda.grand-challenge.org), SegPath (https://zenodo.org/record/7412731) and AGGC (https://zenodo.org/record/6460100). TCGA, CPTAC, HunCRC and TCGA-TILS can also be accessed using The Cancer Imaging Archive175. Links for all datasets are also listed in Supplementary Table 73. We note that data from AGGC were obtained from a public grand challenge (of the same name (https://aggc22.grand-challenge.org)) with a pending publication101, with permission granted by the challenge organizers to present results from this dataset. No internal patient data were specifically collected for this study. This study relies on retrospective analysis of anonymized whole-slide images. Following institution policies, all requests for data collected or curated in-house will be evaluated on a case-by-case basis to determine whether the data requested and the use case comply with intellectual property or patient privacy obligations.


