Abstract
The Holliday junction (HJ) is a central intermediate in various genetic processes including homologous and site-specific recombination and DNA replication. Branch migration allows the exchange between homologous DNA regions, but the detailed mechanism for this key step of DNA recombination is unidentified. Here, we report direct real-time detection of branch migration in individual molecules. Using appropriately designed HJ constructs we were able to follow junction branch migration at the single-molecule level. Branch migration is detected as a stepwise random process with the overall kinetics dependent on Mg2+ concentration. We developed a theoretical approach to analyze the mechanism of HJ branch migration. The data show steps in which the junction flips between conformations favorable to branch migration and conformations unfavorable to it. In the favorable conformation (the extended HJ geometry), the branch can migrate over several base pairs detected, usually as a single large step. Mg2+ cations stabilize folded conformations and stall branch migration for a period considerably longer than the hopping step. The conformational flip and the variable base pair hopping step provide insights into the regulatory mechanism of genetic processes involving HJs.
Keywords: FRET, recombination DNA, four-way junctions, fluorescence microscopy
The Holliday junction (HJ), suggested in 1964 by Robin Holliday (1), is a central intermediate in homologous and site-specific recombination (2). Movement of the crossover junction along the DNA allowing for length extension of the heteroduplex is termed branch migration (Fig. 1a). Branch migration, whether spontaneous or mediated by proteins, is a key step in various genetic processes involving the HJ. Recent data show that HJs are critical intermediates during pauses of DNA replication (3–5). Various models for the HJ intermediate have been used to unravel the structural basis for branch migration and resolution of the junction. The immobile four-way junction was a primary model system, and a great deal of information on the structure of HJs was obtained. Numerous techniques (6–11), including very recent x-ray crystallography analysis (12, 13), show that in the presence of multivalent cations the junction adopts an antiparallel orientation in which the four helices stack in pairs to form two double-helical domains. Single-molecule FRET studies showed that immobile HJs adopt two conformational states undergoing transitions between them by an extended conformation acting as a transient state (14). This important work nicely demonstrated the power of single-molecule FRET microscopy as a tool for dynamic studies of a complex system such as a four-way DNA junction.
Fig. 1.
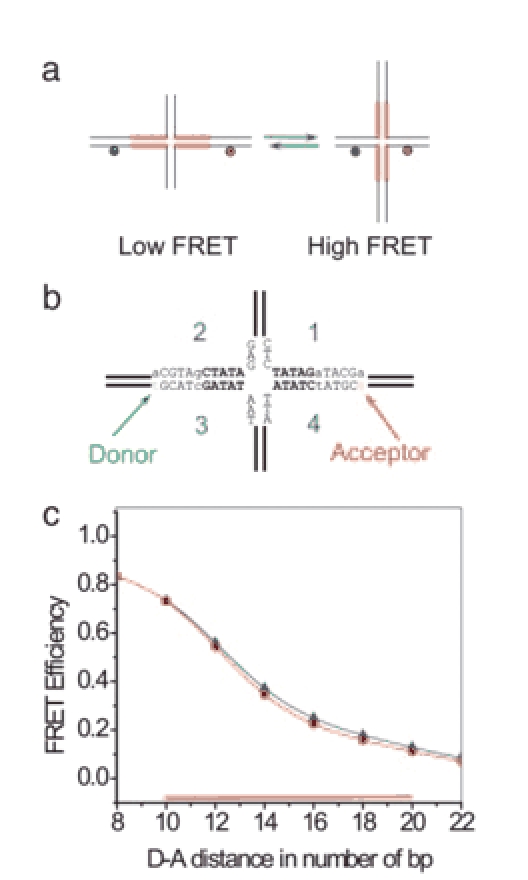
Mobile HJ capable of branch migration. (a) Schematic illustrating the rationale for the mobile HJ design used for the spFRET studies. Homologous regions capable of branch migration are indicated by thick red lines. (b) Nucleotide sequences of the central part of the junction including the donor and acceptor sites are shown. The bases of the homologous region are in bold. Dye locations are indicated by arrows and corresponding nucleotides are in lowercase. (c) Calculated dependence of FRET efficiency on the number of base pairs between the donor and the acceptor. The red line with filled circles corresponds to extended conformation (as in b) and the blue line with triangles corresponds to the folded conformation. The region of HJ mobility among the six possible configurations shown as red or blue dots is indicated by the red horizontal bar.
The majority of available data were obtained from immobile HJs and thus do not provide direct insights into the mechanism of branch migration. An exception is a series of papers by Hsieh and coworkers (15–17) in which the kinetics of branch migration were studied. A model suggested 30 years ago (18) relied on a parallel orientation of exchanging strands. The studies of Hsieh and coworkers (15–17) challenged this view and suggested an alternative model in which an extended configuration of the junction is more appropriate for spontaneous movement of the HJ. We (19) have recently tested these models by using a HJ undergoing branch migration and time-lapse atomic force microscopy, a nanoimaging technique capable of visualizing DNA dynamics. The single-molecule atomic force microscopy experiments support the model for branch migration with the extended conformation of the HJ.
Despite this progress, many questions related to the detailed mechanism of branch migration remain unanswered. Most importantly we still do not know the pathways and step size for branch migration. It has been proposed that branch migration is a stepwise process, but no direct experimental evidence for such a stepwise mechanism has been obtained. We need to know what the step size is and how fast the junction hops between adjacent conformations. Experiments with immobile junctions have shown that divalent cations profoundly affect the junction structure. These cations are physiologically important components, and we must know how they affect branch migration.
In this article, we applied single-pair FRET (spFRET) microscopy to the HJ capable of branch migration (mobile HJ) to answer the questions above. Using appropriately designed HJ constructs (Fig. 1b) we were able to follow junction branch migration at the single-molecule level. Branch migration is detected as a stepwise random process with the overall kinetics dependent on Mg2+ concentration. We developed a theoretical approach for analyzing the mechanism of HJ branch migration. The data obtained paint the following picture. Branch migration is a stepwise process in which the junction flips between an open conformation favorable to branch migration and a folded conformation unfavorable to it. In the favorable conformation (the extended HJ geometry), the branch migrates over several base pairs detected, usually as a single hop. Mg2+ cations facilitate folding the junction in the unfavorable conformation and thus stall branch migration for a period considerably longer than the hopping rate in the activated (open) state. These findings suggest a dynamic model for regulation of the branch migration rate by stabilizing specific conformations; this article provides insight into regulation of recombination events within cells.
Methods
HJ Assembly. Two sets of four oligonucleotides for assembly of mobile (m) and immobile (i) versions of HJs were synthesized (IDT, Coralville, IA). The oligonucleotides were internally amino-labeled for attaching the dyes. The sequences are as follows: 1m, biotin-5′-TCTTTTGATAAGCTTGCAAGCATAGATATCTCGTAATTTCCGGTTAGGT; 2m, 5′-ACCTAACCGGAAATTACGAGATATCGATGCATGCAAGCTTCACA; 3m, 5′-TGTGAAGCTTGCA/iAmMC6T/GCATCGATATAATACGTGAGGCCTAGGATC; 4m, 5′-GATCCTAGGCCTCACGTATTATATCTATGC/iAmMC6T/TGCAAGCTTATCA; 1i, biotin-5′-TCTTTTGATAAGCTTGCAAGCATAGAGATCTCGTAATTTCCGGTTAGGT; 2i: 5′-ACCTAACCGGAAATTACGAGTCAACGATGCATGCAAGCTTCACA; 3i, 5′-TGTGAAGCTTGCA/iAmMC6T/GCATCGTTGAAATACGTGAGGCCTAGGATC; and 4i, 5′-GATCCTAGGCCTCACGTATTATCTCTATGC/iAmMC6T/TGCAAGCTTATCA.
The 3m, 3i, 4m, and 4i oligonucleotides were labeled with succinimide esters of Cy3 or Cy5 dyes (Amersham Pharmacia Biosciences) according to the protocol provided. Labeled oligonucleotides were purified by RP-HPLC using a Matrix Polystyrene/divinyl benzene column (5RPC ST 4.6/150, Amersham Pharmacia Biotech). Purification quality was controlled by measuring the absorption spectra. The molar ratios of DNA and attached dyes in the sample were close to 1:1 for all samples. Left and right parts of the junctions were assembled and then annealed together to form four-way designs. Two tubes with mixtures of oligonucleotides 1 and 4 (tube 1) and oligonucleotides 2 and 3 in concentrations of 3 μM in annealing buffer containing 50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, and 100 mM NaCl, were placed in ≈250 ml of boiling water for 5 min and cooled to ambient temperature for ≈2 h. Four-way junctions (Fig. 1b) were prepared by mixing 1 μM solutions of the two halves in 10 mM Tris (pH 7.5), 50 mM NaCl, and 10 mM MgCl2 buffer, heating at 50°C for 3 min and cooling to 4°C for 30 min. The yield of the annealing reaction was ≈80–90% determined by gel electrophoresis.
A similar protocol was used for the preparation of the mobile HJ with alternative arrangements of the donor and acceptor (see Fig. 5, which is published as supporting information on the PNAS web site).
Measurements and Experimental Setup. Glass coverslips were cleaned with 1:1 nitric acid/hydrogen peroxide mixture and stored in deionized water before use. The coverslips were treated sequentially with 1 mg/ml of biotinylated BSA (Sigma) in pH 7.5 TES buffer (10 mM Tris·HCl/150 mM NaCl/1 mM EDTA) for 10 min, rinsed with TES buffer and a 0.5 μM solution of streptavidin in the same buffer for 10 min, and rinsed with TES buffer. A 250-pM solution of biotinylated HJs in TES buffer was added and incubated for ≈30 min. Coverslips were rinsed with TES buffer. Measurements were performed in imaging buffer containing oxygen-scavenging system based on glucose oxidase and catalase (20) and different concentrations of sodium and magnesium ions to maintain the ionic strength constant (150 mM).
Single-molecule fluorescence measurements were carried out on a home-built confocal microscope built around an Olympus IX70 inverted microscope body equipped with a motorized stage. Specifics are given in Supporting Text, which is published as supporting information on the PNAS web site.
Data Analysis. The single-molecule time trajectories of fluorescence changes well above background were analyzed. Averaged data were obtained by using the following algorithm. The HJ is considered to remain on the same migration step in adjacent time bins 1 and 2 if the FRET efficiency difference |E1 - E2| < 0.12. The mean plateau efficiency is calculated as Ē = (E1 + E2)/2. The plateau continues on the third time bin if |E3 - Ē| < 0.12. This procedure repeats itself for subsequent bins where the new mean efficiency is determined according to general formula
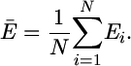 |
If the FRET efficiency at a later time bin differs >0.12 from the mean efficiency of the plateau (|Ei+1 - Ē| > 0.12) the algorithm will set the mean efficiency for all previous points on the plateau, determine the state duration from the total number of time bins, and then move on to a new plateau. The threshold value of 0.12 was chosen as a minimal discernable step size, which is confidentially larger than the background noise. The noise level was estimated from the SD of the data points on the long plateaus for mobile and immobile four-way junctions and does not exceed ≈0.06, half of the chosen threshold. For a Gaussian distribution, only 5% of the data will be scattered beyond a ±2-SD interval from the mean. If we apply this step-averaging algorithm to an immobile HJ (Fig. 2d, red line) we observe just a few false steps, which is evidence that the chosen criteria work well.
Fig. 2.
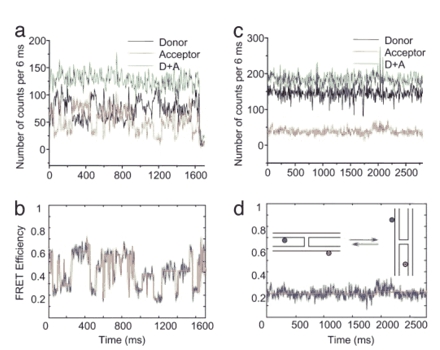
Time traces of the fluorescence intensities and FRET efficiencies of mobile and immobile HJs in 10 mM MgCl2 buffer. (a) The time dependences of single-molecule fluorescence intensities of donor (black), acceptor (red), and the summed intensity [donor plus acceptor (D+A), green] for the mobile HJ. Intensities are taken over a 6-ms binning interval. (b) The time dependence of the FRET efficiency (blue line) of the data shown in a. The red line is the result of an averaging procedure described in Methods, which better reveals plateaus. (c) Immobile HJ time traces of the single-molecule fluorescence intensities of the donor (black) and acceptor (red) and the summed intensity [donor plus acceptor (D+A), green]. (d) The time dependence of the FRET efficiency obtained from the data shown in c (blue line). (Inset) Two folded conformations of immobile HJ are shown schematically.
Calculation of FRET Efficiency. Theoretical FRET efficiencies for different junction locations were estimated from the model where dyes are located on the opposite sides of the junction point and opposite strands are considered aligned across the junction point as in Fig. 1b. The computed curve in Fig. 1c was determined by (21–23):
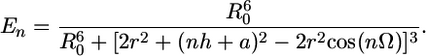 |
Here R0 is the Forster critical distance (in nm) between donor and acceptor at which the energy transfer efficiency is 50%, n is the number of base pairs separating the donor and the acceptor, h is the distance between DNA base pairs, a is the distance through the four-way junction, Ω is the twist angle between adjacent base pairs, and r is the distance to the dye center from the helix axis. The distance across the junction point is estimated to be 1.7 ± 0.1 nm according to crystallographic data (24, 25). The change of the distance between the dyes (0.68 nm per 1-bp hop) includes in addition to translational movement a rotational component (69° per step).
Results and Discussion
The Design of the HJ. Schematics for the HJ design for the spFRET studies are shown in Fig. 1. The junction was assembled from four single-stranded oligonucleotides as described in Methods. The homology between the arms of the junction is interrupted by incorporation of a nonhomology base pair that is sufficient to block spontaneous branch migration (17). Therefore, branch migration of the junction is limited to two 5-bp regions on both sides of the junction indicated in Fig. 1a by thick lines. The sequence for the central part of the construct is shown in Fig. 1b. Nucleotides in the homologous region undergoing branch migration are in bold in Fig. 1b. The donor and acceptor dyes were attached to modified nucleotides indicated by arrows in Fig. 1b. During a 1-bp hop the distance between the donor and acceptor changes by 2 bp, which leads to a measurable change in FRET intensity. Fig. 1c shows the calculated dependence of the FRET intensity based on the known value for R0 (6 nm) for the Cy3 (donor) and Cy5 (acceptor) dyes (22, 23, 26, 27) and the crystallographic data for the unfolded conformation of the junction (24, 25). The highest value of the FRET efficiency corresponds to the smallest donor-acceptor distance (Fig. 1a Right) and the efficiency drops upon moving the dyes apart as the junction migrates. The dependence of the FRET efficiency on the intervening number of base pairs (Fig. 1c, circles) has been calculated assuming extended unfolded arms at 90° (Fig. 1a) and B-form geometry for the helical region. According to the estimates provided by Fig. 1c, the range of FRET values for the design with two 5-bp homologous regions (10 bp for the entire branch migration range) is shown as a horizontal red bar along the x axis and varies between ≈0.7 and 0.1. We estimate from Fig. 1c values of the efficiency Ei of 0.73, 054, 0.35, 022, 0.16, and 0.11 for each of the six branch migration configurations i within the branch migration region. A similar analysis for the folded HJ geometry corresponding to the crystallographic structure of the junction (12) produces the blue curve with triangles in Fig. 1c. This curve almost coincides with the dependence obtained for the unfolded conformation of the junction, suggesting that the selected method of labeling the sample is sensitive to branch migration steps and is much less sensitive to conformational transitions of the junction between unfolded and folded states.
To test our design and its estimated FRET efficiencies, we performed ensemble FRET efficiency measurements for an immobile HJ corresponding to the longest donor-acceptor distance of our design and find the value 0.15. This value is in good agreement with the calculated value of 0.11 given above. Additionally, we perform ensemble FRET experiments of our designed mobile HJ and obtain 0.35 ± 0.2, in good agreement with the mean value of 0.35 obtained by averaging Ei over the six values indicated above.
Branch Migration at the Single-Molecule Level. The results of the spFRET experiments with mobile HJ are shown in Fig. 2a. One end of the HJ molecules was anchored to the glass surface by biotin-streptavidin links. The buffer containing 10 mM Mg2+ cations (TNM: 10 mM Tris·HCl, 100 mM NaCl, and 10 mM MgCl2) was added to the sample in this experiment. The time-dependent signals from the donor, acceptor, and total fluorescence values for both dyes (donor plus acceptor) are shown in Fig. 2a. Correlated changes in the donor and acceptor fluorescence intensity are clearly seen. The total donor plus acceptor fluorescence (Fig. 2a, green line) remains constant during these observations. The FRET efficiency time trajectories calculated from these data are shown in Fig. 2b. They show plateaus with stepwise changes of the FRET intensity as a function of time. This pattern is seen more clearly after averaging the original data by using the approach described in Methods. These data show that FRET efficiencies vary in the range of ≈0.6 to 0.15. These experimental values are in remarkably good agreement with calculated Ei values (Fig. 1c), suggesting that the observed FRET data dynamically follows the HJ undergoing branch migration. Control data for an immobile HJ with a specific positioning of the donor and acceptor molecules showed a constant fluorescence signal in both donor (black) and acceptor channels (red) (Fig. 2c) and a constant FRET efficiency ≈0.2 (Fig. 2d), indicating that the labeling of opposite arms is not sensitive to folding of the HJ in the two different stacked antiparallel conformers. Schematically these conformers are shown in Fig. 2d Inset. This finding supports our assumption that our design is not sensitive to the particular stacking conformation or unfolding of the arms (Fig. 1c), but only to the HJ branch location.
The data in Fig. 2b suggest that mobile HJ undergoes a series of sharp transitions between the states with reasonably well defined FRET values. For example, during the time range between 250 and 450 ms the molecule exists in a state with high FRET (≈0.6) and then drops to the FRET value 0.25. After that, the molecule jumps to another high FRET state (≈0.5), then undergoes a series of fast transitions with lower FRET efficiency values but with a clear preference to return to a high FRET state (during ≈500 and ≈900 ms). After staying at the low FRET state (≈50 ms) the molecule adopts the state with a slightly higher FRET (≈0.38) and after 100 ms adopts the state with a very low FRET (≈0.15). It stays there for another 100 ms followed by a jump to a higher FRET state (0.45) where it stays as long as 400 ms except for a short time interval, ≈1,400 ms, when the transition to a state with a low FRET occurs. Finally, the molecule jumps into the state with a very high FRET value (≈0.6) and after 20 ms quickly adopts the conformation with a low FRET (≈0.25). These long-live states are most easily seen after averaging the data (Fig. 2b, red line). Additional time trajectories for averaged FRET values illustrating a stepwise change of FRET intensities are shown in Fig. 6a, which is published as supporting information on the PNAS web site. Importantly, there is a good correlation in the FRET values for the plateaus in independent experiments. When the FRET plateau values are combined in one histogram, discrete peaks are resolved (Fig. 6b), suggesting that branch migration occurs through a selected number of discrete states along the branch migration path.
A critical issue for this interpretation of the FRET data is that the dye-labeling method used is very sensitive to branch migration and much less sensitive to conformational transitions of the junction between unfolded and folded states (Fig. 2d Inset). There is good evidence supporting this interpretation. First, the estimated FRET values for the unfolded conformation of the HJ and the folded geometry corresponding to the recent crystallographic data of ref. 12 are virtually identical (compare triangles and circles in Fig. 1c). Second, there is no temporal change in the FRET efficiency for the immobile junction (Fig. 2d). If folding and opening of the junction led to changes in the FRET efficiency, we should be able to observe the FRET variation. The results in Fig. 2d show no jumps of FRET values. This finding is consistent with the recent publication from Ha's group (28) indicating that similar labeling arrangement is insensitive to changes of global junction conformation.
Effect of Mg2+ Cations on the HJ Dynamic. The data described above were obtained in the presence of 10 mM Mg2+. To analyze the effect of Mg cations on the branch migration rate the same samples were prepared in different concentrations of Mg2+ and spFRET experiments were performed. The time dependence of the FRET efficiencies for 0.3 mM Mg2+ and 2 mM Mg2+ are shown in Fig. 3. As in Fig. 2b, blue lines are the raw efficiency data (binning interval 6 ms) and red lines are the averaged data. The data for 0.3 mM Mg2+ (Fig. 3a, blue line) are “noisy,” showing a statistical variation about a mean value. The variations of the FRET efficiency are considerably larger than statistical noise detected from an immobile junction (Fig. 2d), suggesting that observed variations are caused by the dynamics of the mobile HJ. The analysis of the data revealed the peaks in the FRET efficiency distribution correspond to peak values obtained for the data at 10 mM Mg2+ (see Supporting Text).
Fig. 3.
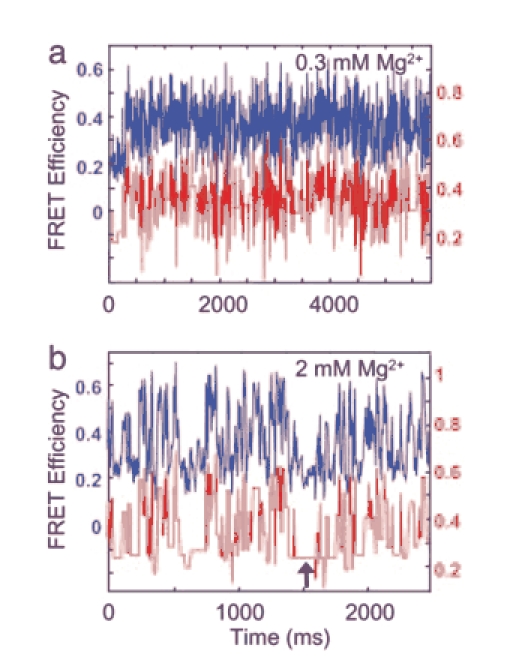
The spFRET data for mobile HJs obtained at different concentrations of Mg2+ cations. (a) Measured time dependence of the FRET efficiency in a 0.3 mM MgCl2 buffer (upper blue curve, left axis). Red line (shifted for clarity, right axis) shows the same data but averaged as described in Methods. (b) Time dependence of the FRET efficiency in a 2 mM MgCl2 buffer. Raw (blue) and averaged data (red) are presented as in a. All efficiency data were binned with 6-ms time intervals.
Data obtained with 2 mM Mg2+ is shown in Fig. 3b. A stepwise dynamics is seen in this spectrum, so the states with a lifetime as large as ≈100 ms, indicated by arrows in Fig. 3b, could be observed. This rare long lifetime at lower Mg2+ concentration and the low rate of branch migration at the elevated 10 mM Mg2+ shows that Mg2+ produces an increase of the residence time of nonmigrating states of the junction. Additional analysis of the data obtained at different concentrations of Mg2+ and with alternative label positions is given in Supporting Text.
Branch Migration and Global Conformations of HJs. To correlate the conformations of HJs with branch migration we performed experiments with the same HJ design, but labeled in a way that allows us to detect transitions between folded conformations of the junction (14). The labels are placed at adjacent arms to detect the transition between two folded conformations (Fig. 5a; see also details in Supporting Text). The time dependence of FRET obtained in the presence of 10 mM Mg2+ (Fig. 5b) shows a step-like pattern in which FRET values change between low (≈0.4) and high values (≈0.8). This observation is similar to the data obtained by Ha's group (14) for immobile HJs, suggesting that the mobile junction also flips between two conformations upon branch migration. There is some variability in the low and high FRET values in Fig. 5b, reflecting the change in the donor-acceptor distance caused by branch migration during the time trajectories. However, the FRET variability is considerably less than observed for the same HJ design, but labeled differently (compare Fig. 2). At low Mg2+ concentration (0.3 mM), the transition is so fast that individual steps cannot be clearly resolved (Fig. 5c). Both data sets are fully consistent with the stepwise dynamics of HJs branch migration observed by the alternative labeling (Fig. 2 b and c) where the two dyes do not stack together. Importantly, we have measured the residence times corresponding to the plateaus lengths for the data obtained in the presence of 10 mM Mg2+. The value 74 ± 5 ms calculated for a set of time trajectories similar to one shown in Fig. 5b is very close to 71 ± 5.4 ms obtained for the design with alternate labeling (Table 1, which is published as supporting information on the PNAS web site). This correlation suggests that plateaus in the branch migration time trajectories (Figs. 2b and 5a) correspond to folded conformations of the junction. Therefore, we conclude that folded conformations of HJs stall branch migration. In summary, these data show that various labeling strategies in spFRET experiments need to be applied, depending on whether global conformational changes or branch migration dynamics of HJs are analyzed.
Theoretical Model for the HJ Branch Migration. We model the single-molecule FRET efficiency E(t) by using a stochastic Monte Carlo hopping process among Nsites sites (here 6), where the average hopping time is τ between nearest-neighbor sites. Further details are provided in Fig. 7, which is published as supporting information on the PNAS web site, and Supporting Text.
We start with a single-step hopping model in which nearest-neighbor hops occur from site i to i + 1 or from site i to site i - 1 with equal rates k = 1/τ. End configurations i = 1 and i = 6 can only hop one way and not two directions as for configurations in the interior. The only parameter is τ, which is adjustable with the Mg2+ concentration (Supporting Text gives further details). The residence time, τR, is the average time that the HJ resides in a specific configuration for a site not at the end, τR = τ/2. An end site has only one hopping direction and the average residence time is twice as long.
The characteristic behavior of E(t) is quite different in the slow and rapid hopping limits. Slow hopping is when the residence time is much larger than the binning time (tbin), and rapid hopping is when it is much shorter. An example of the slow hopping limit is shown in Fig. 4a. The simulation produces plateaus connected by steps, and the qualitative features of the simulated single-molecule FRET efficiency data are quite similar to those observed in the experiments (e.g., Fig. 2b) at high Mg2+ concentrations. An example of the rapid hopping limit is shown in Fig. 4b. The simulation produces rapid fluctuations about the average efficiency and is qualitatively similar to the experimental FRET efficiency data at low Mg2+ concentrations such as those in Fig. 3a.
Fig. 4.
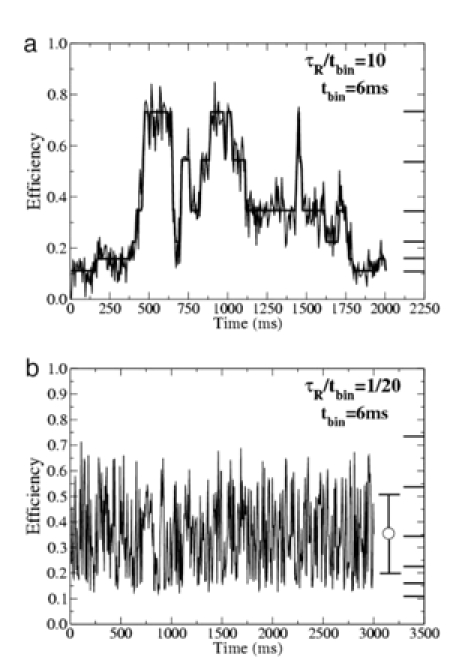
Theoretical Monte Carlo simulations of single-molecule FRET efficiencies of a six-site migrating HJ. The binning time is 6 ms. The central efficiency at each of the six sites is shown as a heavy line on the rightmost axis. (a) The FRET efficiency for long residence time (slow hopping), τR = 10 tbin. The segmented line is the same data without Gaussian noise. (b) Same as a but for a short residence time (rapid hopping), τR = 1/20 tbin. The error bar on the right shows the time-averaged efficiency 〈E〉t and its deviation δE.
The Branch Migration Step Size. The FRET time trajectories similar to one shown in Fig. 2b (see also Fig. 5a) show that the FRET step sizes in the data vary over a broad range. For example, steps corresponding to changes of FRET between 0.2 and 0.6 can be found in addition to FRET changes <0.1. Such variability of the step size indicates that branch migration is not a single equidistant hop, but rather hops occurring over different distances. To extract the hopping distance from the FRET data we must convert the steps in FRET efficiencies into changes of the donor-acceptor distance and eventually into the hopping step size as a number of base pairs. As a rough estimate, the theoretical dependence of the FRET efficiency on the donor-acceptor distance shown in Fig. 1c can be used to translate steps in FRET efficiencies to base pair steps. This dependence predicts possible stepwise change of FRET intensities between 0.11 and 0.71. Experimental data for FRET step values are in the range of 0.15 to 0.6 (Fig. 5b); this result is in satisfactory coincidence, justifying the use of the calibration plot in Fig. 1c for a qualitative analysis of the branch migration data. As examples of calibration using this curve, FRET changes between 0.54 and 0.16 ideally correspond to hopping over 3 bp, whereas FRET changes as small as 0.1 can correspond to a 1-bp hop. Based on this qualitative analysis, we conclude that branch migration over a homology region >1 bp can occur.
Conclusions
The single-molecule, real-time observations of the dynamics of HJs and the analysis of these data provide the following mechanism of branch migration. The junction flips between extended and folded conformations that favor and block branch migration, respectively. Branch migration occurs while the junction is in an unfolded state and it stops when the molecule adopts a folded conformation. The stability of the folded state determines how long the junction stalls. Therefore, the entire branch migration process appears as a random overall stepwise process, with an effective rate that depends on the ratio between lifetimes of folded and unfolded states. This ratio depends on environmental conditions, especially on the concentration of divalent cations. The estimated hopping rate is ≈100 μs, and in the presence of 10 mM Mg2+ folded states with the lifetimes as long as 100 ms were observed, suggesting that the rate of branch migration can drop 1,000-fold upon increase of the Mg2+ concentration. This prediction is in perfect agreement with papers (15, 16) in which the rate of branch migration was measured. Another critical finding of our work is that the step size between adjacent conformations is not necessarily 1 bp as previously hypothesized (15, 16, 29). Rather, the junction migration over the large homology region was observed, suggesting that at favorable conditions one step can cover a large DNA region including several base pairs. This finding explains our recent atomic force microscopy study in which a dissociation of homologous HJs caused by migration over ≈100-bp region in the absence of Mg2+ cations observed as a fast event occurred between two consecutive scans (19).
The HJ is an intermediate state for various genetic processes and it is targeted by a number of structure specific proteins. Some of them (RuvA protein of Escherichia coli) promote branch migration (3), whereas other proteins such as the hMSH4–hMSH5 complex (30) prefer folded conformations of the HJ and act as a clamp for branch migration. In cases where branch migration is required, keeping the junction in an unfolded conformation is sufficient for providing a long-range spontaneous migration of the junction. At the same time, folding the junction is an effective brake that immediately blocks its movement. Therefore, various structural proteins competing for binding to the HJ can play roles of molecular switches turning on or off branch migration.
Supplementary Material
Acknowledgments
We thank L. Shlyakhtenko, A. Lushnikov, and other members of Y.L.L.'s laboratory for discussion of the results at various stages of manuscript preparation and R. Clegg and P. Hsieh for fruitful discussion of the article and valuable suggestions. This research was supported by National Institutes of Health Grant GM0062235 (to Y.L.L.).
Author contributions: Y.L.L. designed research; M.K., D.D., O.F.S., and Y.L.L. performed research; M.K., D.D., O.F.S., and Y.L.L. analyzed data; and M.K., D.D., O.F.S., and Y.L.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HJ, Holliday junction; spFRET, single-pair FRET.
References
- 1.Holliday, R. (1964) Genet. Res. 5, 282-304. [Google Scholar]
- 2.Leach, D. R. F. (1996) Genetic Recombination (Blackwell, Oxford), pp. 15-59.
- 3.McGlynn, P. & Lloyd, R. G. (2001) J. Biol. Chem. 276, 41938-41944. [DOI] [PubMed] [Google Scholar]
- 4.Postow, L., Ullsperger, C., Keller, R. W., Bustamante, C., Vologodskii, A. V. & Cozzarelli, N. R. (2001) J. Biol. Chem. 276, 2790-2796. [DOI] [PubMed] [Google Scholar]
- 5.Grompone, G., Seigneur, M., Ehrlich, S. D. & Michel, B. (2002) Mol. Microbiol. 44, 1331-1339. [DOI] [PubMed] [Google Scholar]
- 6.Churchill, M. E., Tullius, T. D., Kallenbach, N. R. & Seeman, N. C. (1988) Proc. Natl. Acad. Sci. USA 85, 4653-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lilley, D. M. (1997) Proc. Natl. Acad. Sci. USA 94, 9513-9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lilley, D. M. & Norman, D. G. (1999) Nat. Struct. Biol 6, 897-899. [DOI] [PubMed] [Google Scholar]
- 9.Sha, R., Liu, F. & Seeman, N. C. (2002) Biochemistry 41, 5950-5955. [DOI] [PubMed] [Google Scholar]
- 10.Sha, R., Liu, F. & Seeman, N. C. (2000) Biochemistry 39, 11514-11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogg, J. M., Schofield, M. J., Declais, A. C. & Lilley, D. M. (2000) Biochemistry 39, 4082-4089. [DOI] [PubMed] [Google Scholar]
- 12.Ho, P. S. & Eichman, B. F. (2001) Curr. Opin. Struct. Biol. 11, 302-308. [DOI] [PubMed] [Google Scholar]
- 13.Eichman, B. F., Ortiz-Lombardia, M., Aymami, J., Coll, M. & Ho, P. S. (2002) J. Mol. Biol. 320, 1037-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinney, S. A., Declais, A. C., Lilley, D. M. & Ha, T. (2003) Nat. Struct. Biol 10, 93-97. [DOI] [PubMed] [Google Scholar]
- 15.Panyutin, I. G., Biswas, I. & Hsieh, P. (1995) EMBO J. 14, 1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panyutin, I. G. & Hsieh, P. (1994) Proc. Natl. Acad. Sci. USA 91, 2021-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas, I., Yamamoto, A. & Hsieh, P. (1998) J. Mol. Biol. 279, 795-806. [DOI] [PubMed] [Google Scholar]
- 18.Sigal, N. & Alberts, B. (1972) J. Mol. Biol. 71, 789-793. [DOI] [PubMed] [Google Scholar]
- 19.Lushnikov, A. Y., Bogdanov, A. & Lyubchenko, Y. L. (2003) J. Biol. Chem. 278, 43130-43134. [DOI] [PubMed] [Google Scholar]
- 20.Harada, Y., Sakurada, K., Aoki, T., Thomas, D. D. & Yanagida, T. (1990) J. Mol. Biol. 216, 49-68. [DOI] [PubMed] [Google Scholar]
- 21.Clegg, R. M., Murchie, A. I., Zechel, A. & Lilley, D. M. (1993) Proc. Natl. Acad. Sci. USA 90, 2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deniz, A. A., Dahan, M., Grunwell, J. R., Ha, T., Faulhaber, A. E., Chemla, D. S., Weiss, S. & Schultz, P. G. (1999) Proc. Natl. Acad. Sci. USA 96, 3670-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao, J. & Singleton, S. F. (2002) J. Mol. Biol. 320, 529-558. [DOI] [PubMed] [Google Scholar]
- 24.Gopaul, D. N. & Duyne, G. D. (1999) Curr. Opin. Struct. Biol. 9, 14-20. [DOI] [PubMed] [Google Scholar]
- 25.Guo, F., Gopaul, D. N. & van Duyne, G. D. (1997) Nature 389, 40-46. [DOI] [PubMed] [Google Scholar]
- 26.Clegg, R. M., Murchie, A. I. & Lilley, D. M. (1994) Biophys. J. 66, 99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha, T., Rasnik, I., Cheng, W., Babcock, H. P., Gauss, G. H., Lohman, T. M. & Chu, S. (2002) Nature 419, 638-641. [DOI] [PubMed] [Google Scholar]
- 28.Joo, C., McKinney, S. A., Lilley, D. M. & Ha, T. (2004) J. Mol. Biol. 341, 739-751. [DOI] [PubMed] [Google Scholar]
- 29.Bruist, M. F. & Myers, E. (2003) J. Theor. Biol. 220, 139-156. [DOI] [PubMed] [Google Scholar]
- 30.Snowden, T., Acharya, S., Butz, C., Berardini, M. & Fishel, R. (2004) Mol. Cell 15, 437-451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


