Abstract
The production of wild-type-free stocks of recombinant parvovirus minute virus of mice [MVM(p)] is difficult due to the presence of homologous sequences in vector and helper genomes that cannot easily be eliminated from the overlapping coding sequences. We have therefore cloned and sequenced spontaneously occurring defective particles of MVM(p) with very small genomes to identify the minimal cis-acting sequences required for DNA amplification and virus production. One of them has lost all capsid-coding sequences but is still able to replicate in permissive cells when nonstructural proteins are provided in trans by a helper plasmid. Vectors derived from this particle produce stocks with no detectable wild-type MVM after cotransfection with new, matched, helper plasmids that present no homology downstream from the transgene.
The autonomous parvovirus minute virus of mice [MVM(p)] is a lytic virus that replicates as an episome, preferentially in transformed cells. Given its oncotropism but also its innocuousness in the adult host and its resistance to extreme pH and temperatures, MVM(p) has been proposed as a vector for the gene therapy of cancer (34). The 5-kb single-stranded DNA genome of MVM(p) is organized into two overlapping transcription units. The early P4 promoter controls transcription of the nonstructural proteins, NS1 and NS2. The pleiotropic NS1 protein is responsible for the cytotoxic activity of MVM(p) in transformed cells (6, 10), is required for viral DNA amplification, and can positively or negatively affect the activity of homologous or heterologous promoters (26). NS1 also transactivates the second promoter, P38, thus allowing the synthesis of capsid proteins VP1 and VP2 at the end of the viral cycle (15).
We have previously derived a recombinant parvovirus from MVM(p) that transfers the cDNA for human interleukin-2 (IL-2). This virus is defective since part of the genes coding for capsid proteins (VP) is deleted. Transducing recombinant virus can, however, be produced if VP proteins are provided in trans by a cotransfected helper plasmid (35) or from genes integrated into the host chromosome (packaging cells) (7, 17). However, these stocks are contaminated by wild-type (WT) virus, which renders impossible their amplification through serial infections. The WT virus is generated through homologous recombination between vector and helper DNAs. Ideally, removing all sequence homology between these DNA sequences should prevent the formation of WT virus. However, data from Astell et al. suggest that a cis-acting element, necessary for DNA replication, overlaps the capsid-coding genes (2). On the other hand, data obtained from the study of defective particles indicate that particles as small as 15% of the size of WT MVM(p) are able to replicate. These defective particles are generated spontaneously during high-multiplicity infections (19) and are selectively amplified during serial infections. They vary in size from 15 to 70% of that of WT MVM(p), but they always retain the two terminal palindromes of 115 and 206 nucleotides. The stop codon for VP being located 566 nucleotides from the end of the right-hand palindrome, the smallest particles (500 to 600 nucleotides) must have lost all capsid-coding sequences. We have therefore amplified and cloned several of these defective particles in order to identify minimal sequences that are required for parvovirus MVM(p) amplification. Based on the structure of two of the smallest particles, we constructed and characterized IL-2-transducing vectors.
MATERIALS AND METHODS
Production and cloning of defective viral particles of MVM(p).
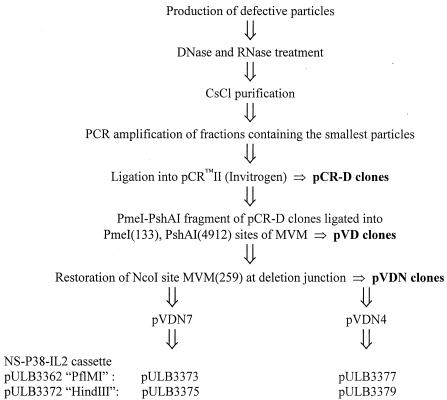
Defective viral particles were produced as described elsewhere (19) with the following modifications. A9 cells were infected with WT virus at a multiplicity of 5 PFU/cell in 2.5 ml of phosphate-buffered saline (PBS) supplemented with 0.5 mM (each) CaCl2 and MgCl2. Adsorption was allowed for 1 h, and then cells were washed with PBS and fed with fresh medium (Dulbecco modified Eagle medium [Life Technologies/Gibco-BRL], 10% fetal calf serum [PAA]). They were harvested after 48 h by trypsinization and centrifugation, resuspended in 250 μl of sterile TE 8.7 buffer (50 mM Tris, 1 mM EDTA, pH 8.7), and lysed by three cycles of freeze-thawing. Lysates were then centrifuged at 13,000 rpm in an Eppendorf centrifuge to eliminate cell debris and used to infect fresh A9 cells. Six successive rounds of infection were performed as described above, except that cells were harvested 24 h after infection. The final lysates were treated with DNase and RNase (20 μg/ml) for 30 min at 37°C and purified on CsCl gradients (38,000 rpm for 20 h at 18°C in a Beckman SW41 rotor). About 20 fractions were recovered. DNAs of relevant fractions, containing defective genomes, were amplified by PCR (see below) and cloned into pCRII (Invitrogen) to generate pCR-D plasmids. Defective particle sequences were subcloned into pUC-MVM to regenerate the palindromes that had been lost during the PCR amplification step (see Results). Some of the resulting pVD plasmids, namely, pVD4 and pVD7, lacked the MVM NcoI (259) site (1) at the deletion junction. The different steps of the cloning of defective particles and the generation of vectors are summarized in Fig. 1.
FIG. 1.
Different steps in the cloning of defective particles of MVM(p).
PCR amplifications and sequencing.
DNA from CsCl fractions was amplified by using the Expand Long Template PCR System (Boehringer Mannheim) in a 50-μl reaction volume (350 μM deoxynucleoside triphosphate, 300 nM [each] primer, buffer 1 with 1.75 mM MgCl2, and 2.6 U of enzyme mix). Conditions were as follows: first denaturation at 94°C for 2 min 30 s; 35 cycles at 94°C for 30 s, 60°C for 1 min, and 68°C for 4 min; and a terminal polymerization at 68°C for 10 min, in a thermocycler (Perkin-Elmer). The positions of primers are indicated in Fig. 4.
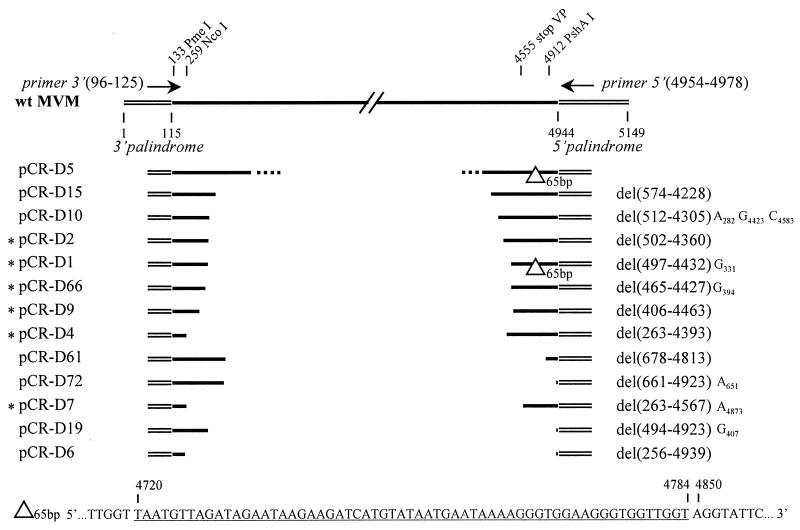
FIG. 4.
Structure of defective particles. The top line indicates the WT MVM genome. Terminal palindromes (thin double lines) are not drawn to scale. The positions of the primers used for amplification of defective particles are indicated by arrows. Nucleotides are numbered according to the description of Astell et al. (1). Asterisks at the left mark inserts that were subcloned into pUC-MVM. Numbers between parentheses indicate the MVM coordinates of the start and the end of the deletion. Letters at the right of the deletion indicate single-base substitutions with their position. The sequence of the 65-bp deletion observed in two clones is given at the bottom of the figure.
Sequences were determined with the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer).
Plasmids.
pUC-MVM was constructed by subcloning the entire genome of MVM(p) as a BamHI restriction fragment from pMM984 (28) into pUC18. Vector and helper plasmids are described in Fig. 2. MVM(p) coordinates are given according to the description of Astell et al. (1).
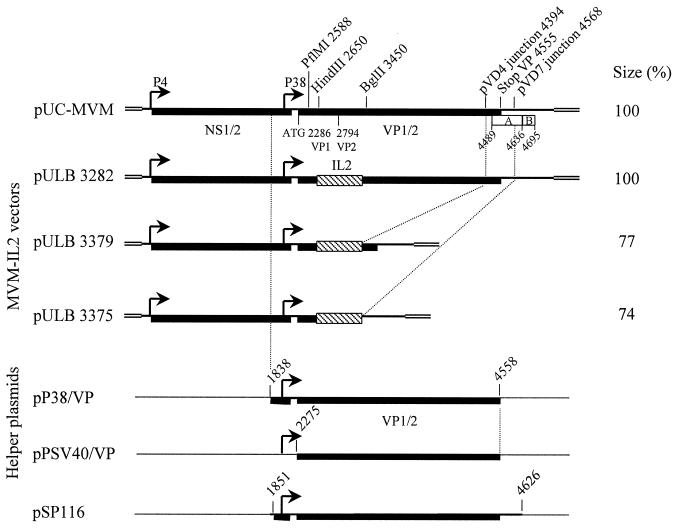
FIG. 2.
Vectors derived from defective particles. The top line represents the WT MVM genome; only relevant restriction sites and MVM coordinates are indicated. Elements A and B (39) are represented as open boxes. Plasmids pULB3373 and pULB3377 (not depicted) are identical to pULB3375 and pULB3379, respectively, downstream from the IL-2 gene. They differ by the insertion site of IL-2 upstream from the transgene, which is the restriction site PflMI instead of HindIII.
(i) Vectors.
pULB3282 was previously described as pMVM100a (8).
The NcoI restriction site of MVM was restored in both pVD7 and pVD4 at the deletion junction by PCR amplification with the 5′ primer (see Fig. 4) used for the initial amplification of defective particles and either primer 5′GTAACCAACTAACCATGGTTTTTCTTTC3′ or 5′GTAACCAACTAACCATGGGAGCAAAAG3′, which overlap the deletion junction in pVD7 or pVD4, respectively. The mismatched nucleotide in each primer is underlined. The Expand Long Template PCR System (Boehringer Mannheim) was used according to the manufacturer's instructions. Agarose gel-purified PCR products were digested with restriction enzymes NcoI and PshAI and cloned into the corresponding sites of plasmid pUC-MVM: NcoI (259)-PshAI (4912). In the resulting molecular clones, pVDN7 and pVDN4, the presence of the NcoI site was confirmed by restriction and by sequencing.
The IL-2 cDNA was cloned as a blunt-ended HindIII-EcoRI fragment from pUC-IL-2 (35) into the PflMI (2588) or HindIII (2650) site of MVM, generating plasmid pULB3362 or pULB3372, respectively. These plasmids provided the NS–P38–IL-2 cassettes that were used to derive vectors from the defective molecular clones. These PflMI and HindIII cassettes were excised as PmeI (133 in MVM)-SmaI (in pUC-IL-2) and inserted into the PmeI-NcoI (blunt-ended) site of pVDN7 or pVDN4. pVDN7-derived vectors are called pULB3373 (PflMI cassette) and pULB3375 (HindIII cassette). pVDN4-derived vectors are pULB3377 (PflMI cassette) and pULB3379 (HindIII cassette).
All plasmids were amplified in the DL795 bacterial strain to avoid deletions in the 5′ palindrome (25).
(ii) Helper plasmids.
The NS1 helper pULB3321 was obtained by inserting the NS-coding region of MVM(p) (restriction fragment NcoI [259]-StuI [2371]) under control of the simian virus 40 (SV40) promoter in plasmid pSBC2 (14) (SmaI site of the polylinker).
The first-generation helper plasmid, pSP116, carries the MVM(p) transcription unit coding for capsid proteins, followed by the SV40 poly(A) sequence (Fig. 2) (17).
To construct pP38-VP and pPSV40-VP, a fragment encompassing the capsid coding genes of MVM(p) with or without their promoter was amplified from pMM984 under the following conditions: 94°C for 2 min 30 s, followed by 35 cycles (94°C for 45 min, 50°C for 1 min, and 68°C for 2 min) and a final polymerization at 68°C for 10 min. Forward primers hybridized to nucleotides 1838 to 1859 (primer 19: 5′TGACAAAAATCGATGGCCCATG3′) or to nucleotides 2275 to 2295 (primer 20: 5′ACTAAGGTACGATGGCGCCTC3′), respectively. Primer 19 introduced a ClaI restriction site at nucleotide 1847. A common reverse primer, used for both amplifications, hybridized to nucleotides 4537 to 4558 (primer 21: 5′GTTAGTAAGTATTTCTAGCAAC3′). PCR products were purified on agarose gels, phosphorylated by T4 polynucleotide kinase, rendered blunt by T4 DNA polymerase, and ligated into the dephosphorylated SmaI site of plasmid pSBC2 (14). Plasmids pPSV40/P38-VP and pPSV40-VP were purified after transformation into TOP10 bacteria. Plasmid pP38-VP was derived from pPSV40/P38-VP by removing the SV40 promoter on a ClaI fragment. Clones were partially sequenced.
Transfections were performed by the calcium phosphate coprecipitation method. Hirt's extractions and Southern blot analyses were done 2 days after transfection. IL-2 production was measured by enzyme-linked immunosorbent assay, in culture supernatants, 3 days after transfection or infection. These methods have been described elsewhere (7).
Serial infections of NBK cells were done as previously published (17).
RESULTS
Production and cloning of defective viral particles of MVM(p).
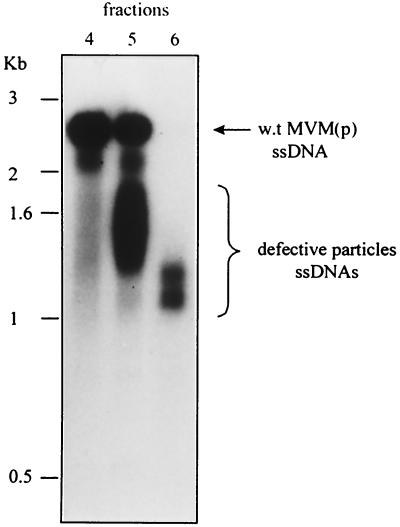
To generate defective particles, A9 cells were first infected at a high multiplicity of infection and then reinfected through six successive rounds of infection with the cell lysates obtained from each of these infections. Proteinase-treated aliquots of CsCl gradient fractions from the final lysate were hybridized on Southern blots with a 32P-labeled MVM(p) probe. Each fraction contained a population of molecules, smaller than WT MVM and migrating as smears mainly between 1 and 2 kb (Fig. 3). The amount of defective particles present in each fraction was estimated by comparing the intensity of the smears with that of the band of titratable WT virus in the same lane. DNA of approximately 107 to 108 defective viral particles was extracted from fractions 5 and 6, containing the smallest particles, and amplified by PCR. The primers used for this reaction hybridized just inboard of the two palindromes, thus allowing the amplification of viral genomes from nucleotides 96 to 4978 (Fig. 4).
FIG. 3.
CsCl gradient fractionation of defective particles. Relevant fractions of one representative gradient are shown. They were revealed with an MVM probe on Southern blots. ssDNAs, single-stranded DNAs.
The amplified products migrated as smears in a 1% agarose gel, mainly around 1 and 2 kb (data not shown). They were ligated into the pCR II plasmid (Invitrogen) to give a series of pCR-D clones. The sizes of inserts were analyzed for 96 clones by digestion with restriction enzymes ApaI and BamHI, which cut the polylinker at either side of the insert but do not cleave the MVM sequence. Sizes of inserts ranged from approximately 300 bp to 3 kb, with a majority of clones, 62 of 96, larger than 1,000 bp and only 13 of 96 smaller than 500 bp.
Sequence analysis of defective particles.
The entire sequence of 15 pCR-D inserts was established. Since the aim of our study was to identify the smallest replication-proficient particle, we selectively analyzed the shortest clones with inserts comprising between 300 and 1,200 bp, deliberately dismissing genomes longer than 2 kb. Only one large clone, pCR-D5, with an insert of approximately 2,500 nucleotides was partially sequenced (Fig. 4).
Three single-nucleotide differences were detected in all the clones compared to the published sequence of MVM(p) (1). Two of them resulted in the loss of a restriction site: BstEII (4486), transition T→C, and SnaBI (4634), transition G→A. The third one created a restriction site: NdeI (4359), transition C→T. Their presence was confirmed by restriction analysis in our molecular clone of MVM(p). Since the defective genomes were not generated from the molecular clone, these nucleotide changes reflect probable variations of the published sequence. Two other groups of nucleotide changes were found in the sequence of the left-hand (3′) palindrome in pMM984 after subcloning of the defective viral particles, as discussed below. (i) Our sequence analysis reveals CA at positions 69 to 70, clarifying the ambiguity (AC or CA) reported by Astell et al. (1). This sequence is consistent with the absence of the AatII restriction site in pMM984. (ii) An insertion of one nucleotide, T, at position 61 was found by J. C. Ramirez (unpublished observation). Other point mutations identified in some defective clones are indicated in Fig. 4. These mutations could have been generated during the PCR amplification.
The smallest sequence retained at the left end of the genome ends at coordinate 256 (Fig. 4, clone pCR-D6) and comprises the 3′ palindrome (1 to 115) and the P4 promoter (ending at coordinate 201) (21). Two other clones, pCR-D4 and pCR-D7, had a short left end, while all the others extended further than nucleotide 400 into the genome of MVM(p). The four smallest sequences at the right end were found in pCR-D6, pCR-D19, pCR-D72, and pCR-D61, with sizes of 210, 226, 226, and 333 nucleotides, respectively, including the 171 terminal nucleotides of the palindrome, which were not amplified by PCR. The right end of clone pCR-D6, starting at nucleotide 4939, retains just the 5′ palindrome (4944 to 5149).
The sequences of pCR-D1 and pCR-D5 contain a second deletion in the right terminal region of the genome, just inboard of the 5′ palindrome (Fig. 4). This deletion results from the removal of one copy of a tandemly repeated sequence of 65 bp between nucleotides 4720 and 4849. We cannot exclude the possibility that this deletion occurred during the PCR amplification.
Deletion junctions.
Two types of deletion junctions could be identified: (i) those involving short direct repeats and (ii) those occurring between nonhomologous regions of the genome. In the genomes of eight defective viruses, pCR-D15, pCR-D10, pCR-D1, pCR-D66, pCR-D4, pCR-D61, pCR-D7, and pCR-D6, the internal deletion occurs through recombination between short direct repeats located at the left and right, resulting in the loss of one copy of the repeat. These short direct repeats are scattered over the viral genome, but in the majority of sequenced clones, they mapped between nucleotides 465 and 574 at the left and nucleotides 4228 to 4567 in the right part (Fig. 5). For clones pCR-D61 and pCR-D66, the two repeats involved in recombination differ by 1 nucleotide. In two groups of clones, the same repeat on one side recombined with a repeat located at a different position on the other side, i.e., clones pCR-D4 and pCR-D7 have the same left extremity whereas clones pCR-D72 and pCR-D19 have identical right ends. It is possible that, by choosing to study mainly short genomes, we have selected for recombination events involving these repeats. Four junctions, in pCR-D2, pCR-D9, pCR-D72, and pCR-D19, could not be ascribed to recombination between short direct repeats. These deletions seemed to occur by directly joining right and left parts of the genome.
FIG. 5.
Sequence of deletion junctions. The sequences correspond to the plus strand of the genome (5′-to-3′ orientation). Coordinates of the internal deletion are listed on the right. The remaining copy of the short direct repeats is underlined. For pCR-D66 and pCR-D61, recombination involved repeats with one mismatched nucleotide; the sequence of the deleted copy is shown on top.
Replication assay of defective viral particles.
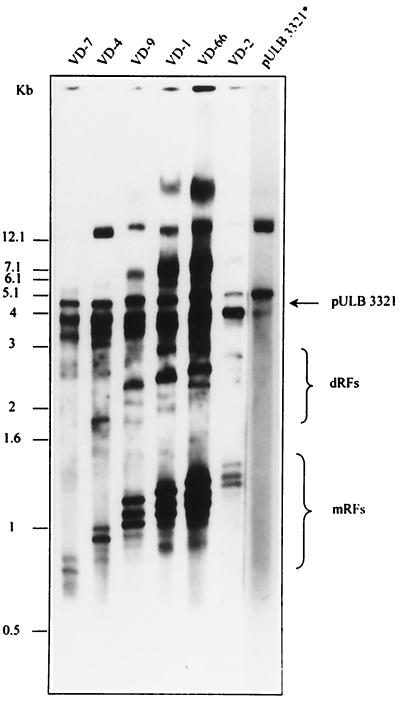
To check if minimal cis-acting sequences for DNA amplification were maintained in the defective genomes, the palindromic termini that had been lost during the PCR amplification step were restored. This was done for 6 of the 13 sequenced pCR-D clones, pCR-D2, pCR-D1, pCR-D66, pCR-D9, pCR-D4, and pCR-D7. They were chosen for the presence or absence of different sequences that have been reported as being involved in replication (2) but also for the presence of convenient restriction sites for subcloning into pUC-MVM. Inserts of clones pCR-D6, pCR-D19, and pCR-D72, with the smallest sequences at the right terminus, were not subcloned for lack of restriction sites. Each insert was extracted from the pCR-D vector by digestion at the MVM restriction sites PmeI (133) and PshAI (4912) and ligated into the same restriction sites of pUC-MVM. The resulting plasmids, pVD2, pVD1, pVD66, pVD9, pVD4, and pVD7, were cotransfected into NBE cells with the helper plasmid pULB3321, which expresses the NS1 protein under control of the SV40 promoter-enhancer. Replicative forms of viral DNA were extracted in Hirt's extracts and revealed by Southern blotting with a 32P-labeled MVM(p) probe (35). For each plasmid, a band of the expected size corresponding to the excised double-stranded genome was detected (Fig. 6). Other bands corresponding to differently processed hairpin ends of the replicative forms have already been described (39). The pULB3321 band represents input DNA; this plasmid does not replicate and was therefore considered an internal standard to evaluate the relative replication efficiency of the different VD clones. VD7 and VD4 genomes replicate, indicating that the first 263 nucleotides are sufficient for DNA replication in the presence of the NS1 protein. This sequence encompasses the minimal active origin of replication (39, 13). VD7, however, seems to replicate less efficiently than VD4. This clone, but not VD4, has lost a large part of the right-hand regulatory element A (4489 to 4636) but retains element B (4636 to 4695), both described as being relevant for replication (39, 40).
FIG. 6.
Replication of defective particles. pCR-D plasmids (3 μg/6-cm-diameter dish), carrying defective particles, were cotransfected into NBE cells at a molar ratio of 1 with plasmid pULB3321, which provides NS1 protein in trans, allowing the excision and amplification of MVM DNA. Hirt's extracts prepared 2 days after transfection were separated by electrophoresis, blotted, and revealed with an MVM probe. The names of the defective particle clones are indicated above each lane. The lane marked with an asterisk shows a shorter exposure time from the same Southern blot. It contains an extract of cells transfected only with pULB3321.
The deletion of one copy of the 65-bp tandem repeat present in the right-hand part of the VD1 genome did not affect the replication efficiency (Fig. 6), in agreement with published results (39).
Replication of vectors derived from defective particles.
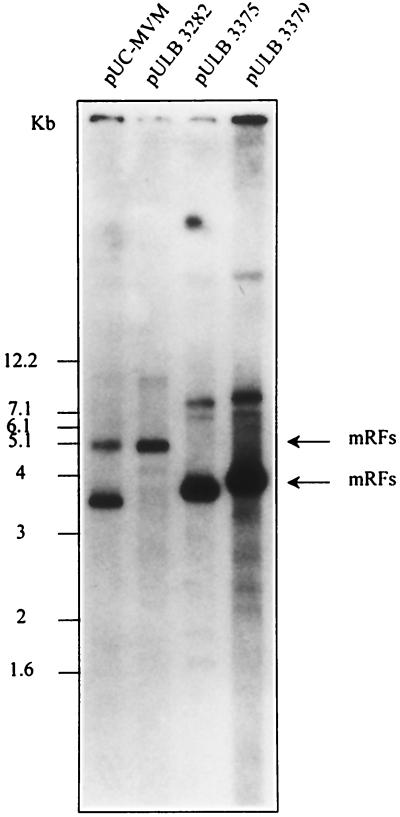
New transducing vectors were derived from the small replication-competent defective particles pVD7 and pVD4 by the addition of the NS transcription unit and the human IL-2 cDNA under the control of the P38 promoter of MVM. The insertion of the IL-2 cDNA was done at two different restriction sites behind P38, PflMI and HindIII, generating plasmids pULB3373 and pULB3375 for pVD7 and pULB3377 and pULB3379 for pVD4 (Fig. 1). The former reduces the homology of the vector with helper plasmids upstream of the IL-2 gene. Excision from their respective plasmids and their replication were compared to those of WT MVM (pUC-MVM) and of the first-generation vector (pULB3282) using Hirt's extracts prepared 2 days after transfection of NBE cells. Replicative monomeric forms were observed for all DNAs with equal efficiencies, except for pULB3373, which replicated less efficiently (Fig. 7 and results not shown for pULB3373 and pULB3377).
FIG. 7.
Replication of vectors derived from defective particles. The NS transcription unit of MVM(p) and the human IL-2 cDNA under the control of the P38 promoter were cloned into defective particles to generate vectors that were transfected into NBE cells (1 μg/6-cm-diameter dish). Control cells were transfected with pUC-MVM. The name of the transfected plasmid is indicated on top of each lane.
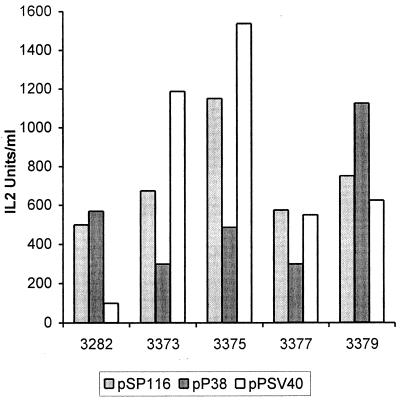
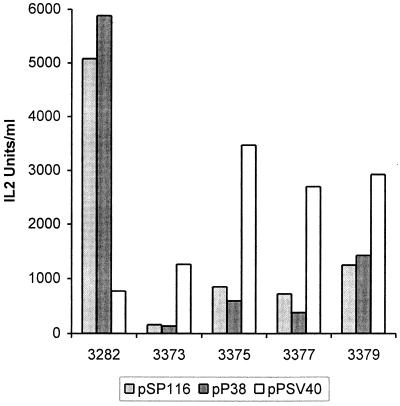
New helper plasmids that express capsid proteins under the control of their natural promoter, P38 (pP38-VP), or the SV40 early promoter (pPSVA40-VP) were designed. Both have little homology (165 nucleotides) with vectors pULB3377 and pULB3379 and no homology with the smallest vectors, pULB3373 and pULB3375, downstream from the transgene. The homology upstream from the transgene is greatly reduced, 817 or 756 nucleotides for helper pP38-VP and 380 or 319 nucleotides for helper pPSV40-VP with vectors in which the IL-2 gene is inserted into the PflMI or HindIII restriction site, respectively. To produce stocks of these vectors, the different plasmids were cotransfected with either the pP38-VP or the pSV40-VP helper. IL-2 production of transfected cells was measured as an indication of the functionality of the P38–IL-2 expression cassette. One- to threefold variations were observed between vectors cotransfected with P38 helpers pSP116 and pP38-VP. These variations were more important with the helper pPSV40-VP: up to 15-fold between pULB3375 and pULB3282 (Fig. 8). Lysates prepared 3 days after transfection were used to infect NBK cells, and IL-2 production was measured to give an estimation of the relative concentration of recombinant virus stocks. Titration of recombinant virus or WT MVM was done by in situ hybridization with an IL-2 or VP probe, respectively. The most efficient vector production was observed by cotransfection of the pULB3282 vector with the pP38 helpers. The new vectors described here, however, yielded the best titers when produced with the pPSV40-VP helper. These stocks were equivalent to or better than those obtained for pULB3282 with the same helper, except for pULB3373 (see Fig. 10A). Although the titers of pULB3375/ pPSV40-VP stocks are 1 order of magnitude lower than those produced with pULB3282/pSP116, they produce approximately the same amount of IL-2 after transduction of NBK cells (70%) (Fig. 9). The ratio of IL-2 produced over transducing units varied between experiments, but it was in general higher for pVD7-derived vectors, pULB3373 and pULB3375, than for pULB3282- and pVD4-derived vectors, pULB3377 and pULB3379 (Fig. 10B). Stocks produced by cotransfection with pSP116 contained WT virus except for pULB3373. However, the titers of this vector were probably too low (around 100 IU/ml) to detect the WT. The reduction of homology upstream from the transgene in pULB3377 compared to pULB3379 did not significantly reduce the proportion of WT virus (Fig. 10C). On the other hand, eliminating all homology downstream from the transgene in pULB3375 completely abolished WT virus formation (<0.03%) after cotransfection with pP38-VP and pPSV40-VP (Fig. 10C). Serial infections of NBK cells with pULB3375/pP38-VP stocks, however, revealed the presence of WT in the first or second round of infection, depending on the experiment (results not shown). In contrast, the absence of WT virus in pULB3375/ pPSV40-VP stocks was confirmed by three cycles of infection of NBK cells.
FIG. 8.
IL-2 production after transfection of NBE cells with vectors derived from defective particles. Different vectors (3 μg/6-cm-diameter dish) were cotransfected with either of three helper plasmids (threefold molar excess), pSP116, pP38-VP, or pPSV40-VP. The first two helpers express capsid proteins under the control of the parvoviral P38 promoter, and the third uses the SV40 promoter. IL-2 levels were titrated 3 days after transfection in total lysates. Results of one representative experiment are shown.
FIG. 10.
Transducing particles of recombinant and WT infectious virus were titrated by in situ hybridization with IL-2- and VP-specific probes, respectively. WT virus production is shown for two experiments that produced similar amounts of recombinant virus; results of the first experiment are shown in Fig. 8. The ratio of IL-2 produced after infection to transducing units was calculated for that experiment as well.
FIG. 9.
Transducing activity of vectors. Cells from the experiments described for Fig. 8 were lysed 3 days after transfection. The transducing activity of these lysates was measured as IL-2 expression of NBK cells, 3 days after infection. Results are from the same experiment as in Fig. 8.
DISCUSSION
The production of recombinant MVM(p) and autonomous parvovirus vectors in general is hampered by the low efficiency of recombinant virus production after transfection of permissive cells and by the generation of WT virus that prevents amplification in permissive cells (11). The WT virus is generated through homologous recombination between vector and helper DNAs. Eliminating all homology between vector and helper genomes is rendered difficult by the compactness of the parvoviral genome. Indeed, transcription units for nonstructural and structural proteins are overlapping, one of the isoforms of NS2 overlaps the start of the structural protein VP1, and the P38 promoter is contained in the 3′ coding region of NS. Moreover, different elements have been reported to be required for DNA replication (2) and for encapsidation (24).
Since defective particles of autonomous parvoviruses are able to replicate and to be propagated in cells infected at a high multiplicity of infection, we have postulated that these particles must possess all cis-acting sequences required for these functions. Moreover, they should lack all VP sequences, as some of these particles are only 500 to 600 nucleotides long and retain the 115 and 206 nucleotides of the 3′ and 5′ palindromes. We have therefore isolated, amplified, and sequenced some of these defective particles to identify the minimal cis-acting sequences that should be retained in vectors based on MVM(p). The genomes that we cloned are consistent with a structure of type I defective particles, since they retain both right and left palindromes with an internal deletion of NS and VP coding regions (19). No rearrangements, i.e., repeats or insertions like those described for H1 (32), were observed in this study. Such rearrangements seem to be rare for MVM: one insertion was described by Hogan and Faust (22). In a majority of clones, the internal deletion occurred between short direct repeats, localized at different sites in the genome. The role of short direct repeats in the generation of defective particles of MVM(p) has already been studied. We could not associate the location of the direct repeats with a nearby consensus motif like the one, CTA/TTTC/T, identified in this earlier study (22). Also, some of the clones did not involve recombination in direct repeats. Although the molecular mechanisms of illegitimate recombination are not fully understood, two models are presently proposed: a copy choice model via short direct repeats or a strictly nonhomologous, cut-and-join process (16). The two mechanisms have been proposed to be mediated through different enzymes, such as exonuclease V or topoisomerases and gyrases, respectively (5, 38). Recombination in direct repeats can also result from replication slippage following DNA polymerase arrest at the first sequence, mainly during rolling-circle replication (30). The generation of deletions through illegitimate recombination mediated by short direct repeats has been described for numerous prokaryote or eukaryote genomes (16, 29). In two recent studies, defective interfering particles of RNA viruses, Toscana virus and tomato spotted wilt virus, were shown to be generated by this mechanism (27, 23). Recombination through copy choice can also occur during PCR amplification by Thermus aquaticus DNA polymerase I (42). However, it is very unlikely that our defective genomes were generated during PCR amplification. Indeed, the heterogeneous population of short viral DNAs that we cloned after PCR is DNase resistant, suggesting that they were encapsidated. Moreover, similar structures were isolated without PCR amplification (19). Illegitimate recombination by the cut-and-join mechanism has not been described for MVM(p) before, maybe because it occurs less frequently than recombination through direct repeats, as suggested from our results. Recombination through direct repeats is also consistent with the rolling-circle type of replication of parvoviruses (30, 41). In defective interfering particles of tomato spotted wilt virus, the two types of junctions have been observed, although with a predominance of cut joining sites (23).
All the defective particles that we have cloned were able to replicate when NS proteins were provided in trans from the cotransfected plasmid, pULB3321. Two clones with a deletion of a 65-bp tandem repeat replicated similarly to WT virus. This 65-bp sequence is not duplicated in the genome of the closely related lymphotropic variant of MVM(p), MVM(i) (1, 36). An analogous, tandemly repeated region of 55 bp has been described in the right-hand region of H1, which is also very closely related to MVM (33). Interestingly, defective interfering particles of H1 are mainly characterized by integral reiterations of this 55-bp sequence (20, 33). A variant of H1, H1-dr, that was shown to have three tandemly repeated copies of the 55-bp sequence has a clear selective advantage over standard H1 (18). The 55- to 65-bp repeat has been described for a variety of autonomous parvoviruses, but its role remains unclear (3, 4, 18, 33, 37, 39).
The shortest defective particle, pVD7, replicated less efficiently than the others. This particle has lost part of the regulatory element A but has retained element B. Replication was not impaired for pVD4, which has the same sequence at the 3′ palindrome but has retained both elements A and B. These cis-acting regulatory elements may constitute a portion of the origin of replication at the right terminus and may also be important for the packaging of single-stranded DNA (12). Moreover, these sites apparently bind a host cell protein in a sequence-specific manner (9). Deletion of element A alone abolishes replication in mouse A9 cells while reducing it in simian Cos7 cells. Our results, obtained in human NBE cells, are similar to those observed in Cos7 cells. However, one vector derived from defective particle pVD7, pULB3375, replicates as well as WT virus and as vectors derived from pVD4, pULB3377, and pULB3379. Despite an efficient replication of vectors derived from pULB3375, pULB3377, and pULB3379, they yielded 10-, 20-, and 50-fold fewer transducing particles than the reference vector pULB3282 with helpers expressing capsid proteins from the P38 promoter, pSP116 and pP38-VP. With the pPSV40-VP helper this reduction was much less marked for pULB3375 or not observed at all for pULB3377 and pULB3379. Recently it has been suggested that sequences in the 3′ part of the VP transcription unit of parvovirus H1 (nucleotides 3591 to 4012) could contain cis-acting stimulatory elements for recombinant virus encapsidation (24). The corresponding sequences of MVM (nucleotides 3636 to 4002) are missing in both pULB3375 and pULB3379, which could explain the approximately 10-fold reduction in transducing particle formation of pULB3379 compared to pULB3282 with pP38 helpers. The further reduction in transducing particle production for pULB3375 might be due to a deficit in replication, given the absence of part of the regulatory region A, although no difference in DNA amplification was detected in our experiments for this vector. The role of the putative packaging potentiating region is, however, difficult to explain when pPSV40-VP is used as a helper plasmid. Indeed, the production of defective particle-based vectors was better with the pPSV40-VP helper than with the two others, whereas the reverse was true for vector pULB3282. The above-described regulatory elements at the right extremity of MVM might contain binding sites for factors that are necessary for the replication and/or expression from the SV40 origin-promoter region. They would have to be located in the region that is lacking in both pULB3375 and pULB3379, upstream from elements A and B, since both vectors are produced better with the pPSV40-VP helper than with pULB3282. An effect of the NS1 protein on the SV40 origin-promoter can be excluded since vector pULB3282 also expresses NS1. A definitive explanation for the differences in recombinant virus yields associated with the different helper plasmids can thus not yet be proposed.
Elimination of all homology between helper pP38-VP or pPSV40-VP and the vector to the right of the transgene, greatly reduces recombination, so that no WT virus is detected in stocks of pULB3373 and pULB3375. In vectors pULB3377 and pULB3379, the 165-base homology that remains downstream from the IL-2 gene with the same helpers is sufficient to produce WT virus after cotransfection. A sequence homology as small as 105 nucleotides in the NS coding region has been shown to be sufficient to generate WT NS1 through recombination between cotransfected mutant plasmids (31).
The best recombinant virus stocks that contained no WT virus were produced with vector pULB3375 and helper pPSV40-VP. Although crude lysates contained only about 3 × 103 transducing particles/ml, 10-fold fewer than pULB3282 produced with pSP116, vector pULB3375 should be characterized further. Indeed, first, transgene expression from this vector is more efficient than that of pULB3282/pSP116, given the higher ratio of IL-2/transducing particles. Second, its yield can likely be improved through modifications of its structure, i.e., increasing its size to 100% of that of WT MVM (8) and/or restoring its resolution site at the viral left-hand terminus (24). Third, vector stocks can hopefully be amplified by serial infections of packaging cells, since they do not contain any detectable WT virus (17).
ACKNOWLEDGMENTS
N. Clément and K. El Bakkouri were supported by grants from the FNRS/Télévie. We benefited from a grant given by the Fédération contre le Cancer from Belgium and the Grand Duchy of Luxembourg.
REFERENCES
- 1.Astell C R, Gardiner E M, Tattersall P. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J Virol. 1986;57:656–669. doi: 10.1128/jvi.57.2.656-669.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astell C R, Liu G, Brunstein J, Jindal H K, Tam P. Minute virus of mice cis-acting sequences required for genome replication and the role of the trans-acting viral protein NS1. Prog Nucleic Acids Res Mol Biol. 1996;55:245–285. doi: 10.1016/s0079-6603(08)60196-8. [DOI] [PubMed] [Google Scholar]
- 3.Ball-Goodrich L J, Johnson E. Molecular characterization of a newly recognized mouse parvovirus. J Virol. 1994;68:6476–6486. doi: 10.1128/jvi.68.10.6476-6486.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besselsen D G, Pintel D J, Purdy G A, Besch-Williford C L, Franklin C L, Hook R R, Jr, Riley L K. Molecular characterization of newly recognized rodent parvoviruses. J Gen Virol. 1996;77:899–911. doi: 10.1099/0022-1317-77-5-899. [DOI] [PubMed] [Google Scholar]
- 5.Bierne H, Ehrlich S D, Michel B. Deletions at stalled replication forks occur by two different pathways. EMBO J. 1997;16:3332–3340. doi: 10.1093/emboj/16.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandenburger A, Legendre D, Avalosse B, Rommelaere J. NS-1 and NS-2 proteins may act synergistically in the cytopathogenicity of parvovirus MVMp. Virology. 1990;174:576–584. doi: 10.1016/0042-6822(90)90110-d. [DOI] [PubMed] [Google Scholar]
- 7.Brandenburger A, Russell S. A novel packaging system for the generation of helper-free oncolytic MVM vector stocks. Gene Ther. 1996;3:927–931. [PubMed] [Google Scholar]
- 8.Brandenburger A, Coessens E, El Bakkouri K, Velu T. Influence of sequence and size of DNA on packaging efficiency of parvovirus MVM-based vectors. Hum Gene Ther. 1999;10:1229–1238. doi: 10.1089/10430349950018210. [DOI] [PubMed] [Google Scholar]
- 9.Brunstein J, Astell C R. Analysis of the internal replication sequence indicates that there are three elements required for efficient replication of minute virus of mice minigenomes. J Virol. 1997;71:9087–9095. doi: 10.1128/jvi.71.12.9087-9095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corsini J, Afanasiev B, Maxwell I H, Carlson J O. Autonomous parvovirus and densovirus gene vectors. Adv Virus Res. 1996;47:303–351. doi: 10.1016/s0065-3527(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 12.Cossons N, Zannis-Hadjopoulos M, Tam P, Astell C R, Faust E A. The effect of regulatory sequence elements upon the initiation of DNA replication of the minute virus of mice. Virology. 1996;224:320–325. doi: 10.1006/viro.1996.0535. [DOI] [PubMed] [Google Scholar]
- 13.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dirks W, Wirth M, Hauser H. Dicistronic transcription units for gene expression in mammalian cells. Gene. 1993;128:247–249. doi: 10.1016/0378-1119(93)90569-o. [DOI] [PubMed] [Google Scholar]
- 15.Doerig C, Hirt B, Beard P, Antonietti J P. Minute virus of mice nonstructural protein NS-1 is necessary and sufficient for transactivation of the viral P39 promoter. J Gen Virol. 1988;69:2563–2573. doi: 10.1099/0022-1317-69-10-2563. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich S D, Bierne H, d'Alençon E, Vilette D, Petranovic M, Noirot P, Michel B. Mechanisms of illegitimate recombination. Gene. 1993;135:161–166. doi: 10.1016/0378-1119(93)90061-7. [DOI] [PubMed] [Google Scholar]
- 17.El Bakkouri K, Clément N, Velu T, Brandenburger A. Amplification of MVM(p) vectors through serial infection of a new packaging cell line. Tumor Targeting. 1999;4:210–217. [Google Scholar]
- 18.Faisst S, Faisst S R, Dupressoir T, Plaza S, Pujol A, Jauniaux J C, Rhode S L, Rommelaere J. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J Virol. 1995;69:4538–4543. doi: 10.1128/jvi.69.7.4538-4543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faust E A, Ward D. Incomplete genomes of the parvovirus minute virus of mice selective conservation of genome termini, including the origin for DNA replication. J Virol. 1979;32:276–292. doi: 10.1128/jvi.32.1.276-292.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faust E A, Hogan A. Defective interfering particles. In: Tijssen P, editor. Handbook of parvoviruses. Boca Raton, Fla: CRC Press; 1990. pp. 91–107. [Google Scholar]
- 21.Fuks F, Deleu L, Dinsart C, Rommelaere J, Faisst S. ras oncogene-dependent activation of the P4 promoter of minute virus of mice through a proximal P4 element interacting with the Ets family of transcription factors. J Virol. 1996;70:1331–1339. doi: 10.1128/jvi.70.3.1331-1339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogan A, Faust E A. Nonhomologous recombination in the parvovirus chromosome: role for a CTATTTCT motif. Mol Cell Biol. 1986;6:3005–3009. doi: 10.1128/mcb.6.8.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue-Nagata A K, Kormelink R, Sgro J-Y, Nagata T, Kitajima E W, Golbach R, Peters D. Molecular characterization of tomato spotted wilt virus defective interfering RNAs and detection of truncated L proteins. Virology. 1998;248:342–356. doi: 10.1006/viro.1998.9271. [DOI] [PubMed] [Google Scholar]
- 24.Kestler J, Neeb B, Struyf S, Van Damme J, Cotmore S F, D'Abramo A, Tattersall P, Rommelaere J, Dinsart C, Cornelis J J. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum Gene Ther. 1999;10:1619–1632. doi: 10.1089/10430349950017626. [DOI] [PubMed] [Google Scholar]
- 25.Leach D. Cloning and characterization of DNAs with palindromic sequences. Genet Eng (New York) 1996;18:1–11. doi: 10.1007/978-1-4899-1766-9_1. [DOI] [PubMed] [Google Scholar]
- 26.Legendre D, Rommelaere J. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J Virol. 1992;66:5705–5713. doi: 10.1128/jvi.66.10.5705-5713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchi A, Nicoletti L, Accardi L, Di Bonito P, Giorgi C. Characterization of Toscana virus-defective interfering particles generated in vivo. Virology. 1998;246:125–133. doi: 10.1006/viro.1998.9195. [DOI] [PubMed] [Google Scholar]
- 28.Merchlinsky M J, Tattersall P J, Leary J J, Cotmore S F, Gardiner E M, Ward D C. Construction of an infective molecular clone of the autonomous parvovirus minute virus of mice. J Virol. 1983;47:227–232. doi: 10.1128/jvi.47.1.227-232.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meuth M. Illegitimate recombination in mammalian cells. In: Berg D E, Hove M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 833–860. [Google Scholar]
- 30.Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 31.Pearson J L, Pintel D J. Recombination within the nonstructural genes of the parvovirus minute virus of mice (MVM) generates functional levels of wild-type NS1, which can be detected in the absence of selective pressure following transfection of nonreplicating plasmids. Virology. 2000;269:128–136. doi: 10.1006/viro.2000.0202. [DOI] [PubMed] [Google Scholar]
- 32.Rhode S L., III Defective interfering particle of parvovirus H-1. J Virol. 1978;27:347–356. doi: 10.1128/jvi.27.2.347-356.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhode S L, III, Klaassen B. DNA sequence of the 5′ terminus containing the replication origin of parvovirus replicative form DNA. J Virol. 1982;41:990–999. doi: 10.1128/jvi.41.3.990-999.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell S J. Lymphokine gene therapy for cancer. Immunol Today. 1990;11:196–200. doi: 10.1016/0167-5699(90)90081-j. [DOI] [PubMed] [Google Scholar]
- 35.Russell S J, Brandenburger A, Flemming C L, Collins M K L, Rommelaere J. Transformation-dependent expression of interleukin genes delivered by a recombinant parvovirus. J Virol. 1992;66:2821–2828. doi: 10.1128/jvi.66.5.2821-2828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahli R, McMaster G K, Hirt B. DNA sequence comparison between two tissue-specific variants of the autonomous parvovirus, minute virus of mice. Nucleic Acids Res. 1985;13:3617–3633. doi: 10.1093/nar/13.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salvino R, Skiadopoulos M, Faust E A, Tam P, Shade R O, Astell C R. Two spatially distinct genetic elements constitute a bipartite DNA replication origin in the minute virus of mice genome. J Virol. 1991;65:1352–1363. doi: 10.1128/jvi.65.3.1352-1363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu H, Yamaguchi H, Ashizawa Y, Kono Y, Asami M, Kato J, Ikeda H. Short-homology-independent illegitimate recombination in Escherichia coli: distinct mechanism from short-homology dependent illegitimate recombination. J Mol Biol. 1997;266:297–305. doi: 10.1006/jmbi.1996.0794. [DOI] [PubMed] [Google Scholar]
- 39.Tam P, Astell C R. Replication of minute virus of mice minigenomes: novel replication elements required for MVM DNA replication. Virology. 1993;193:812–814. doi: 10.1006/viro.1993.1190. [DOI] [PubMed] [Google Scholar]
- 40.Tam P, Astell C R. Multiple cellular factors bind to cis-regulatory elements found inboard of the 5′ palindrome of minute virus of mice. J Virol. 1994;68:2840–2848. doi: 10.1128/jvi.68.5.2840-2848.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tattersall P, Ward D C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976;263:106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 42.Zaphiropoulos P G. Non-homologous recombination mediated by Thermus aquaticus DNA polymerase I. Evidence supporting a copy choice mechanism. Nucleic Acids Res. 1998;26:2843–2848. doi: 10.1093/nar/26.12.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]