Abstract
Background and Aims
The regenerative capacity of the pancreas diminishes with age. Understanding acinar cell responses to injury and the resolution of regenerative processes is crucial for tissue homeostasis. However, knowledge about the impact of aging on these processes remains limited.
Methods
To investigate the influence of aging on pancreas regeneration, we established a cohort of young (7–14 weeks) and old (18 months) C57bl/6 mice. Experimental pancreatitis was induced using caerulein, and pancreas samples were collected at various time points after induction, covering acute damage response, inflammation, peak proliferation, and inflammation resolution. Our analysis involved immunohistochemistry, quantitative imaging, and gene expression analyses.
Results
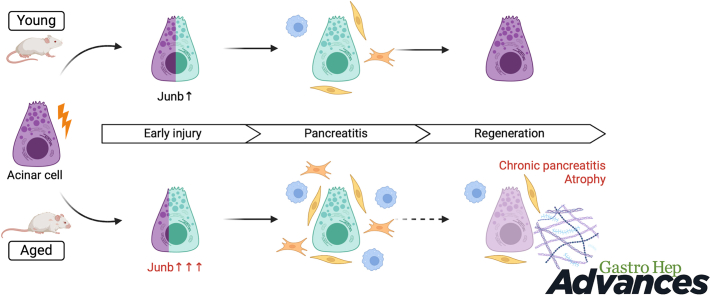
Our study revealed a significant decline in the regenerative capacity of the pancreas in old mice. Despite similar morphology and transcriptional profiles between the pancreas of young and old mice under homeostasis, the aged pancreas is primed to generate an exacerbated proinflammatory reaction in response to injury. Specifically, we observed notable upregulation of Junb expression in acinar cells and aberrant myofibroblast activation in the aged pancreas.
Conclusion
The response of acinar cells to injury in the pancreas of aged mice is characterized by an increased susceptibility to inflammation and stromal reactions. Our findings uncover a pre-existing proinflammatory state in aged acinar cells, offering insights into potential strategies to prevent the onset of pancreatic insufficiency and the development of inflammatory conditions. These insights hold implications for preventing conditions such as chronic pancreatitis and pancreatic ductal adenocarcinoma.
Keywords: ADM, Plasticity, Aging, Junb
Graphical abstract
Introduction
Acute pancreatitis is one of the most prevalent gastrointestinal causes for hospitalization, with its global incidence steadily increasing.1 In contrast to the young population, elderly individuals frequently manifest severe acute pancreatitis.2 The clinical trajectory in this age group tends to be less predictable, leading to more adverse outcomes that often culminate in multi-organ failure and elevated mortality rates.2, 3, 4 Aging impacts the homeostatic mechanisms necessary for tissue function and regeneration.5 Notably, the pancreas of older individuals displays morphological alterations, associated with increased systemic inflammation and thrombosis, including atrophy, fibrosis, and lymphocyte infiltration.6,7 While our understanding of tissue homeostasis and adaptation to injury in the pancreas is expanding, the effects of aging on these processes remain less understood.
Pancreatic injury triggers acinar cells within the exocrine tissue to undergo acinar-to-ductal metaplasia (ADM), a complex process involving the cessation of their dedicated secretory program and the acquisition of traits similar to ductal cells and embryonic progenitors.8,9 ADM provides acinar cells the ability to re-enter the cell cycle while simultaneously coordinating tissue remodeling by attracting immune cells, activating fibroblasts, and inducing modifications in the extracellular matrix to facilitate efficient tissue repair.10 The maladaptation of acinar cells to injury or the failure to resolve the regenerative response results in an exacerbated activation of fibroblasts, deviations in the composition of infiltrating immune cells, fibrosis, and ultimately a decline in tissue regeneration and loss of function.11, 12, 13 The activator protein (AP)-1 family of transcription factors regulates the transcriptional shift in acinar cells, transitioning them from their differentiated secretory function to a proinflammatory response.14 This shift plays a crucial role in regulating the activation of resident tissue fibroblasts and the recruitment of immune cells to the injury site, thereby fostering an environment conducive to tissue repair.15 Furthermore, activating mutations in the Kirsten rat sarcoma proto-oncogene (KRAS) impede the resolution of the regenerative process, leading to an irreversible state of acinar injury.16 This process is mediated by the irreversible deposition of Junb, a member of the AP-1 family of transcription factors, at regulatory elements of ADM drivers and inflammatory cytokines. Similarly, analysis of chromatin accessibility in sorted acinar cells has identified newly accessible chromatin domains in proinflammatory cytokines during the progression from homeostasis to ADM to KRAS-induced neoplastic transformation.17 It is important to note that the molecular analysis of chronic pancreatitis and early neoplastic lesions demonstrates that they share molecular signatures and cellular composition.17, 18, 19, 20 These studies highlight a progressive dysregulation of acinar adaptation to injury, a proinflammatory regulatory network, and an exacerbated response of the tissue microenvironment as determinants of disease severity. Overall, although we are gaining extensive knowledge in the mechanisms regulating tissue homeostasis in the pancreas, the effect of aging on acinar adaptation to injury and the capacity to resolve the regenerative program remains to be elucidated in old individuals.
Materials and Methods
In Vivo Mouse Experiments
Young (7–14 weeks of age) and aged (18 months of age) C57Bl/6J mice were administered with various bouts of caerulein (CAE) (Sigma-Aldrich #C9026). Acute pancreatitis was induced by 8 hourly intraperitoneal injections of CAE (125 μg/kg per injection) on 2 consecutive days, and mice were sacrificed immediately after the last injection (D0), 2 days (D2), or 14 days later (D14). To investigate the early damage response, mice were sacrificed after four-hourly injections (5H). Control mice were non-injected or injected with phosphate-buffered saline (PBS). Animals were housed in accordance with the best animal husbandry guideline recommendations of the European Union Directive (2010/63/EU). The Danish Animal Experiments Inspectorate reviewed and approved all animal experiments.
Immunohistochemistry and Immunofluorescence
Pancreata were fixed in 4% paraformaldehyde (VWR Chemicals # 9713-1000) for 24 hours (for paraffin) or 3 hours (for O.C.T.) and subsequently dehydrated in 70% ethanol (VWR Chemicals # 20824.365) at room temperature until paraffin embedding or 30% sucrose for cryoprotection in O.C.T.
Tissues were cut into 4 μm representing different depths of the tissue sections and placed on Superfrost Plus slide (10149870; Fisher Scientific). Antigen retrieval was performed in Tris-ethylenediaminetetraacetic acid buffer (pH 9). Sections were washed in PBS-Tween 0.5% and blocked with 1% donkey serum before incubation with primary antibodies overnight at 4 degrees. Secondary antibodies + 4′,6-diamidino-2-phenylindole were incubated for 1 hour at room temperature. Sections were mounted with Vectashield Mounting medium (H-1000; Vector Laboratories). Images were acquired using ScanR (Olympus) or a confocal microscope (Leica SP8) using the same laser intensities and parameters.
For immunohistochemistry, after overnight incubation with the primary antibody, the sections were washed in PBST and endogenous peroxidase activity was suppressed using Peroxidase Suppressor (Thermo Scientific # 35000). This was followed by incubation with the relevant horseradish peroxidase-conjugated secondary antibody for 1 hour and a 15-minute incubation with Metal enhanced 3,3′-diaminobenzidine tetra hydrochloride (DAB) substrate working buffer (Metal Enhanced DAB Substrate Kit, Thermo Scientific, USA). Slides were counterstained with hematoxylin (3 min) and blueing agent (H&E Staining Kit, Abcam # ab245880). Finally, the sections were dehydrated in 100% ethanol (1 minute) and xylene (1 minute) before mounting with Entellan (Sigma-Aldrich, Germany # 107960) with coverslips (Thermo Scientific #15747592). Whole section imaging was acquired using NanoZoomer-XR Digital slide scanner C12000-01 (Hamamatsu). For antibodies applied, see Table A1.
For Fast Green and Sirius Red (FGSR) stain, sections were deparaffinized in xylene and rehydrated through 100%, 70%, and 50% ethanol to water. Sections were covered with 0.1% Fast Green (Sigma #F7258) and 0.1% Sirius Red (Sigma #365548) in saturated picric acid solution (Merck #P6744) and incubated for 1 hour at room temperature. The sections were dehydrated in 100% ethanol, cleared in xylene before mounting with Entellan (Sigma-Aldrich, Germany # 107960). Whole section imaging was acquired using NanoZoomer-XR Digital slide scanner C12000-01 (Hamamatsu).
Image Processing
For quantification of acinar and tissue area in H&E-stained sections, QuPath (open-source software for bioimage analysis) was applied. The polygon tool was used to loosely define the area of interest, that is, excluding spleen and artefacts. The thresholder settings were adapted accordingly and saved as a classifier to be applied across all images. Similarly, a threshold was created within the tissue annotation to measure the acinar area with parameters adjusted for eosin stain. Non-acinar regions such as ducts, veins, and arteries were manually removed. Similar to H&E quantification, quantification of FGSR stain was done using QuPath Pixel Thresholder. A tissue annotation was created based on the threshold for average channels, including interlobular spaces clearly within the tissue area while excluding spleen and artefacts. Within the tissue annotation, an annotation was created based on the Sirius Red Pixel threshold, yielding area measurements of both annotations. For F4/80 quantification, a tissue annotation was defined as for FGSR. However, F4/80 positive cells within this annotation were quantified using Positive Cell Detection based on an appropriate threshold for “DAB optical density mean” intensity. Similarly, cleaved caspase 3 quantification was done using Positive Cell Detection within a stricter Pixel threshold-created tissue annotation to restrict the detection of positive nuclei to being within pancreatic lobules.
Cell detection, classification, and intensity measurement of immunofluorescence using QuPath
Quantification of Vimentin (Vim+), alpha-smooth muscle actin (αSMA+), Junb+, Ecad+ cells were based on up to 25 images per section, obtained from ScanR microscope, and quantified using QuPath. Two sections were analyzed per tissue. Cell detection was adjusted to the 4′,6-diamidino-2-phenylindole intensity and applied to all images with the same parameters. Cell classification was trained in a random image. The “Points” tool was used to manually annotate a number of cells positive for a given channel, and the Train object classifier was used for automatic annotation. The training was repeated in other channels when needed. The single classifier or merged multiple classifiers were run for each image yielding a total number of detections and the number of classified detections for each channel per image. Junb intensity was measured in QuPath using the Set cell intensity classification tool on top of already run Cell Detection and Object Classification. Here pixel value thresholds (arbitrary units) of “Nucleus: Junb mean” were set as follows: low (<1500), medium (1500–2500), and high (>2500). This yielded the number of Ecad-positive cells falling within each of the Junb mean intensity ranges. The threshold for “low” was set based on control tissues (young and aged) to exclude background staining.
Intensity measurement using ImageJ
To measure Mist1 intensity, Mist1 channel images acquired from a confocal microscope were opened in ImageJ. The threshold was set automatically on a random image of the 5H time point since this was expected to be lower in intensity. This threshold was applied to all images for quantification. For background normalization, a rectangle was drawn in the background of each image, and the mean fluorescence intensity (MFI) within the rectangle was measured. Then the background MFI was subtracted from the MFI of the region of interest, yielding the final MFI value.
Gene Expression Analyses
RNA was extracted using the RNeasy Mini Kit following manufacturer instructions (Qiagen #74106). DNA removal was performed with the DNase I Amplification Grade kit (Thermo Fisher Scientific, USA) and stored at −20 °C. Expression levels were calculated using the comparative ddCT method of relative quantitation, with ribosomal protein L5 (Rpl5) and ribosomal protein S2 (Rps29) as housekeeping genes. For primer sequences, see Table A1.
RNA-sequencing and downstream analyses
Bulk RNA from control and 5H time points of young and aged mice were sequenced in Illumina NovaSeq 6000 Sequencing System with paired-end 150 bp read length and output of ≥ 20 million read pairs per sample (Novogene). Bioinformatic analysis provided by Novogene unless otherwise indicated. Visualization of differentially expressed genes by volcano plot was done in R Studio (v2023.06.1 + 524) using the packages; tidyverse, ggplot, and ggrepel. Gene set enrichment analysis (GSEA) was performed on count data derived from RNA-seq data against Hallmark signatures in the MSigDB database (GSEA v4.3.2, Broad Institute). The default settings were used except for collapse to gene symbols and permutation type: gene_set. For all analyses, the metric for ranking genes was Signal2Noise except for the comparisons including A_control where Diff_of_Classes was used. This was due to an n = 2 which was not applicable for Signal2Noise.
Motif analysis
HOMER was used to do motif analysis with a standard setting.21 We tested either up-regulated genes (log2FC > 0.5 and FDR < 0.05) or downregulated genes (log2FC < 0.5 and FDR < 0.05) vs a background of non-changing genes (−0.5 > log2FC < 0.5, FDR > 0.05), based on the DESeq2 analysis from Table A2. For each gene ID in each such gene set, HOMERs in-built sequence feature retrieval function was used to identify the −1000 to +100 region around the ENSEMBL-annotated TSS (mm 10 assembly). Only known motif over-representation was reported in this analysis.
Statistical Analyses
Plots and statistical analyses were done using GraphPad Prism (v.9.5.1). For quantitative polymerase chain reaction, the statistical significance of differences between 2 experimental groups was assessed by one-way analysis of variance (P value < .05). For multiple conditions, one-way analysis of variance was used with correction for multiple comparisons using Tukey (P value < .05). The significance of gene sets from GSEA was based on a nominal P value < .05 and an FDR q-value < 0.25.
Pancreas regeneration is compromised in old mice
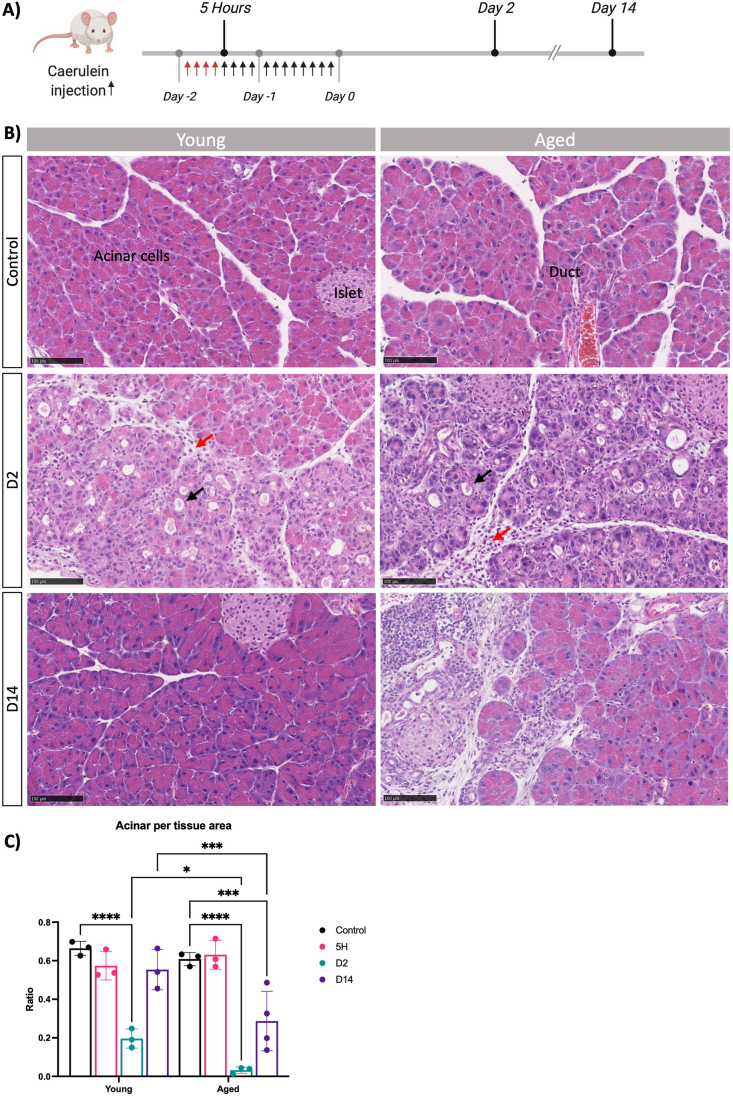
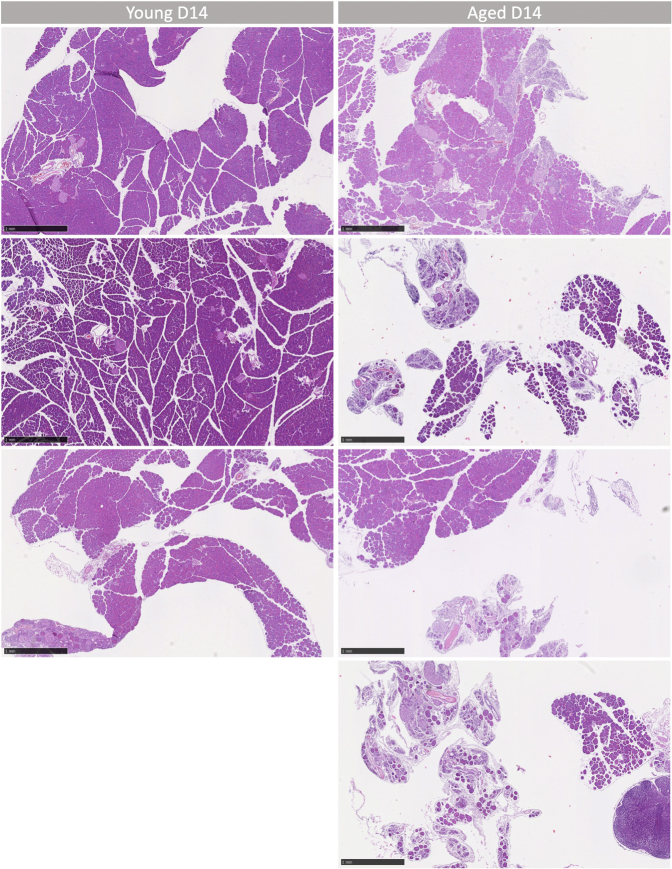
In this study, we aimed to discern the impact of aging on the adaptive behavior of pancreatic acinar cells and the regenerative potential of the pancreas following acute pancreatitis. To achieve this objective, we employed a mouse model of pancreatic injury through repeated administration of supraphysiological levels of a cholecystokinin analogue, CAE.8,22 This model was applied to both young (7–14 weeks) and old (18 months) C57Bl/6 mice (Figure 1A). Histological examinations of the pancreas in both young and old mice revealed normal morphology, characterized by overall normal architecture, well-defined lobules, and the absence of discernible immune infiltration within the interlobular space. The acinar cells displayed their characteristic pyramidical shape, and the ducts exhibited a single layer of cuboidal epithelial cells (Figure 1B). In the group subjected to CAE-induced injury, we observed evident significant alterations in pancreatic morphology. Both young and old mice displayed signs of acute inflammation with granulocytes and lymphocytes. Oedematous acini, focally dilated and enlarged intralobular space, obvious immune infiltration, and acinar to ductal metaplasia, indicative of inflammatory tissue response (Figure 1B). Remarkably, while the young mice demonstrated complete recovery within a 14-day period, the old mice displayed notable pancreatic atrophy and morphological signs of chronic inflammation with fibrosis around ducts and infiltration of lymphocytes (Figure 1B). We quantified this observation by whole-slide imaging through quantification of acinar area relative to tissue area, which highlighted a substantial reduction in acinar tissue in old mice when comparing the 2 age groups (Figure 1C). This ratio is based on measurements of eosin-dense areas, where a decrease indicates a loss of the protein-rich acinar cell phenotype. Moreover, pathological evaluation by expert pathologists who were blinded to the experimental groups revealed significant differences: all 4 pancreases from the old mice exhibited atrophy, loss of acini, and focal loss of lobular architecture, in contrast to only one of the 3 young mice, where the damage was more localized (Table A3 and Figure A1). Collectively, while the aged pancreas exhibited comparable tissue morphology and responses to tissue damage as observed in the young pancreas, the regenerative capability was significantly impaired in the older mice by day 14. These findings underscore a marked diminishment in pancreas recovery following experimental pancreatitis with the progression of age.
Figure 1.
Pancreas regeneration is affected with aging. (A) Schematic representation of the experimental model to induce acute pancreatitis in young and old mice. (B) Representative H&E images of indicated conditions of young (left) and aged (right) mouse pancreas. Scale bars represent 100 μm. Arrows indicate infiltration (red) and apparent ADMs (black). (C) Quantification of ratio acinar area per tissue area in young and aged mice. At least 3 mice per group. Statistical analysis was determined by two-way analysis of variance. Error bars represent SD. Values of significance: ns P > .05 is omitted, ∗P ≤ .05, ∗∗∗P ≤ .001, ∗∗∗∗P ≤ .0001.
Acinar cells retain the proliferative capacity in old mice
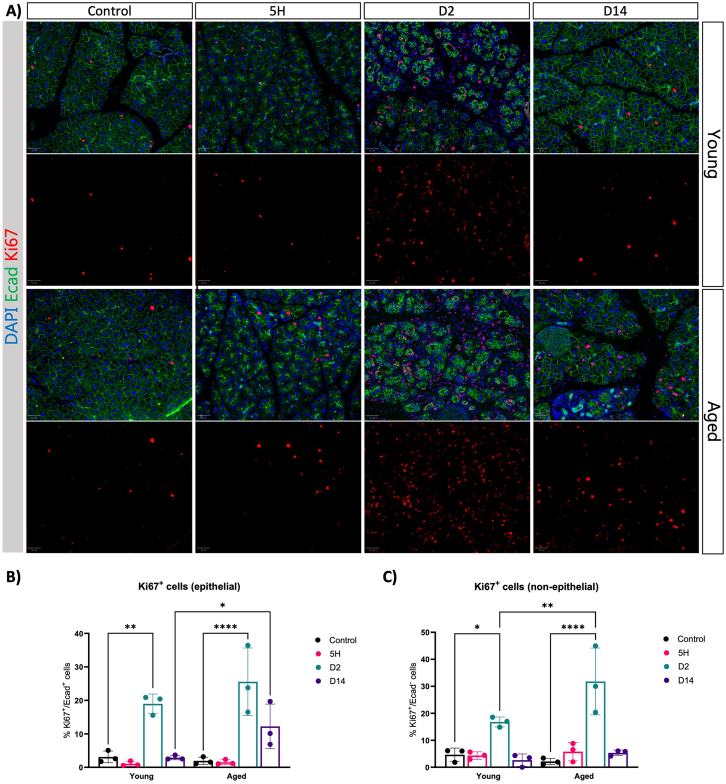
Next, we sought to determine whether acinar proliferation and apoptosis were altered during pancreas regeneration in both young and old mice. We examined the proliferation index of epithelial and non-epithelial cells by immunofluorescence. Under basal conditions, the pancreas was mostly quiescent in young mice (Figure 2A), as expected.23 Following the cessation of CAE treatment, the number of proliferating epithelial cells (marked by Ki67+) increased in both young and old mice, precisely 2 days post treatment (Figure 2A and B). Interestingly, in the context of aged mice, we did not observe any discernible differences compared to the young (Figure 2A and B). Moreover, although acinar cell proliferation reverted to basal levels 14 days after injury in the young, the residual acinar cell population within the elderly mice demonstrated a sustained capacity for initiating proliferation (Figure 2B). Hence, the impaired regenerative capacity in the aged pancreas cannot be attributed to a defect in acinar proliferation. We detected an increase in cleaved caspase 3 positive cells on day 2 that was marginally, yet significantly, higher in older mice (Figure A2). Our observations also unveiled an elevated number of proliferating cells within the non-epithelial compartment, exclusively at day 2, across both age groups (Figure 2C). The proportion of Ki67+ cells was higher in old mice compared to young individuals. These findings suggest that the increased proliferation among non-epithelial cells may be indicative of an exaggerated stromal reaction in aged mice compared to young individuals.
Figure 2.
Aging does not affect the capacity of acinar cells to enter the cell cycle. (A) Immunofluorescence of Ecad and Ki67 at given time points after caerulein treatment in young and aged mice. DAPI was used as a counterstain to visualize nuclei. Images were acquired using ScanR, and representative images were shown (n = 3). Scale bars represent 50 μm. (B) Percentage of epithelial cells (Ecad+) that are proliferating (Ki67+). (C) Percentage of non-epithelial cells (Ecad-) that are proliferating (Ki67+). Statistical analysis was determined by two-way analysis of variance. Error bars represent SD. Values of significance: ns P > .05 is omitted, ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001, ∗∗∗∗P ≤ .0001. DAPI, 4′,6-diamidino-2-phenylindole.
Old mice showed an increased proinflammatory response upon tissue injury
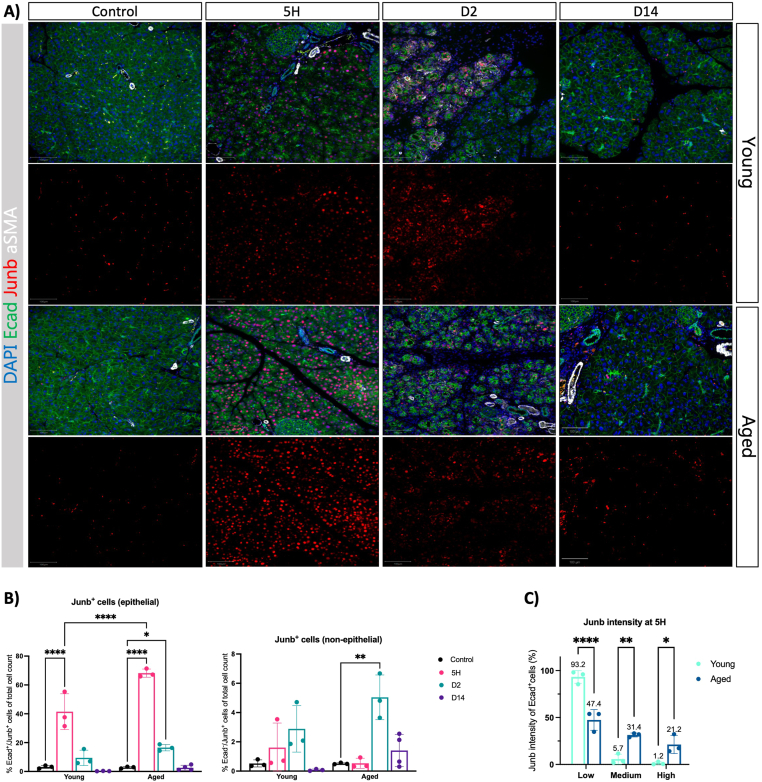
To obtain a more precise understanding of the acinar adaptation to injury in old and young mice, we performed Junb immunofluorescence analysis. We observed Junb upregulation as early as 5 hours of initiating the CAE injections, which was maintained after 2 days of the last CAE injection in young and old mice (Figure 3A). We observed an increase in the number of Junb + cells predominantly of epithelial origin in old mice (Figure 3B) and an increased intensity of Junb staining per cell (Figure 3C). Notably, at the early stages of tissue injury, the pancreas showed a normal histology with no evidence of immune infiltration or fibroblast activation (Figure A3A). We did not observe Junb upregulation in islets or ductal cells (Figure A3B). Indicating that Junb expression following CAE injury is restricted to the acinar cells. Despite regeneration being severely compromised in old mice, the expression of Junb returned to basal conditions in both age groups. Overall, our data suggest that the dynamics of Junb expression in acinar cells is similar in young and old mice. Indicating that acinar cells respond to the injury and can exit the regenerative program in young and old mice. However, acinar cells have an exacerbated response to tissue injury in old mice.
Figure 3.
Junb expression is increased with aging. (A) Immunofluorescence of Ecad, Junb, and aSMA at given time points after caerulein treatment in young and aged mice. DAPI was used as a counterstain to visualize nuclei. Images were acquired using ScanR, and representative images were shown (n = 3). Scale bars represent 100 μm. (B) We quantified Junb + cells in the epithelial (Ecad+) and non-epithelial (Ecad-) compartments as a percentage of total cells in young and old mice at the time points indicated. (C) We quantified the number of epithelial (Ecad+) Junb + cells according to the immunofluorescent intensity of Junb in the 5-hour time point. Statistical analysis was determined by two-way analysis of variance. Error bars represent SD. Values of significance: ns P > .05 is omitted, ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗∗P ≤ .0001. DAPI, 4′,6-diamidino-2-phenylindole.
Acinar plasticity precedes tissue remodeling
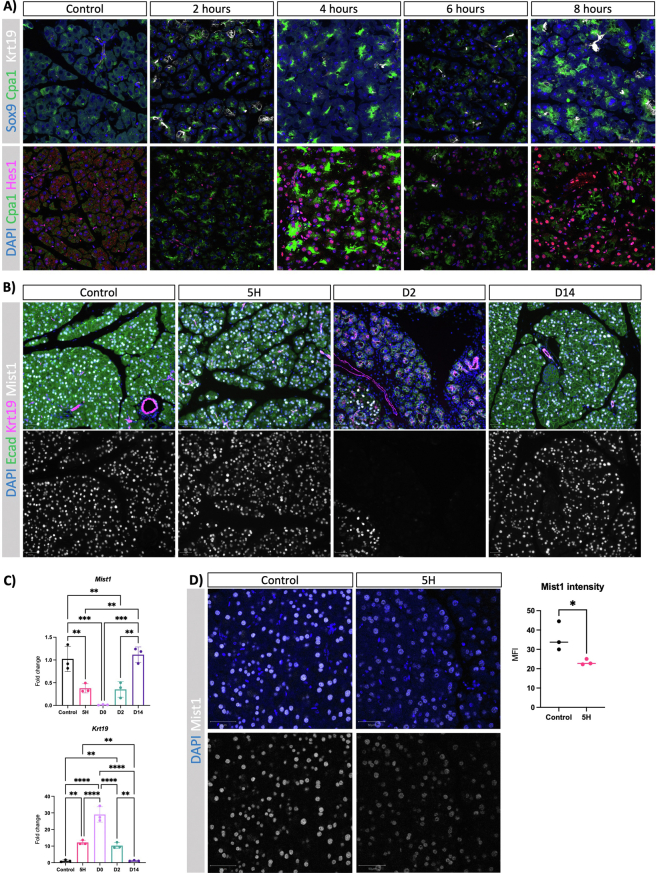
Our data indicate that acinar response to injury is independent of changes in the morphology and cellular composition of the pancreas. This suggests that aging affects cell-autonomous processes in the acinar cells that contribute to the diminished regenerative capacity of the pancreas in old mice. In order to explore whether the acinar response to tissue injury during the early phases is cell autonomous, we conducted a comprehensive time course experiment employing both quantitative polymerase chain reaction and histopathological analysis. First, we focused on young mice to avoid cofounding effects associated with aging. Our findings revealed a swift increase in Sox9 expression, a marker of ADM, emerging as early as 2 hours after CAE injection (Figure A4A). In addition to the upregulation of ductal markers, acinar adaptation to injury is associated with the upregulation of pancreas progenitor markers and the concomitant downregulation of the master regulator of the acinar program.8,24 We observed upregulation of Krt19 and Notch signaling (Hes1 transcription factor) as early as 2 hours after CAE injection (Figure A4A). Furthermore, while the abundance of Mist1+ cells, the master regulator of acinar differentiation (also called Bhlha15), remained unchanged at the 5-hour mark (Figure A4B), Mist1 expression was significantly downregulated (Figure A4C). To assess potential alterations in Mist1 protein expression during the initial response to damage, we acquired confocal microscopy images under uniform laser intensities (Figure A4D). Our observations show a decrease in Mist1 protein per cell at the 5-hour interval, indicating an ongoing process of acinar dedifferentiation at this stage. Cumulatively, our data indicate that acinar dedifferentiation and plasticity begin early in the process of tissue adaptation to the injury.
Aging is associated with an exacerbated activation of myofibroblasts
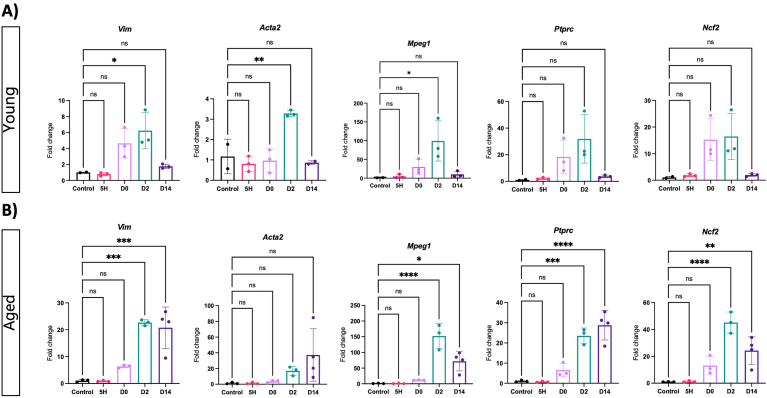
Next, we investigated the dynamics of acinar dedifferentiation and tissue remodeling in young and old mice during pancreas regeneration. We assessed the expression of Vim, Acta2, which encodes αSMA—a marker of activated fibroblasts—as well as macrophage-expressed gene (Mpeg1), protein tyrosine phosphatase (Ptprc), and neutrophil cytosolic factor 2 (Ncf2), which are markers for macrophages, leukocytes, and neutrophils, respectively, and general indicators of myeloid cells. In young mice, an upregulation of αSMA and Mpeg1 was observed, indicative of fibroblast activation and macrophage infiltration on day 2 following CAE treatment (Figure 4A). The values reverted to basal levels on day 14, in conjunction with the resolution of tissue injury. No discernible gene expression changes were observed between the control and the 5-hour time point, indicating that acinar dedifferentiation predates the activation of pancreatic fibroblasts and the infiltration of immune cells.
Figure 4.
Aging exacerbates the remodeling of the tissue microenvironment in acute pancreatitis. Quantitative real-time polymerase chain reaction of microenvironment markers at given time points following caerulein treatment in young (A) and aged (B) mice. Fibroblast and fibroblast activation was determined by Vim and Acta2 expression, respectively. Expression of immune cell markers was determined: Mpeg1 for macrophages, Ptprc for leukocytes and Ncf2 for neutrophils. Fold changes are shown relative to control and normalized with the housekeeping genes Rpl5 and Rps29. Statistical analysis was determined by unpaired one-way analysis of variance. n = 3 mice per group, except n = 2 for young controls and young D14 for Acta2 and n = 4 for aged D14. Error bars represent SD. Values of significance: P > .05 is ns, ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001, ∗∗∗∗P ≤ .0001.
Subsequently, we assessed the dynamics of gene expression in the cohort of aged mice. Similar to young mice, the early phase of acinar dedifferentiation occurred independently of changes in the expression of tissue remodeling markers in old mice (Figure 4B). However, we noted an exacerbated increase in markers associated with activated fibroblasts and immune infiltration on day 2, an increased response that persisted even 14 days after CAE treatment, indicating a compromised acinar regeneration within the context of aging.
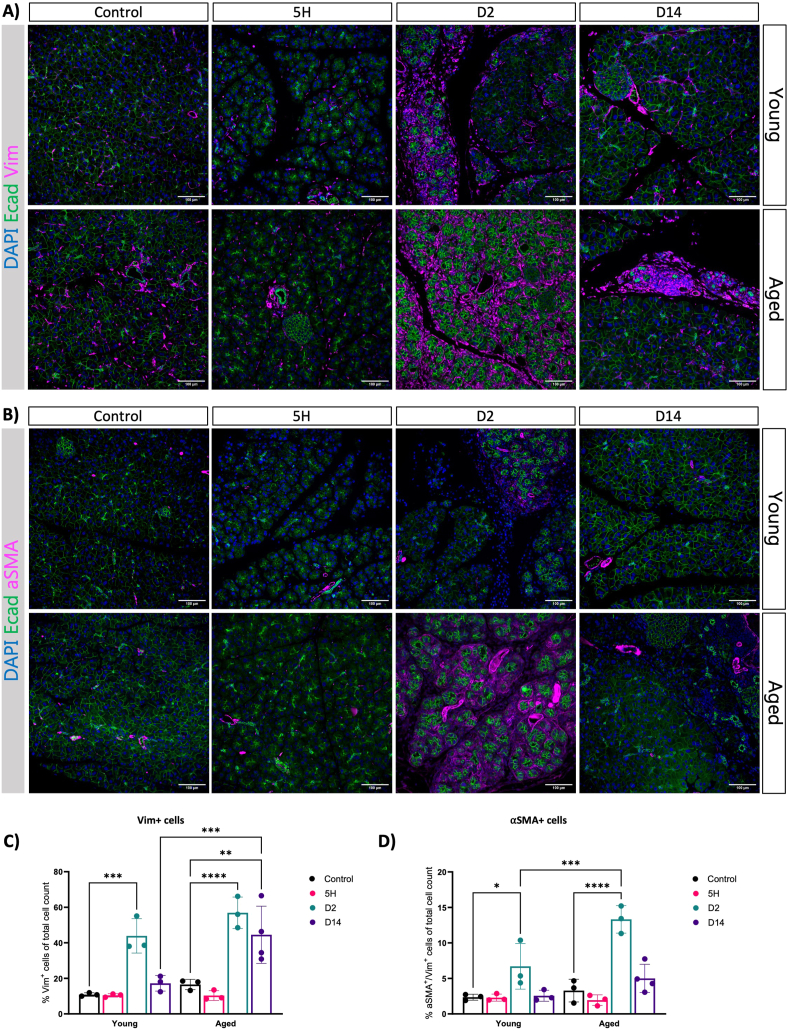
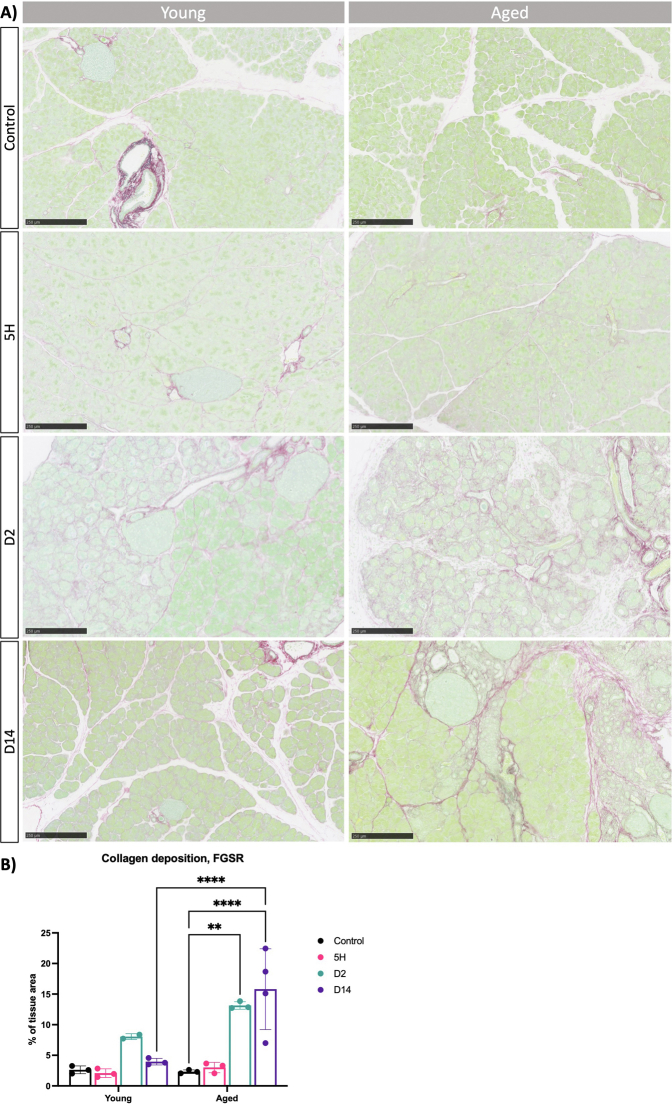
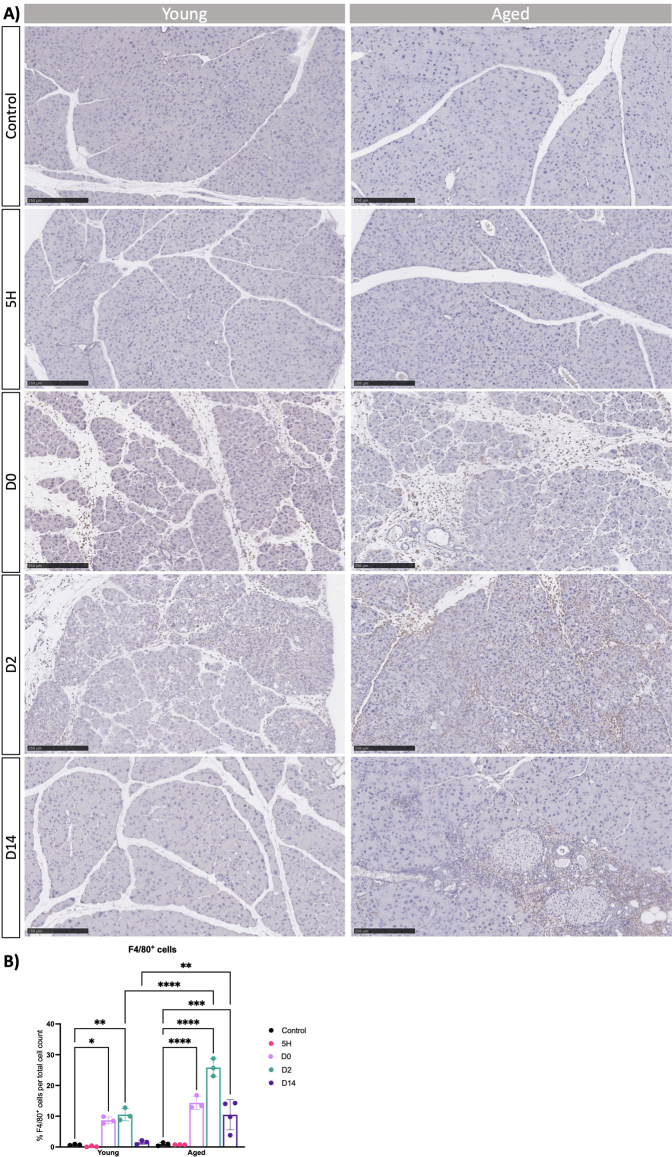
To support our observations, we conducted immunofluorescence analysis for Vim and αSMA in both young and aged mice. In alignment with the gene expression data, an increased number of Vim + cells and activated fibroblasts emerged on the second day within both age groups, especially enriched around presumed ADM lesions (Figure 5A). Interestingly, the proportion of αSMA + cells was notably higher in the older group (Figure 5B). Supporting these findings, we observed an increase in the deposition of collagens as determined by FGSR staining (Figure A5). Next, we performed immunohistochemistry for F4/80 to assess macrophages presence in old and young mice. We observed an increased presence of macrophages in the pancreas on days 0 and 2 in young mice, with higher levels persisting in old mice even 14 days after injury induction (Figure A6). Our data suggest that the dynamics of tissue remodeling are similar in young and adult mice. Like young mice, the pancreas from old mice retained the capacity to acquire alternative cellular states in response to injury. However, the old mice exhibit an exaggerated response to injury, leading to an amplified activation of myofibroblasts, deposition of collagen, and infiltration of macrophages. Furthermore, the pancreas remains in a state of injury even 14 days after the cessation of CAE treatment.
Figure 5.
Fibroblast activation is increased in response to injury in old mice. (A and B) Immunofluorescence of Ecad and Vim (A) and Ecad and aSMA (B) at given time points after caerulein treatment in young and aged mice. DAPI was used as a nuclear counterstain. Images were acquired using ScanR, and scale bars represent 50 μm. DAPI was used as a nuclear counterstain. Images were acquired using ScanR, and scale bars represent 50 μm. (C and D) Quantification of IF showing the percentage of Vim+ (C) and Vim+αSMA+ (D) of total cell count. Statistical analysis was determined by two-way analysis of variance. At least 3 mice per group. Error bars represent SD. Values of significance: P > .05 is omitted, ∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001, ∗∗∗∗P ≤ .0001. DAPI, 4′,6-diamidino-2-phenylindole.
Transcriptome analysis identify Junb dysregulation in old mice
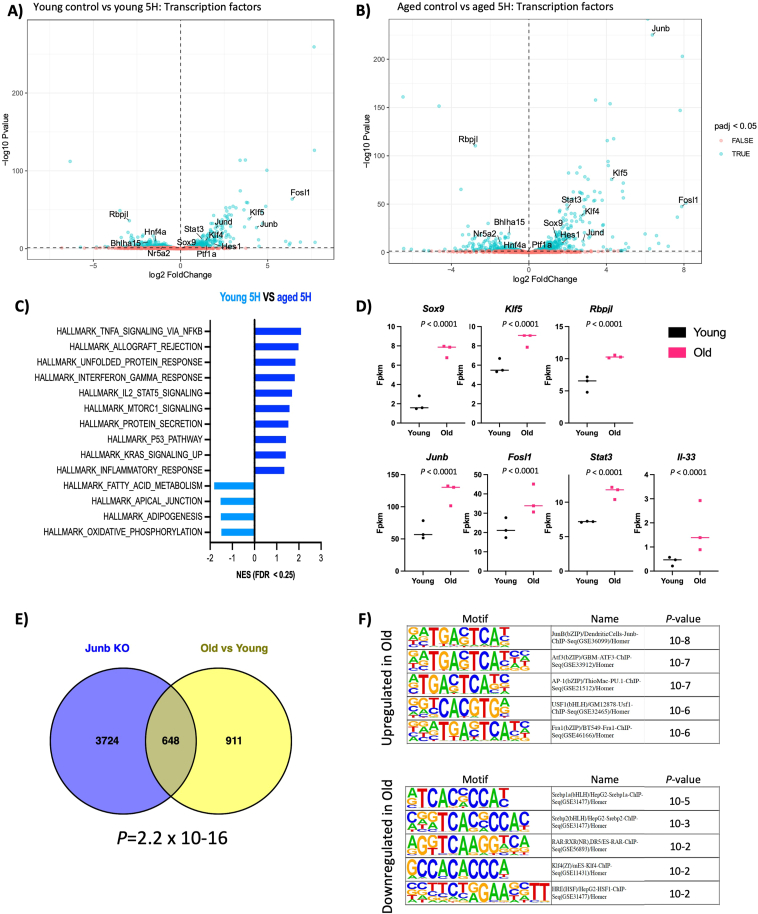
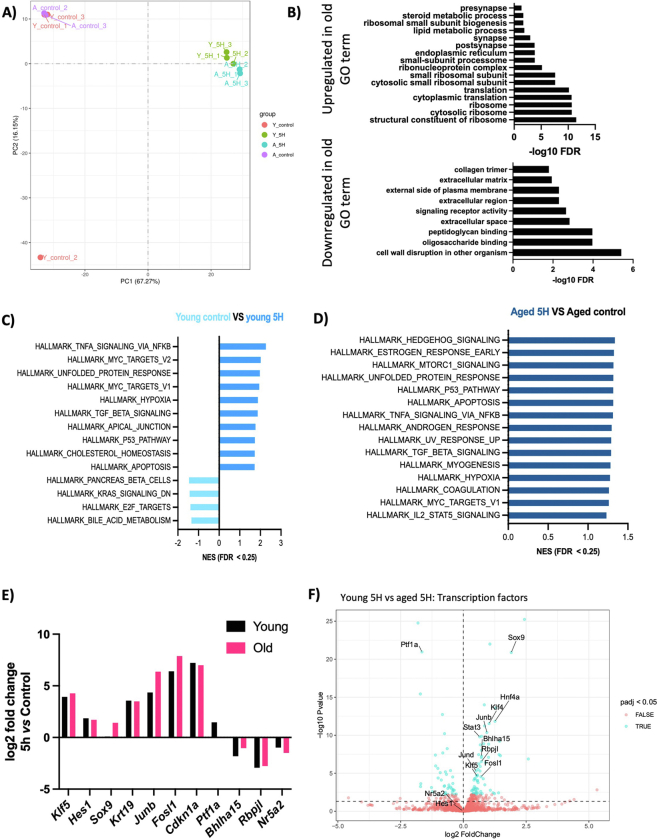
To analyze the effects of aging and pancreatic injury at the transcriptional level, we performed RNAseq on the pancreata of young (7 weeks) and old (18 months) mice, both in homeostasis and 5 hours after CAE administration. Principal component analysis revealed that 67% of the variance among all groups could be explained by the administration of CAE, causing groups to cluster according to treatment conditions (Figure A7A). This infers that the transcriptional regulation of acinar cell function remains remarkably robust and is maintained in old mice. These results are consistent with our previous histological analysis of the pancreas in both young and old mice.
Next, we conducted a differential gene expression analysis between age groups in the homeostatic state and identified a list of significantly differentially expressed genes (n = 34, padj < 0.05) (Table A4). Gene ontology analysis indicated increased lipid metabolism and decreased collagen deposition in the old group (Figure A7B).25 Moreover, among the top downregulated and upregulated genes in the older pancreas were members of the regenerative (REG) protein family (Reg3a, Reg2, and Reg3b) and ribosomal proteins transcripts (Rpl21, Rpl36), respectively (Table A4). This finding aligns with the recently identified populations of regenerative (Acinar-REG+) and secretory acinar cells (Acinar-S).20
To investigate the different responses to injury, we analyzed the effects of CAE treatment and identified many genes differentially expressed upon treatment in young and old mice (n = 6060-young, n = 6642-old, padj < 0.05) (Figure 6A and B; Tables A5 and A6). Despite the limited morphological alterations at this early time point, these results suggest that the pancreas has initiated an extensive transcriptional reprogramming in response to injury. As anticipated, GSEA analysis revealed enrichment of pathways associated with pancreatic injury (Figure A7C and D). We observed the upregulation of genes linked to acinar plasticity and the downregulation of markers of acinar differentiation in both age groups. Furthermore, we observed upregulation of the senescent marker Cdkn1a (Figure A7E). However, the increase was similar in both aging groups, and there were no differences in homeostatic conditions (Table A4). This implies that an increase in the population of senescent cells may not be the cause of the altered stromal response and regenerative capacity associated with aging.
Figure 6.
Acinar cells show a proinflammatory response to injury in aged mice. (A and B) Volcano plots showing statistical significance (−log10 P value) on the y-axis versus magnitude of fold change (log2 FC) on the x-axis. Each dot represents a gene. The genes are color-coded by adjusted P value (padj) < 0.05 (blue) and > 0.05 (red). (C) Gene set enrichment analysis of hallmark signatures gene sets between 5H old and young mice. Significantly enriched hallmark signature gene sets were determined by FDR q-value < 0.25 and nominal P value < .05. (D) Differential gene expression analysis of selected genes as determined by RNAseq and DESEq2 analysis comparing old and young mice at the 5H time point. We observed upregulation of markers of pancreas development and proinflammatory signals in old mice compared to young. n = 3 mice/group. (E) Venn diagram depicting the overlap between the following datasets. Junb KO: differentially expressed genes in Junb KO vs WT control cell line derived from an autochthonous model of PDAC. Old vs Young: differentially expressed genes in old and young mice at the 5H time point. (F) We analyzed the transcription factor binding sites in the promoter region of differentially expressed genes in old and young mice at the 5H time point by HOMER. The binding site for Junb is the top enriched in the set of upregulated genes.
Further examination focused on the differences between age groups in the early response to tissue damage. Differential gene expression analysis identified 1558 differentially expressed genes (padj < 0.05, Table A2), highlighting differences in the response of acinar cells to injury between the old and young mice. Gene set enrichment analysis identified pathways associated with pancreas development, the unfolded protein response, and inflammatory processes enriched in the aged group (Figure 6C). We confirmed the upregulation of transcription factors linked to pancreas development and inflammatory processes, indicating an aggravated response of acinar cells to tissue injury in the aged pancreas. We observed the upregulation of transcription factors promoting an inflammatory response in the pancreas (Figure 6D; Figure A7F). Notably, Junb was among the top upregulated genes in the old group. Furthermore, there was an increased expression of interleukin 33 (IL-33), a cytokine involved in promoting the remodeling of the tumor microenvironment and epithelial transformation in preneoplastic lesions, particularly in the presence of Kras mutation. Consistent with this finding, KRAS UP signaling pathways were also enriched in old mice. Next, we ask whether Junb expression is associated with the difference on gene expression in old mice upon CAE treatment. Intriguingly, transcriptional analysis of Junb-depleted tumors derived from an autochthonous model of pancreatic ductal adenocarcinoma (PDAC) exhibited a significant overlap with the differentially expressed genes in old mice after 5 hours of CAE treatment (Figure 6E). It is important to acknowledge that these comparisons involve different stages of the disease. Nonetheless, our findings revealed a significant overlap in genes differentially expressed between these conditions. These data suggest that Junb mediates, at least in part, the transcriptional changes observed in response to injury in old mice. Supporting this observation, the transcription factor binding motif of Junb was the most enriched at the promoter regions of differentially expressed genes upon CAE treatment in both age groups (Figure 6F). This indicates a potential connection between the AP-1 targets and the decline in the regenerative capacity of the pancreas observed in old mice. Further exploration of this overlap could provide valuable insights into the mechanisms underlying pancreatic healing and severe acute pancreatitis associated with aging.
Discussion
The process of pancreas recovery following tissue damage requires a coordinated interplay between transcriptional programs of acinar adaptation to injury and various cell types within the regenerative microenvironment. While the impact of aging on tissue repair is well-established across diverse tissues, its precise mechanisms within the pancreas remain largely elusive. Our findings reveal that the pancreas from aged mice lacks the capacity to recover from acute pancreatitis, showing evidence of tissue atrophy and fibroinflammatory stroma. We showed that acinar cells initiate a substantial transcriptional reprogramming even prior to the discernible morphological indicators of tissue remodeling, and even more strongly in aged mice. The exacerbated response of acinar cells to the injury is associated with an increased activation of αSMA + myofibroblasts in old mice, which have been shown to contribute to fibrosis and inflammation.26,27 Therefore, our findings show that aging affects the homeostatic mechanism of acinar cells and the stromal remodeling regulating pancreas regeneration.
The reprogramming of acinar cells is crucial to orchestrate the fibroinflammatory response of the pancreas during tissue regeneration.28 Molecular analyses of lesions derived from pancreatitis as well as precursor lesions of cancer, underscore an increasing proinflammatory transcriptional network in the acinar cells associated with disease progression.19,29,30 Analysis of chromatin accessibility in sorted acinar cells has identified newly accessible chromatin domains in proinflammatory cytokines during the progression from homeostasis to ADM to KRAS-induced neoplastic transformation.17 These studies emphasize the leading role of acinar cells in pancreas regeneration and the implications in disease progression. In our study, we observed an increased number of acinar cells that express the AP-1 family member Junb in old mice. Junb expression in acinar cells during disease progression prevents the resolution of tissue damage and the progression toward precursor lesions of cancer.16 Mechanistically, JunB co-opts a regulatory network of enhancers activated downstream of KRAS signaling perpetuating an inflammatory environment. Among the secreted factors upregulated in old mice, we observed the cytokine IL-33, associated with a fibroinflammatory response, to be exclusively upregulated upon tissue injury in old mice.17,31 Notably, transcriptional analysis of sorted acinar cells identifies IL-33 as a mediator of tissue remodeling and promoting disease progression in precursor lesions of PDAC. This indicates that IL-33 may be responsible for the increased number of myofibroblast in old mice after injury. Furthermore, we observed enhanced unfolded protein response in older mice, indicating potential defects in maintaining endoplasmic reticulum homeostasis and a heightened susceptibility to stress in the acinar cells of these mice. Overall, our data suggest that aging exerts changes in the acinar cells that make them prone to a proinflammatory response that is only seen in precursor lesions of PDAC.
Conclusion
Our study reveals that despite the significant differences in the response to injury, the transcriptome of young and old individuals remained remarkably comparable under homeostatic conditions. This suggests that while the transcriptional regulation of acinar function remains robust and resilient to aging processes, acinar cells become sensitized to mounting an inflammatory response following injury. Remarkably, the aged pancreas has notable differences in the methylation patterns of regulatory elements. However, these differences do not necessarily correlate with changes in the transcription of associated genes.32 These insights indicate that variations in the accessibility of regulatory elements could potentially underlie the heightened response to injury associated with aging. In forthcoming experiments, it would be valuable to focus on the cell-autonomous regulatory elements that prime acinar cells toward a proinflammatory response to injury in aged mice.
Recent studies demonstrate the presence of heterogeneous populations of acinar cells characterized by their varying proliferative capacities, possibly contributing to the decline in regenerative potential observed in aged mice. Specifically, aging is associated with the accumulation of metabolically active and cell cycle-arrested senescent cells.33 While there is increasing evidence of senescent cell buildup in the context of KRAS-induced neoplastic transformation,34 the extent to which senescent cells accumulate with age remains uncertain. Our transcriptome analysis indicated comparable expression levels of senescence markers in young and old mice, with their expression increasing in response to injury. Interestingly, similar to our findings, skeletal muscles in aged mice harbor a minimal population of senescent cells that increases following injury.35 Collectively, our data suggest that senescent cells in the pancreas are not a direct consequence of the aging process but rather stem from oncogene-induced senescence. Furthermore, our study did not observe alterations in the expression of telomerase reverse transcriptase (Tert), a marker associated with a rare subpopulation of acinar cells possessing regenerative potential.36 We identified a reduction in the expression of markers associated with the Acinar-reg subpopulation, accompanied by an increase in markers characterizing the Acinar-s population in elderly mice. These findings suggest the possibility of clonal amplification among distinct acinar cell populations, each endowed with varying regenerative capacities or susceptibility to damage, which may contribute to the diminished regenerative capability of the pancreas that is observed with aging.
Our study has certain limitations as we used mice that have been housed under sterile conditions with a strictly controlled diet and light/dark cycles. While such conditions are unfeasible to replicate in human subjects, they are indispensable for isolating the effects of aging on pancreas physiology from potential confounding influences of lifestyle-related alterations in transcriptional and cellular composition. Notably, recent functional analyses of genomic variations identified in genome-wide association studies have revealed a connection between the transcriptional dysregulation of the AP-1 complex in acinar cells and an underlying proinflammatory state within the pancreas aggravating the progression of experimental pancreatitis in mice.14 Thereby underscoring the critical importance of comprehending the impact of aging on the proinflammatory status of acinar cells and the acinar-specific regulatory mechanisms governing the remodeling of the tissue microenvironment and disease progression.
Acknowledgments:
We are sincerely grateful for the Histology and Microscopy, Light Microscopy and Bioinformatics core facilities at the Biotech Research and Innovation Centre (BRIC), University of Copenhagen. Especially, the invaluable support and expertise of Mia Kristine Grønning Høg, Yasuko Antoku and Juliana Assis from the core facilities have significantly contributed to the success of this research. The graphical abstract was created with BioRender.com.
Authors’ Contributions:
Acquisition of data: all authors. Drafting of the manuscript: Kristina Høj and Luis Arnes. Critical revision of the manuscript for important intellectual content: all authors. Obtained funding, Study supervision, Study concept and design: Luis Arnes.
Footnotes
Conflicts of Interest: The authors disclose no conflicts.
Funding: Luis Arnes laboratory is supported by core funding of the Biotech Research and Innovation Center, the Danish Cancer Society (R302-A17481, R322-A17.350), The Novo Nordisk Foundation (NNF21OC0070884) and The Innovation Fund (Eurostars 2807). The Novo Nordisk Foundation Center for Stem Cell Biology was supported by Novo Nordisk Foundation grants NNF17CC0027852.
Ethical Statement: All the animals were treated ethically in accordance with best animal husbandry guideline recommendations of the European Union Directive (2010/63/EU). All the protocols and procedures were approved by the Danish Animal Experiments Inspectorate (2024-15-0201-01673).
Data Transparency Statement: Study materials will be made available upon request. The transcriptomic and sequencing data discussed in this publication have been deposited in the Gene Expression Omnibus archive (GSE268278).
Reporting Guidelines: The study was conducted in accordance with ARRIVE guidelines.
Material associated with this article can be found, in the online version, at https://doi.org/10.1016/j.gastha.2024.07.002.
Supplementary Materials
Figure A1.
(A) Low magnification H&E images of young and aged D14 mouse pancreas. Scale bars represent 1 mm.
Figure A2.
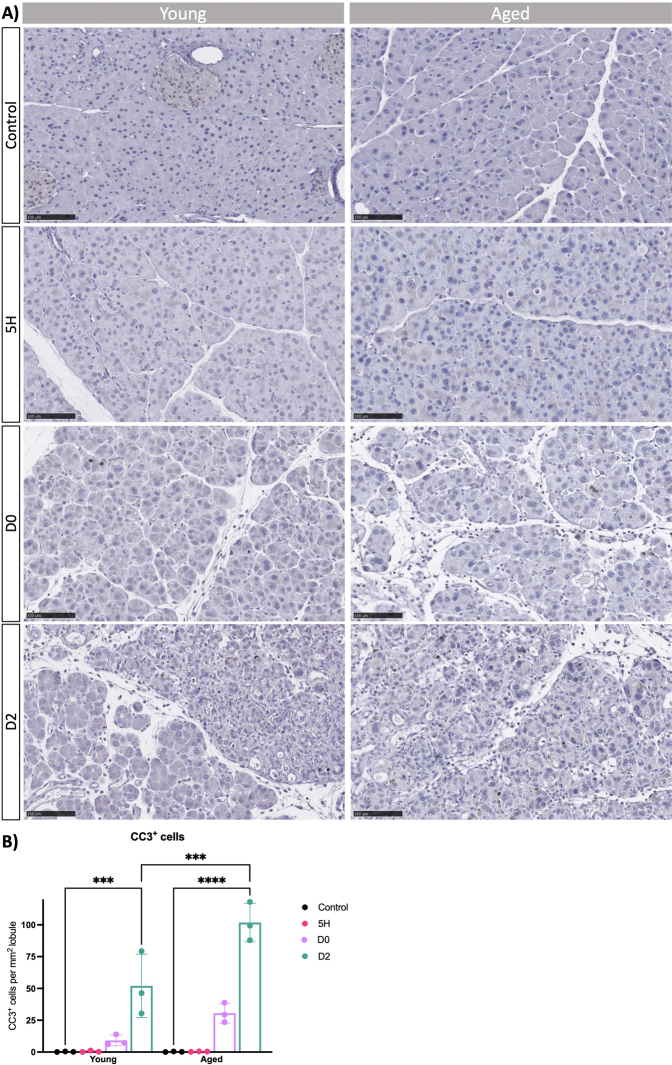
(A) Cell death was investigated by IHC stain against cleaved caspase-3 (CC3) in young and aged mice at indicated time points. Representative images of n ≥ 3 mice are shown. Scale bars represent 100 μm. (B) Quantification of CC3+ cells per mmˆ2 lobule (excluding interlobular space). Statistical analysis was determined by two-way ANOVA. Error bars represent SD. Values of significance: ns P > .05 is omitted, ∗P ≤ .05, ∗∗∗ P ≤ .001 and ∗∗∗∗ P ≤ .0001.
Figure A3.
(A) Representative H&E images of 5-hour timepoint in young (left) and aged (right) mouse pancreata (n = 3). Scale bars represent 100 μm. (B) Immunofluorescence images of Ecad and Junb in pancreata from young 5H and D2 mice (related to Figure 3). Representative images are shown with zoom in on islets and ducts, demonstrating that neither express Junb. DAPI was used as a counterstain to visualise nuclei. Images were acquired using ScanR. Scale bars represent 50 μm.
Figure A4.
(A) Immunofluorescence of Sox9, Cpa1 and Krt19 (top panel) and Cpa1, Hes1 and Krt19 (bottom panel) at given time points following caerulein treatment of young mice (n = 3). Images were acquired using confocal. (B) Immunofluorescence of Ecad, Krt19 and Mist1 at given timepoints after caerulein treatment of young mice. DAPI was used as a counterstain to visualise nuclei. Images were acquired using ScanR, and scale bars represent 50 μm. (C) Quantitative real-time PCR of markers associated with acinar plasticity at given time following caerulein treatment in young mice. Mist1 is acinar-specific, whereas Krt19 is expressed in mature ductal cells and ADM. Fold changes are shown relative to control and normalised with the housekeeping genes Rpl5 and Rps29. Statistical analysis was determined by unpaired one-way ANOVA. n = 3 mice per group. Error bars represent SD. Values of significance: ns P > .05 is omitted, ∗∗ P ≤ .01, ∗∗∗ P ≤ .001, ∗∗∗∗ P ≤ .0001. (D) Immunofluorescence of Mist1 in young control and following 4 injections with caerulein. Nuclei were counterstained by DAPI. Images were acquired using confocal microscopy, and scale bars represent 50 μm. Quantification of IF showing mean fluorescent intensity (MFI) of Mist1 in arbitrary units. Statistical analysis was determined by an unpaired t-test. n = 3 mice per group. Error bars represent SD. Values of significance: ns P > .05, ∗ P ≤ .05.
Figure A5.
(A) We examined the collagen deposition in the extracellular matrix by Fast Green and Sirius Red (FGSR) staining of young and aged mice at the indicated time points. Scale bars represent 250 μm. (B) Collagen deposition, visualized by Sirius Red stain, was quantified as percentage positive area of total tissue area. Statistical analysis was determined by two-way ANOVA. Error bars represent SD. Values of significance: ns P > .05 is omitted, ∗∗P ≤ .01, ∗∗∗∗ P ≤ .0001.
Figure A6.
(A) Macrophage presence was investigated by IHC stain against F4/80 in young and aged mice at indicated time points. Representative images are shown. Scale bars represent 250 μm. (B) Quantification of F4/80+ cells per total cell count. Statistical analysis was determined by two-way ANOVA. Error bars represent SD. Values of significance: ns P > .05 is omitted, ∗P ≤ .05, ∗∗ P ≤ .01, ∗∗∗ P ≤ .001 and ∗∗∗∗ P ≤ .0001.
Figure A7.
(A) Principal component analysis (PCA) of RNA-seq data reveal clustering of each condition, regardless of age. PCA score plot of PC1 and PC2 illustrates 87.42 % of the total variance in transcriptomic data between young control (red), young 5H (green), aged 5H (blue) and aged control (purple). PCA was done by Novogene. (B) Gene ontology analyses of differentially expressed genes in the young and aged control tissues. (C and D) GSEA of hallmark signatures gene sets between 5H and control in young mice (C) and aged versus young 5H (D). Significantly enriched hallmark signature gene sets were determined by FDR q-value < 0.25 and nominal P-value < .05. (E) Differentially expressed genes related to acinar plasticity and differentiation compared between the 5-hour timepoint and control for each age group. (F) Volcano plots showing statistical significance (−log10 P-value) on the y-axis versus magnitude of fold change (log2 FC) on the x-axis. Each dot represents a gene. The genes are colour-coded by adjusted P-value (padj) < .05 (blue) and > .05 (red).
References
- 1.Baeza-Zapata A.A., Garcia-Compean D., Jaquez-Quintana J.O., et al. Acute pancreatitis in elderly patients. Gastroenterology. 2021;161:1736–1740. doi: 10.1053/j.gastro.2021.06.081. [DOI] [PubMed] [Google Scholar]
- 2.Quero G., Covino M., Fiorillo C., et al. Acute pancreatitis in elderly patients: a single-center retrospective evaluation of clinical outcomes. Scand J Gastroenterol. 2019;54:492–498. doi: 10.1080/00365521.2019.1588369. [DOI] [PubMed] [Google Scholar]
- 3.Yadav D., Lowenfels A.B. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–1261. doi: 10.1053/j.gastro.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beger H.G., Rau B.M. Severe acute pancreatitis: clinical course and management. World J Gastroenterol. 2007;13:5043–5051. doi: 10.3748/wjg.v13.i38.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodell M.A., Rando T.A. Stem cells and healthy aging. Science. 2015;350:1199–1204. doi: 10.1126/science.aab3388. [DOI] [PubMed] [Google Scholar]
- 6.Okamura D., Starr M.E., Lee E.Y., et al. Age-dependent vulnerability to experimental acute pancreatitis is associated with increased systemic inflammation and thrombosis. Aging Cell. 2012;11:760–769. doi: 10.1111/j.1474-9726.2012.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda Y. Age-related morphological changes in the pancreas and their association with pancreatic carcinogenesis. Pathol Int. 2019;69:450–462. doi: 10.1111/pin.12837. [DOI] [PubMed] [Google Scholar]
- 8.Jensen J.N., Cameron E., Garay M.V., et al. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Pinho A.V., Rooman I., Real F.X. p53-dependent regulation of growth, epithelial-mesenchymal transition and stemness in normal pancreatic epithelial cells. Cell Cycle. 2011;10:1312–1321. doi: 10.4161/cc.10.8.15363. [DOI] [PubMed] [Google Scholar]
- 10.Brown J.W., Cho C.J., Mills J.C. Paligenosis: cellular remodeling during tissue repair. Annu Rev Physiol. 2022;84:461–483. doi: 10.1146/annurev-physiol-061121-035954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baer J.M., Zuo C., Kang L.I., et al. Fibrosis induced by resident macrophages has divergent roles in pancreas inflammatory injury and PDAC. Nat Immunol. 2023;24:1443–1457. doi: 10.1038/s41590-023-01579-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velez-Delgado A., Donahue K.L., Brown K.L., et al. Extrinsic KRAS signaling shapes the pancreatic microenvironment through fibroblast reprogramming. Cell Mol Gastroenterol Hepatol. 2022;13:1673–1699. doi: 10.1016/j.jcmgh.2022.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter E.S., Elhossiny A.M., Kadiyala P., et al. Analysis of donor pancreata defines the transcriptomic signature and microenvironment of early neoplastic lesions. Cancer Discov. 2023;13:1324–1345. doi: 10.1158/2159-8290.CD-23-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobo I., Martinelli P., Flandez M., et al. Transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas. Nature. 2018;554:533–537. doi: 10.1038/nature25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burdziak C., Alonso-Curbelo D., Walle T., et al. Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science. 2023;380 doi: 10.1126/science.add5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., He Y., Peng J., et al. Mutant Kras co-opts a proto-oncogenic enhancer network in inflammation-induced metaplastic progenitor cells to initiate pancreatic cancer. Nat Cancer. 2021;2:49–65. doi: 10.1038/s43018-020-00134-z. [DOI] [PubMed] [Google Scholar]
- 17.Alonso-Curbelo D., Ho Y.J., Burdziak C., et al. A gene-environment-induced epigenetic program initiates tumorigenesis. Nature. 2021;590:642–648. doi: 10.1038/s41586-020-03147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlesinger Y., Yosefov-Levi O., Kolodkin-Gal D., et al. Single-cell transcriptomes of pancreatic preinvasive lesions and cancer reveal acinar metaplastic cells' heterogeneity. Nat Commun. 2020;11:4516. doi: 10.1038/s41467-020-18207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Z., Lytle N.K., Chen B., et al. Single-cell transcriptomics reveals a conserved metaplasia program in pancreatic injury. Gastroenterology. 2022;162:604–620.e20. doi: 10.1053/j.gastro.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tosti L., Hang Y., Debnath O., et al. Single-nucleus and in Situ RNA-sequencing reveal cell topographies in the human pancreas. Gastroenterology. 2021;160:1330–1344.e11. doi: 10.1053/j.gastro.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Heinz S., Benner C., Spann N., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keefe M.D., Wang H., De La O.J., et al. beta-catenin is selectively required for the expansion and regeneration of mature pancreatic acinar cells in mice. Dis Model Mech. 2012;5:503–514. doi: 10.1242/dmm.007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong B., Bruns P., Behler N.A., et al. Dynamic landscape of pancreatic carcinogenesis reveals early molecular networks of malignancy. Gut. 2018;67:146–156. doi: 10.1136/gutjnl-2015-310913. [DOI] [PubMed] [Google Scholar]
- 24.Pin C.L., Rukstalis J.M., Johnson C., et al. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol. 2001;155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Otin C., Blasco M.A., Partridge L., et al. Hallmarks of aging: an expanding universe. Cell. 2023;186:243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Ozdemir B.C., Pentcheva-Hoang T., Carstens J.L., et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2015;28:831–833. doi: 10.1016/j.ccell.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Rhim A.D., Oberstein P.E., Thomas D.H., et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storz P., Crawford H.C. Carcinogenesis of pancreatic ductal adenocarcinoma. Gastroenterology. 2020;158:2072–2081. doi: 10.1053/j.gastro.2020.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DelGiorno K.E., Chung C.Y., Vavinskaya V., et al. Tuft cells inhibit pancreatic tumorigenesis in mice by producing prostaglandin D(2) Gastroenterology. 2020;159:1866–1881.e8. doi: 10.1053/j.gastro.2020.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Poggetto E., Ho I.L., Balestrieri C., et al. Epithelial memory of inflammation limits tissue damage while promoting pancreatic tumorigenesis. Science. 2021;373 doi: 10.1126/science.abj0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liew F.Y., Girard J.P., Turnquist H.R. Interleukin-33 in health and disease. Nat Rev Immunol. 2016;16:676–689. doi: 10.1038/nri.2016.95. [DOI] [PubMed] [Google Scholar]
- 32.Chondronasiou D., Gill D., Mosteiro L., et al. Multi-omic rejuvenation of naturally aged tissues by a single cycle of transient reprogramming. Aging Cell. 2022;21 doi: 10.1111/acel.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Micco R., Krizhanovsky V., Baker D., et al. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat Rev Mol Cell Biol. 2021;22:75–95. doi: 10.1038/s41580-020-00314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amor C., Feucht J., Leibold J., et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature. 2020;583:127–132. doi: 10.1038/s41586-020-2403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moiseeva V., Cisneros A., Sica V., et al. Senescence atlas reveals an aged-like inflamed niche that blunts muscle regeneration. Nature. 2023;613:169–178. doi: 10.1038/s41586-022-05535-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuhofer P., Roake C.M., Kim S.J., et al. Acinar cell clonal expansion in pancreas homeostasis and carcinogenesis. Nature. 2021;597:715–719. doi: 10.1038/s41586-021-03916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.