Abstract
The ubiquitin-proteasome system (UPS), which involves E3 ligases and deubiquitinates (DUBs), is critical for protein homeostasis. The epigenetic reader ZMYND8 (zinc finger MYND-type containing 8) has emerged as an oncoprotein, and its protein levels are elevated in various types of cancer, including breast cancer. However, the mechanism by which ZMYND8 protein levels are increased in cancer remains elusive. Although ZMYND8 has been reported to be regulated by the E3 ligase FBXW7, it is still unknown whether ZMYND8 could be modulated by DUBs. Here, we identified USP7 (ubiquitin carboxyl-terminal hydrolase 7) as a bona fide DUB for ZMYND8. Mechanically, USP7 directly binds to the PBP (PHD-BRD-PWWP) domain of ZMYND8 via its TRAF (tumor necrosis factor receptor-associated factor) domain and UBL (ubiquitin-like) domain and removes F-box and WD repeat domain containing 7 (FBXW7)-catalyzed poly-ubiquitin chains on lysine residue 1034 (K1034) within ZMYND8, thereby stabilizing ZMYND8 and stimulating the transcription of ZMYND8 target genes ZEB1 (zinc finger E-box binding homeobox 1) and VEGFA (Vascular Endothelial Growth Factor A). Consequently, USP7 enhances the capacity of breast cancer cells for migration and invasion through antagonizing FBXW7-mediated ZMYND8 degradation. Importantly, the protein levels of USP7 positively correlates with those of ZMYND8 in breast cancer tissues. These findings delineate an important layer of migration and invasion regulation by the USP7-ZMYND8 axis in breast cancer cells.
Keywords: ZMYND8, USP7, cell migration, cell invasion, breast cancer cells
ZMYND8 (Zinc finger MYND-type containing 8), also known as RACK7 and PRKCBP1, is an epigenetic reader that contains a PHD (plant homeodomain) domain, a BRD (bromodomain) domain, a PWWP (Pro-Trp-Trp-Pro) chromatin-binding domain, and a MYND (Myeloid, Nervy and DEAF-1) domain (1, 2). The PBP (PHD-BRD-PWWP) domain recognizes modified chromatin and mediates protein-protein interaction, while the MYND domain mainly bridges the association between ZMYND8 and other nonhistone proteins (1, 3, 4, 5, 6). It is well documented that ZMYND8 can activate or repress gene expression in cancer cells. ZMYND8 stimulates gene expression by interacting with HIF (hypoxia-inducible factor) (1, 7), BRD4 (bromodomain-containing protein 4) (1, 8, 9), NRF2 (nuclear factor erythroid 2-related factor 2) (9), SREBP2 (sterol regulatory element-binding protein 2) (10), and EZH2 (enhanced zeste homolog 2) (11), whereas repressing gene transcription by associating with KDM5C (lysine demethylase 5C) (12), p65 (13), and NuRD (nucleosome remodeling and deacetylase) complex (3, 14). Our recent findings have demonstrated that ZMYND8 expression is upregulated in human breast cancer tissues and promotes breast tumor growth and metastasis by controlling HIF signaling (1), cGAS-STING-NF-κB axis (15), and 27-hydroxycholesterol metabolism (8). In addition to breast cancer, ZMYND8 expression is also increased in other types of cancer, including intestinal cancer (10), glioblastoma (16), clear cell renal cell carcinoma (11), and bladder cancer (17) and consequently provoke tumor growth and metastasis. Despite ZMYND8 transcription is regulated by HIF (1), NRF2 (9) and YAP (yes-associated protein) (10), the role of posttranslational modification (PTM) on ZMYND8 remain elusive. Recently, ubiquitination and E3 ligase FBXW7 (F-box and WD repeat domain containing 7) have been demonstrated to modulate ZMYND8 stability (17, 18). However, whether deubiquitinases (DUBs) modulate ZMYND8 stability remains unknown.
DUBs are critical for protein homeostasis (19, 20, 21). USP7 is a multifunctional DUB containing TRAF (tumor necrosis factor receptor-associated factor) domain, catalytic domain, and UBL (ubiquitin-like) domain (22, 23). Accumulating evidence suggests that USP7 is overexpressed in a plethora of cancer types and participates in diverse processes of cancer, including genome stability, inflammation, proliferation, metastasis, angiogenesis and cell death (24). Intriguingly, USP7 functions as either a tumor suppressor or an oncoprotein, depending on its predominant targets in different cancer types (24). USP7 stabilizes HIF-1α to induce the expression of SLC2A1 which encodes GLUT1 (glucose transporter type 1) in triple negative breast cancer (TNBC) cells, thereby promoting TNBC growth (25). USP7 enhances the ability of breast cancer cells in metastasis by stabilizing EZH2 and LSD1 (lysine demethylase 1) (26, 27). Moreover, USP7 contributes to chemoresistance of TNBC cells by increasing ABC1 (ATP-binding cassette sub-family B member 1) stability (28). Hence, USP7’s function is highly dependent on its substrates in breast cancer cells. Identification of novel USP7 substrates is critical for elucidating USP7’s function in breast cancer cells.
In the present study, we identified USP7 as a bona fide DUB for ZMYND8 by performing a DUB complementary (cDNA) library binding screen. USP7 directly binds to the PBP domain of ZMYND8 via its TRAF and UBL domains to deubiquitinate ZMYND8 and subsequently increases ZMYND8 stability and the transcription of ZMYND8 target genes ZEB1 (zinc finger E-box binding homeobox 1) and VEGFA (Vascular Endothelial Growth Factor A), which are pivotal for cancer cell metastasis. Ultimately, USP7 promotes breast cancer cell migration and invasion. Moreover, the protein levels of USP7 are positively correlated with that of ZMYND8 in human breast tumors. These results uncover the functional role and regulatory mechanism of USP7-ZMYND8 axis in breast cancer cells.
Results
ZMYND8 associates with USP7 in breast cancer cells
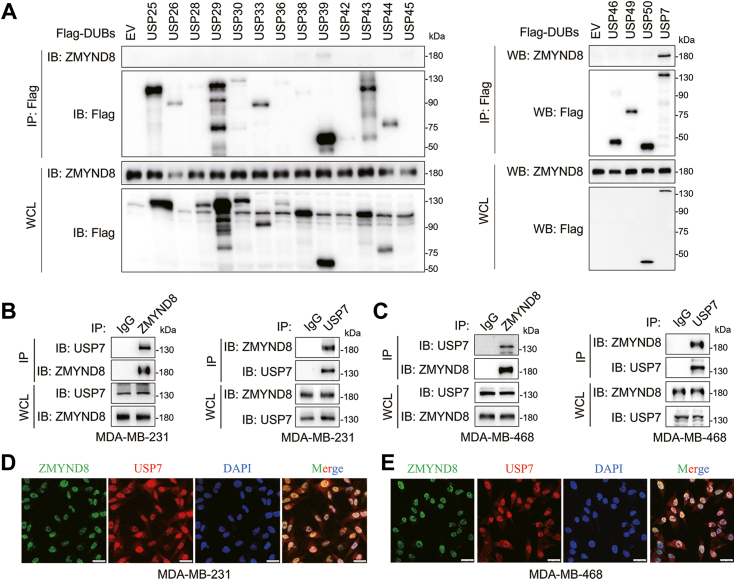
To delineate the mechanism underlying the upregulation of ZMYND8 protein levels in breast cancer, we sought to identify potential ZMYND8-interacting DUBs by scrutinizing the Biological General Repository for Interaction Datasets (BioGRID), a publicly accessible database of physical and genetic interactions available at http://www.thebiogrid.org (29). Our analysis revealed USP7 as the sole DUB among the 193 potential ZMYND8-interacting proteins (Table S1). To further explore DUBs potentially regulating ZMYND8, we transfected HEK293T cells with 17 Flag-tagged DUBs, as described previously (30). Coimmunoprecipitation (co-IP) assays were conducted using anti-Flag antibody-conjugated magnetic beads. USP7 displayed the most robust association with endogenous ZMYND8 among these DUBs, while the interaction with USP39 was weak (Fig. 1A). Given that ZMYND8 is a primary driver of metastasis in breast cancer (1), we investigated the ZMYND8-USP7 interaction in metastatic breast cancer cells, including MDA-MB-231 and MDA-MB-468. Reciprocal interactions between endogenous ZMYND8 and USP7 were observed in both MDA-MB-231 cells and MDA-MB-468 cells (Fig. 1, B and C). To further confirm their interaction, immunofluorescence assays were performed to evaluate the subcellular colocalization of endogenous ZMYND8 and USP7, both of which predominantly localize in the nucleus (3, 31). As expected, ZMYND8 and USP7 displayed an excellent colocalization in the nuclei of MDA-MB-231 cells and MDA-MB-468 cells (Fig. 1, D and E). Together, ZMYND8 interacts with USP7 in breast cancer cells.
Figure 1.
ZMYND8 associates with USP7.A, HEK293T cells transfected with constructs encoding Flag tagged deubiquitinases (DUBs) were subjected to co-immunoprecipitation (co-IP) assays using the anti-Flag magnetic beads followed by western blotting using anti-ZMYND8 antibody. B and C, Co-IP assay of endogenous ZMYND8 with USP7 and vice versa in MDA-MB-231 cells or MDA-MB-468 cells. WCL, whole cell lysate. D and E, MDA-MB-231 cells (D) or MDA-MB-468 cells (E) were co-stained with anti-ZMYND8 and anti-USP7 antibodies using immunofluorescence staining to determine the colocalization of ZMYND8 and USP7. Nuclei were counterstained with DAPI. Scale bar: 25 μm.
ZMYND8 directly binds to the TRAF and UBL domains of USP7 via its PBP domain
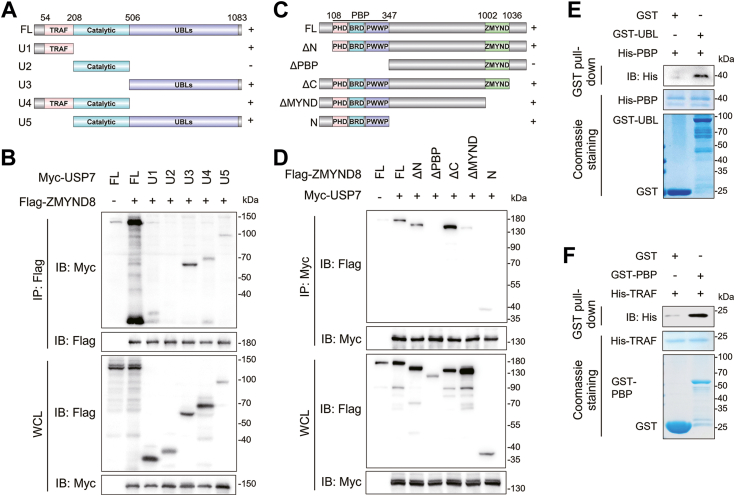
Since USP7 physically associates with ZMYND8, we next aimed to identify the specific domains mediating their interaction. USP7 comprises a catalytic domain flanked by an N-terminal TRAF domain and a UBL domain at the C-terminus, both the TRAF domain and UBL domain are critical for substrate binding (24, 32, 33). Therefore, we constructed truncated mutants expressing Myc-tagged TRAF domain, catalytic domain, and UBL domain, respectively (Fig. 2A). Full-length Myc-USP7 and these mutants were transiently transfected into HEK293T cells followed by co-IP assays. The results of co-IP assays demonstrated that both the TRAF domain and UBL domain of USP7 associated with ZMYND8, whereas the catalytic domain of USP7 failed to interact with ZMYND8 (Fig. 2B). Thus, the TRAF domain and UBL domain of USP7 both mediate the interaction of USP7 and ZMYND8. We also generated truncated Flag-ZMYND8 lacking the PBP domain or MYND domain, which are pivotal for protein-protein interaction (Fig. 2C) (1, 9). Full-length and truncated Flag-ZMYND8 were transiently transfected into HEK293T cells for co-IP assays and we found that only truncations lacking the MYND domain had no interaction with USP7, suggesting that the MYND domain is essential for USP7-ZMYND8 association (Fig. 2D). To investigate whether USP7 directly interacts with ZMYND8, we performed GST pull-down assays using purified GST- and His-fusion proteins. The results indicated that GST-UBL, but not GST alone, interacted with His-PBP (Fig. 2E). Moreover, His-TRAF was pulled down by GST-PBP, but not GST (Fig. 2F). Taken together, ZMYND8 directly binds to the TRAF and UBL domains of USP7 through its PBP domain.
Figure 2.
ZMYND8 directly binds to the TRAF and UBL domains of USP7 via its PBP domain.A, schematic depiction of full length (FL) and truncated USP7. Plus (+) and minus (−) indicates binding and no binding with ZMYND8, respectively. B, Co-IP assay of Flag-ZMYND8 and FL or truncated Myc-USP7 in HEK293T cells. C, schematic depiction of FL ZMYND8 and its truncates. Plus (+) and minus (−) indicates binding and no binding with USP7, respectively. D, Co-IP assay of Myc-USP7 and FL or truncated Flag-ZMYND8 in HEK293T cells. E and F, the direct interaction of ZMYND8 and USP7 was examined by in vitro GST pull-down assay using purified GST-UBL and His-PBP (E) or GST-PBP and His-TRAF (F).
USP7 stabilizes ZMYND8 in breast cancer cells
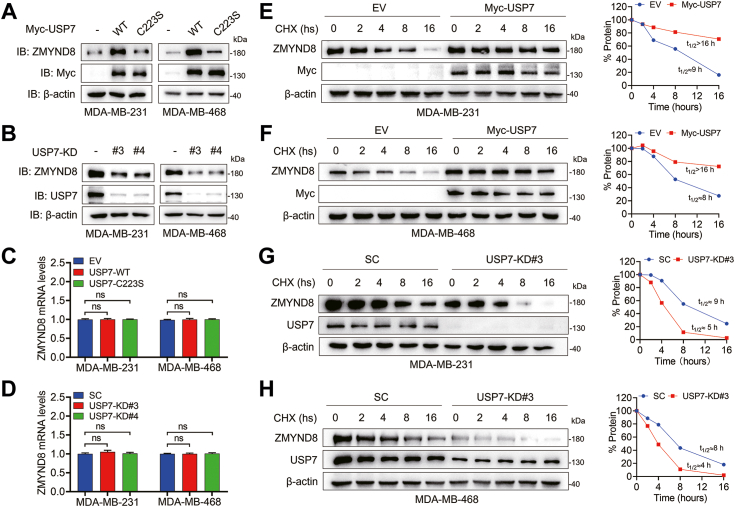
USP7 is a well-known DUB that has been reported to stabilize several substrates, including MDM2 (Mouse double minute 2) (34), ABCB1 (28), LSD1 (27), and MLL5 (Mixed Lineage Leukemia 5) (35). ZMYND8 functions as an oncoprotein in breast cancer cells (1, 8, 9, 15). Since USP7 directly associates with ZMYND8, we attempted to examine the effect of USP7 on ZMYND8 stability in breast cancer cells. To this end, wild-type (WT) and catalytically inactive (C223S) Myc-USP7 were ectopically expressed in MDA-MB-231 and MDA-MB-468 breast cancer cells (36). WT, but not C223S, Myc-USP7 significantly increased the protein levels of endogenous ZMYND8 in these breast cancer cells, indicating that USP7 controls ZMYND8 protein levels in an enzymatically active manner (Fig. 3A). Next, USP7 was depleted via two independent shRNAs (shUSP7#3 and shUSP7#4) in MDA-MB-231 and MDA-MB-468 cells. USP7 ablation in these cells remarkably downregulated endogenous ZMYND8 protein levels (Fig. 3B). Notably, USP7 participates in gene expression by modulating DNA or histone modification (37, 38, 39). Moreover, USP7 stabilizes HIF-1α to upregulates the expression of glycolysis genes, and HIF-1α promotes the transcription of ZMYND8 in triple negative breast cancer (TNBC) cells (1, 25). Therefore, we performed RT-qPCR in the aforementioned MDA-MB-231 and MDA-MB-468 cells to determine whether ZMYND8 mRNA levels were regulated by USP7. Our findings indicated that USP7 has no effect on ZMYND8 mRNA levels (Fig. 3, C and D), suggesting USP7 did not affect ZMYND8 expression at the transcription level. To further examine the impact of USP7 on ZMYND8 stability, we conducted cycloheximide (CHX) chasing assays in both MDA-MB-231 and MDA-MB-468 cells ectopically expressing Myc-USP7 to evaluate the protein half-life of endogenous ZMYND8. In contrast to the control, cells stably expressing Myc-USP7 exhibited a prolonged half-life of ZMYND8 (Fig. 3, E and F). Conversely, USP7 knockdown in MDA-MB-231 and MDA-MB-468 cells markedly decreased the half-life of ZMYND8 (Fig. 3, G and H). In summary, USP7 regulates ZMYND8 protein stability in breast cancer cells.
Figure 3.
USP7 modulates ZMYND8 stability.A, protein levels of endogenous ZMYND8 were analyzed in MDA-MB-231 cells and MDA-MB-468 cells stably expressing wild type (WT) or catalytically inactive (C223S) Myc-USP7. B, protein levels of endogenous ZMYND8 in control or USP7-knockdown (USP7-KD) MDA-MB-231 cells and MDA-MB-468 cells. C, mRNA levels of ZMYND8 were analyzed by RT-qPCR in MDA-MB-231 cells and MDA-MB-468 cells stably WT or C223S Myc-USP7. ns, no significance, by 1-way ANOVA Tukey’s multiple comparisons test. D, mRNA levels of ZMYND8 were analyzed by RT-qPCR in MDA-MB-231 cells and MDA-MB-468 cells with USP7 ablation. ns, no significance, by 1-way ANOVA Tukey’s multiple comparisons test. E and F, the half-life of ZMYND8 protein in MDA-MB-231 (E) or MDA-MB-468 (F) cells expressing Myc-USP7. Cells were treated with 10 μg/ml CHX for the indicated time. G and H, the half-life of ZMYND protein were analyzed in scrambled shRNA (SC) or USP7-KD#3 MDA-MB-231 (G) or MDA-MB-468 (H) cells. Cells were treated with 10 μg/ml CHX for the indicated time.
USP7 deubiquitinates ZMYND8 in vivo and in vitro
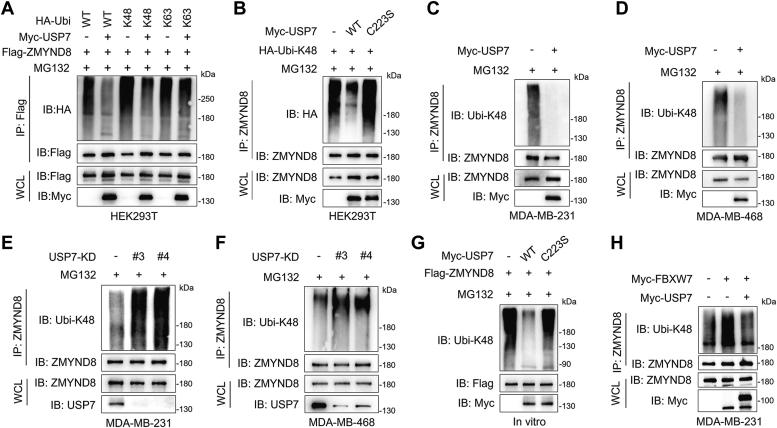
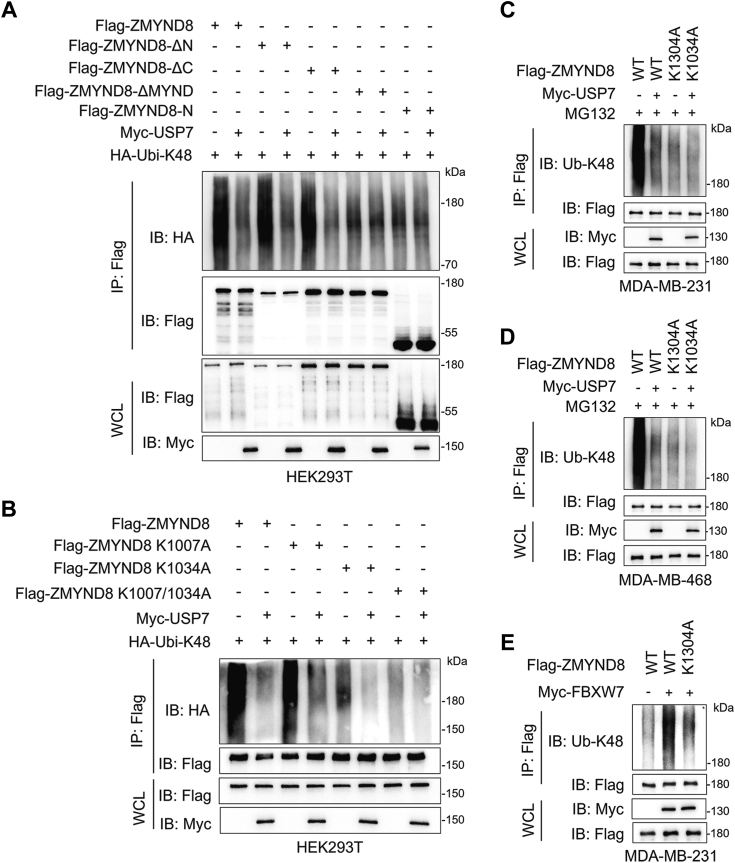
Since USP7 is a well-known DUB and governs ZMYND8 stability (Fig. 2, K and L), we performed in vivo ubiquitination assays to determine whether USP7 governs ZMYND8 turnover through ubiquitination. Given that USP7 cleaves both K48- and K63-linked ubiquitination chains from substrates (40), we first examined which kind of ubiquitination chains were removed from ZMYND8 by USP7. HEK293T cells were transfected with vectors encoding Flag-ZMYND8, WT or single-lysine only HA-ubiquitin (Ubi) together with or without Myc-USP7 followed by treatment with MG231. Expectedly, USP7 significantly decreased the degrative K48 linkage of ZMYND8, but had no effect on non-degrative K63 linkage (Fig. 4A). To demonstrate the enzymatic activity of USP7 is indispensable for ZMYND8 deubiquitination, HA-Ubi-K48 together WT or C223S Myc-USP7 were ectopically expressed in HEK293T cells treated with MG132 and the ubiquitination levels of endogenous ZMYND8 were detected. Consistent with the effect of USP7 on ZMYND8 stability (Fig. 3A), WT but not C223S Myc-USP7 dramatically decreased the K48-linked ubiquitination of endogenous ZMYND8 (Fig. 4B). Investigation in MDA-MB-231 and MDA-MB-468 cells expressing WT or C223S Myc-USP7 revealed a substantial decrease in K48-linked polyubiquitination of ZMYND8 by WT but not C223S Myc-USP7 (Fig. 4, C and D). Furthermore, both USP7-KD#3 and USP7-KD#4 dramatically elevated K48-linked polyubiquitination of ZMYND8 in MDA-MB-231 and MDA-MB-468 cells (Fig. 5, E and F). Notably, the protein levels of ZMYND8 were comparable in these MG132-treated MDA-MB-231 and MDA-MB-468 cells, suggesting that USP7 hinders proteasomal degradation of ZMYND8 (Fig. 4, C–F). To further strengthen our conclusion about the regulation of USP7 on ZMYND8 ubiquitination, we also performed an in vitro ubiquitination assay. Ubiquitinated Flag-ZMYND8 was immunoprecipitated and incubated with purified WT and C223S Myc-USP7, respectively. The results indicated that Flag-ZMYND8 was only deubiquitinated by purified WT but not C223S Myc-USP7 (Fig. 4G). Together, USP7 deubiquitinates ZMYND8 in breast cancer cells. Since the E3 ligase FBXW7 ubiquitinates ZMYND8 in bladder cancer cells (17), we next explored whether ZMYND8 ubiquitination is coordinately controlled by USP7 and FBXW7 in breast cancer cells. Our results indicated that FBXW7 overexpression dramatically increased ZMYND8 ubiquitination, which was almost completely reversed by ectopically expressed USP7 in MDA-MB-231 cells (Fig. 4H), suggesting that USP7 antagonizes FBXW7-mediated ZMYND8 ubiquitination in breast cancer cells.
Figure 4.
USP7 deubiquitinates ZMYND8.A, immunoblots of WCL and co-IP from HEK293T cells transfected with Flag-ZMYND8 vector, WT or single-lysine only HA-ubiquitin (Ubi) vector, or Myc-USP7 vector and treated with 10 μM MG132 for 6 h before harvest. B, ubiquitination of endogenous ZMYND8 in HEK293T ectopically expressing WT or C223S Myc-USP7. C and D, ubiquitination of endogenous ZMYND8 in MDA-MB-231 cells (C) and MDA-MB-231 cells (D) stably expressing Myc-USP7. E and F, ubiquitination of endogenous ZMYND8 in SC and USP7-KD MDA-MB-231 cells (E) and MDA-MB-231 cells (F). G, ubiquitinated Flag-ZMYND8 was precipitated from HEK293T cells transfected with constructs encoding Flag-ZMYND8 and HA-Ubi-K48 and incubated with purified WT or C223S Myc-USP7 in deubiquitylation buffer for examination of ZMYND8 ubiquitination. H, ubiquitination of endogenous ZMYND8 in MDA-MB-231 cells stably expressing Myc-FBXW7 with or without Myc-USP7.
Figure 5.
K1034 is the key deubiquitination site for ZMYND8 by USP7.A, ubiquitination of full length and truncated ZMYND8 in HEK293T cells treated with 10 μM MG132 for 6 h before harvest. B, ubiquitination of WT and mutant ZMYND8 in HEK293T cells treated with 10 μM MG132 for 6 h before harvest. C and D, ubiquitination of ectopic WT and K1034A mutant Flag-ZMYND8 in MDA-MB-231 (C) and MDA-MB-468 (D) cells treated with 10 μM MG132 for 6 h before harvest. E, ubiquitination of endogenous ZMYND8 in MDA-MB-231 cells stably expressing WT or K1034A mutant Flag-ZMYND8 together with Myc-FBXW7.
K1034 is the key (de)ubiquitination site for ZMYND8 in breast cancer cells
To identify the putative deubiquitination sites of ZMYND8 by USP7, we first detected the ubiquitination levels of full-length and truncated ZMYND8 proteins that associate with USP7 in HEK293T cells with or without the ectopic expression of USP7. These ZMYND8 proteins containing the MYND domain were robustly ubiquitinated, and USP7 dramatically decreased their ubiquitination (Fig. 5A). In contrast, the ubiquitination levels of ZMYND8 proteins lacking MYND domain were very weak and not influenced by USP7 (Fig. 5A). All these findings indicated that the key ubiquitination sites of ZMYND8, including those affected by USP7, exist in the MYND domain. Previous research showed that there were two evolutionary conserved lysine (K) residues in the MYND domain, namely K1007 and K1034 (1, 7). Therefore, we constructed single and double lysine-to-alanine (A) mutant ZMYND8 for ubiquitination assays. Compared with K1007A, K1034A remarkably downregulated ZMYND8 ubiquitination to a very low levels, and USP7 completely abolished the ubiquitination of ZMYND8-K1034A in HEK293T cells (Fig. 5B). However, the K1007/1034A double mutation totally abrogated ZMYND8 ubiquitination (Fig. 5B). Collectively, K1007 and K1034 are ubiquitination sites in ZMYND8, and K1034 plays a predominant role in ZMYND8 ubiquitination. Moreover, we demonstrated that K1034 is the key deubiquitination site for ZMYND8 by USP7 in breast cancer cells (Fig. 5, C and D). As mentioned above, FBXW7 is an E3 ligase for ZMYND8 (17). We further investigated whether FBXW7 accounts for ZMYND8 ubiquitination at K1034. In line with our prediction, K1034A significantly shielded ZMYND8 from FBXW7-mediated ubiquitination (Fig. 5E), suggesting K1034 as a key ubiquitination site for ZMYND8 by FBXW7. Taken together, USP7 antagonizes FBXW7-mediated ZMYND8 ubiquitination.
USP7 enhances breast cancer cell migration and invasion through ZMYND8
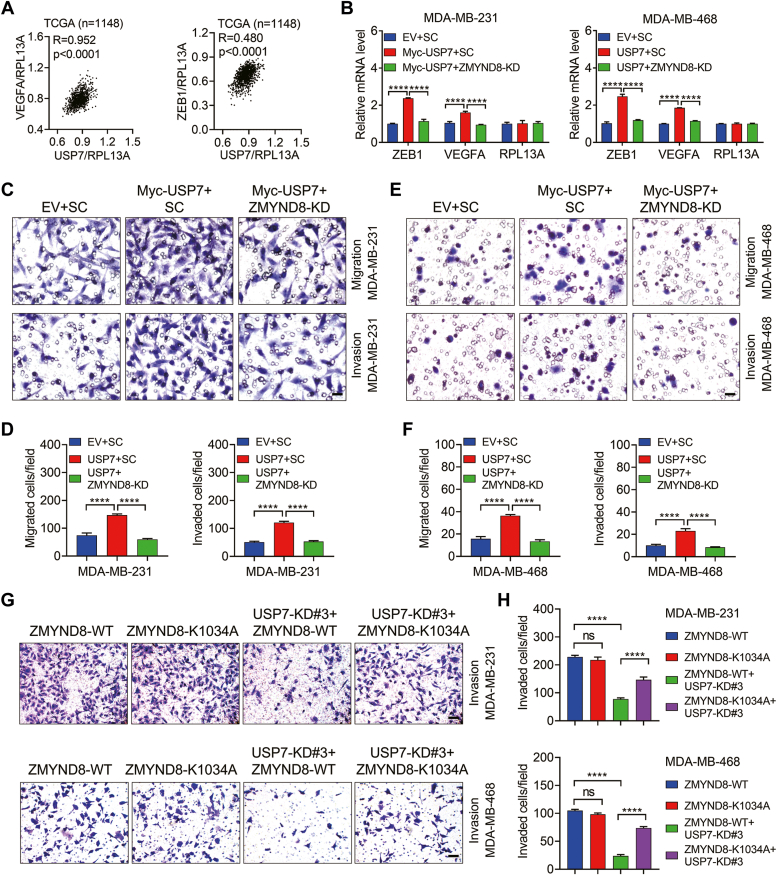
Metastasis is a leading cause of breast cancer mortality and recurrence. ZMYND8 promotes breast cancer metastasis by stimulating the expression of several pro-metastatic genes, such as VEGFA, ZEB1, ANGPTL4 (angiopoietin-like 4), and AGR2 (Anterior Gradient 2) (1, 8). Motivated by the finding that USP7 deubiquitinates and stabilizes ZMYND8, we sought to examine whether USP7 promotes ZMYND8-mediated breast cancer cell migration and invasion. To this end, we first determined the relationship between USP7 and these metastatic genes by gene correlation analysis across The Cancer Genome Atlas (TCGA) cohort of breast cancer tissues. Intriguingly, USP7 expression was positively associated with ZEB1 and VEGFA (Fig. 6A), but not ANGPTL4 or AGR2 (data not shown). To verify that USP7 modulates the transcription of ZEB1 and VEGFA via ZMYND8 in breast cancer cells, ZMYND8 was depleted in MDA-MB-231 or MDA-MB-468 cells stably expressing Myc-USP7. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) assays indicated that USP7 indeed upregulated the transcription of ZEB1 and VEGFA and this induction was eliminated by ZMYND8 ablation in both MDA-MB-231 and MDA-MB-468 cells (Fig. 6B). In contrast, the mRNA levels of the negative control RPL13A were not influenced by ZMYND8 or USP7 (Fig. 6B). All these findings indicate that USP7 may augments the capacity of breast cancer cells in migration and invasion via ZMYND8. To confirm this prediction, Boyden chamber assays were performed in the aforementioned breast cancer cells. As expected, Myc-USP7 overexpression increased the ability of MDA-MB-231 and MDA-MB-468 cells in migration and invasion and ZMYND8 depletion effectively ameliorated the inductive effect of Myc-USP7 (Fig. 6, C–F). Together, USP7 enhances breast cancer cell migration and invasion through ZMYND8. To address the significance of USP7-mediated ZMYND8 deubiquitination on breast cancer cell invasion, MDA-MB-231 or MDA-MB-468 stable cell lines expressing comparable levels of ZMYND8-WT and ZMYND8-K1034A were generated, followed by transfection with vector encoding shRNA designed to knockdown USP7 (hereafter referred to as ZMYND8-WT + USP7-KD and ZMYND8-K1034A + USP7-KD, respectively). As the Boyden chamber assays showed, USP7 depletion remarkably attenuated ZMYND8-WT-mediated invasion of MDA-MB-231 and MBA-MB-468 cells but barely affected the invasion of MDA-MB-231 and MBA-MB-468 cells expressing ZMYND8-K1034A (Fig. 6, G and H). Collectively, USP7 promotes breast cancer cell migration and invasion through deubiquitinating ZMYND8.
Figure 6.
USP7 promotes breast cancer cell migration and invasion via ZMYND8.A, Pearson correlation analysis of USP7 mRNA with ZEB1 and VEGFA mRNAs in human breast cancer tissues from the TCGA dataset. B, the mRNA levels of ZEB1 and VEGFA in control, Myc-USP7, and USP7+ZMYND8-KD MDA-MB-231 cells and MDA-MB-468 cells. ∗∗∗∗p < 0.0001, by 1-way ANOVA Tukey’s multiple comparisons test. RPL13A was negative control. C–F, the ability of control, Myc-USP7, and USP7+ZMYND8-KD MDA-MB-231 cells (C and D) and MDA-MB-468 (E and F) cells in migration and invasion, respectively. G and H, Invasion of MDA-MB-231 and MDA-MB-468 cells expressing wild-type ZMYND8 (ZMYND8-WT) or ubiquitination site-mutated ZMYND8 (ZMYND8-K1034A) together with USP7 depletion (USP7-KD#3). ∗∗∗∗p < 0.0001, by 1-way ANOVA Tukey’s multiple comparisons test. Scale bar: 20 μm.
USP7 expression is positively correlated with ZMYND8 expression in human breast tumors
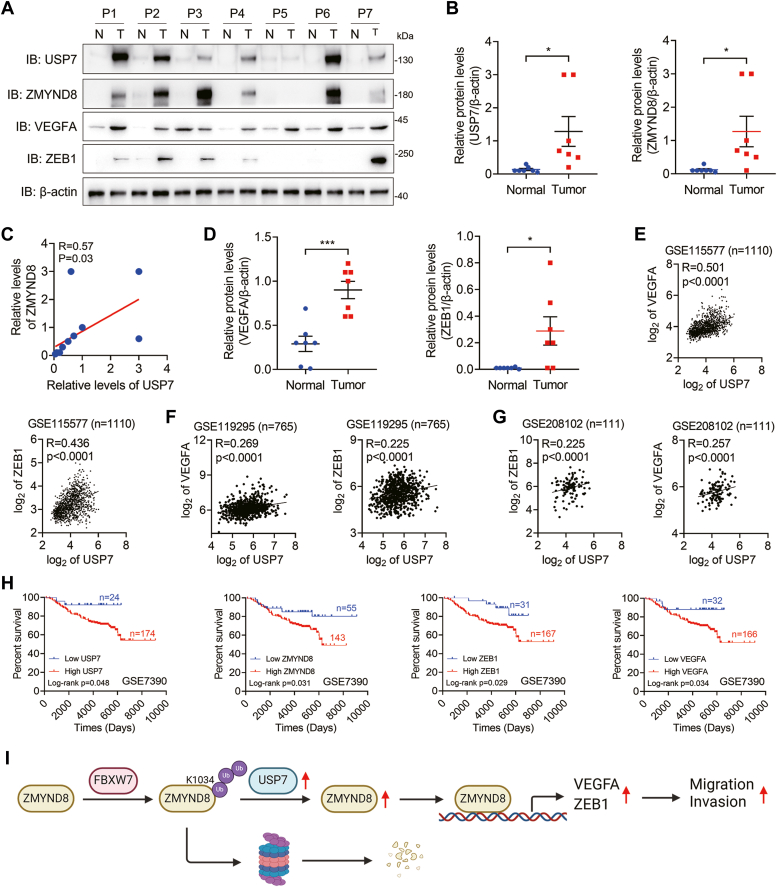
We have illustrated that USP7 upregulates ZMYND8 protein levels in breast cancer cells. Furthermore, to explore the potential relevance of USP7 and ZMYND8 in human breast cancer, we extracted cell lysates from breast tumors and adjacent normal breast tissues for Western blotting. It was shown that the protein levels of USP7 were significantly increased in breast tumors coincided with upregulation of ZMYND8 protein levels (Fig. 7, A and B), suggesting a positive correlation between USP7 and ZMYND8 in breast cancer. Indeed, the correlation analysis confirmed that USP7 expression was positively associated with ZMYND8 expression in breast tumors (Fig. 7C). Additionally, the protein levels of VEGFA and ZEB1 were also increased in breast tumors (Fig. 7, A and D), and their mRNA levels positively correlated with those of USP7 in breast cancer tissues (Fig. 7, E–G). These findings, together with those in Figure 6, A and B, supported the conclusion that USP7 modulates the expression of VEGFA and ZEB1 via ZMYND8 in breast tumors. Importantly, high levels of USP7, ZMYND8, VEGFA, or ZEB1 decreased the overall survival rates of patients with breast cancer (Fig. 7H). Taken together, USP7 expression is positively correlated with ZMYND8 expression in human breast tumors.
Figure 7.
.USP7 expression is positively correlated with ZMYND8 expression in human breast tumors.A–D, protein levels of USP7, ZMYND8, VEGFA, and ZEB1 in human breast tumors (n = 7) and adjacent normal breast tissues (n = 7) (A, B and D) and the correlation of USP7 expression and ZMYND8 expression in human breast tumors and normal breast tissues (B). ∗p < 0.05, by two-tailed t test. P, patient. B, the correlation of USP7 expression and ZMYND8 expression in human breast tumors and normal breast tissues. E–G, Pearson correlation analysis of USP7 mRNA with ZEB1 and VEGFA mRNAs in human breast cancer tissues. The data were retrieved from the GSE115577 (E), GSE119295 (F), and GSE208102 (G) databases. H, Kaplan-Meier survival analysis for patients with breast cancer using datasets downloaded from the PrognoScan database. I, a proposed model of USP7-mediated ZMYND8 stability in breast cancer. USP7 directly interacts with ZMYND8 and antagonizes FBXW7-mediated ZMYND8 ubiquitination and degradation, thereby promoting breast cancer cell migration and invasion by stimulating the transcription of ZMYND8 target genes, such as VEGFA and ZEB1.
In summary, FBXW7 promotes ZMYND8 polyubiquitination at K1034, leading to ZMYND8 proteasomal degradation in breast cancer cells. However, USP7 is highly expressed in breast tumors and antagonizes FBXW7-mediated ZMYND8 turnover, thereby stimulating the expression of ZMYND8 target genes, including VEGFA and ZEB1. Consequently, USP7 enhances breast cancer cell migration and invasion (Fig. 7I).
Discussion
In the present study, we identified USP7 as a bona fide DUB for ZMYND8 in breast tumors. Mechanically, USP7 directly interacts with the PBP domain of ZMYND8 via its TRAF and UBL domains, promoting the deubiquitylation of ZMYND8 at K1034 and subsequent stability. Consequently, the transcription of ZMYND8 target genes VEGFA and ZEB1 is activated to enhance breast cancer cell migration and invasion. Furthermore, the protein levels of USP7 and ZMYND8 are positively correlated in breast tumors. Therefore, the USP7-ZMYND8 axis emerges as a potential therapeutic target in breast cancer.
ZMYND8 has been well characterized as an oncoprotein, and its protein levels are pretty high in multiple cancer types, including breast cancer (1), intestinal cancer (10), glioblastoma (16), clear cell renal cell carcinoma (11) and bladder cancer (17). Importantly, high levels of ZMYND8 lead to poor clinical outcomes in these cancers. Hence, ZMYND8 is an attractive diagnostic biomarker and therapeutic target. However, the mechanism by which ZMYND8 expression is increased in cancers remains elusive. Although some studies have demonstrated that ZMYND8 expression is fine-tuned at the transcription level by transcriptional factors HIF (1), NRF2 (9), and YAP (10), and at the posttranslational modification (PTM) levels by ubiquitination and E3 ligase FBXW7 (17, 18), the effect of DUBs on ZMYND8 has not been reported. In this study, we found that USP7 interacts with ZMYND8 and promotes ZMYND8 stability. There are about 100 DUBs in humans (19, 21). In this study, we only verified the interaction between ZMYND8 and 17 DUBs and found a robust interaction between USP7 and ZMYND8. Whether other DUBs regulate ZMYND8 expression needs to be investigated in the future. What needs to be emphasized is how E3 ligases and DUBs coordinately modulate ZMYND8, given that ZMYND8 turnover is controlled by both E3 ligase FBXW7 and USP7. Here, we showed that USP7 specifically deubiquitinates ZMYND8 at K1034, where polyubiquitin chains are conjugated by FBXW7, suggesting that USP7 antagonizes FBXW7-mediated ZMYND8 ubiquitination and proteasomal degradation. How the precise ubiquitination status of ZMYND8 is fine-tuned by FBXW7 and USP7 is an important question to be answered in future investigations. FBXW7/USP7 ubiquitinates/deubiquitinates ZMYND8 at K1007 and K1034, which are also acetylated by p300 (1), the mechanism by which ZMYND8 ubiquitination and acetylation are balanced by FBXW7, USP7, and p300 needs to be deciphered in the future. Notably, ZMYND8 is phosphorylated by the mammalian MST1/2 (STE20-like protein kinases 1 and 2) to impede ZMYND8-dependent DNA double-stand break repair (41), indicating that other PTMs may be involved in ZMYND8 expression regulation. Further investigations are needed to comprehensively analyze how PTMs regulate ZMYND8 expression.
USP7 acts as an oncogenic driver in most cancer types and binds to approximately 200 substrate proteins (24). However, the predominant targets of USP7 differ in various cancers (22). In breast cancer cells, the known USP7 substrates are LSD1 (27), ABCB1 (28), EZH2 (26), and p53 (42). Identification of novel USP7 substrates will strengthen the understanding of USP7’s fundamental function and regulatory mechanisms. We demonstrated that ZMYND8 is a novel USP7 substrate in breast cancer cells. USP7 usually binds to the (P/A/E)-X-X-S or K-X-X-X-K motif of substrates via its TRAF or UBL domains, but only 25% of the substrate binding domains or motifs have been identified (24, 43). Our results revealed that USP7 directly binds to the PBP domain of ZMYND8, which possesses five (P/A/E)-X-X-S and K-X-X-X-K motifs (data not shown). Thus, the motif in the PBP domain of ZMYND8 that mediates the USP7-ZMYND8 interaction should be identified in the future research. Although USP7 has been demonstrated to remove K48- but not K63-linked polyubiquitin chains from ZMYND8, whether USP7 affects other linkage types on ZMYND8 should be determined, because USP7 also efficiently cleaves K6-, K11-, and K33-linked chains from substrates (40). In addition to the epigenetic reader ZMYND8, USP7 modulates the stability of several other epigenetic regulators, such as the epigenetic writers PHF8 (36), UHRF1 (37), and SUV39H1 (44); and the epigenetic erasers LSD1 (27) and KDM5B (45). How USP7 spatially and temporally regulates these epigenetic regulators remains elusive. It is worth noting that ZMYND8 recognizes and binds to acetylated H3K14 (H3K14ac) and H4K16ac (1), whereas the aforementioned writers and erasers mainly modulate histone methylation or ubiquitination, suggesting that USP7 may control various epigenetic events by regulating different epigenetic regulators. Accumulating evidence indicates that USP7 is a promising therapeutic target in cancer and more than 25 USP7 inhibitors have been identified (22). However, most USP7 inhibitors lack selectivity due to their cross-reaction with other UDBs (22). In addition, these inhibitors have poor potency or solubility (22). Therefore, identifying highly efficient and selective USP7 inhibitors is an urgent need for cancer treatment. Targeting protein-protein interaction is an ideal strategy for developing inhibitors. ZMYND8 and USP7 are attractive therapeutic targets, and since USP7 upregulates ZMYND8 stability, screening for small molecules that disrupt the ZMYND8-USP7 interaction provides a promising avenue for future research.
Experimental procedures
Cell culture
MDA-MB-231, MDA-MB-468, and HEK293T cells were obtained from the American Type Culture Collection (ATCC) and cultured at 37°C with 5% CO2 in DMEM (CM15019, Macgene) medium supplemented with 10% FBS (FSP500, ExCell), 1% penicillin, and 1% streptomycin. All cells are mycoplasma-free and have been authenticated by STR DNA profiling analysis.
Plasmids
cDNA encoding full length and truncated human USP7, ZMYND8 or FBXW7 gene was amplified by PCR and inserted into lentiviral vector pLVX-IRES-Flag or pLVX-IRES-Myc. For protein purification, pGEX-6P-1 or pET28a (+) was used as a carrier vector for truncated human USP7 or ZMYND8 genes. USP7 catalytically dead mutant (C223S) was constructed by site-directed mutagenesis PCR. shRNA sequences (shRNA#3 primer: 5′-CCAGCTAAGTATCAAAGGAAA-3’; shRNA#4 primer: 5′-CCTGGATTTGTGGTTACGTTA-3′) targeting human USP7 were cloned into the pLKO.1 lentiviral vector. All plasmids were confirmed by DNA sequencing. Other plasmids were described previously (30, 46).
Antibodies and reagents
MG132 (ab141003), anti-ubiquitin antibody (linkage-specific K63, ab179424), and anti-ubiquitin antibody (linkage-specific K48, ab140601) were obtained from Abcam. Normal mouse IgG antibody (sc-2025) was obtained from Santa Cruz. Anti-Flag-M2 antibody (F1804) was obtained from Sigma. Anti-USP7 antibody (66514-1-IG), anti-HA antibody (51064-2-AP), anti-β-actin antibody (66009-1-Ig), anti-Myc antibody (16286-1-AP), anti-VEGFA antobody (19003-1-AP), anti-ZEB1 antibody (21544-1-AP), goat anti-mouse secondary antibody (SA00001-1), and HRP-conjugated affinipure goat anti-rabbit secondary antibody (SA00001-2) were obtained from Proteintech. Anti-ZMYND8 (A302-089A) antibody was obtained from Bethyl. Cycloheximide (CHX, 2112S), anti-ubiquitin antibody (3936S), and normal rabbit IgG antibody (2729) were obtained from Cell Signaling Technology. Alexa Fluor 488-conjugated donkey anti-rabbit secondary antibody (A21206) and Alexa Fluor 594-conjugated donkey anti-mouse secondary antibody (A21203) were obtained from Thermo Fisher.
Lentivirus production and establishment of stable cell lines
Lentivirus were generated according to a previous study (30, 46, 47). Briefly, HEK293T cells were co-transfected with the transducing vector scrambled control shRNA (shSC), shUSP7#3, or shUSP7#4 and packaging vectors pMD2.G and psPAX2 using Polyjet (SignaGen) according to the manufacture’s protocol. After 48 h post-transfection, virus particles were harvested and filtered using a 0.45 μm syringe filter. Breast cancer cells were co-cultured with lentivirus for 48 h followed by selection using puromycin (1.0 μg/ml) for another 72 h.
Immunofluorescence staining
Immunofluorescence staining was conducted according to previous research (48, 49, 50). In brief, MDA-MB-231 or MDA-MB-468 cells were fixed with 4% paraformaldehyde at room temperature for 20 min followed by permeabilization with 0.2% Triton X-100 solution for 20 min. Cells were washed with PBS three times and blocked with 3% BSA at room temperature for 30 min. Subsequently, cells were co-stained overnight with anti-ZMYND8 antibody (1:2000) and anti-USP7 antibody (1:1000) at 4°C followed by incubation with Alexa Fluor 488-conjugated donkey anti-rabbit secondary antibody and Alexa Fluor 594-conjugated donkey anti-mouse secondary antibody for 1 h at room temperature. Immunofluorescence images were captured with Leica TCS SPE confocal microscope (Leica).
Boyden chamber assay
5 × 104 breast cancer cells were resuspended in serum-free medium and seeded into the upper chamber for migration or in a Matrigel-coated upper chamber for invasion. The bottom chamber was filled with medium supplemented with 10% FBS. After 16 h (migration) or 24 h (invasion), cells that migrated or invaded to the lower surface of the transwell insert were fixed with methanol, stained with 0.5% crystal violet, and counted using Image J.
Immunoprecipitation and western blotting
Cells were sonicated in NETN lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, pH7.4, and protease inhibitor cocktail) followed by centrifugation. The supernatant was collected for immunoprecipitation overnight with the indicated primary antibodies and protein G/A magnetic beads (Bio-Rad) at 4 °C. Subsequently, the beads were gently washed with NETN lysis buffer three times, and immunoprecipitates were resuspended in one x Laemmli buffer and boiled for 6 minutes. The protein-protein interactions were then detected by SDS-PAGE.
Protein purification and pull-down assay
GST- and His-fusion proteins were purified according to the previous description (51, 52). Briefly, GST-UBL, GST-PBP, six x His-PBP, or six x His-TRAF were expressed in BL21 competent E. Coli cells and purified with glutathione-Sepharose 4B (GE Healthcare) or Ni-NTA resin (GE Healthcare), respectively, following standard protocols. Purified GST or GST-fusion proteins were incubated with purified His-fusion proteins and glutathione-Sepharose 4B beads. Subsequently, beads were washed extensively five times before being boiled, and western blotting was performed to analyze the proteins bound to the beads.
In vivo deubiquitylation assay
In vivo deubiquitylation assays were performed as described previously (30, 47). Briefly, cells with Myc-USP7 overexpression or USP7 depletion were treated with 10 μM MG132 for 6 h before harvest. Endogenous ZMYND8 was precipitated by anti-ZMYND8 antibody and then subjected to an immunoblotting assay to determine its ubiquitination levels using anti-ubiquitin (linkage-specific K48, ab140601) antibody.
In vitro deubiquitylation assay
In vitro deubiquitylation assays were carried out according to previous research (53). Briefly, HEK293T cells were co-transfected with constructs encoding Flag-ZMYND8 and HA-Ubi-K48 and lysed in NETN lysis buffer for immunoprecipitation using anti-Flag magnetic beads. Meanwhile, wide type (WT) or catalytically mutant (C223S) Myc-USP7 expressed in HEK293T cells was immunoprecipitated with anti-Myc magnetic beads and subsequently eluted by Myc peptide. Ubiquitinated Flag-ZMYND8 was incubated with WT or C223S Myc-USP7 in deubiquitylation buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA, pH 8.0, 10 mM dithiothreitol, and 5% glycerol) at 37 °C for 2 h. Then, anti-ubiquitin (linkage-specific K48, ab140601) antibody was used to evaluate the ubiquitination level of Flag-ZMYND8.
RT-qPCR
Total RNA was isolated from cultured cells using Trizol reagent (Invitrogen) and treated with DNase I (Ambion). RT-qPCR assays were performed according to the standard protocol (46, 54, 55). The following primer pair was used for RT-qPCR: ZMYND8 (Forward: 5′-GGGTTTATCACGCTAAGTGTCTG-3′, Reverse: 5′- GGCTTTACTCTGGGTCTCGATG-3′), VEGFA (Forward: 5′-CTTGCCTTGCTGCTCTAC-3′, Reverse: 5′-TGGCTTGAAGATGTACTCG-3′), ZEB1 (Forward: 5′-CAGTCCCACACGACCACA-3′, Reverse: 5′-GTAATGGGCCACCACCAG-3′), PRL13A (Forward: 5′- CTCAAGGTCGTGCGTCTG-3′, Reverse: 5′-TGGCTTTCTCTTTCCTCTTCTC-3′), and 18s rRNA (Forward: 5′-CGGCGACGACCCATTCGAAC-3′, Reverse: 5′- GAATCGAACCCTGATTCCCCGTC-3′).
Statistical analysis
At least three samples prepared from independent experiments were used to ensure adequate power for all studies. Statistical analysis was carried out by one-way ANOVA with multiple testing correlation within multiple groups or two-tailed t test within two groups. Data were presented as mean ± SD p < 0.05 is considered significant.
Data availability
USP7, VEGFA and ZEB1 expression data in human breast tumors in the TCGA and GEO datasets were downloaded from the UCSC Cancer Browser (http://genome-cancer.ucsc.edu) and the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl), respectively. Kaplan–Meier survival analysis for patients with breast cancer was performed using datasets downloaded from the PrognoScan database (http://www.prognoscan.org/). All data generated or analyzed during this study are included in this article and its supporting information files. Any additional data presented in this paper are available from the corresponding author upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
Y. W. and Y. C. funding acquisition; Y. W. and Y. C. supervision; Y. W. and Y. C. conceptualization; Y. W. and Y. C. formal analysis; Y. W. and Y. C. writing-reviewing and editing; K. T., T. Y., M. W., and B. D. methodology; K. T., T. Y., and B. D. investigation; K. T., T. Y., M. W., Z. R., and B. D. data curation; K. T., T. Y., M. W., and B. D. conceptualization; K. T., T. Y., M. W., and B. D. software; K. T., T. Y., and B. D. writing–original draft; K. T., T. Y., B. D., Z. R., S. W., X. L., K. T., H. L., J. W., and Y. D. formal analysis; S. W., X. L., K. T., H. L., J. W., and Y. D. validation; S. W., X. L., K. T., H. L., J. W., and Y. D. software; J. Z. methodology; J. Z. writing–reviewing and editing.
Funding and additional information
This work was supported by grants from the National Natural Science Foundation of China (82203297 and 81902691), Taishan Scholar Young Expert Program of Shandong Province (tsqn202306154 and tsqn202306155), Shandong Excellent Young Scientists Fund Program (Overseas) (2022HWYQ-077), Shandong Provincial Natural Science Foundation (ZR2023MH252), Guangdong Basic and Applied Basic Research Foundation (2022A1515011738 and 2021A1515011224), Medical Joint Fund of Jinan University (YXJC2022002), Science Foundation of AMHT Group (2023YK07), and Science Foundation of Human Aerospace Hospital (2023YJ02).
Reviewed by members of the JBC Editorial Board. Edited by Brian D. Strahl
Contributor Information
Yan Chen, Email: yanchen@sdnu.edu.cn.
Yijie Wang, Email: yijiewang@sdnu.edu.cn.
Supporting information
References
- 1.Chen Y., Zhang B., Bao L., Jin L., Yang M., Peng Y., et al. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J. Clin. Invest. 2018;128:1937–1955. doi: 10.1172/JCI95089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Tsai Y.H., Tseng S.H. Regulation of ZMYND8 to treat cancer. Molecules. 2021;26:1083. doi: 10.3390/molecules26041083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29:197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savitsky P., Krojer T., Fujisawa T., Lambert J.P., Picaud S., Wang C.Y., et al. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 2016;17:2724–2737. doi: 10.1016/j.celrep.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li N., Li Y., Lv J., Zheng X., Wen H., Shen H., et al. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Mol. Cell. 2016;63:470–484. doi: 10.1016/j.molcel.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adhikary S., Sanyal S., Basu M., Sengupta I., Sen S., Srivastava D.K., et al. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 2016;291:2664–2681. doi: 10.1074/jbc.M115.679985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Wang Y., Luo W. ZMYND8 is a primary HIF coactivator that mediates breast cancer progression. Mol. Cell Oncol. 2018;5 doi: 10.1080/23723556.2018.1479619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo M., Bao L., Chen Y., Xue Y., Wang Y., Zhang B., et al. ZMYND8 is a master regulator of 27-hydroxycholesterol that promotes tumorigenicity of breast cancer stem cells. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abn5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo M., Bao L., Xue Y., Zhu M., Kumar A., Xing C., et al. ZMYND8 protects breast cancer stem cells against oxidative stress and ferroptosis through activation of NRF2. J. Clin. Invest. 2024;134 doi: 10.1172/JCI171166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Q., Zhong S., Wang H., Wang X., Li N., Li Y., et al. The ZMYND8-regulated mevalonate pathway endows YAP-high intestinal cancer with metabolic vulnerability. Mol. Cell. 2021;81:2736–2751.e8. doi: 10.1016/j.molcel.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Tang B., Sun R., Wang D., Sheng H., Wei T., Wang L., et al. ZMYND8 preferentially binds phosphorylated EZH2 to promote a PRC2-dependent to -independent function switch in hypoxia-inducible factor-activated cancer. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2019052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen H.F., Zhang W.J., Huang Y., He Y.H., Hu G.S., Wang L., et al. The dual function of KDM5C in both gene transcriptional activation and repression promotes breast cancer cell growth and tumorigenesis. Adv. Sci. (Weinh.) 2021;8 doi: 10.1002/advs.202004635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia P., Li X., Wang X., Yao L., Xu Y., Hu Y., et al. ZMYND8 mediated liquid condensates spatiotemporally decommission the latent super-enhancers during macrophage polarization. Nat. Commun. 2021;12:6535. doi: 10.1038/s41467-021-26864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong F., Clouaire T., Aguirrebengoa M., Legube G., Miller K.M. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 2017;216:1959–1974. doi: 10.1083/jcb.201611135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Luo M., Chen Y., Wang Y., Zhang B., Ren Z., et al. ZMYND8 expression in breast cancer cells blocks T-lymphocyte surveillance to promote tumor growth. Cancer Res. 2021;81:174–186. doi: 10.1158/0008-5472.CAN-20-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao F., Li Z., He C., Xu W., Yang G., Liu T., et al. RACK7 recognizes H3.3G34R mutation to suppress expression of MHC class II complex components and their delivery pathway in pediatric glioblastoma. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu F., Jin Y., Pu J., Huang Y., Hou J., Zhao X., et al. Aberrant FBXW7-mediated ubiquitination and degradation of ZMYND8 enhances tumor progression and stemness in bladder cancer. Exp. Cell Res. 2021;407 doi: 10.1016/j.yexcr.2021.112807. [DOI] [PubMed] [Google Scholar]
- 18.Jin X., Xu X.E., Jiang Y.Z., Liu Y.R., Sun W., Guo Y.J., et al. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aat9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Liu X., Huang W., Liang J., Chen Y. The intricate interplay between HIFs, ROS, and the ubiquitin system in the tumor hypoxic microenvironment. Pharmacol. Ther. 2022;240 doi: 10.1016/j.pharmthera.2022.108303. [DOI] [PubMed] [Google Scholar]
- 20.Dikic I., Schulman B.A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 2023;24:273–287. doi: 10.1038/s41580-022-00543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dewson G., Eichhorn P.J.A., Komander D. Deubiquitinases in cancer. Nat. Rev. Cancer. 2023;23:842–862. doi: 10.1038/s41568-023-00633-y. [DOI] [PubMed] [Google Scholar]
- 22.Saha G., Roy S., Basu M., Ghosh M.K. USP7 - a crucial regulator of cancer hallmarks. Biochim. Biophys. Acta Rev. Cancer. 2023;1878 doi: 10.1016/j.bbcan.2023.188903. [DOI] [PubMed] [Google Scholar]
- 23.Guo N.J., Wang B., Zhang Y., Kang H.Q., Nie H.Q., Feng M.K., et al. USP7 as an emerging therapeutic target: a key regulator of protein homeostasis. Int. J. Biol. Macromol. 2024;263 doi: 10.1016/j.ijbiomac.2024.130309. [DOI] [PubMed] [Google Scholar]
- 24.Park H.B., Baek K.H. Current and future directions of USP7 interactome in cancer study. Biochim. Biophys. Acta Rev. Cancer. 2023;1878 doi: 10.1016/j.bbcan.2023.188992. [DOI] [PubMed] [Google Scholar]
- 25.He J., Li C.F., Lee H.J., Shin D.H., Chern Y.J., Pereira De Carvalho B., et al. MIG-6 is essential for promoting glucose metabolic reprogramming and tumor growth in triple-negative breast cancer. EMBO Rep. 2021;22 doi: 10.15252/embr.202050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan D., Shang M., Han Y., Liu J., Liu J., Kong S.H., et al. EZH2-CCF-cGAS Axis promotes breast cancer metastasis. Int. J. Mol. Sci. 2022;23:1788. doi: 10.3390/ijms23031788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J., Feng J., Li L., Lin L., Ji J., Lin C., et al. Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep. 2020;21 doi: 10.15252/embr.201948597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y.T., Lin J., Liu Y.E., Chen Y.C., Liu S.T., Hsu K.W., et al. USP7 induces chemoresistance in triple-negative breast cancer via deubiquitination and stabilization of ABCB1. Cells. 2022;11:3294. doi: 10.3390/cells11203294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W., Liu X., Zhang Y., Deng M., Li G., Chen G., et al. USP5 promotes breast cancer cell proliferation and metastasis by stabilizing HIF2alpha. J. Cell Physiol. 2022;237:2211–2219. doi: 10.1002/jcp.30686. [DOI] [PubMed] [Google Scholar]
- 31.Cheng J., Yang H., Fang J., Ma L., Gong R., Wang P., et al. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat. Commun. 2015;6:7023. doi: 10.1038/ncomms8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkari F., La Delfa A., Arrowsmith C.H., Frappier L., Sheng Y., Saridakis V. Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. J. Mol. Biol. 2010;402:825–837. doi: 10.1016/j.jmb.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 33.Faesen A.C., Dirac A.M., Shanmugham A., Ovaa H., Perrakis A., Sixma T.K. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 34.Sheng Y., Saridakis V., Sarkari F., Duan S., Wu T., Arrowsmith C.H., et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 35.Ding X., Jiang W., Zhou P., Liu L., Wan X., Yuan X., et al. Mixed lineage leukemia 5 (MLL5) protein stability is cooperatively regulated by O-GlcNac transferase (OGT) and ubiquitin specific protease 7 (USP7) PLoS One. 2015;10 doi: 10.1371/journal.pone.0145023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q., Ma S., Song N., Li X., Liu L., Yang S., et al. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J. Clin. Invest. 2016;126:2205–2220. doi: 10.1172/JCI85747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felle M., Joppien S., Nemeth A., Diermeier S., Thalhammer V., Dobner T., et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin W., Leonhardt H., Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell Biochem. 2011;112:439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 39.Maertens G.N., El Messaoudi-Aubert S., Elderkin S., Hiom K., Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faesen A.C., Luna-Vargas M.P., Geurink P.P., Clerici M., Merkx R., van Dijk W.J., et al. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 41.An L., Cao Z., Nie P., Zhang H., Tong Z., Chen F., et al. Combinatorial targeting of Hippo-STRIPAK and PARP elicits synthetic lethality in gastrointestinal cancers. J. Clin. Invest. 2022;132:e155468. doi: 10.1172/JCI155468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Epping M.T., Meijer L.A., Krijgsman O., Bos J.L., Pandolfi P.P., Bernards R. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat. Cell Biol. 2011;13:102–108. doi: 10.1038/ncb2142. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z., Kang W., You Y., Pang J., Ren H., Suo Z., et al. USP7: novel drug target in cancer therapy. Front. Pharmacol. 2019;10:427. doi: 10.3389/fphar.2019.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mungamuri S.K., Qiao R.F., Yao S., Manfredi J.J., Gu W., Aaronson S.A. USP7 enforces heterochromatinization of p53 target promoters by protecting SUV39H1 from MDM2-mediated degradation. Cell Rep. 2016;14:2528–2537. doi: 10.1016/j.celrep.2016.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang B., Li J., Wang Y., Liu X., Yang X., Liao Z., et al. Deubiquitinase USP7 stabilizes KDM5B and promotes tumor progression and cisplatin resistance in nasopharyngeal carcinoma through the ZBTB16/TOP2A axis. Cell Death Differ. 2024;31:309–321. doi: 10.1038/s41418-024-01257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Liu X., Wang M., Wang Y., Wang S., Jin L., et al. UBE3B promotes breast cancer progression by antagonizing HIF-2alpha degradation. Oncogene. 2023;42:3394–3406. doi: 10.1038/s41388-023-02842-z. [DOI] [PubMed] [Google Scholar]
- 47.Guan T., Yang X., Liang H., Chen J., Chen Y., Zhu Y., et al. Deubiquitinating enzyme USP9X regulates metastasis and chemoresistance in triple-negative breast cancer by stabilizing Snail1. J. Cell Physiol. 2022;237:2992–3000. doi: 10.1002/jcp.30763. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Chen Y., Wang C., Yang M., Wang Y., Bao L., et al. MIF is a 3' flap nuclease that facilitates DNA replication and promotes tumor growth. Nat. Commun. 2021;12:2954. doi: 10.1038/s41467-021-23264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie W., Gao S., Yang Y., Li H., Zhou J., Chen M., et al. CYLD deubiquitinates plakoglobin to promote Cx43 membrane targeting and gap junction assembly in the heart. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111864. [DOI] [PubMed] [Google Scholar]
- 50.Hong R., Tan Y., Tian X., Huang Z., Wang J., Ni H., et al. XIAP-mediated degradation of IFT88 disrupts HSC cilia to stimulate HSC activation and liver fibrosis. EMBO Rep. 2024;25:1055–1074. doi: 10.1038/s44319-024-00092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma B., Chen Y., Chen L., Cheng H., Mu C., Li J., et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 2015;17:95–103. doi: 10.1038/ncb3073. [DOI] [PubMed] [Google Scholar]
- 52.Wang S., Li H., Liu X., Yin T., Li T., Zheng M., et al. VHL suppresses UBE3B-mediated breast tumor growth and metastasis. Cell Death Dis. 2024;15:446. doi: 10.1038/s41419-024-06844-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang T., Periz G., Lu Y.N., Wang J. USP7 regulates ALS-associated proteotoxicity and quality control through the NEDD4L-SMAD pathway. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28114–28125. doi: 10.1073/pnas.2014349117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Chen Y., Bao L., Zhang B., Wang J.E., Kumar A., et al. CHD4 promotes breast cancer progression as a coactivator of hypoxia-inducible factors. Cancer Res. 2020;80:3880–3891. doi: 10.1158/0008-5472.CAN-20-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao Y., Zhang X., Hu M., Yang S., Li X., Han R., et al. CYLD inhibits osteoclastogenesis to ameliorate alveolar bone loss in mice with periodontitis. J. Cell Physiol. 2023;238:1036–1045. doi: 10.1002/jcp.30990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
USP7, VEGFA and ZEB1 expression data in human breast tumors in the TCGA and GEO datasets were downloaded from the UCSC Cancer Browser (http://genome-cancer.ucsc.edu) and the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl), respectively. Kaplan–Meier survival analysis for patients with breast cancer was performed using datasets downloaded from the PrognoScan database (http://www.prognoscan.org/). All data generated or analyzed during this study are included in this article and its supporting information files. Any additional data presented in this paper are available from the corresponding author upon request.