Abstract
Snake envenomation poses a significant risk to Malaysians and country visitors. Malaysia witnesses an estimated 650 snake bites per 100,000 population annually. The primary treatment for snake envenomation involves administering antivenom derived from horses, despite its drawbacks, such as anaphylactic reactions and serum sickness. Identifying the venom proteome is crucial for understanding and predicting the clinical implications of envenomation and developing effective treatments targeting specific venom proteins. In this study, we employ an immunoprecipitation assay followed by LC-MS/MS to identify antigenic proteins in five common venomous snakes in Malaysia compassing of two families which are pit vipers, (Calloselasma rhodostoma and Cryptelytrops purpureomaculatus) and cobras (Ophiophagus hannah, Naja kaouthia, and Naja sumatrana). The immunoprecipitation assay utilises a 2 % agarose gel, allowing antigenic proteins to diffuse and bind with antibodies in the antivenom. The antivenom utilised in this research was procured from the Queen Saovabha Memorial Institute (QSMI), Thailand, including king cobra antivenom (KCAV), cobra antivenom (CAV), Malayan pit viper antivenom (MPAV), Russell's viper antivenom (RPAV), hematopolyvalent antivenom (HPAV), neuropolyvalent antivenom (NPAV), banded krait antivenom (BKAV), and Malayan krait antivenom (MKAV). The protein identified through these interactions which are exclusive to the cobras are three-finger toxins (3FTXs) while snake C-type lectins (Snaclecs) are unique to the pit vipers. Common protein that are present in both families are L-amino acid oxidase (LAAO), Phospholipase A2 (PLA2), and snake venom metalloproteinase (SVMP). Identifying these proteins is vital for formulating a broad-spectrum antivenom applicable across multiple species.
Keywords: Antivenom, Proteomics, Malaysian venomous snake, Elapidae, Crotalinae
Graphical abstract
Highlights
-
•
Unique cobra proteins include 3FTXs, while pit vipers have Snaclecs.
-
•
Common venom proteins detected across both snake families are LAAO, PLA2, and SVMP.
-
•
The study reveals key proteins that could enable broader antivenom development.
-
•
Novel findings offer potential improvements in treating diverse snakebites.
-
•
These insights pave the way for more effective universal antivenom.
1. Introduction
Venomous snakes pose a significant threat to human health worldwide, with the World Health Organization (WHO) reclassifying them as one of the world's neglected tropical diseases in 2017 [1]. WHO reports approximately 81,000 to 138,000 deaths worldwide annually due to snakebite envenomation, with a high incidence of snakebites recorded in the Southeast Asian region [1]. Envenomation can happen when venomous snake bites or when venom is sprayed into the eyes of the victim by snakes that possess the capability to spray their venom as an act of defence [2]. In Malaysia, cobras and pit vipers are recognized as medically significant snakes [2], and these species are commonly found throughout the land region of peninsula Malaysia, including Thailand, Laos, and Myanmar [3]. Cobras' venoms contain potent neurotoxic, cardiotoxic, and cytotoxic activities, attributed to protein families like three-finger toxins, cardiotoxins, and cytotoxins [4]. On the other hand, pit viper venoms can induce potent necrotic and hemotoxic effects through toxins such as L-amino acid oxidase and snake venom metalloproteinase, respectively [5].
The primary treatment for snake envenomation typically relies on administering horse-derived antisera, known as antivenom, to counteract snake venom's clinical and toxic effects [6]. While effective in preventing fatalities, the current antivenom presents several challenges. These include severe anaphylactic reactions and limited efficacy against snake venom from diverse geographical regions due to species-specific formulations [7]. Other factors effecting the efficacy of the antivenom includes the variation of venom caused by diet, sex and ontogenetic factor [8]. Moreover, the current formulation and treatment approach for antivenom has remained stagnant for many decades, highlighting the urgent need for a novel and innovative strategy to address snakebite envenomation. There are different approaches being studied, including small molecule inhibitor, peptide inhibitor and natural products where they target specific proteins [9]. Proteomics technique using tandem mass spectrometry has been the gold standard in profiling and characterising snake venom proteins. This approach enables scientists to accurately identify and profile different protein families within the crude venom of diverse snake species, facilitating the relative measurement of protein abundance and prediction of clinical outcomes following envenomation by these species [10,11]. Applying similar proteomic techniques, antigenic venom proteins from venomous snakes in other regions, such as India [12], South America [13], and African snakes [14], were successfully profiled. This comprehensive understanding of venom composition aids in the design and formulation of antivenom that is effective across various geographical regions and against different snake species, thereby enhancing the treatment outcomes for snakebite victims.
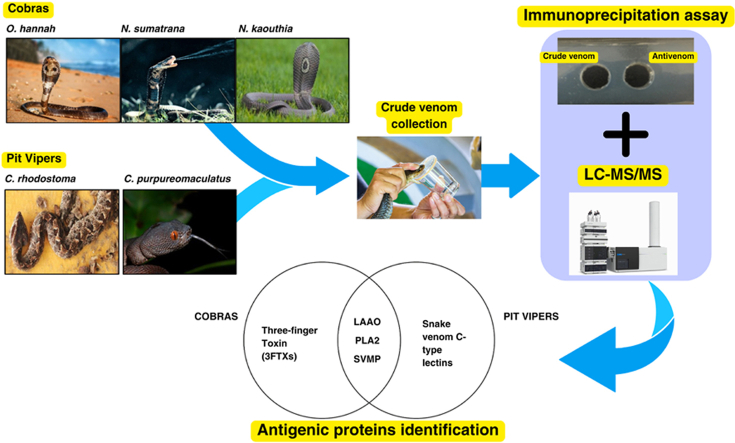
Therefore, in this present study, we investigated the antigenic venom proteins from five of the most common venomous snakes compassing of two families which are pit vipers, (Calloselasma rhodostoma and Cryptelytrops purpureomaculatus) and cobras (Ophiophagus hannah, Naja kaouthia, and Naja sumatrana). Using the agarose immunoprecipitation technique, we assayed the venom against various types of antivenom procured from Thailand (Queen Saovabha Memorial Institute, Bangkok). Subsequently, we employed high-resolution tandem mass spectrometry (LC-MS/MS) to identify the proteins from positive venom-antivenom interactions. These findings offer valuable insights that could contribute to the development of broad range antivenom.
2. Materials and methods
2.1. Crude venom collection
Crude venom of Calloselasma rhodostoma and Cryptelytrops purpureomaculatus) and cobras (Ophiophagus hannah, Naja kaouthia, and Naja sumatrana was purchased and collected from local and licensed venomous snake enthusiasts, Mr. Zainuddin Ismail (Bukit Bintang Enterprises Sdn. Bhd.) in Malaysia. The adult snakes are native to northern Malaysia (Perlis) and they are of both sex. For each species, the venom of 3–4 snakes were collected by gently placing the venom's fangs on a container wrapped with parafilm. The container was then sealed, stored in a cool box, and transported back to Monash Malaysia. Crude venom was then stored at −20 °C, followed by freeze-drying to preserve the venom activity. Freeze-dried crude venom was dissolved in double-distilled (Milli-Q) water before usage.
2.2. Antivenom
Antivenom was acquired from the Queen Saovabha Memorial Institute (QSMI) of the Thai Red Cross Society in Bangkok, Thailand, were used, i.e., king cobra antivenom (KCAV; LH00118), cobra antivenom (CAV; NK00117), Malayan pit viper antivenom (MPAV; NK00117), Russell's viper antivenom (RPAV; WR00308), hematopolyvalent antivenom (HPAV; HP00416), neuropolyvalent antivenom (NPAV; NP00116), banded krait antivenom (BKAV; BK00114) and Malayan krait antivenom (MKAV). The antivenom was diluted according to the information leaflet using the normal saline (0.9 % NaCl) provided.
2.3. Immunoprecipitation agarose gel assay
Agarose gel medium (2 g agarose in 100 mL MilliQ water) was pipetted on the glass slides and then allowed to polymerise. Two wells were made in the agarose gel with a distance of at least 1 cm between the wells and approximately 1 cm in diameter. Hundred microlitre of venom and antivenom were pipetted into one of each well and allowed to diffuse through the gel for 24–48 h. The venom-antivenom complex appeared as a white precipitate (white band) in the middle of the wells and proceeded to in-gel digestion and LC-MS/MS analysis.
2.4. In-gel tryptic digestion
After the excision of the gel band, an in-gel tryptic digestion protocol was conducted to digest the proteins on the band. The gel was prepared for digestion with 200 μL of a reduction buffer (3.1 mg of 10 mM dithiothreitol (DTT) in 2 mL of 50 mM ammonium bicarbonate) incubated at 56 °C for 60 min. The gel pieces were then centrifuged briefly before removing all liquid in the Eppendorf tube. Then, 200 μL of an alkylation buffer (20.4 mg of 55 mM iodoacetamide (IAM) in 2 mL of 50 mM ammonium bicarbonate) was added to the gel piece for incubation in the dark for 30 min, and all liquid was discarded after. The gel piece was then washed with 200 μL of 50 mM ammonium bicarbonate for 15 min before discarding all liquid and repeated with 200 μL of 50 mM ammonium bicarbonate in 50 % acetonitrile (ACN). The gel piece was washed with 200 μL of 100 % ACN for 15 min at 37 °C before being centrifuged briefly and all liquid contents discarded. 1 μL of Trypsin/Lys-C was added with 50 μL of 40 mM ammonium bicarbonate in 9 % ACN and left overnight at 37 °C. The digested sample was then centrifuged briefly, and its supernatant was collected in a separate collection tube. 50 μL of 5 % formic acid (FA) was added to the gel piece, vortexed briefly and incubated at 37 °C for 15 min. This process was repeated with 5 % FA in 50 % ACN and then with pure ACN. The supernatant was recovered into the collection tube at the end of each incubation. The gel piece was discarded, and the sample in the collection tube was subjected and spun to dry overnight in a vacuum concentrator before LC-MS/MS analysis.
2.5. LC-MS/MS analysis
The analysis was performed on Agilent 6550 Quadrupole Time-of-Flight (QTOF). Digested peptides (100 μg/ml) from in-solution tryptic digestion were loaded into an AdvancedBio Peptide Mapping, 2.1 × 250mm, 7 μm (pn 651750-902). Peptides were eluted with an increasing gradient, 5–100 % of 90 % acetonitrile in 0.1 % formic acid in water. The LCMS-QTOF parameters were set as positive polarity with the capillary voltage set at 2050 V and 300 V, respectively and 5 L/min of gas flow with a temperature of 300 °C. The spectrum was analysed in auto-MS mode, ranging from 110 to 3000 m/z for MS scan and 50–3000 m/z for MS/MS scan.
2.6. Data analysis using PEAKS Studio X plus
Antigenic proteins were identified and analysed using the UniProt database (Species: Serpentes and Equus caballus) through the PEAKS Studio X Plus software. The identification of the antigenic proteins was run against the Serpentes database, while the identification of the antibodies was run against the Equus caballus database. The profiling parameters include false discovery rate (FDR) at 0.1 %, −10logP values (>20), and a minimum of 1 unique protein identified for each result.
3. Results
3.1. Immunoprecipitation assay
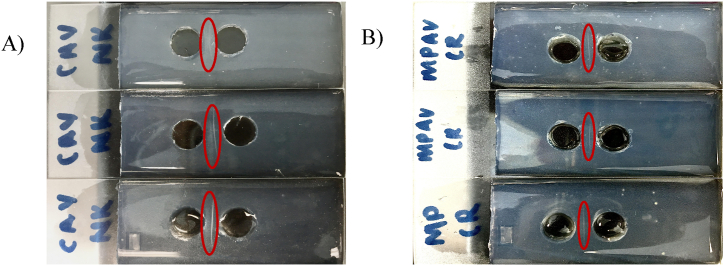
The immunoprecipitation assay was conducted to investigate the interaction between venom and antivenom [king cobra antivenom (KCAV), cobra antivenom (CAV), Malayan pit viper antivenom (MPAV), Russell's viper antivenom (RPAV), hematopolyvalent antivenom (HPAV), neuropolyvalent antivenom (NPAV), banded krait antivenom (BKAV), and Malayan krait antivenom (MKAV)]. Positive interaction was indicated by the formation of white precipitate between the wells. Fig. 1A shows the positive interactions of the immunoprecipitation assay of Malayan pit viper antivenom against C. rhodostoma (Malayan pit viper) while Fig. 1B shows the positive interactions of the immunoprecipitation assay of cobra antivenom against N. kaouthia (Monocled cobra). Bands extracted from the immunoprecipitation assay underwent LC-MS/MS analysis to identify antigenic venom proteins. These proteins were then analysed against the UniProt database (Species: Serpentes) using PEAKS Studio X Plus software. Confirmation of venom protein binding with antivenoms was achieved by analysing against the Equus caballus database (Refer to supplementary documents).
Fig. 1.
A). Positive interactions of the immunoprecipitation assay of cobra antivenom against N. kaouthia. B) Positive interactions of the immunoprecipitation assay of malayan pit viper antivenom against C. rhodostoma. The red circle highlights the precipitation between the venom and antivenom indication a positive interaction. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
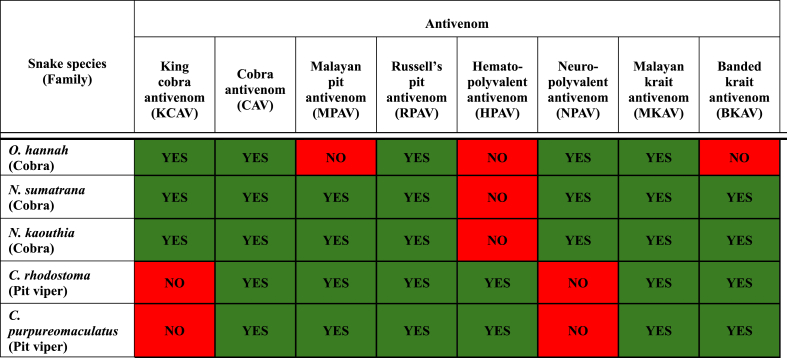
The results of the immunoprecipitation assay were summarised in Table 1. Both Naja species showed same interactions with all the eight antivenom while O. hannah exhibited 5 positive interactions. Both the pit vipers presented with the same interactions across all the antivenom used.
Table 1.
Results of immunoprecipitation assay for all 5 crude venom against 8 antivenom. The green highlighted boxes indicate positive interaction, and the red highlighted boxes indicates a negative interaction between the venom and antivenom. King cobra antivenom (KCAV), Cobra antivenom (CAV), Malayan pit antivenom (MPAV), Russell's pit antivenom (RPAV), Hemato-polyvalent antivenom (HPAV), Neuro-polyvalent antivenom (NPAV), Malayan krait antivenom (MKAV), Banded krait antivenom (BKAV).
3.2. O. hannah antigenic proteins
Table 2 summarised all the proteins identified from the immunoprecipitation assay from Ophiophagus hannah. Five proteins were identified in the interaction between O. hannah venom and KCAV: cysteine-rich venom proteins (CRVP), L-amino-acid oxidase (LAAO), beta-cardiotoxin, ShKT domain-containing protein, and alpha-elapitoxin. In the case of RPAV, seven proteins were identified: LAAO, amine oxidase, ophiophagus venom factor, SVMP-disintegrin, CRVP, ShKT domain-containing protein, and beta-cardiotoxin. Interaction with NPAV led to the identification of five proteins: LAAO, ophiophagus venom factor, SVMP-disintegrin, CRVP, and beta-cardiotoxin. Finally, interaction with MKAV revealed five proteins: CRVP, amine oxidase, LAAO, ShKT domain-containing protein, and beta-cardiotoxin. CRVP, LAAO, and amine oxidase were common proteins identified in interactions with all five antivenoms, while beta-cardiotoxin was present in all interactions except with CAV. Alpha-elapitoxin was only detected in interactions with elapid antivenom. Beta-cardiotoxin and cysteine-rich venom proteins exhibited the highest coverage among the identified proteins. Each protein contained at least one unique peptide in its sequence, with all −10logP values exceeding 40. All identified proteins were specific to the O. hannah species.
Table 2.
Proteins identified via LC-MS/MS from the O. hannah crude venom against KCAV, CAV, RPAV, NPAV. MKAV, and BKAV. King cobra antivenom (KCAV), Cobra antivenom (CAV), Neuro-polyvalent antivenom (NPAV), Malayan krait antivenom (MKAV).
| Accession | −10lgP | Coverage (%) | Unique | Avg. Mass | Description (OS) |
|---|---|---|---|---|---|
|
O. hannah crude venom against KCAV | |||||
| Q7ZT98 | 197.6 | 31 | 10 | 26869 | Cysteine-rich venom protein ophanin (O. hannah) |
| P81383 | 152.54 | 21 | 5 | 55977 | L-amino-acid oxidase (O. hannah) |
| Q2VBN8 | 85.13 | 27 | 3 | 9380 | Beta-cardiotoxin CTX9 (O. hannah) |
| V8N8B4 | 163.58 | 12 | 1 | 24779 | ShKT domain-containing protein (O. hannah) |
|
Q53B58 |
57.06 |
20 |
1 |
10210 |
Alpha-elapitoxin-Oh3a (O. hannah) |
|
O. hannah crude venom against CAV | |||||
| P81383 | 215.81 | 40 | 5 | 55977 | L-amino-acid oxidase (O. hannah) |
| V8N3Q9 | 207.35 | 43 | 2 | 55593 | Amine oxidase (Fragment) (O. hannah) |
| I2C090 | 191.18 | 18 | 17 | 183927 | Ophiophagus venom factor (O. hannah) |
| A3R0T9 | 189.13 | 28 | 6 | 69049 | Zinc metalloproteinase-disintegrin-like ohanin (O. hannah) |
| Q7ZT98 | 185.82 | 51 | 14 | 26869 | Cysteine-rich venom protein ophanin (O. hannah) |
|
Q53B58 |
174.67 |
59 |
7 |
10210 |
Alpha-elapitoxin-Oh3a (O. hannah) |
|
O. hannah crude venom against RPAV | |||||
| P81383 | 243.32 | 43 | 3 | 55977 | L-amino-acid oxidase (O. hannah) |
| V8N3Q9 | 238.19 | 46 | 3 | 55593 | Amine oxidase (Fragment) (O. hannah) |
| I2C090 | 206.75 | 15 | 11 | 183927 | Ophiophagus venom factor (O. hannah) |
| A3R0T9 | 197.39 | 25 | 14 | 69049 | Zinc metalloproteinase-disintegrin-like ohanin (O. hannah) |
| Q7ZT98 | 180.33 | 55 | 17 | 26869 | Cysteine-rich venom protein ophanin (O. hannah) |
| tV8N8B4 | 170.3 | 48 | 2 | 24779 | ShKT domain-containing protein (O. hannah) |
|
Q53B46 |
110.47 |
61 |
6 |
9352 |
Beta-cardiotoxin CTX15 (O. hannah) |
|
O. hannah crude venom against NPAV | |||||
| I2C090 | 182.32 | 21 | 13 | 183927 | Ophiophagus venom factor (O. hannah) |
| Q7ZT98 | 158.23 | 46 | 4 | 26869 | Cysteine-rich venom protein ophanin (O. hannah) |
| A3R0T9 | 139.75 | 61 | 12 | 69049 | Zinc metalloproteinase-disintegrin-like ohanin (O. hannah) |
| P81383 | 135.33 | 32 | 15 | 55977 | L-amino-acid oxidase (O. hannah) |
|
Q53B46 |
104.21 |
61 |
6 |
9352 |
Beta-cardiotoxin CTX15 (O. hannah) |
|
O. hannah crude venom against MKAV | |||||
| Q7ZT98 | 199.17 | 33 | 5 | 26869 | Cysteine-rich venom protein ophanin (O. hannah) |
| V8N3Q9 | 146.06 | 14 | 6 | 55593 | Amine oxidase (Fragment) (O. hannah) |
| P81383 | 146.06 | 14 | 6 | 55977 | L-amino-acid oxidase (O. hannah) |
| V8N8B4 | 188.08 | 16 | 1 | 24779 | ShKT domain-containing protein (O. hannah) |
| Q2VBN8 | 45.61 | 27 | 2 | 9380 | Beta-cardiotoxin CTX9 (O. hannah) |
3.3. N. sumatrana antigenic proteins
Table 3 summarised the proteins identified from all interactions involving Naja sumatrana. Predominantly, cytotoxins were identified as the common antigenic proteins across interactions with seven antivenoms. These include cytotoxin isoforms 4b, 4a, 3a, KJC3, 2, 1, and 3. Notably, cytotoxin KJC3 and cytotoxin 4b from Naja sputatrix were consistently identified across all interactions. In the interaction with KCAV, seven proteins were identified: cytotoxin 4b, 4a, 3a, KJC3, 2, 1, and 3. Similarly, six proteins were determined from the interaction with CAV, including cytotoxin KJC3, 4b, 4a, 5, and 2, along with neutral phospholipase A2. MPAV interaction revealed cytotoxin KJC3, 4b, 4a, 1, and 2, alongside weak neurotoxin 6 and weak neurotoxin 8. RPAV interaction identified cytotoxin KJC3, 4b, 3a, 4a, and 1, along with neutral phospholipase A2. Interaction with NPAV unveiled cytotoxin KJC3, 4b, 4a, 1, and 3a, along with weak neurotoxin 6. BKAV interaction resulted in the identification of five proteins: cytotoxin 4b, 3a, KJC3, 2, and 7. Lastly, MKAV interaction identified cytotoxin 4b, KJC3, 2, 1, and 3. All proteins contain at least one unique peptide sequence, with −10logP values exceeding 40. The proteins identified from the database belong to the Naja genus.
Table 3.
Proteins identified via LC-MS/MS from the N. sumatrana crude venom against KCAV, CAV, MPAV, RPAV, NPAV. MKAV, and BKAV. King cobra antivenom (KCAV), Cobra antivenom (CAV), Malayan pit antivenom (MPAV), Russell's pit antivenom (RPAV), Neuro-polyvalent antivenom (NPAV), Malayan krait antivenom (MKAV), Banded krait antivenom (BKAV).
| Accession | −10lgP | Coverage (%) | Unique | Avg. Mass | Description (OS) |
|---|---|---|---|---|---|
|
N. sumatrana crude venom against KCAV | |||||
| O73856 | 211.24 | 74 | 2 | 9084 | Cytotoxin 4b (N. sputatrix) |
| Q98959 | 201.16 | 74 | 1 | 9065 | Cytotoxin 3a (N. atra) |
| O93473 | 209.07 | 74 | 1 | 9068 | Cytotoxin 4a (N. sputatrix) |
| P60311 | 203.85 | 100 | 3 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| P01440 | 196.07 | 100 | 2 | 6763 | Cytotoxin 2 (N. naja) |
| P0CH80 | 195.54 | 87 | 1 | 6807 | Cytotoxin 1 (N. kaouthia) |
|
P01459 |
162.87 |
70 |
1 |
6839 |
Cytotoxin 3 (N. annulifera) |
|
N. sumatrana crude venom against CAV | |||||
| P60311 | 213.52 | 100 | 5 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| O73856 | 206.02 | 74 | 2 | 9084 | Cytotoxin 4b (N. sputatrix) |
| O93473 | 203.82 | 74 | 2 | 9068 | Cytotoxin 4a (N. sputatrix) |
| P24779 | 143.27 | 92 | 1 | 6654 | Cytotoxin 5 (N. kaouthia) |
| P01440 | 221.91 | 100 | 1 | 6763 | Cytotoxin 2 (N. naja) |
|
Q92084 |
147.73 |
44 |
7 |
16189 |
Neutral phospholipase A2 muscarinic inhibitor (N. sputatrix) |
|
N. sumatrana crude venom against MPAV | |||||
| O73856 | 211.08 | 74 | 2 | 9084 | Cytotoxin 4b (N. sputatrix) |
| O93473 | 208.52 | 74 | 2 | 9068 | Cytotoxin 4a (N. sputatrix) |
| P60311 | 206.01 | 100 | 4 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| P01440 | 212.59 | 100 | 1 | 6763 | Cytotoxin 2 (N. naja) |
| P0CH80 | 207.96 | 100 | 2 | 6807 | Cytotoxin 1 (N. kaouthia) |
| O42256 | 205.16 | 71 | 3 | 9807 | Weak neurotoxin 6 (N. sputatrix) |
|
Q802B3 |
204.9 |
71 |
1 |
9809 |
Weak neurotoxin 8 (N. sputatrix) |
|
N. sumatrana crude venom against RPAV | |||||
| A0A7T7DMY7 | 199.41 | 74 | 2 | 9054 | Cytotoxin 1 (N. sumatrana) |
| O73856 | 194.33 | 69 | 1 | 9084 | Cytotoxin 4b (N. sputatrix) |
| P60311 | 192.51 | 97 | 1 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| Q98959 | 200.03 | 74 | 2 | 9065 | Cytotoxin 3a (N. atra) |
| O93473 | 187.72 | 74 | 1 | 9068 | Cytotoxin 4a (N. sputatrix) |
|
Q92084 |
142.3 |
50 |
6 |
16189 |
Neutral phospholipase A2 muscarinic inhibitor (N. sputatrix) |
|
N. sumatrana crude venom against NPAV | |||||
| P60311 | 204.07 | 88 | 2 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| O73856 | 210.07 | 72 | 1 | 9084 | Cytotoxin 4b (N. sputatrix) |
| O93473 | 208.53 | 77 | 2 | 9068 | Cytotoxin 4a (N. sputatrix) |
| P0CH80 | 161.65 | 82 | 1 | 6807 | Cytotoxin 1 (N. kaouthia) |
| Q98959 | 229.39 | 77 | 2 | 9065 | Cytotoxin 3a (N. atra) |
|
O42256 |
190.64 |
66 |
12 |
9807 |
Weak neurotoxin 6 (N. sputatrix) |
|
N. sumatrana crude venom against MKAV | |||||
| A0A7T7DMY7 | 221.01 | 74 | 4 | 9054 | Cytotoxin 1 (N. sumatrana) |
| O73856 | 217.17 | 74 | 1 | 9084 | Cytotoxin 4b (N. sputatrix) |
| P60311 | 208.21 | 90 | 2 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| P01459 | 159.08 | 75 | 3 | 6839 | Cytotoxin 3 (N. annulifera) |
|
P01440 |
188.22 |
100 |
1 |
6763 |
Cytotoxin 2 (N. naja) |
|
N. sumatrana crude venom against BKAV | |||||
| O73856 | 266.94 | 74 | 1 | 9084 | Cytotoxin 4b (N. sputatrix) |
| P60311 | 251.44 | 100 | 3 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| P01440 | 247.06 | 100 | 1 | 6763 | Cytotoxin 2 (N. naja) |
| Q98959 | 222.82 | 74 | 1 | 9065 | Cytotoxin 3a (N. atra) |
| O73859 | 267.43 | 97 | 1 | 7062 | Cytotoxin 7 (Fragment) (N. sputatrix) |
3.4. N. kaouthia antigenic proteins
The proteins identified from all N. kaouthia interactions were summarised in Table 4. In the interaction with KCAV, the identified proteins include cytotoxin 3, cytotoxin 2, cytotoxin 2a, cytotoxin 1d/1e, and cytotoxin 2c. Interaction with CAV revealed cytotoxin KJC3, cytotoxin 3, cytotoxin 10, cytotoxin 1d/1e, and cytotoxin 1. MPAV interaction resulted in the identification of cytotoxin 5, cytotoxin 2, cytotoxin 1d/1e, cytotoxin 1a, and tryptophan-containing weak neurotoxin. RPAV interaction led to the identification of cytotoxin KJC3, cytotoxin 3, cytotoxin 4a, cytotoxin 1d/1e, and cytotoxin 1a. Interaction with NPAV identified cytotoxin 1d/1e, cytotoxin 2, venom phosphodiesterase, and CRVP. MKAV interaction identified cytotoxin 3, cytotoxin 10, cytotoxin 1d/1e, cytotoxin 1, and acidic phospholipase A2. Lastly, interaction with BKAV resulted in the identification of cytotoxin VC-1, cytotoxin 3, cytotoxin 1d/1e, cytotoxin 10, and tryptophan-containing weak neurotoxin. All identified proteins contain at least one unique peptide sequence, with −10logP values exceeding 40. These proteins belong to the Naja genus as identified from the sample using the database.
Table 4.
Proteins identified via LC-MS/MS from N. kaouthia crude venom against KCAV, CAV, MPAV, RPAV, NPAV, MKAV and BKAV. King cobra antivenom (KCAV), Cobra antivenom (CAV), Malayan pit antivenom (MPAV), Russell's pit antivenom (RPAV), Neuro-polyvalent antivenom (NPAV), Malayan krait antivenom (MKAV), Banded krait antivenom (BKAV).
| Accession | −10lgP | Coverage (%) | Unique | Avg. Mass | Description (OS) |
|---|---|---|---|---|---|
|
N. kaouthia crude venom against KCAV | |||||
| P01446 | 162.15 | 100 | 3 | 6717 | Cytotoxin 3 (N. kaouthia) |
| P01440 | 153.53 | 100 | 1 | 6763 | Cytotoxin 2 (N. naja) |
| P86538 | 115.11 | 77 | 1 | 6711 | Cytotoxin 2a (N. naja) |
| Q98958 | 180.79 | 73 | 2 | 8992 | Cytotoxin 1d/1e (N. atra) |
|
O93472 |
153.56 |
74 |
1 |
9128 |
Cytotoxin 2c (N. sputatrix) |
|
N. kaouthia crude venom against CAV | |||||
| Q98958 | 226.56 | 72 | 3 | 8992 | Cytotoxin 1d/1e (N. atra) |
| P60311 | 204.82 | 98 | 1 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| P86541 | 217.5 | 100 | 1 | 6764 | Cytotoxin 10 (N. naja) |
| P01446 | 236.57 | 100 | 2 | 6717 | Cytotoxin 3 (N. kaouthia) |
| P01451 | 191.98 | 92 | 1 | 6821 | Cytotoxin 1 (N. oxiana) |
|
N. kaouthia crude venom against MPAV | |||||
| Q98958 | 200.51 | 73 | 3 | 8992 | Cytotoxin 1d/1e (N. atra) |
| Q98957 | 190.66 | 72 | 1 | 8976 | Cytotoxin 1a (N. atra) |
| P07525 | 170.94 | 87 | 1 | 6810 | Cytotoxin 5 (N. atra) |
| P01440 | 163.56 | 80 | 1 | 6763 | Cytotoxin 2 (N. naja) |
|
P82935 |
200.59 |
67 |
15 |
9915 |
Tryptophan-containing weak neurotoxin (N. kaouthia) |
|
N. kaouthia crude venom against RPAV | |||||
| Q98958 | 173.3 | 65 | 1 | 8992 | Cytotoxin 1d/1e (N. atra) |
| P01446 | 142.21 | 87 | 2 | 6717 | Cytotoxin 3 (N. kaouthia) |
| P60311 | 145.61 | 78 | 1 | 6753 | Cytotoxin KJC3 (N. sputatrix) |
| Q98957 | 164.24 | 58 | 1 | 8976 | Cytotoxin 1a (N. atra) |
|
O93473 |
137.84 |
58 |
1 |
9068 |
Cytotoxin 4a (N. sputatrix) |
|
N. kaouthia crude venom against NPAV | |||||
| A0A2D0TC04 | 183.48 | 23 | 17 | 94616 | Venom phosphodiesterase (N. atra) |
| P84805 | 160.04 | 29 | 3 | 26846 | Cysteine-rich venom protein kaouthin-1 (N. kaouthia) |
| P01445 | 159.76 | 87 | 1 | 6745 | Cytotoxin 2 (N. kaouthia) |
|
Q98958 |
161.89 |
64 |
1 |
8992 |
Cytotoxin 1d/1e (N. atra) |
|
N. kaouthia crude venom against MKAV | |||||
| Q98958 | 157.76 | 65 | 1 | 8992 | Cytotoxin 1d/1e (N. atra) |
| P01446 | 163 | 97 | 3 | 6717 | Cytotoxin 3 (N. kaouthia) |
| P01451 | 171.51 | 82 | 1 | 6821 | Cytotoxin 1 (N. oxiana) |
| P00597 | 172.32 | 69 | 1 | 16016 | Acidic phospholipase A2 (N. kaouthia) |
|
P86541 |
134.67 |
62 |
1 |
6764 |
Cytotoxin 10 (N. naja) |
|
N. kaouthia crude venom against BKAV | |||||
| Q98958 | 184.06 | 72 | 1 | 8992 | Cytotoxin 1d/1e (N. atra) |
| Q9PS33 | 186.73 | 92 | 1 | 6724 | VC-1 cytotoxin (N. oxiana) |
| P01446 | 171.49 | 97 | 4 | 6717 | Cytotoxin 3 (N. kaouthia) |
| P86541 | 148.45 | 80 | 2 | 6764 | Cytotoxin 10 (N. naja) |
| P82935 | 169.17 | 66 | 9 | 9915 | Tryptophan-containing weak neurotoxin (N. kaouthia) |
3.5. C. rhodostoma antigenic proteins
The proteins identified from all C. rhodostoma interactions were summarised in Table 5. The common antigenic proteins identified across all interactions of C. rhodostoma venom against six antivenoms were PLA2. Snaclec rhodocetin subunit beta was consistently identified in every interaction except with CAV, while snaclec rhodocetin subunit alpha was present in almost every interaction except with MKAV and BKAV. Zinc metalloproteinase/disintegrin was identified in all interactions except against HPAV. Snake venom metalloproteinase kistomin was identified in interactions with CAV, HPAV, MKAV, and BKAV. Lastly, LAAO was detected in the interaction with MPAV, HPAV, MKAV, and BKAV. All identified proteins contain at least one unique peptide sequence, with −10logP values exceeding 40. These proteins were matched with the Calloselasma rhodostoma species in the database.
Table 5.
Proteins identified via LC-MS/MS from C. rhodostoma crude venom against CAV, MPAV, RPAV, HPAV, MKAV and BKAV. Cobra antivenom (CAV), Malayan pit antivenom (MPAV), Russell's pit antivenom (RPAV), Hemato-polyvalent antivenom (HPAV), Malayan krait antivenom (MKAV), Banded krait antivenom (BKAV).
| Accession | −10lgP | Coverage (%) | Unique | Avg. Mass | Description (OS) |
|---|---|---|---|---|---|
|
C. rhodostoma crude venom against CAV | |||||
| P30403 | 185.86 | 17 | 14 | 54006 | Zinc metalloproteinase/disintegrin (C. rhodostoma) |
| P81398 | 160.51 | 59 | 15 | 15190 | Snaclec rhodocetin subunit beta (C. rhodostoma) |
| A0A0H3U266 | 160.8 | 68 | 5 | 15486 | Phospholipase A2 (C. rhodostoma) |
|
P0CB14 |
133.27 |
20 |
7 |
47446 |
Snake venom metalloproteinase kistomin (C. rhodostoma) |
|
C. rhodostoma crude venom against MPAV | |||||
| P81397 | 137.25 | 83 | 8 | 15962 | Snaclec rhodocetin subunit alpha (C. rhodostoma) |
| P81382 | 166.86 | 46 | 16 | 58221 | L-amino-acid oxidase (C. rhodostoma) |
| P81398 | 150.91 | 59 | 10 | 15190 | Snaclec rhodocetin subunit beta (C. rhodostoma) |
| A0A0H3U266 | 150 | 58 | 5 | 15486 | Phospholipase A2 (C. rhodostoma) |
|
P30403 |
181.83 |
22 |
11 |
54006 |
Zinc metalloproteinase/disintegrin (C. rhodostoma) |
|
C. rhodostoma crude venom against RPAV | |||||
| P81397 | 182.81 | 80 | 9 | 15962 | Snaclec rhodocetin subunit alpha (C. rhodostoma) |
| P81398 | 171.56 | 59 | 14 | 15190 | Snaclec rhodocetin subunit beta (C. rhodostoma) |
| A0A0H3U266 | 149.25 | 43 | 4 | 15486 | Phospholipase A2 (C. rhodostoma) |
| Q9PVF4 | 143.51 | 42 | 2 | 15457 | Basic phospholipase A2 homolog W6D49 (C. rhodostoma) |
|
P30403 |
136.12 |
10 |
7 |
54006 |
Zinc metalloproteinase/disintegrin (C. rhodostoma) |
|
C. rhodostoma crude venom against HPAV | |||||
| P81382 | 219 | 70 | 25 | 58221 | L-amino-acid oxidase (C. rhodostoma) |
| P81398 | 159.43 | 70 | 12 | 15190 | Snaclec rhodocetin subunit beta (C. rhodostoma) |
| A0A0H3U266 | 138.8 | 43 | 11 | 15486 | Phospholipase A2 (C. rhodostoma) |
| P81397 | 179.33 | 80 | 11 | 15962 | Snaclec rhodocetin subunit alpha (C. rhodostoma) |
|
P0CB14 |
218.61 |
37 |
24 |
47446 |
Snake venom metalloproteinase kistomin (C. rhodostoma) |
|
C. rhodostoma crude venom against MKAV | |||||
| P81382 | 181.91 | 60 | 19 | 58221 | L-amino-acid oxidase (C. rhodostoma) |
| P81398 | 135.22 | 58 | 8 | 15190 | Snaclec rhodocetin subunit beta (C. rhodostoma) |
| P30403 | 134.28 | 13 | 5 | 54006 | Zinc metalloproteinase/disintegrin (C. rhodostoma) |
| A0A0H3U266 | 132.5 | 43 | 4 | 15486 | Phospholipase A2 (C. rhodostoma) |
| P0CB14 | 127.08 | 21 | 10 | 47446 | Snake venom metalloproteinase kistomin (C. rhodostoma) |
|
Q9PVF4 |
124.81 |
42 |
2 |
15457 |
Basic phospholipase A2 homolog W6D49 (C. rhodostoma) |
|
C. rhodostoma crude venom against BKAV | |||||
| A0A0H3U266 | 204.27 | 52 | 3 | 15486 | Phospholipase A2 (C. rhodostoma) |
| P81398 | 199.58 | 59 | 7 | 15190 | Snaclec rhodocetin subunit beta (C. rhodostoma) |
| P30403 | 149.41 | 13 | 5 | 54006 | Zinc metalloproteinase/disintegrin (C. rhodostoma) |
| P0CB14 | 148.7 | 15 | 9 | 47446 | Snake venom metalloproteinase kistomin (C. rhodostoma) |
| P81382 | 134.69 | 27 | 8 | 58221 | L-amino-acid oxidase (C. rhodostoma) |
3.6. C. purpureomaculatus antigenic proteins
The proteins identified from all C. purpureomaculatus interactions were summarised in Table 6. PLA2 was identified as the common antigenic protein across all interactions of C. purpureomaculatus venom with six antivenoms. Snaclec rhodocetin subunit beta was consistently identified in every interaction except with RPAV and BKAV, while snaclec rhodocetin subunit alpha was present in three interactions: CAV, MPAV, and MKAV. Snaclec coagulation factor was detected in the interaction of the crude venom with CAV, MPAV, and RPAV. LAAO was only identified in interactions with MKAV and HPAV. Zinc metalloproteinase/disintegrin was solely identified in interactions with HPAV. All identified proteins contain at least one unique peptide sequence, with −10logP values exceeding 40. These proteins were matched with the Cryptelytrops species, a subset of the Trimeresurus family, in the database.
Table 6.
Proteins identified via LC-MS/MS from the C. purpureomaculatus crude venom against, CAV, MPAV, RPAV, HPAV. MKAV, and BKAV. Cobra antivenom (CAV), Malayan pit antivenom (MPAV), Russells pit antivenom (RPAV), Hemato-polyvalent antivenom (HPAV), Malayan krait antivenom (MKAV), Banded krait antivenom (BKAV).
| Accession | −10lgP | Coverage (%) | Unique | Avg. Mass | Description (OS) |
|---|---|---|---|---|---|
|
C. purpureomaculatus crude venom against CAV | |||||
| P0DJL3 | 155.05 | 85 | 9 | 14498 | Snaclec purpureotin subunit beta (T. purpureomaculatus) |
| A0A0H3U1W4 | 137.69 | 57 | 1 | 15895 | Phospholipase A2 (T. albolabris) |
|
Q71RR4 |
131.02 |
42 |
4 |
17092 |
Snaclec coagulation factor IX/factor X-binding protein subunit A (T. stejnegeri) |
|
C. purpureomaculatus crude venom against MPAV | |||||
| A0A0H3U245 | 166.9 | 51 | 3 | 15197 | Phospholipase (T. albolabris) |
| P0DJL3 | 129.87 | 53 | 7 | 14498 | Snaclec purpureotin subunit beta (T. purpureomaculatus) |
| Q71RR4 | 115.37 | 43 | 5 | 17092 | Snaclec coagulation factor IX/factor X-binding protein subunit A (T. stejnegeri) |
|
P0DJL2 |
102.04 |
52 |
3 |
15613 |
Snaclec purpureotin subunit alpha (T. purpureomaculatus) |
|
C. purpureomaculatus crude venom against RPAV | |||||
| A0A0H3U1W4 | 141.04 | 51 | 1 | 15895 | Phospholipase A2 (T. albolabris) |
|
Q71RR4 |
146.93 |
55 |
6 |
17092 |
Snaclec coagulation factor IX/factor X-binding protein subunit A (T. stejnegeri) |
|
C. purpureomaculatus crude venom against HPAV | |||||
| A0A0H3U267 | 132.93 | 36 | 5 | 15239 | Phospholipase A2 (T. albolabris) |
| P0DJL3 | 128.15 | 60 | 1 | 14498 | Snaclec purpureotin subunit beta (T. purpureomaculatus) |
| Q6WP39 | 120.49 | 15 | 2 | 58601 | L-amino-acid oxidase (T. stejnegeri) |
|
P0C6E8 |
121.69 |
14 |
9 |
48204 |
Zinc metalloproteinase/disintegrin (Fragment) (T. gramineus) |
|
C. purpureomaculatus crude venom against MKAV | |||||
| A0A0H3U245 | 161.1 | 59 | 3 | 15197 | Phospholipase A2 (T. albolabris) |
| P0DJL3 | 112.96 | 37 | 5 | 14498 | Snaclec purpureotin subunit beta (T. purpureomaculatus) |
| Q6WP39 | 111.82 | 14 | 1 | 58601 | L-amino-acid oxidase (T. stejnegeri) |
|
P0DJL2 |
118.9 |
52 |
5 |
15613 |
Snaclec purpureotin subunit alpha (T. purpureomaculatus) |
|
C. purpureomaculatus crude venom against BKAV | |||||
| A0A0H3U239 | 133.98 | 17 | 3 | 15816 | Phospholipase A2 (T. erythrurus) |
4. Discussion
The current study focuses on the identification of antigenic proteins from five medically significant snakes in Malaysia using immunoprecipitation assay and LC-MS/MS. It is essential to analyse the venom composition to further understand the effects of envenomation and how can the treatment of snakebite be improved. The current antivenom that are available are either species specific (monovalent: raised against one crude venom) or syndromic antivenom (polyvalent: raised against snake venom with similar envenomation symptoms). Studying the venom composition allows us to identify and categorise them into the correct family and genus while also providing insights into the evolutionary history and adaptation of the snakes [15]. In this case, the information gathered from venom analysis was used to identify the antigenic proteins that are present in the five medically significant Malaysian venomous snakes (Ophiophagus hannah, Naja sumatrana, Naja kaouthia, Calloselasma rhodostoma, and Cryptelytrops purpureomaculatus). The immunoprecipitation assay is a technique utilised to study protein-protein interactions, based on the principle of antibody binding to the protein of interest. It involves the use of a medium, such as agarose, through which proteins and antibodies can diffuse. This facilitates the binding of both proteins and antibodies to form a precipitate, typically observed as a white band [16]. This approach enables the detection of protein binding, as well as the screening and purification of the protein complex that forms the band [16]. One of the main advantages of this approach is its simplicity and cost-effectiveness. Moreover, it is reproducible, as demonstrated by its repeated use with various venom and antivenom combinations in this study.
After observing the interaction on the agarose assay, LC-MS/MS was employed to identify the proteins bound to the antibodies from the antivenom. This approach was chosen due to its superior sensitivity compared to other methods such as gel electrophoresis or enzyme-linked immunosorbent assay (ELISA) [17]. LC-MS/MS operates with high-speed, high-throughput, and automated processing, facilitating the deep detection of trace protein components [18,19], crucial for identifying all proteins digested from the gel. Numerous studies have utilised the LC-MS/MS approach to profile snake venoms and determine the relative abundance of protein compositions [10,20]. For example, proteins from two pit vipers, Tropidolaemus wagleri and Cryptelytrops purpureomaculatus, were identified, and relative abundance of the venom proteins was detected using LC-MS/MS [10]. Prior to that, the venom proteome of Naja haje, a cobra species, was documented using chromatographic fractionation and Nano ESI-liquid chromatography and tandem mass spectrometry (nano-ESI-LCMS/MS) [18].
4.1. Crude venom and antivenom cross reactivity
The white band (Fig. 1) formed in the immunoprecipitation assay confirms there are interactions between the protein from the venom and the antibodies from the antivenom. The formation of the white band in the immunoprecipitation assay represents visual confirmation of interactions between the venom protein and the antibodies from the antivenom [21]. This assay demonstrates antivenom cross-reactivity, where the antivenom recognises a linear epitope on the protein originating from multiple snake species. A linear epitopic element may cover either the entire epitope sequence or a partial sequence that potentially folds into a structure mimicking the conformational epitope, fitting the paratopes of the antibodies. In polyclonal antivenom development, cross-reactivity refers to the antivenom's ability to effectively bind with antigens from various snake species, achieved through immunisation with venoms from diverse snake species [22].
Proposed theories of cross-reactivity suggest that monoclonal antibodies may possess cross-binding abilities toward multiple proteins due to antibody tolerance regarding antigen variation [23] or that polyclonal antibodies constitute a pool of monoclonal antibodies capable of binding to all toxins present in the venom [22]. While other studies on cross-reactivity have been conducted [[24], [25], [26]], the precise mechanism contributing to these observations remains unknown. Despite the undetermined mechanism, the concept of cross-reactivity holds potential benefits for antivenom production.
Among the eight antivenoms used in this study, six of them are monovalent (monospecific) antivenom; KCAV, CAV, MPAV, RPAV, MKAV, and BKAV, while the remaining two are polyvalent (polyspecific) antivenoms; NPAV and HPAV. Monospecific antivenoms target venom from a single species, whereas polyvalent antivenoms are effective against multiple species [27,28]. CAV, RPAV, and MKAV exhibited positive interactions across all venoms, potentially driven by a short motif of conserved residues present across the identified proteins of all five venoms, regardless of their families [29]. KCAV showed positive interaction with venoms from O. hannah, N. sumatrana, and N. kaouthia. The monospecific antibodies in KCAV are specific to proteins in O. hannah, as it was raised against its crude venom. Interestingly, KCAV also demonstrated positive interactions with two other vipers, due to the presence of heterologous proteins with similar antigenic properties as the venom used during immunisation [30]. These heterologous venom proteins may fold to fit the structure of the paratope, leading to neutralisation by the antibodies [21].
MPAV and BKAV exhibited positive interactions for the immunoprecipitation assay, except with O. hannah crude venom. Although these antivenoms did show positive interactions with the other two cobras, N. sumatrana and N. kaouthia, they failed to form a white precipitate with O. hannah. These absence of may be due to the lack of epitope recognition by the antibodies, as Ophiophagus hannah is not considered a "true cobra" [31]. Even though O. hannah comes under the cobra family, they may not possess similar proteins to the other two cobras in this study. Hence why MKAV and BKAV showed positive interactions with both the cobras but not O. hannah. Due to evident taxonomic differences, they possess distinct venom proteins leading to differing results from those observed with the other two cobras. King cobra anatomically differs from members of the Naja genus and is more closely related to the mambas [32]. It is expected that KCAV would exhibit a positive interaction with all the cobras as it was raised against the king cobra crude venom.
NPAV and HPAV, are syndromic polyvalent antivenom designed to address snake envenomation symptoms manifested by the victims [28,33]. NPAV comprises immunogens from Naja kaouthia, Ophiophagus hannah, and Bungarus fasiatus [34] and Bungarus candidus. NPAV demonstrated a positive interaction with all three elapids and negative interaction with the vipers. NPAV targets envenomation by elapids, whose venom primarily consists of neurotoxins and cytotoxins, aligning with the assay results from our present study [28,34]. The antibodies in NPAV can bind to the heterologous antigenic venom proteins in the cobras [28]. Due to the nature of the NPAV and the venom from elapids, the findings obtained from the immunoprecipitation assay corresponds to the function of NPAV.
HPAV, produced using venoms from Daboia russellii, Calloselasma rhodostoma, and Trimeresurus albolabris, targets hematologic disorders typically presented in viper envenomation victims, characterised by incoagulable blood and prolonged clotting time [35]. This is due to the composition of viper venom that are predominantly hemotoxic. The HPAV interaction from our present study showed positive reaction with both the vipers used in this study and the positive reactions are anticipated, as C. rhodostoma venom was used in its production. Since C. purpureomaculatus was formerly classified as Trimeresurus purpureomaculatus, it shares similar proteins with the crude venom used to produce hematopolyvalent antivenom. The proteins in C. purpureomaculatus have much similar to proteins found within the Trimeresurus family venom such as SVMP and PLA2 [10]. While cross-reactivity between venoms and antivenoms was observed in our present study, identifying the proteins responsible for these interactions is crucial for venomics research. This information could be instrumental in fortifying plasma-derived antivenoms with monoclonal antibodies, potentially broadening neutralisation capacity or enhancing efficacy against key toxins. Improving antivenom specificity can reduce adverse effects and have broader applicability across different demographics.
4.2. Antigenic protein across cobra and pit viper venom
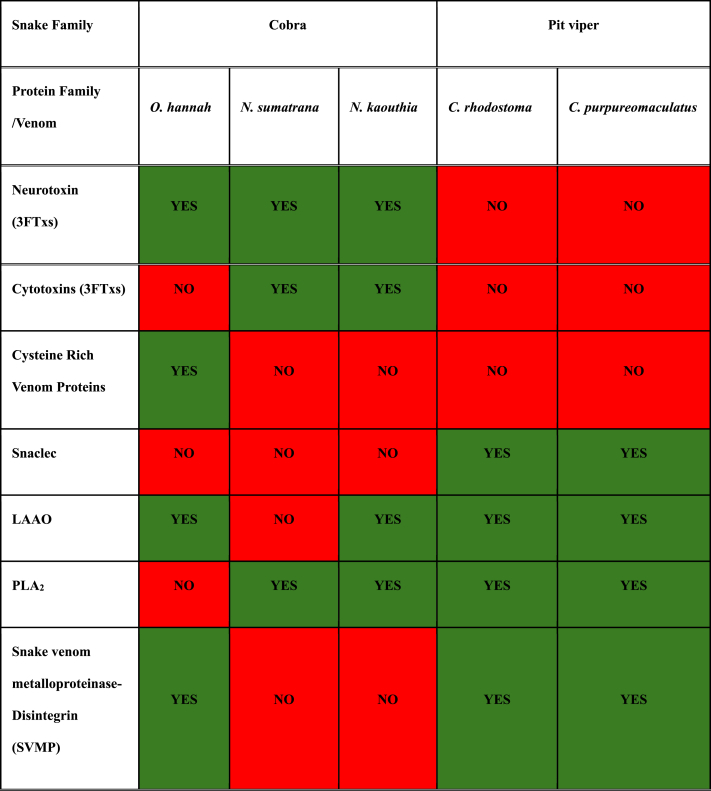
Snake envenomation leads to various pathological effects, including neurotoxicity, hemotoxicity, and cytotoxicity at the bite site [36]. These diverse clinical manifestations are due to the variations in the toxin constituents within the venom. Our LC-MS/MS analysis of the protein bands from the immunoprecipitation assay demonstrated antigenic proteins specific to certain snake families and species (see Table 7). We identified eight antigenic protein families across the five species studied. However, distinct differences were identified in the antigenic proteins found in cobras and pit vipers alongside shared proteins. 3FTx (neurotoxins) and CRVP are antigenic in elapids, reflecting the prominent neurotoxic effects of cobra envenomation [37]. Although elapids exhibit similar symptoms, CRVP was only found to be antigenic in king cobra interactions. Conversely, proteins specific to pit vipers, such as Snaclec and SVMP (P-I and P-II class), were identified. However, despite varying post-envenomation effects, PLA2 and LAAO emerge as common antigenic proteins in both cobras and pit vipers. Despite the similarity in names, venom proteins can exhibit diverse immunogenicity, affecting their ability to bind to antibodies. Hence, identifying the antigenic properties in venom holds promise for developing effective, broad-spectrum antivenoms.
Table 7.
Common antigenic proteins across all venom and antivenom interactions. The green highlighted boxes indicate presence of the antigenic protein, and the red highlighted boxes indicate an absence of the antigenic protein from the interactions. King cobra antivenom (KCAV), Cobra antivenom (CAV), Malayan pit antivenom (MPAV), Russells pit antivenom (RPAV), Hemato-polyvalent antivenom (HPAV), Neuro-polyvalent antivenom (NPAV), Malayan krait antivenom (MKAV), Banded krait antivenom (BKAV).
4.3. Common antigenic proteins in cobra
Our study identified 3FTXs and cytotoxins as the common antigenic protein across the three cobras. The clinical manifestations of cobra envenomation include severe neurotoxic effects such as limb weakness, respiratory muscle paralysis and paraesthesia [38]. Major venom families found in king cobra include LAAO, metalloprotease, CRVP, PLA2 and 3FTxs (Chang, Tsai, and Tsai 2013). In the Naja genus, the key toxins are PLA2, neurotoxins, and cytotoxins/cardiotoxins (Yap et al., 2014). These cobra proteins are responsible for the above biological symptoms onto the victim.
4.3.1. Three-finger toxins (3FTxs)
3FTxs comprises of three parallel β-sheets, with multiple disulphide bonds. The specific biological effects induced by 3FTxs heavily depend on the toxin subtype and the target receptor or ion channel [39] inducing neurotoxicity, cardiotoxicity anticoagulation and cytotoxicity [40]. Additionally, the interaction of 3FTx with the lipid bilayer of the cell leads to the disruption of the cell membrane and subsequent physiological changes in cell metabolism [41,42]. The three main types of 3FTX are short-chain, long-chain and nonconventional 3FTx [43]. On average, 3FTx constitutes approximately 51.3 % of the venom composition in elapids [39].
4.3.1.1. Neurotoxin
There are two kinds of neurotoxins that are particularly harmful to the human body: A) α-neurotoxins, which acts as a non-enzymatic acetylcholine receptor (AchrRs) blockers and B) β-neurotoxins, which functions as a pre-synaptic phospholipase A2 [44]. α-neurotoxins, classified into three main groups-long chain, short chain, and non-conventional α-neurotoxins are post-synaptically active neurotoxins that bind to the post-synaptic muscle nAChRs. Generally, α-neurotoxins’ binding are irreversibly even when administering of antivenom or acetylcholinesterase inhibitors (AChEIs) [45]. Neurotoxin proteins in snake venom target the nervous system, leading to muscle paralysis, respiratory failure, and neurological symptoms [45].
4.3.1.2. Cytotoxin
Various isoforms of cytotoxins were identified from the interaction of Naja venom against all the antivenom. Cytotoxins constitute approximately 40–70 % of Naja venom and they are highly amphipathic proteins. These cytotoxins mediate a range of biological processes, including depolarizing excitable membranes of heart cells and neurons, modulating the activity of membrane-bound enzymes, inhibiting platelet aggregation, inducing haemolytic and cytotoxic activity, and triggering a cardiac arrest [44,46,47]. The majority cytotoxin-mediated toxicity arises from their ability to bind to cell membranes, subsequently altering the structure and function of the lipid bilayer [44]. Cytotoxins act on various cell types, such as red blood cells, lymphocytes, tumour cells, spleen cells and cardiac myocytes. However, the pathological effects depend on the proteins found on the cell membrane and the phospholipids on the outer part of the plasma membrane. Generally, cytotoxins kill cells by disrupting the cell membranes [48]. The predominant antigenic proteins in the Naja venom are cytotoxins of various isoforms causing effects such as oedema, severe blistering, and necrosis. Furthermore, these effects can lead to secondary injuries including limb loss due to severe local tissue damage [44,49]. Targeting these proteins while formulating an antivenom may help prevent rapid swelling, enlargement of lymph nodes, or necrosis at the bite site.
4.3.1.3. Β-cardiotoxin
β-cardiotoxin from O. hannah is a relatively new protein identified in 2007, exhibiting a distinct structure and function compared to other cardiotoxins [50]. Despite sharing similar sequences with conventional cytotoxins, β-cardiotoxin as an individual protein is less harmful than cytotoxins. β-cardiotoxin has been shown to decrease heart rate while acting by directly acting on cardiac tissue [50]. The combined action of various 3FTxs has been postulated to produce synergistic antagonising effects, leading to neuromuscular paralysis along respiratory failure in snakebite victims [51]. Inhibitors of 3FTx include polyphenols present in plants, which can prevent the activity of 3FTx by binding to the toxin itself, thus blocking their interaction with their respective receptors [52]. Epigallocatechin-3-gallate (EGCG), a polyphenol, successfully blocked the activity of alpha cobra toxin by binding to the toxin, thereby preventing adherence to the acetylcholine receptor [52]. When the toxin fails to bind to the receptor, minimal neurotoxicity, cardiotoxicity, anticoagulation, and cytotoxicity occurs. Hence, by identifying 3FTx as one of the antigenic proteins, we can target this protein family in developing therapeutics for snake envenomation.
4.4. Common antigenic proteins in pit vipers
The common antigenic proteins identified across both pit vipers in this study are SVMP and snaclecs, which align with previous investigations on viper envenomation effects [53,54]. Viper venom induces hemotoxic and myotoxic effects in the victims. The major proteins responsible for these clinical manifestations include PLA2, which causes local inflammation; metalloprotease, leading to haemorrhaging and degradation of fibrinogen; and snaclecs, inhibiting platelet activation [55]. Viperid venoms typically have a relatively long half-life, potentially prolonging patient recovery [54,56]. These proteins are crucial in causing the symptoms observed in victims’ post-envenomation.
4.4.1. Snake C-type lectins (Snaclecs)
Snaclecs have a basic heterodimeric structure with two subunits α and β [57], such as rhodocetin, and both subunits were separately identified in the LC-MS/MS analysis. In our present study, snaclecs were exclusively identified in the viper samples of C. rhodostoma and C. purpureomaculatus. In the C. rhodostoma sample, only snaclec rhodocetin subunit beta was identified in the interaction against CAV, MKAV and BKAV. In contrast, both subunits were identified in the interactions against MPAV, RPAV and HPAV. The snaclec protein from C. rhodostoma has dual effects on platelet aggregation. Rhodocetin (four subunits) is a platelet aggregation inhibitor, while rhodocytin (two subunits) induces it. For C. purpureomaculatus samples, the interaction against CAV and HPAV only identified snaclec rhodocetin subunit beta. In contrast, both subunits were found to be antigenic in the reaction against MPAV and MKAV. This protein is a potent inhibitor of collagen-induced platelet aggregation; it binds to an integrin α2A domain, blocking collagen from binding to the integrin [58]. It is possible that some antibodies could only bind to the beta subunits of this protein, but antibodies from other antivenom could bind to the alpha subunit of Snaclecs. This could be due to difference the structure of the subunit or the position of the subunits itself.
Currently, there are no small molecules or drugs specifically targeting snaclec rhodocetin or snaclec rhodocytin. Theoretically, an inhibitor for snaclec rhodocetin could allow platelets to adhere to each other and form a haemostatic plug, thereby preventing bleeding that may occur during envenomation. This inhibitor could target the protein or its binding site, integrin alpha2A domain. Similarly, a rhodocytin inhibitor could prevent platelet aggregation by potentially binding to the protein or its specific binding site, C-type lectin domain family 1 member B (CLEC1B/CLEC2). Inhibiting the activities of rhodocytin could prevent the formation of blood clots during envenomation.
4.5. Common antigenic proteins in cobras and pit vipers
In this study, we examined the crude venom from two distinct families, each associated with different clinical manifestations following envenomation. Despite this divergence, we have successfully identified common antigenic proteins across both families, notably LAAO and PLA2. While LAAO is more prevalent in cobras, viper venom also contains trace amounts of this enzyme [59]. Conversely, PLA2 constitutes a sizeable portion of cobra venom composition. Despite the difference in symptoms, pit vipers and cobras share common antigenic proteins, this could be because snake venoms' composition and function can differ between inter and intra-species [31]. Recognising these shared antigenic proteins is crucial, as it provides insights that could assist the development of novel treatment approaches for envenomation cases.
4.5.1. L-amino acid oxidase (LAAO)
Snake venom LAAO poses significant harm to the human body, inducing red blood cell haemolysis and extensive bleeding by compromising blood vessel integrity, leading to haemorrhage [60]. LAAO are also capable of inducing programmed cell death in various cells and tissues resulting in severe organ damage. LAAOs also induce programmed cell death in various tissues, resulting in severe organ damage and increased vascular permeability, leading to fluid leakage into interstitial spaces [60]. Variants like TM-LAAO from Cryptelytrops mucrosquamatus and LAAO from O. hannah have been reported to be edematogenic [61]. Although the precise mechanism of LAAO-induced oedema remains unclear, it likely differs from typical toxin-mediated inflammatory mediator release, as antihistamines do not alleviate its effects [62]. LAAO triggers local effects such as swelling and necrosis and systemic effects like disseminated intravascular coagulation (DIC) and haemorrhaging, which are consistent with king cobra envenomation symptoms.
Given the conserved structure of LAAO, inhibitors targeting its binding site can be designed using its protein sequence. Aristolochic acid from Aristolochia indica has shown inhibitory effects on LAAO from Russell's viper venom by inducing cell genotoxicity [63]. Subsequently, derivatives of aristolochic acid were synthesised to reduce toxicity while retaining inhibitory activity against LAAO. These derivatives significantly decreased reactive oxygen species induced by LAAO without affecting DNA [64]. Therefore, Inhibiting LAAO holds significant promise in antivenom development due to its crucial role in venom-induced pathogenesis. By targeting LAAO, it's possible to mitigate the destructive effects of snake envenomation, such as haemolysis, bleeding, tissue necrosis, and systemic complications like DIC.
4.5.2. Phospholipase A2 (PLA2)
PLA2 was identified in two of N. sumatrana interactions (RPAV and CAV), and in all the pit viper samples. However, the PLA2 found in the elapids is a neutral PLA2 muscarinic inhibitor. This specific snake venom PLA2 inhibits the muscarinic acetylcholine receptors (mAChR) and catalyses the calcium-dependent hydrolysis of 2-acyl groups (amides and esters) in 3-sn-phosphoglyceride [65]. While this protein shares high homology with PLA2 from the Naja family, it is the first to be identified as a muscarinic inhibitor [65]. In contrast, the functional role of PLA2 identified in pit vipers remains to be elucidated. Nevertheless, PLA2 generally exerts diverse toxic effects, which can be categorized into three main groups: neurotoxins, myotoxins, and haemostasis-impairing toxins [66].
Given the wide range of toxic effects attributed to PLA2, targeting this compound in snake venom is crucial for developing effective treatments. PLA2 inhibitors have been investigated extensively, including compounds derived from plant extracts, steroids, synthetic molecules, and phenolic compounds [67]. For example, the aqueous extract of Casearia sylvestris has demonstrated protective effects against PLA2-induced damage from several snake species, including Bothrops moojeni, B. pirajai, B. neuwiedi, and B. jararacussu [68,69]. Additionally, synthetic inhibitors like Edunol, a derivative of isoflavonoids, have exhibited anti-PLA2 activities against B. jararacussu venom [70,71]. Further research in this area holds promise for developing potent PLA2 inhibitors for treating snake envenomation.
4.5.3. Snake venom metalloproteinase (SVMP)
SVMP are zinc-dependent enzymes predominantly found in vipers but also present in elapids, play a crucial role in snake venom toxicity. This study identified SVMP as antigenic in both vipers and king cobra. These enzymes can be classified into three classes based on size and structure [72]. The P-I class has a single catalytic metalloproteinase domain and exhibits less severe toxic effects [73]. In contrast, the P-II class has an additional disintegrin domain, influencing its integrin-binding motif and domain composition [72]. The effects of the P-II proteins heavily depend on the integrin-binding motif and domain composition [72]. Lastly, the P-III class contains metalloproteinase, disintegrin-like, and cysteine-rich domains, contributing to potent haemorrhagic activities. The variety of structure in P-III class is due to proteolytic cleavage, occurrence of other ancillary domains and continual domain loss [72,74].
The LC-MS/MS analysis from the present study (Table 2, Table 5, Table 6) revealed the presence of SVMP with a disintegrin, (Zinc metalloproteinase-disintegrin-like ohanin, Zinc metalloproteinase/disintegrin, Snake venom metalloproteinase kistomin) suggesting membership in the SVMP P-II subclass. Zinc metalloproteinase-disintegrin-like ohanin inhibits adenosine diphosphate (ADP), disrupting the platelet aggregation [75]. Zinc metalloproteinase/disintegrin from C. rhodostoma is also known as snake venom metalloproteinase rhodostoxin and it impairs the envenomed victims’ homeostasis [76]. Snake venom metalloproteinase kistomin from C. rhodostoma also inhibits the platelet aggregation by blocking the adhesion of platelets immobilised collagen [77].
Neutralising the haemorrhagic effects of SVMPs is crucial in snakebite treatment. Antibodies, small molecules, or protease inhibitors targeting these proteins can help mitigate their effects. Matrix metalloprotease inhibitors (MMPIs), such as marimastat and batimastat, have shown promise in reducing haemorrhage, necrosis, and oedema induced by SVMPs [78]. Marimastat inhibits lethal and haemorrhagic effects from E. ocellatus venom, while batimastat potentially treats local haemorrhage and dermonecrosis post-Bothrops asper envenomation [79,80]. Further research in this area is essential for developing effective treatments for snake envenomation.
5. Conclusion
In this study, we successfully identified the antigenic proteins in the venoms of five snake species By using immunoprecipitation coupled with LC-MS/MS, we identified key antigenic proteins from pit vipers and cobras, revealing distinct venom components such as 3FTXs in cobras and Snaclecs in pit vipers. The presence of common proteins, including LAAO, PLA2, and SVMP, across both families suggests potential targets for developing broader spectrum antivenoms. Current findings from the conventional antivenoms primarily target antigenic toxins; however, proteins not recognized by these antivenoms pose a challenge for developing new treatments. It is also important to note that several antigenic proteins do not necessarily cause clinical manifestations, adding complexity to the development of effective therapeutic interventions. The immunoprecipitation assay, despite its advantages in reproducibility and cost-efficiency, has potential cross-contamination risks, emphasizing the need for careful dissection and sample handling. Nonetheless, by understanding the specific toxic antigenic proteins involved in snake envenomation, we can potentially enhance the efficacy and safety of snakebite therapy. Further research is essential for advancing snakebite management and reducing the global burden of snakebite-related morbidity and mortality.
Funding
This work was supported by theMalaysia Ministry of Higher Education, Fundamental Research Grant Scheme (FRGS) 2022 (Ref: FRGS/1/2022/SKK10/MUSM/03/2), the Royal Society of Tropical Medicine and Hygiene (RSTMH) Early Career Grant Award 2020 and Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, 2020 Seed Grant.
Data availability statement
The data has not been deposited into a publicly available repository. The data is included in the article/supplementary material/referenced in the article.
CRediT authorship contribution statement
Preetha Rajendiran: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Rakesh Naidu: Writing – review & editing, Validation, Supervision. Iekhsan Othman: Validation, Supervision. Syafiq Asnawi Zainal Abidin: Writing – review & editing, Validation, Supervision, Project administration, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Syafiq Asnawi Zainal Abidin reports financial support was provided byMalaysia Ministry of Higher Education. Syafiq Asnawi Zainal Abidin reports financial support was provided by Royal Society of Tropical Medicine and Hygiene. Syafiq Asnawi Zainal Abidin reports financial support was provided by Monash University Malaysia Jeffrey Cheah School of Medicine and Health Sciences. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Mr Zainuddin Ismail (Bukit Bintang Enterprise, Perlis) for the snake venom and the Monash University Malaysia Proteomics and Metabolomics Platform staff, Ms Aliaa Idrus, for running the samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37243.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Snakebite Envenoming. Health Topics; Available from: https://www.who.int/health-topics/snakebite#tab=tab_1.
- 2.Ismail A.K. Snakebite and envenomation management in Malaysia. Toxinology: Clinical Toxinology in Asia Pacific and Africa. 2015;2:71–102. [Google Scholar]
- 3.Chippaux J.-P. Guidelines for the production, control and regulation of snake antivenom immunoglobulins. Biologie aujourd'hui. 2010;204(1):87–91. doi: 10.1051/jbio/2009043. [DOI] [PubMed] [Google Scholar]
- 4.Kalita B., Utkin Y.N., Mukherjee A.K. Current insights in the mechanisms of cobra venom cytotoxins and their complexes in inducing toxicity: implications in antivenom therapy. Toxins. 2022;14(12) doi: 10.3390/toxins14120839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adisakwattana P., et al. Venom-gland transcriptomics of the Malayan pit viper (Calloselasma rhodostoma) for identification, classification, and characterization of venom proteins. Heliyon. 2023;9(5) doi: 10.1016/j.heliyon.2023.e15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Silva H.A., Ryan N.M., de Silva H.J. Adverse reactions to snake antivenom, and their prevention and treatment. Br. J. Clin. Pharmacol. 2016;81(3):446–452. doi: 10.1111/bcp.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sriapha C., et al. Early adverse reactions to snake antivenom: poison center data analysis. Toxins. 2022;14(10):694. doi: 10.3390/toxins14100694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patiño R.S.P., et al. Bothrops atrox from Ecuadorian Amazon: initial analyses of venoms from individuals. Toxicon. 2021;193:63–72. doi: 10.1016/j.toxicon.2021.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Alangode A., Rajan K., Nair B.G. Snake antivenom: challenges and alternate approaches. Biochem. Pharmacol. 2020;181 doi: 10.1016/j.bcp.2020.114135. [DOI] [PubMed] [Google Scholar]
- 10.Abidin S.Z., et al. Proteomic characterization and comparison of Malaysian Tropidolaemus wagleri and Cryptelytrops purpureomaculatus venom using shotgunproteomics. Toxins. 2016;8(2016) doi: 10.3390/toxins8100299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunalan S., et al. Proteomic characterization of two medically important Malaysian snake venoms, Calloselasma rhodostoma (malayan pit viper) and ophiophagus hannah (king cobra) Toxins. 2018;10(11):434. doi: 10.3390/toxins10110434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhury M., et al. Comparison of proteomic profiles of the venoms of two of the ‘Big Four’ snakes of India, the Indian cobra (Naja naja) and the common krait (Bungarus caeruleus), and analyses of their toxins. Toxicon. 2017;135:33–42. doi: 10.1016/j.toxicon.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Sanz L., et al. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J. Proteonomics. 2008;71(1):46–60. doi: 10.1016/j.jprot.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Dingwoke E.J., et al. Venom proteomic analysis of medically important Nigerian viper Echis ocellatus and Bitis arietans snake species. Biochem Biophys Rep. 2021;28 doi: 10.1016/j.bbrep.2021.101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan C.H. Snake venomics: fundamentals, recent updates, and a look to the next decade. Toxins. 2022;14(4) doi: 10.3390/toxins14040247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alhabbab R.Y. In: Basic Serological Testing. Alhabbab R.Y., editor. Springer International Publishing; Cham: 2018. Precipitation and agglutination reactions; pp. 23–30. [Google Scholar]
- 17.Lomonte B., Calvete J.J. Strategies in ‘snake venomics’ aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017;23:26. doi: 10.1186/s40409-017-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malih I., et al. Proteomic analysis of Moroccan cobra Naja haje legionis venom using tandem mass spectrometry. J. Proteonomics. 2014;96:240–252. doi: 10.1016/j.jprot.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Sharma M., et al. Unveiling the complexities of Daboia russelii venom, a medically important snake of India, by tandem mass spectrometry. Toxicon. 2015;107:266–281. doi: 10.1016/j.toxicon.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 20.Li S., et al. Proteomic characterization of two snake venoms: Naja naja atra and Agkistrodon halys. Biochem. J. 2004;384(Pt 1):119–127. doi: 10.1042/BJ20040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledsgaard L., et al. Antibody cross-reactivity in antivenom research. Toxins. 2018;10(10) doi: 10.3390/toxins10100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Leary M.A., et al. Cross-neutralisation of Australian brown and tiger snake venoms with commercial antivenoms: cross-reactivity or antivenom mixtures? Toxicon. 2007;50(2):206–213. doi: 10.1016/j.toxicon.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Wu X., et al. Maturation and diversity of the VRC01-antibody lineage over 15 years of chronic HIV-1 infection. Cell. 2015;161(3):470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engmark M., et al. High-density peptide microarray exploration of the antibody response in a rabbit immunized with a neurotoxic venom fraction. Toxicon. 2017;138:151–158. doi: 10.1016/j.toxicon.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 25.Lauridsen L.P., et al. Exploring the venom of the forest cobra snake: toxicovenomics and antivenom profiling of Naja melanoleuca. J. Proteonomics. 2017;150:98–108. doi: 10.1016/j.jprot.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Tan C.H., et al. Venom proteome of the yellow-lipped sea krait, Laticauda colubrina from Bali: insights into subvenomic diversity, venom antigenicity and cross-neutralization by antivenom. J. Proteonomics. 2017;166:48–58. doi: 10.1016/j.jprot.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Ganthavorn S. Toxicities of Thailand snake venoms and neutralization capacity of antivenin. Toxicon. 1969;7(3):239–241. doi: 10.1016/0041-0101(69)90012-9. [DOI] [PubMed] [Google Scholar]
- 28.Ratanabanangkoon K. Polyvalent snake antivenoms: production strategy and their therapeutic benefits. Toxins. 2023;15(9):517. doi: 10.3390/toxins15090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peggion C., Tonello F. Short linear motifs characterizing snake venom and mammalian phospholipases A2. Toxins. 2021;13(4) doi: 10.3390/toxins13040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.León G., et al. Current technology for the industrial manufacture of snake antivenoms. Toxicon. 2018;151:63–73. doi: 10.1016/j.toxicon.2018.06.084. [DOI] [PubMed] [Google Scholar]
- 31.Tan C.H., Bourges A., Tan K.Y. King Cobra and snakebite envenomation: on the natural history, human-snake relationship and medical importance of Ophiophagus hannah. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021;27 doi: 10.1590/1678-9199-JVATITD-2021-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueroa A., et al. A species-level phylogeny of extant snakes with description of a new colubrid subfamily and genus. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams D.J., et al. Ending the drought: new strategies for improving the flow of affordable, effective antivenoms in Asia and Africa. J. Proteonomics. 2011;74(9):1735–1767. doi: 10.1016/j.jprot.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 34.Chotwiwatthanakun C., et al. Production of potent polyvalent antivenom against three elapid venoms using a low dose, low volume, multi-site immunization protocol. Toxicon. 2001;39(10):1487–1494. doi: 10.1016/s0041-0101(01)00108-8. [DOI] [PubMed] [Google Scholar]
- 35.Sapsutthipas S., et al. Effective equine immunization protocol for production of potent poly-specific antisera against Calloselasma rhodostoma, Cryptelytrops albolabris and Daboia siamensis. PLoS Neglected Trop. Dis. 2015;9(3) doi: 10.1371/journal.pntd.0003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casewell N.R., et al. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020;41(8):570–581. doi: 10.1016/j.tips.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan C.H., et al. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah) BMC Genom. 2015;16(1):687. doi: 10.1186/s12864-015-1828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kakaria A., Agarwa S., Bagul J. A study of cobra envenomation: clinical features and management. J. Evol. Med. Dent. Sci. 2014;3:12394+. [Google Scholar]
- 39.Hiremath K., et al. Three finger toxins of elapids: structure, function, clinical applications and its inhibitors. Mol. Divers. 2023 doi: 10.1007/s11030-023-10734-3. [DOI] [PubMed] [Google Scholar]
- 40.Srodawa K., et al. Evolution of three-finger toxin genes in neotropical colubrine snakes (colubridae) Toxins. 2023;15(9):523. doi: 10.3390/toxins15090523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peetla C., Stine A., Labhasetwar V. Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery. Mol. Pharm. 2009;6(5):1264–1276. doi: 10.1021/mp9000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyba B., et al. Effects of 3FTx protein fraction from Naja ashei venom on the model and native membranes: recognition and implications for the mechanisms of toxicity. Molecules. 2021;26(8) doi: 10.3390/molecules26082164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gulsevin A., Meiler J. An investigation of three-finger toxin—nAChR Interactions through Rosetta protein docking. Toxins. 2020;12(9):598. doi: 10.3390/toxins12090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasanov S.E., Dagda R.K., Rael E.D. Snake venom cytotoxins, phospholipase A(2)s, and Zn(2+)-dependent metalloproteinases: mechanisms of action and pharmacological relevance. J. Clin. Toxicol. 2014;4(1) doi: 10.4172/2161-0495.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferraz C.R., et al. Multifunctional toxins in snake venoms and therapeutic implications: from pain to hemorrhage and necrosis. Frontiers in ecology and evolution. 2019;7:218. [Google Scholar]
- 46.Harvey A. Shier WT, Mebs D; 1990. Cytolytic Toxins. Handook of Toxicology. [Google Scholar]
- 47.Harvey A. vol. 5. Marcel Dekker; New York: 1991. pp. 85–106. (Handbook of Natural Toxins). [Google Scholar]
- 48.Feofanov Alexei V., et al. Cancer cell injury by cytotoxins from cobra venom is mediated through lysosomal damage. Biochem. J. 2005;390(1):11–18. doi: 10.1042/BJ20041892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dufton M., Hider R. Structure and pharmacology of elapid cytotoxins. Pharmacology & therapeutics. 1988;36(1):1–40. doi: 10.1016/0163-7258(88)90111-8. [DOI] [PubMed] [Google Scholar]
- 50.Rajagopalan N., et al. β‐Cardiotoxin: a new three‐finger toxin from Ophiophagus hannah (king cobra) venom with beta‐blocker activity. Faseb. J. 2007;21(13):3685–3695. doi: 10.1096/fj.07-8658com. [DOI] [PubMed] [Google Scholar]
- 51.Gutiérrez J.M., et al. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017;3(1) doi: 10.1038/nrdp.2017.79. [DOI] [PubMed] [Google Scholar]
- 52.Pithayanukul P., et al. Inhibition of Naja kaouthia venom activities by plant polyphenols. J. Ethnopharmacol. 2005;97(3):527–533. doi: 10.1016/j.jep.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 53.Reid H., et al. 1963. Clinical Effects of Bites by Malayan Viper (Ancistrodon Rhodostomd) [DOI] [PubMed] [Google Scholar]
- 54.Tang E.L.H., et al. Venomics of Calloselasma rhodostoma, the Malayan pit viper: a complex toxin arsenal unraveled. J. Proteonomics. 2016;148:44–56. doi: 10.1016/j.jprot.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Thornton S.L. In: Encyclopedia of Toxicology. third ed. Wexler P., editor. Academic Press; Oxford: 2014. Snakes; pp. 310–312. [Google Scholar]
- 56.Reid H.A., Chan K., Thean P. Prolonged coagulation defect (defibrination syndrome) in Malayan viper bite. Lancet. 1963;281(7282):621–626. doi: 10.1016/s0140-6736(63)91269-8. [DOI] [PubMed] [Google Scholar]
- 57.Clemetson K.J. Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon. 2010;56(7):1236–1246. doi: 10.1016/j.toxicon.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Eble J.A., et al. Alpha 2beta 1 integrin is not recognized by rhodocytin but is the specific, high affinity target of rhodocetin, an RGD-independent disintegrin and potent inhibitor of cell adhesion to collagen. J. Biol. Chem. 2001;276(15):12274–12284. doi: 10.1074/jbc.M009338200. [DOI] [PubMed] [Google Scholar]
- 59.Gutiérrez J.M., et al. The search for natural and synthetic inhibitors that would complement antivenoms as therapeutics for snakebite envenoming. Toxins. 2021;13(7):451. doi: 10.3390/toxins13070451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Izidoro L.F., et al. Snake venom L-amino acid oxidases: trends in pharmacology and biochemistry. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/196754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahn M.Y., Lee B.M., Kim Y.S. Characterization and cytotoxicity of L-amino acid oxidase from the venom of king cobra (Ophiophagus hannah) Int. J. Biochem. Cell Biol. 1997;29(6):911–919. doi: 10.1016/s1357-2725(97)00024-1. [DOI] [PubMed] [Google Scholar]
- 62.Ali S.A., et al. Isolation, structural, and functional characterization of an apoptosis-inducing L-amino acid oxidase from leaf-nosed viper (Eristocophis macmahoni) snake venom. Arch. Biochem. Biophys. 2000;384(2):216–226. doi: 10.1006/abbi.2000.2130. [DOI] [PubMed] [Google Scholar]
- 63.Bhattacharjee P., Bhattacharyya D. Characterization of the aqueous extract of the root of Aristolochia indica: evaluation of its traditional use as an antidote for snake bites. J. Ethnopharmacol. 2013;145(1):220–226. doi: 10.1016/j.jep.2012.10.056. [DOI] [PubMed] [Google Scholar]
- 64.Bhattacharjee P., et al. Aristolochic acid and its derivatives as inhibitors of snake venom L-amino acid oxidase. Toxicon. 2017;138:1–17. doi: 10.1016/j.toxicon.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 65.Miyoshi S.-i., Tu A.T. Phospholipase A2fromNaja naja sputatrixVenom is a muscarinic acetylcholine receptor inhibitor. Arch. Biochem. Biophys. 1996;328(1):17–25. doi: 10.1006/abbi.1996.0137. [DOI] [PubMed] [Google Scholar]
- 66.Tonello F., Rigoni M. In: Cellular Mechanisms of Action of Snake Phospholipase A2 Toxins. Venoms Snake, Inagaki H., et al., editors. Springer Netherlands; Dordrecht: 2017. pp. 49–65. [Google Scholar]
- 67.Carvalho B.M.A., et al. Snake venom PLA2s inhibitors isolated from Brazilian plants: synthetic and natural molecules. BioMed Res. Int. 2013;2013 doi: 10.1155/2013/153045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Borges M.H., et al. Effects of aqueous extract of Casearia sylvestris (Flacourtiaceae) on actions of snake and bee venoms and on activity of phospholipases A2. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000;127(1):21–30. doi: 10.1016/s0305-0491(00)00237-6. [DOI] [PubMed] [Google Scholar]
- 69.Borges M., et al. Neutralization of proteases from Bothrops snake venoms by the aqueous extract from Casearia sylvestris (Flacourtiaceae) Toxicon. 2001;39(12):1863–1869. doi: 10.1016/s0041-0101(01)00169-6. [DOI] [PubMed] [Google Scholar]
- 70.Da Silva A.J., et al. Synthesis and pharmacological evaluation of prenylated and benzylated pterocarpans against snake venom. Bioorg. Med. Chem. Lett. 2004;14(2):431–435. doi: 10.1016/j.bmcl.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 71.Soares A.M., et al. Medicinal plants with inhibitory properties against snake venoms. Curr. Med. Chem. 2005;12(22):2625–2641. doi: 10.2174/092986705774370655. [DOI] [PubMed] [Google Scholar]
- 72.Olaoba O.T., et al. Snake venom metalloproteinases (SVMPs): a structure-function update. Toxicon X. 2020;7 doi: 10.1016/j.toxcx.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suvilesh K., et al. Snake venom proteinases as toxins and tools. Proteases in Physiology and Pathology. 2017:485–515. [Google Scholar]
- 74.Escalante T., et al. Key events in microvascular damage induced by snake venom hemorrhagic metalloproteinases. J. Proteonomics. 2011;74(9):1781–1794. doi: 10.1016/j.jprot.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 75.Guo X.X., et al. Isolation and cloning of a metalloproteinase from king cobra snake venom. Toxicon. 2007;49(7):954–965. doi: 10.1016/j.toxicon.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 76.Au L.C., et al. A common precursor for a putative hemorrhagic protein and rhodostomin, a platelet aggregation inhibitor of the venom of Calloselasma rhodostoma: molecular cloning and sequence analysis. Biochem. Biophys. Res. Commun. 1991;181(2):585–593. doi: 10.1016/0006-291x(91)91230-a. [DOI] [PubMed] [Google Scholar]
- 77.Huang T.F., et al. A novel alpha-type fibrinogenase from Agkistrodon rhodostoma snake venom. Biochim. Biophys. Acta. 1992;1160(3):262–268. doi: 10.1016/0167-4838(92)90086-s. [DOI] [PubMed] [Google Scholar]
- 78.Gutiérrez J.M., Rucavado A. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie. 2000;82(9–10):841–850. doi: 10.1016/s0300-9084(00)01163-9. [DOI] [PubMed] [Google Scholar]
- 79.Arias A.S., Rucavado A., Gutiérrez J.M. Peptidomimetic hydroxamate metalloproteinase inhibitors abrogate local and systemic toxicity induced by Echis ocellatus (saw-scaled) snake venom. Toxicon. 2017;132:40–49. doi: 10.1016/j.toxicon.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 80.Rucavado A., Escalante T., Gutiérrez J.M.a. Effect of the metalloproteinase inhibitor batimastat in the systemic toxicity induced by Bothrops asper snake venom: understanding the role of metalloproteinases in envenomation. Toxicon. 2004;43(4):417–424. doi: 10.1016/j.toxicon.2004.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data has not been deposited into a publicly available repository. The data is included in the article/supplementary material/referenced in the article.