Abstract
Conjugation of the naturally occurring product piplartine (PPT, 1), which is a potent cytotoxic compound and ROS inducer, with a diphenyl sulfonyl-substituted furoxan moiety (namely, 3,4-bis(phenylsulfonyl)-1,2,5-oxadiazole-2-oxide), an important type of NO donor, via an ether linker of different chain lengths is described, characterized and screened for the anticancer potential. The cytotoxicity of the new hybrids was evaluated on a panel of human cancer cell lines (MCF-7, PC3 and OVCAR-3) and two non-cancer human cells (MCF10A and PNT2). In general, the synthesized hybrids were more cytotoxic and selective compared to their furoxan precursors 4–6 and PPT in the above cancer cells. Particularly, PC3 cells are the most sensitive to hybrids 7 and 9 (IC50 values of 240 nM and 50 nM, respectively), while a lower potency was found for the prostate normal cells (IC50 = 17.8 μM and 14.1 μM, respectively), corresponding to selectivity indices of ca. 75 and 280, respectively. NO generation by the PPT–furoxan compounds in PC3 cells was confirmed using the Griess reaction. Furthermore, the cell growth inhibitory effect of 9 was significantly attenuated by the NO scavenger carboxy-PTIO. The intracellular ROS generation by 7 and 9 was also verified, and different assays showed that co-treatment with the antioxidant N-acetyl-l-cysteine (NAC) provided protection against PPT-induced ROS generation. Further mechanistic studies revealed that 7 and 9 had strong cytotoxicity to induce apoptosis in PC3 cells, being mediated, at least in part, by the NO-release and increase in ROS production. Notably, the ability of 9 to induce apoptosis was stronger than that of 7, which may be attributed to higher levels of NO released by 9. Compounds 7 and 9 modulated the expression profiles of critical regulators of cell cycle, such as CDKN1A (p21), c-MYC, and CCND1 (cyclin D1), as well as induced DNA damage. Overall, tethering the furoxan NO-releasing moiety to the cytotoxic natural product PPT had significant impact on the potential anticancer activity and selectivity of the novel hybrid drug candidates, especially 9, as a result of synergistic effects of both furoxan and PPT's ability to release NO, generate ROS, induce DNA damage, and trigger apoptosis.
A novel hybrid integrating piplartine with a furoxan moiety exhibited a sub-micromolar IC50 and extraordinary selectivity for PC3 cells, which was associated with its capacity to release NO, generate ROS, induce DNA damage, and trigger apoptosis.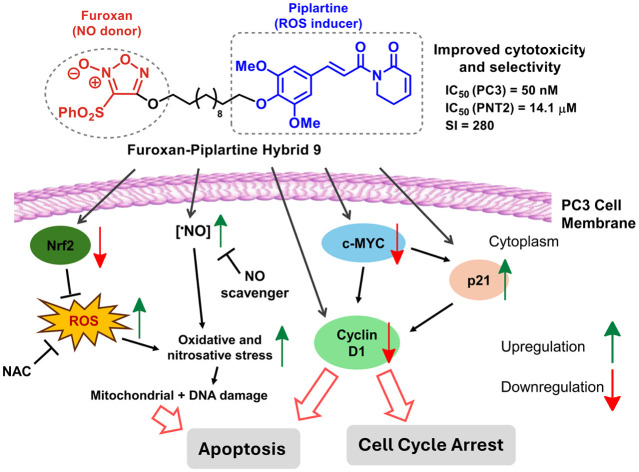
1. Introduction
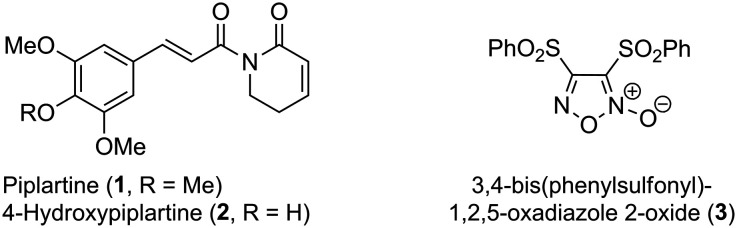
Piplartine (1, PPT, Fig. 1), also known as piperlongumine, is an α,β-unsaturated 2-piperidinone isolated from Piper longum which displays potent cytotoxic activity against several cancer cell lines being highly selective over healthy cells.1,2 Related to PPT (1), its 4-hydroxy analogue (2, 4-HOPPT, Fig. 1) is also a natural product isolated from Piper cenocladum.3
Fig. 1. Chemical structures of natural products piplartine (PPT, 1) and its 4-hydroxy analogue (4-HOPPT, 2), and synthetic furoxan (3).
The antiproliferative activity of piplartine has been extensively reported in the literature,4–10 and it seems to correlate with the increase in reactive oxygen species (ROS) levels and disruption of the cell redox balance.11 An increase in protein glutathionylation by insertion of piplartine between glutathione and a glutathione-binding protein may also be associated with cellular toxicity.12
Several piplartine analogues have been prepared and assessed for their pharmacological profile.12–17 Also, the use of carrier systems to increase piplartine potency and distribution has been described.18–25 To our knowledge, however, few studies have employed the molecular hybridization strategy in order to improve its pharmacological profile.26–30
Nitric oxide (·NO) is a well-known cell-signaling molecule playing a key role in vasodilation and neurotransmission, but its role in cancer is still under debate. It seems that while low levels (pico- to nanomolar range) of nitric oxide enhance tumor proliferation, higher levels (micromolar range) lead not only to cell cycle arrest and apoptosis but also increase tumor sensitivity to chemo-, radio- and immunotherapy in addition to slowing down angiogenesis and metastasis processes.31–41 High levels of NO can also react with ROS to generate reactive nitrogen species (RNS). Some experiments suggest that NO can be used in cancer treatment by adjusting ROS/RNS equilibration, triggering oxidative stress, inducing DNA damage and, ultimately, inducing apoptosis.38–41
Although the use of NO for the treatment of persistent pulmonary hypertension in newborns has been in clinical use for some time, its reactivity and problems associated with the handling of a gaseous species make the use of NO-donors such as organic nitrates, metal–NO complexes and furoxans a better alternative.42–46
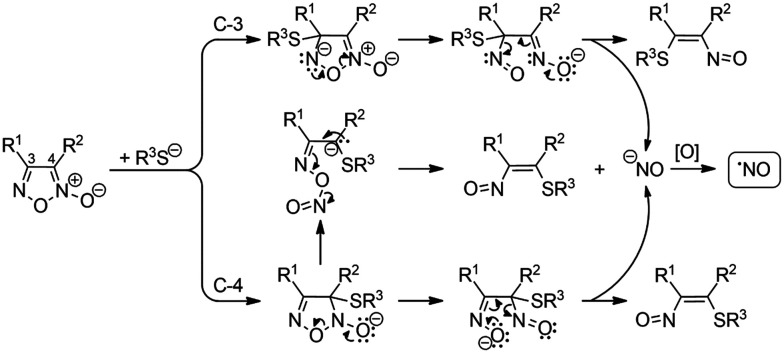
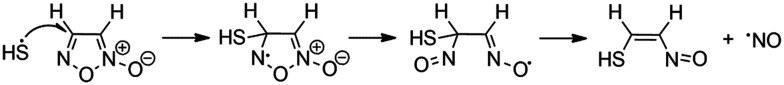
Furoxans (1,2,5-oxadiazole N-oxides) are especially attractive as they liberate NO upon reaction with a nucleophilic thiol group such as the one present in glutathione. Although an ionic mechanism involving nucleophilic addition of a thiolate anion has been proposed to be involved in NO formation from furoxans (Fig. 2), DFT calculation has pointed out that this is a thermodynamically unfavorable process while the radical process is thermodynamically favored (Fig. 3).47,48
Fig. 2. Proposed ionic mechanisms for the formation of NO from furoxans.
Fig. 3. Proposed radical mechanism for the formation of NO from furoxans.
Glutathione (GSH) is an intracellular antioxidant thiol that regulates the redox state preventing cell damage by oxygen and nitrogen reactive species, among others, and is considered to be involved in removing exogenous carcinogens or chemicals formed via oxidative stress. Its elevated concentration in cancer cells has been associated with tumor progression and increased resistance to cancer treatments.49 In the presence of a NO-donor, glutathione may be nitrosylated, and the S-nitrosylglutathione formed may act as a NO reservoir transferring the nitrosyl group to nucleophilic residues of other biomolecules, thus modulating the functioning of redox systems. It can also participate in NO liberation from furoxans.50–52 The high intracellular concentration of NO liberated by furoxans inhibits the expression of P-glycoprotein and, consequently, the efflux of exogenous chemicals as well as the proliferation of multidrug resistant tumor cells.53
Furoxan derivatives known for their ability to release NO have been previously reported for cancer therapy, and a promising strategy to selectively deliver high concentration of NO to tumor cells and reduce damage to healthy cells is to hybridize a thermally stable NO-donor, such as furoxan, with a highly selective antiproliferative molecule.40,54–62
Our previous studies with piplartine (1) and corresponding hybrids and derivatives4,22,28 have led us to consider its hybridization with NO-releasing compounds, particularly 3,4-bis(phenylsulfonyl)-1,2,5-oxadiazole-2-oxide (3, Fig. 1), as a strategy to improve the antiproliferative potency and selectivity of such hybrid structures compared to the natural products against breast (MCF-7), prostate (PC3) and ovary (OVCAR-3) cancer cells. Here, we used non-hydrolysable, conformationally mobile linkers (i.e. an ether linkage with carbon chains of different lengths) to connect the bioactive compounds piplartine (1) and furoxan aiming to retain their corresponding activities in a single molecule.
2. Results and discussion
2.1. Synthesis of the piplartine–furoxan hybrids
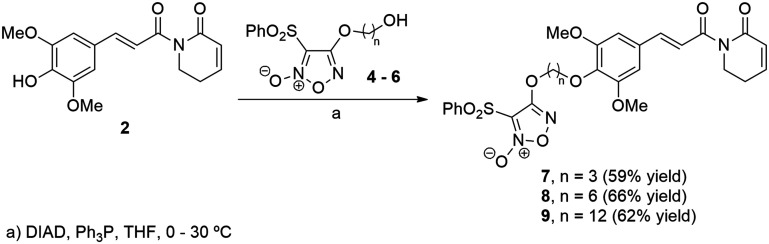
Our approach to the synthesis of PPT–furoxan hybrids was based on using an ether linkage of different chain lengths to isolate the electronic and steric effects of each pharmacophoric group and to allow conformational flexibility for the hybrid structures. For that, we planned to use Mitsunobu reaction, where a primary alcohol derived from furoxan 3 would couple with the phenoxide derived from 4-hydroxypiplartine (2).
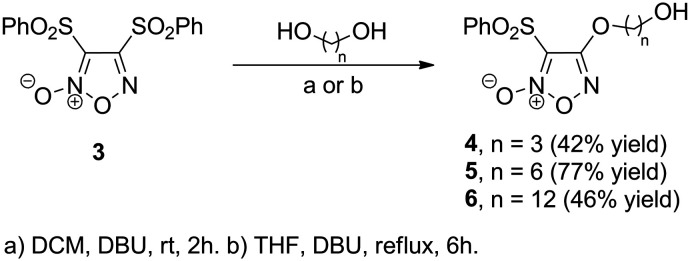
The syntheses of piplartine (1) and 4-hydroxypiplartine (2) through the regioselective C-4 demethylation of 1 have been previously described by us,22,28 while furoxan 3 was obtained according to a literature procedure.63,64 Alcohols 4–6 were obtained from the corresponding diols via aromatic substitution at the C-4 position of the bis-sulfonylfuroxane 3 (Scheme 1).
Scheme 1. Preparation of 4-alkoxyfuroxans 4–6.
The Mitsunobu reaction proved to be a robust methodology to merge the two desired fragments (Scheme 2).65,66 In fact, in the presence of diisopropyl azodicarboxylate (DIAD) and triphenylphosphine (Ph3P), we were able to prepare the hybrid furoxan 7 in 59% yield, after 2 h at 30 °C. The same protocol served well for the synthesis of the hybrid furoxan 8 by using the previously prepared hydroxyfuroxan 5 (Scheme 1). It is worth mentioning that as the regioselective demethylation of the methoxy group at the para position was already reported by our group,28 only phenolic OH in 4-hydroxypiplartine was explored for conjugation chemistry.
Scheme 2. Preparation of hybrid structures of 4-hydroxypiplartine (2) and furoxans 7–9.
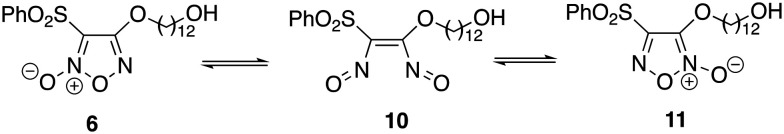
The preparation of PPT-furoxan hybrid 9 containing a 12-carbon tether required hydroxyfuroxan 6 which could not be efficiently prepared under the same reaction conditions as analogues 4 and 5 by using DBU in CH2Cl2 at room temperature (ca. 30% yield). Using such conditions, 6 was isolated together with the regioisomer 11 resulting from the substitution at C-3. This was rationalized by a tautomerization process which involves the formation of the bisnitroso olefin 10 that may undergo cyclization to produce the thermodynamically more stable furoxan 11 (Scheme 3).44,67 Instead, after 6 hours by using THF at reflux compound 6 could be isolated in 46% yield. Then, in THF at 30 °C, hybrid 9 was successfully synthesized in 62% yield.
Scheme 3. Equilibration of furoxans 6 and 11via bisnitroso olefin 10.
2.2. Biological evaluation
2.2.1. In vitro cytotoxicity study
The cytotoxicity of the novel PPT–furoxan hybrids 7–9 was evaluated using the Cell Counting Kit-8 (CCK-8) assay towards three human cancer cell lines (MCF-7: breast adenocarcinoma cells, PC3: prostate adenocarcinoma cells, and OVCAR-3: ovary adenocarcinoma cells) and two human non-cancer cell lines (MCF10A: normal breast cells, and PNT2: normal prostate cells). We have chosen these cell lines considering that breast, ovarian and prostate cancer are very common cancer types and have been the focus of most of our recent studies.28 The cells were incubated with varied concentrations of each compound for 48 h at 37 °C. Cisplatin as a reference compound was selected for comparison, as well as the natural product piplartine (1) and the hydroxyfuroxans 4–6. The obtained results are presented in Table 1.
In vitro cytotoxic activities and selectivity indices (SI) of piplartine–furoxan hybrids 7–9 as well as their precursors hydroxyfuroxans (4–6) and the natural product PPT (1) against different human cell lines using the CCK-8 assay.
| Compounds | IC50a (μM) | SIb | |||||
|---|---|---|---|---|---|---|---|
| MCF-7 | MCF10A | PC3 | PNT2 | OVCAR-3 | SIbreastc | SIprostated | |
| PPT (1) | 5.4 ± 0.4 | 5.3 ± 0.1 | 5.6 ± 0.6 | 8.5 ± 0.3 | 3.5 ± 0.2 | 1.0 | 1.5 |
| 4 | 2.1 ± 0.3 | 1.8 ± 0.7 | 7.2 ± 0.2 | 1.9 ± 0.6 | 3.3 ± 0.5 | 0.9 | 0.3 |
| 5 | 2.0 ± 0.5 | 1.7 ± 0.3 | 3.8 ± 0.6 | 12.9 ± 4.1 | 2.2 ± 0.1 | 0.9 | 3.4 |
| 6 | 4.3 ± 1.0 | 14.8 ± 1.9 | 0.13 ± 0.06 | 17.6 ± 4.0 | 0.04 ± 0.01 | 3.4 | 136.9 |
| 7 | 1.2 ± 0.4 | 3.5 ± 1.3 | 0.24 ± 0.01 | 17.8 ± 3.9 | 0.4 ± 0.1 | 3.0 | 75.5 |
| 8 | 1.1 ± 0.4 | 3.6 ± 0.2 | 0.13 ± 0.06 | 3.3 ± 0.3 | 0.3 ± 0.1 | 3.1 | 26.2 |
| 9 | 7.9 ± 0.4 | 123.3 ± 4.5 | 0.05 ± 0.04 | 14.1 ± 2.2 | 0.12 ± 0.10 | 15.7 | 285.1 |
| Cisplatine | 14.8 ± 4.5 | 5.1 ± 1.3 | 7.5 ± 0.5 | 6.6 ± 0.5 | 14.9 ± 2.1 | 0.3 | 0.9 |
Half maximum inhibitory concentration (IC50) values for each compound were calculated, and the data are expressed as the mean IC50 value (μM) ± SD from two independent experiments performed in triplicate per plate.
SI: selectivity index defined as the ratio between the IC50 value for the normal cell line and the IC50 value for the cancer cell line.
SIbreast = IC50 (MCF10A)/IC50 (MCF-7).
SIprostate = IC50 (PNT2)/IC50 (PC3).
Positive control.
As shown in Table 1, the synthesized hybrid compounds 7–9 had, in general, very high cytotoxicity towards all cancer cells analyzed (MCF-7, PC3, and OVCAR-3 cell lines) with IC50 values ranging from low micromolar to the nanomolar range. Their IC50 values were significantly lower in comparison to that of the reference drug cisplatin (MCF-7: 14.8 ± 4.5 μM, PC3: 7.5 ± 0.5 μM, OVCAR-3: 14.9 ± 2.1 μM) against the three cancer cell lines tested. Significant cytotoxic effects were observed in PC3 cells, and the hybrids 7–9 also showed much higher cytotoxic activity than their corresponding hydroxyfuroxans 4–6 and piplartine (1). More specifically, compound 7 (IC50 = 0.24 ± 0.01 μM) with a 3-carbon linker displayed more potent activity than its precursors hydroxyfuroxan 4 (IC50 = 7.2 ± 0.2 μM) and piplartine (1) (IC50 = 5.6 ± 0.6 μM), which corresponds to a 30- and 23-fold increase in potency toward the PC3 cell line. The PPT–furoxan hybrid 8 (IC50 = 0.13 ± 0.06 μM) with a 6-carbon linker was 29- and 43-fold more cytotoxic than its precursor 5 (IC50 = 3.8 ± 0.6 μM) and piplartine (1), respectively, while hybrid 9 (IC50 = 0.05 ± 0.04 μM) with the longest linker (12-carbon) exhibited approximately 3- and 110-fold higher cytotoxicity in PC3 cells when compared to 6 (IC50 = 0.13 ± 0.06 μM) and piplartine (1).
PPT–furoxan hybrids 7 and 8 also presented higher cytotoxicity than their precursors 4 and 5, respectively, and piplartine (1), against both breast (MCF-7) and ovary (OVCAR-3) cancer cells. A much better cytotoxic profile toward OVCAR-3, however, was observed for hybrids 7 (IC50 = 0.4 ± 0.1 μM) and 8 (IC50 = 0.3 ± 0.1 μM) when compared to 4 (IC50 = 3.3 ± 0.5 μM) and 5 (IC50 = 2.2 ± 0.1 μM), respectively, as well as to piplartine (1) (IC50 = 3.5 ± 0.2 μM). Despite the fact that hydroxyfuroxans 4–6 and piplartine (1) are inherently cytotoxic compounds, the introduction of a piplartine-derived moiety via an ether linker with different chain lengths led, in most cases, to hybrid compounds with enhanced cytotoxic activity. These outcomes suggest that the cytotoxic activity of PPT–furoxan hybrids may be attributed to the synergistic effects of PPT and NO donors in cancer cells.
As a first approximation of the safety profile of the synthesized hybrid compounds 7–9 and aiming to determine the selectivity against cancer cells, the antiproliferative activity in non-cancer cells (MCF10A and PNT2) was also evaluated. The selectivity index (SI) was calculated for the compounds by dividing the IC50 value observed for the non-cancer cell line by the IC50 value obtained for the corresponding cancer cell line. The data in Table 1 revealed that all PPT–furoxan hybrids 7–9 exhibited weaker cytotoxicity on normal MCF10A and PNT2 cells when compared to cancer cells MCF-7 and PC3, respectively, indicating their excellent selectivity against breast and prostate cancer cells, especially in the latter ones. Noteworthily, the exceptional SIprostate of compound 7 (75.5) significantly exceeded the values obtained for the hydroxyfuroxan 4 (0.3) and piplartine (1.5), suggesting a 250- and 50-fold higher selectivity for cancer cells. Moreover, compound 8 (SIprostate = 26.2) was ca. 8-fold more selective for prostate cancer cells than its precursor 5 (SIprostate = 3.4), while compound 9 (SIprostate = 285.1) had ca. 2-fold higher selectivity than 6 (SIprostate = 136.9), and they were much more selective than piplartine (SIprostate = 1.5).
Considering that hybrids 7, 8 and 9 display in common both the furoxan and piplartine moieties, we hypothesized that the high cytotoxicity and exceptional selective activity of hybrid 9 for cancer cells when compared to hybrids 7 and 8 may result from different mechanisms of its transport into the membrane as a consequence of its longest linker containing 12 carbon atoms. Indeed, the main purpose of varying non-cleavable and flexible linkers with carbon chains of three different lengths (more specifically: 3, 6 and 12-carbon atoms for hybrids 7, 8 and 9, respectively) was to modulate the internalization of these compounds in the cancer and non-cancer cells.
Clearly, our findings show that the combination of piplartine (1) and a furoxan moiety attached via an ether linker significantly impacts the cytotoxic activity as well as the selectivity of these hybrid drug candidates compared to their precursors. Hence the designed PPT–furoxan hybrids 7, with the shortest linker (3-carbon atoms), and 9, with the longest linker (12-carbon atoms), were chosen as promising candidates for further investigation.
2.2.2. Detection of in vitro NO release
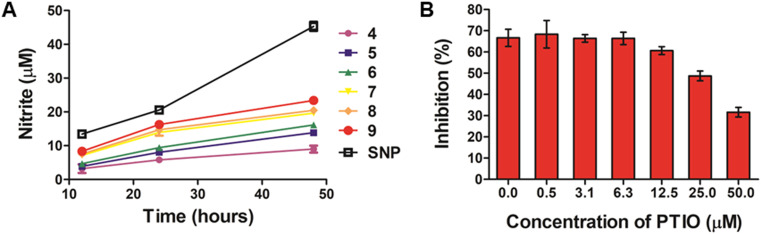
The novel hybrid compounds synthesized in this work contain the furoxan moiety which is well known for its ability to release high amounts of ·NO through a thiol-dependent mechanism.47,48 In this context, the NO release capacity of hybrid compounds 7–9 was examined to indicate whether their high cytotoxicity in PC3 cells could be associated with the levels of NO produced. The furoxan precursors 4–6 were included for comparison, as well as sodium nitroprusside (SNP), used as the positive control. Here, the levels of NO released in the cell supernatants were indirectly quantified using the Griess assay. This method is based on the reaction between the released thiol-induced NO from the compounds tested and oxygen, providing nitrite in the medium, a stable oxidation product of NO in aqueous solution. This nitrite formed is further derivatized after a few steps into an azo compound using the Griess reagent, which consists of 1% sulfanilamide in 2.5% phosphoric acid and 0.1% N-(1-naphthyl)ethylenediamine dihydrochloride. We have studied the release of NO by exposing PC3 cells to 100 μM of each compound for varying durations (12 h, 24 h, and 48 h) at 37 °C. The results, represented as concentration of nitrite (NO2−; μM), are summarized in Fig. 4.
Fig. 4. (A) Cumulative levels of NO (expressed as concentration of nitrite) produced by compounds 4–9 and sodium nitroprusside (SNP). PC3 cells were treated with individual compounds at 100 μM for 12 h, 24 h, and 48 h at 37 °C, and the concentrations of nitrite in the cell supernatants were calculated according to the calibration curve generated with NaNO2 using the Griess assay. Data are expressed as the mean value (μM) ± SD from three independent experiments. (B) Effects of NO scavenger carboxy-PTIO on the antiproliferative activity of 9. PC3 cells were pretreated with the indicated concentrations of carboxy-PTIO for 1 h and then treated with 0.05 μM of compound 9 for another 48 h. The results are expressed as the percentage of cell growth inhibition relative to control cells. Data are the mean value ± SD obtained from three independent determinations.
All the furoxans (4–9) were able to induce nitrite formation with time in the cell supernatant at levels ranging from 3.2 to 23.5 μM. As expected, treatment with SNP, a well-stablished source of NO, resulted in higher levels of nitrite (approximately 45 μM) in the cells. Among the hybrid compounds, 7–9 generated the highest NO levels, while precursors 4–6 exhibited the lowest NO-releasing abilities, which correlate with their relatively poorer cytotoxic activity. Interestingly, considering the chain lengths of the linkers in the two series (hydroxyfuroxans: 4–6 and PPT–furoxan hybrids: 7–9), compounds with longer linker chains (e.g.9) released higher levels of NO than compounds with shorter linker chains (e.g.7).
Subsequently, hybrid 9, which exhibited the highest cytotoxic activity and the highest amount of NO released after 48 h incubation, was investigated for its inhibitory activity toward PC3 cells in the presence and absence of the NO scavenger carboxy-PTIO (PTIO). PC3 cells were pretreated with various concentrations of PTIO (0, 0.50, 3.13, 6.25, 12.50, 25, and 50 μM) for 1 h and then treated with 0.05 μM of 9 for another 48 h, and cell viability was determined by the CCK-8 assay. As shown in Fig. 4b, treatment with 9 alone inhibited ca. 65% of the growth of PC3 cells; on the other hand, the cell growth inhibitory effect of 9 was dose-dependently attenuated by pretreatment with increasing concentrations of PTIO. These results strongly suggested that the high levels of intracellular NO generated by hybrid 9 were associated with its antiproliferative activity.
2.2.3. Evaluation of cell morphology
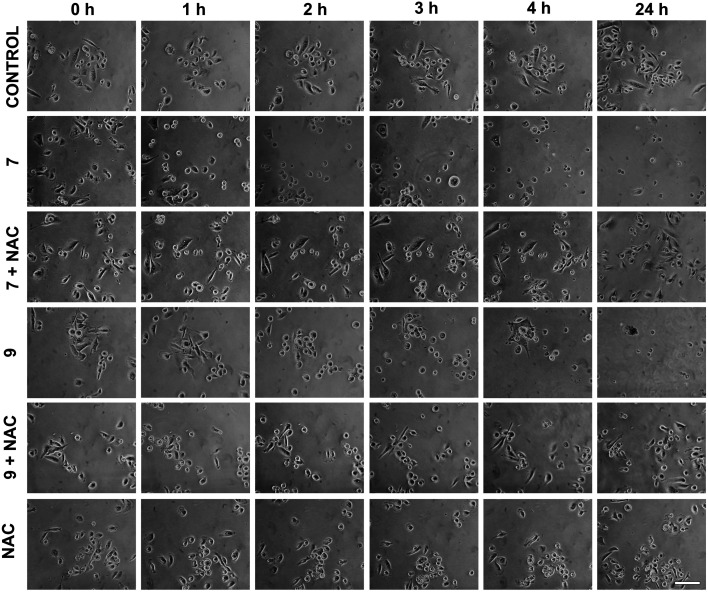
To further elucidate if other possible mechanisms of action would account for the high cytotoxic activity of hybrid compounds 7 and 9, the morphological changes in PC3 cells after treatment with 0.24 μM of 7 and 0.05 μM of 9 at different time points were evaluated and the results are shown in Fig. 5. Monitoring morphological changes over the course of treatment allowed us to observe that in the control group (DMSO), cells did not undergo morphological impairment in the first few hours. After 24 hours, however, there was a noticeable increase in cell density. In contrast, cultures treated with compounds 7 and 9 displayed evident morphological alterations from the early stages of treatment, characterized by cell rounding and partial loss of adhesion. A substantial reduction in cell density was observed after 24 h of treatment, indicating a pronounced cytotoxic effect of these compounds on the prostate cancer cell line. Intriguingly, co-treatment with the antioxidant N-acetyl-l-cysteine (NAC), which acts as a free-radical scavenger and inhibits the production of ROS that can damage cellular structures, mitigated the cytotoxicity of the tested compounds and preserved cellular morphology. These findings suggest that the PPT–furoxan hybrids 7 and 9 may also exert their mechanism of action through ROS generation.
Fig. 5. Representative images obtained by phase contrast microscopy showing morphological effects of PPT–furoxan hybrids 7 and 9 on PC3 cells. Cell cultures were treated with 7 and 9 at 0.24 μM and 0.05 μM, respectively, for 1 h, 2 h, 3 h, 4 h or 24 h in the presence or absence of N-acetyl-l-cysteine (NAC) at 1 mM (scale bar = 100 μm).
2.2.4. ROS production
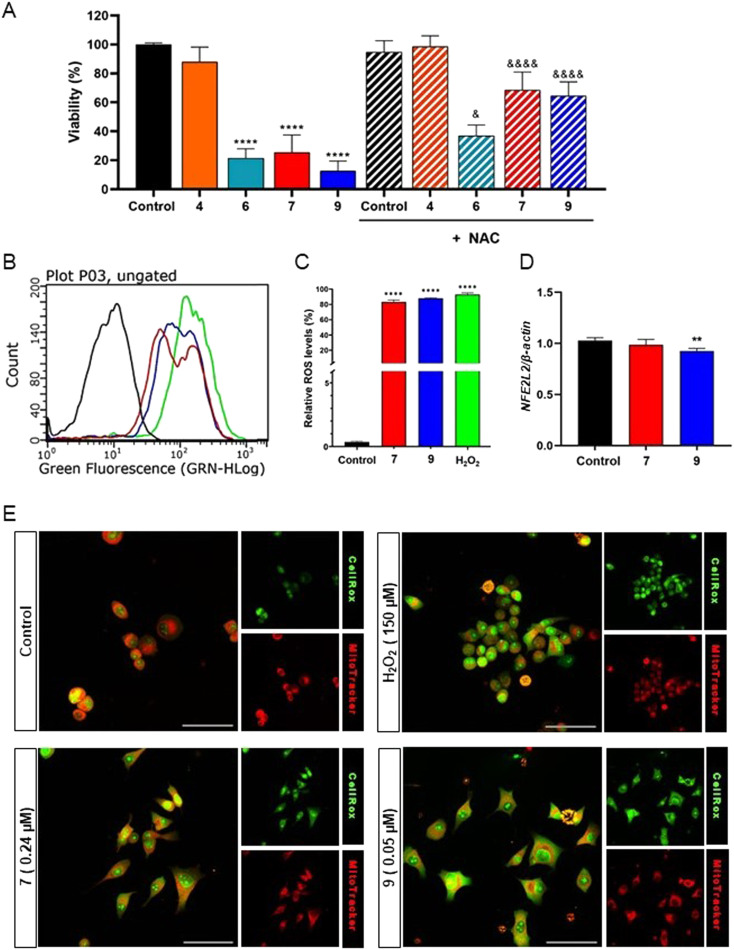
To further clarify the morphological observations, a cell viability assay was conducted through assessing the relative percentage of viable cells after 24 hours of treatment with the PPT–furoxan hybrids 7 and 9, and their respective precursors hydroxyfuroxan 4 and 6, both in the presence and absence of NAC (Fig. 6A). It was possible to observe a reduction in viable cells when cultures were treated only with the NO donor compounds. Notably, co-treatment with NAC led to attenuation in the cytotoxic activities of the furoxan-based compounds, with a more significant decrease in the cytotoxicities occurring in PC3 cells treated with hybrids 7 and 9 when compared to hydroxyfuroxans 4 and 6. Thus, these results provided additional evidence that the cytotoxic mechanism of these molecules may, at least in part, involve ROS-mediated pathways, especially in the case of PPT–furoxan hybrids 7 and 9.
Fig. 6. (A) Cell viability determined by the sulforhodamine B (SRB) assay after 24 hours of treatment. PPT–furoxan hybrid 7 and its precursor hydroxyfuroxan 4 were tested at 0.24 μM, while PPT–furoxan hybrid 9 and its precursor 6 were tested at 0.05 μM. The DMSO group represents the negative control where cells were treated only with the vehicle (0.1% DMSO). The NAC group represents samples treated with N-acetyl-l-cysteine (NAC) at 1 mM. The treated samples in the absence of NAC were compared to the control group ****p < 0.0001 according to ANOVA followed by Tukey's post-test. The effects of hybrids 7 and 9 in the presence of NAC were compared to their precursors. &p < 0.05, &&&&p < 0.0001 according to ANOVA followed by Tukey's post-test. (B and C) Intracellular ROS production determined by flow cytometry, using CellROX Green®, after 30 minutes of treatment with PPT–furoxan hybrids 7 at 0.24 μM (red line) and 9 at 0.05 μM (blue line) in PC3 cells. DMSO (0.1% v/v) was used as a negative control (black line), and H2O2 was used as a positive control at 1 mM (green line). (D) Relative expression of NFE2L2 (NRF2) determined by qPCR after 3 h treatment with hybrids 7 and 9. The analyses were performed by comparing treated samples with control groups. ****p < 0.0001, ***p < 0.001, **p < 0.01 according to ANOVA followed by Tukey's post-test. (E) Confocal imaging of PC3 cells loaded with CellROX-Green® and MitoTracker Red® fluorescent probes. Scale bars: 50 μm.
Recent studies have shown that the cytotoxic activity of piplartine in tumor cells correlates with the increase in intracellular ROS levels, which in turn perturbates the normal cell redox homeostasis.11 Furthermore, NO can easily be converted to RNS in the presence of intracellular oxidative stress factors, such as ROS. Exogenous RNS alter the original balance between RNS/ROS production and scavenging in mitochondria, which leads cells to an imbalance of redox status.38–41 Hence, to verify the capacity of 7 and 9 to trigger ROS accumulation, as well as to determine whether a possible ROS generation would be affected in the presence of NO donors, we used CellROX® green reagent to detect the intracellular levels of ROS induced by 7 (at 0.24 μM) and 9 (at 0.05 μM) in PC3 cells (Fig. 6B and C). Hydrogen peroxide (H2O2) was used as the positive control. Data obtained from flow cytometry showed a notable increase in the proportion of CellROX-positive cells in the cultures treated with 7 and 9 for 30 min relative to non-treated cells (negative control). The intracellular levels of ROS induced by 7 and 9 in PC3 cells were comparable to that of H2O2 at a higher concentration (1 mM) in experiments performed by flow cytometry. We also demonstrated that compound 9 was able to modulate gene expression of NFE2L2 (NRF2) (Fig. 6D), a transcription factor associated with redox metabolism.68 Moreover, illustrative fluorescence microscopy images from CellROX- and MitoTracker-treated PC-3 cells (Fig. 6E) show the effects of hybrids 7 and 9 on cellular redox status and membrane mitochondrial potential, respectively, in comparison with positive (H2O2) and negative (vehicle, DMSO) controls. The CellROX-Green® confocal microscopy images confirm the effects of the hybrids in increasing the levels of intracellular ROS in PC3 cells. We also used MitoTracker Red®, a cationic fluorescent dye that stains intact mitochondria but diffuses out when the membrane potential drops. Prominent perinuclear staining was observed in the control PC3 cells indicating healthy mitochondria. However, in treated samples, especially those treated with compound 9, the fluorescent signals for MitoTracker Red® were diffuse (Fig. 6E). These results indicate that PPT–furoxan hybrids could rapidly generate high levels of ROS in the presence of NO donors and induce oxidative stress in PC3 cells which may influence the observed cytotoxic activity.
2.2.5. Cell proliferation analysis
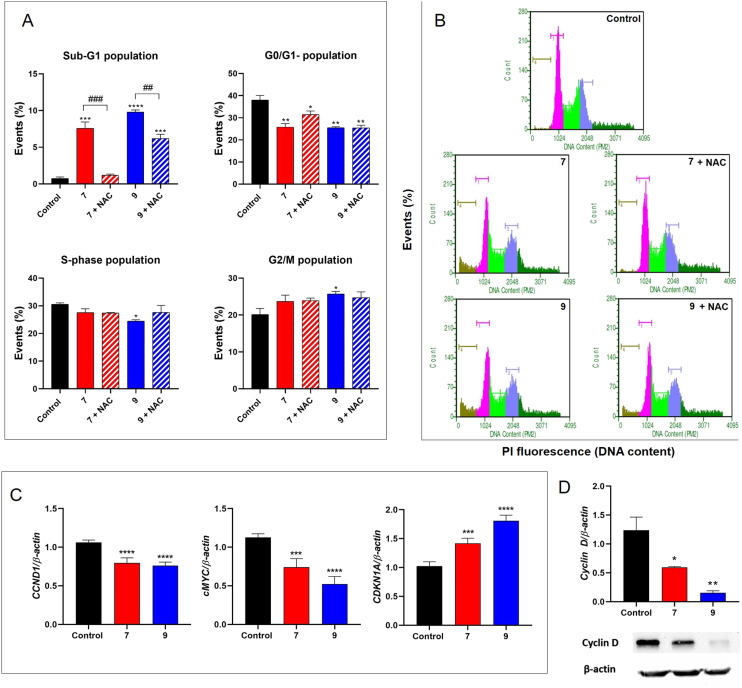
Cell cycle analysis was performed in PC3 cells treated with compound 7 at 0.24 μM and compound 9 at 0.05 μM for 3 hours (Fig. 7A and B). We observed a substantial increase in the percentage of dead cells (Sub-G1 population) compared to vehicle-treated control cells (0.1% DMSO). More specifically, the cell population in the sub-G1 phase increased from 0.75 ± 0.21% (control group) to 7.60 ± 0.85% and 9.80 ± 0.29% after treatment with 7 and 9, respectively. When we pretreated the PC3 cells with the classic antioxidant NAC (at 1 mM) prior to treatment with compounds 7 and 9, induced cell death by 7 was prevented, whereas for 9 it was considerably attenuated. The results indicated that the novel hybrids appear to rapidly induce death in PC3 cells, which, at least in part, is associated with their pro-oxidant effect.
Fig. 7. (A and B) Cell cycle analysis of PPT-hybrids 7 and 9 in PC3 cells. Cells were treated with DMSO (control), 7 (at 0.24 μM) or 9 (at 0.05 μM) for 3 h, in the presence or absence of N-acetyl-l-cysteine (NAC) at 1 mM. The percentages of cells in sub-G1, G0/G1, S and G2/M phases were compared with the control group (0.1% DMSO): *p < 0.05; **p < 0.01 ***p < 0.001 ****p < 0.0001 according to ANOVA followed by Tukey's post-test. ##p < 0.01; ###p < 0.001 represents statistical differences between treated groups in the presence or absence of NAC. (C) Relative expression of CCND1, c-MYC, and CDKN1A determined by qPCR. (D) Protein expression of cyclin D1 determined by western blot. Gene expression analysis at mRNA or protein was conducted after 3 h treatment with hybrid 7 (at 0.24 μM) or 9 (at 0.05 μM). *p < 0.05; **p < 0.01 ***p < 0.001 ****p < 0.0001 according to ANOVA followed by Tukey's post-test.
We also observed that compounds 7 and 9 were able to alter the dynamics of the progression of cell cycle as a significant reduction in G0/G1 population was observed in all treated samples (Fig. 7A and B). Interestingly, the frequency of cells in the S phase was reduced in samples treated with hybrid 9, while the population in G2/M was increased in relation to the control group (Fig. 7A and B). However, pretreatment with NAC did not prevent the effects of hybrids 7 and 9 on cell cycle progression in PC3 cells, indicating that the ability of these compounds in modulating cell cycle progression is not associated with their pro-oxidative activity. In addition, we demonstrated that hybrids 7 and 9 modulated the expression of key regulators of the cell cycle. There was a notable downregulation of CCND1 (encode cyclin D1) and c-Myc gene expression (Fig. 7C) after 3 h treatment with PPT–furoxan hybrids 7 (at 0.24 μM) and 9 (at 0.05 μM). In contrast, the CDKN1A gene (encode p21) was upregulated (Fig. 7C) in response to treatment. In accordance with qPCR data, the protein levels of cyclin D1 were also reduced by hybrid compounds as demonstrated by western blot analysis (Fig. 7D). These findings are very promising considering that c-Myc and cyclin D1 are associated with tumor development and progression in several types of cancer, including prostate cancer.69,70 Cyclin D1 is an important positive regulator of the cell cycle, which may be transcriptionally regulated by c-Myc.71c-Myc plays multiple roles in tumorigenesis, affecting cell proliferation, survival, metabolic adaptation, epithelial mesenchymal transition (EMT), angiogenesis, and metastasis in prostate cancer.70 Overexpression of c-Myc has also been reported in both prostate cancer cells and cancer stem cells.72 Importantly, c-Myc can not only reduce the expression of positive regulators of the cell cycle such as cyclin D1, cyclin-dependent kinase 1 or 4 (CDK1 or CDK4), and cyclin B1, but it can suppress the expression of p21, a pan CDK inhibitor.71,73 In fact, our results showed a significant increase in the expression profile of CDKN1A in PC3 cells in response to treatment with PPT–furoxan hybrids 7 and 9. Further studies will be performed to elucidate the molecular mechanism associated with hybrid-induced p21 upregulation in androgen-independent human prostate cancer cells PC3, which are TP53-null.
2.2.6. Annexin V apoptosis assay
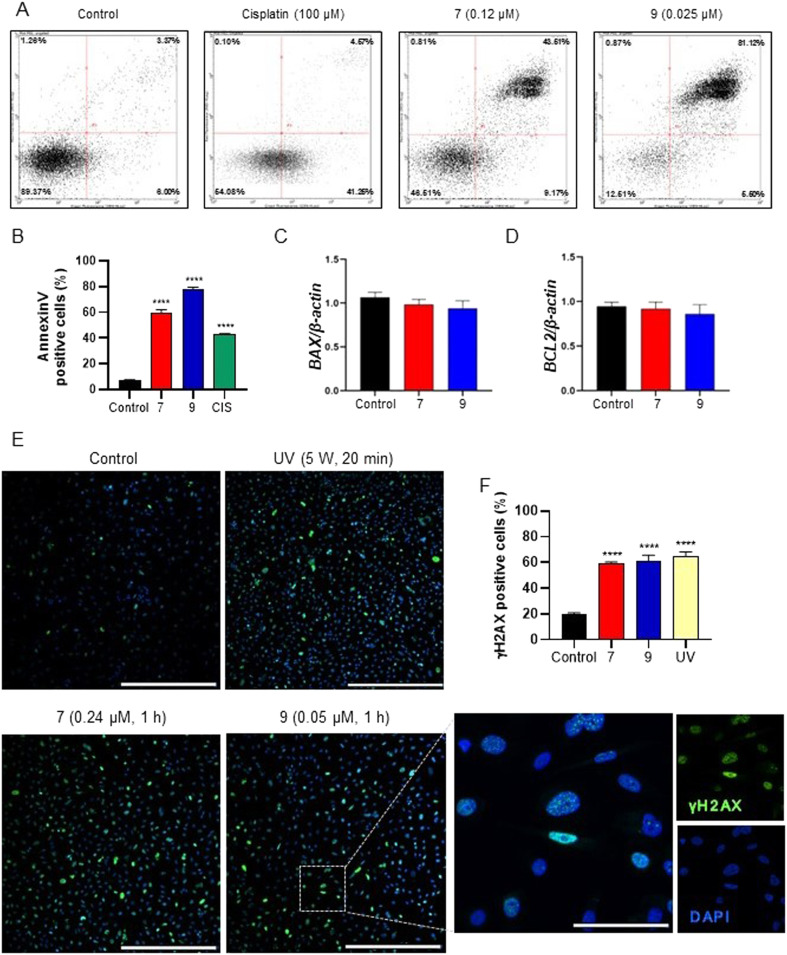
To confirm whether the cytotoxic effects of PPT–furoxan hybrids 7 and 9 in PC3 cells are accompanied by enhanced pro-apoptotic activity, an annexin V-Alexa Fluor 488 and PI apoptosis detection assay was conducted. PC3 cells were incubated with 7 (at 0.24 μM), 9 (at 0.05 μM), cisplatin (at 100 μM, positive control) or vehicle (0.1% DMSO, negative control) for 6 h, and the percentage of apoptotic cells was determined by flow cytometry analysis (Fig. 8A and B). A significant increase in the percentage of annexin V positive cells was found after treatment with both compounds 7 and 9: from 5.52% of the vehicle control to 46.86% (3.54% and 43.32% of early and late apoptotic cells, respectively) for 7 and 82.39% (80.21% and 2.18% of early and late apoptotic cells, respectively) for 9. Notably, the ability of 9 to induce apoptosis was stronger than that of 7 in PC3 cells, which may be attributed to higher levels of NO released by 9. These results corroborate all those previously obtained by morphological, cell viability and cell cycle analyses, indicating the ability of PPT–furoxan hybrids 7 and 9, especially the latter one, to induce tumor cell apoptosis, at least partly, through a ROS- and NO-dependent manner.
Fig. 8. (A and B) Annexin V-Alexa Flour 488 and PI assay conducted after 6 h of treatment with PPT–furoxan hybrid compounds 7 and 9. (A) Representative dot plots obtained by flow cytometry are shown for the vehicle (negative control, 0.1% DMSO), cisplatin (positive control, at 100 μM), 7 (at 0.24 μM), and 9 (at 0.05 μM). Viable cells (lower left quadrant), early apoptosis (lower right quadrant), late apoptosis (upper right quadrant), and necrotic cells (upper left quadrant). (B) The frequency of apoptotic cells (annexin V positive cells) was determined considering both early and late apoptosis data from n = 3. ****p < 0.0001 according to ANOVA followed by Tukey's post-test. (C and D) Relative gene expression of BAX and BCL-2 determined by qPCR. PC3 cells were treated with PPT–furoxan hybrids 7 at 0.24 μM and 9 at 0.05 μM for 3 h. Results are expressed as mRNA expression levels of each gene, normalized according β-actin expression. (E and F) Immunodetection of phospho-H2AX (γ-H2AX), a DNA damage marker, on PC3 cells performed after 6 h of treatment with PPT–furoxan hybrid compounds 7 (at 0.24 μM) and 9 (at 0.05 μM). (E) Illustrative fluorescence images showing the immunoreaction pattern for p-H2AX in PC3 cells. Nuclei were stained with DAPI. Cells irradiated with ultraviolet C light for 20 min were used as positive controls. ****p < 0.0001 according to ANOVA followed by Tukey post-test.
Our results also showed that there was no alteration in the gene expression of BAX and BCL2 after 3 h treatment with compounds 7 and 9 under the experimental conditions evaluated (Fig. 8C and D). However, we observed the pro-apoptotic potential of these hybrids on PC3 cells using the annexin V assay. Bax is a pro-apoptotic member of the Bcl-2 family that under pro-apoptotic conditions induces the permeabilization of the outer mitochondrial membrane, triggering the intrinsic apoptosis pathway. The Bcl-2 protein, in turn, plays a crucial anti-apoptotic role by inhibiting Bax activity.74 It is well-known, however, that the intrinsic apoptosis process also involves other pro-apoptotic (Bim, Puma, Bad, and Bid) and anti-apoptotic (Bcl-xL, Bcl-w, and Mcl-1) members of the Bcl-2 family,75,76 which were not assessed in the present study. Indeed, Bcl-xL appears to represent a crucial survival mechanism in human prostate cancer cells. Overexpression of Bcl-xL has been described in PC3 cells and cell lines exhibiting resistance to multiple drugs,77–79 and this overexpression relationship seems to be directly associated with prostate cancer progression.80 Bcl-xL seems to play a crucial role in resistance to staurosporine-induced apoptosis in PC3 cells, and restoration of apoptosis is achieved through the negative regulation of Bcl-xL.77,79 Poudel and colleagues demonstrated that the treatment of PC3 cells with sulforaphane (3′,4′,6′-trihydroxyaurone) led to the suppression of cyclin D1 expression, causing cell death, similar to the findings of the present study.81 They observed that this outcome occurs in a temporal pattern likewise to the reduction in Bcl-xL expression, suggesting an interconnection between the regulation of cyclin D1 and Bcl-xL-mediated apoptosis.81 Studies have shown that reduced levels of cyclin D1 are associated with increased sensitivity to apoptosis induced by various agents, implying a crucial role of cyclin D1 in the regulation of cell survival.82–84 Thus, further studies will be performed to elucidate the molecular mechanism associated with the pro-apoptotic activity of hybrids 7 and 9 in PC3 cells.
It is well recognized that oxidative stress can induce DNA damage and, in turn, apoptosis.85,86 Thus, we investigated if hybrids 7 and 9 could induce DNA damage in PC3 cells (Fig. 8E and F). The results revealed that there was a significant increase in the frequency of cells positive for phospho-H2AX (γ-H2A.X), a marker of DNA damage, in samples treated with 7 (at 0.24 μM) and 9 (at 0.05 μM) for 6 h. The levels of DNA damage found in treated samples were similar to that observed for the positive control (UVC). Therefore, the data indicate that the mechanism associated with the high cytotoxicity of 7 and 9 in PC3 cells involves their ability to induce ROS generation, DNA damage and, ultimately, apoptosis.
3. Conclusions
Three novel piplartine–furoxan hybrids (compounds 7–9) conjugating the natural product piplartine (1) and a furoxan moiety (3,4-bis(phenylsulfonyl)-1,2,5-oxadiazole-2-oxide) as the NO-donor subunit via an ether linker with different chain lengths were designed, synthesized, characterized, and evaluated for their anticancer potential. In the CCK-8 study, we have shown that the introduction of a furoxan fragment to piplartine led in most cases to hybrid compounds with improved cytotoxic activity toward a panel of human cancer cells (MCF-7, PC3 and OVCAR-3) compared to their precursors hydroxyfuroxans 4–6 and the natural product PPT (1). Particularly, the cytotoxicity of compounds 7–9 toward prostate cancer cells (PC3) was much higher than their activity against normal prostate cells (PNT2), demonstrating significant selectivities in these cancer cell lines. The concentrations of NO released by the compounds correlated well with their cytotoxicity profile against PC3 cells. These outcomes suggested that the cytotoxic activity of PPT–furoxan hybrids in cancer cells may be attributed to the synergistic effects of PPT and NO donor moieties. Furthermore, 7 and 9 could rapidly generate high levels of ROS.
The promising compounds 7 and 9 showed a similar mode of action, significantly reducing cell proliferation, promoting DNA damage, and inducing apoptosis. It could be speculated that the pro-apoptotic activity in PC3 cells induced by 7 and 9 was associated, at least in part, with their capacity of NO-release and ROS generation. Notably, the ability of 9 to induce apoptosis was stronger than that of 7 in PC3 cells, which may be attributed to higher levels of NO released by 9. We have also identified that critical regulators of the cell cycle were modulated by hybrids 7 and 9 in PC3 cells. Thus, the novel PPT–furoxan hybrids described here represent an interesting class of ROS inducers and NO-donor hybrids s treatments.
In particular, our results show that hybrid 9 is a unique dual-function bioactive compound that acts by releasing high levels of NO from its furoxan moiety and generating intracellular ROS due to the presence of piplartine. Besides its very high cytotoxicity toward PC3 cells (IC50 = 50 nM), compound 9 demonstrated an exceptional selectivity (SI = 285.1) for these prostate cancer cells. Although many conventional anticancer drugs are highly cytotoxic, one of the main issues of chemotherapy is the lack of selectivity of these drugs to cancer cells, which severely affects the efficacy of the treatment. Thus, the potent cytotoxic activity and extraordinary selectivity for prostate cancer cells associated with the capacity to release NO, generate ROS, induce DNA damage and trigger apoptosis make hybrid 9 a unique and more advantageous molecule compared with other therapeutic frameworks. Since it is well-known that cancer is a complex disease and its treatment a big challenge, the construction of molecular frameworks with various degrees of complexity and improved pharmacodynamic effects to address the complexities of cancer is highly desirable. Therefore, the biological data provided in our study offers a solid foundation for future research regarding the anticancer potential of compound 9.
4. Experimental section
4.1. Chemistry
4.1.1. General considerations
All reactions were carried out with freshly distilled solvents, using anhydrous conditions unless otherwise noted. DCM and Et3N were distilled over calcium hydride. THF was distilled over metallic Na and benzophenone. DMF was obtained in anhydrous grade and was used without previous treatment. The anhydrous solvents were transferred by using oven-dried syringes. The sealed tubes used in the cross-coupling reactions were previously dried in an oven for at least 3 h. Other reagents were obtained from commercial sources and were used without prior purification. The reactions were monitored by thin-layer chromatography (silica gel 60 F254 in aluminum foil, Merck). Silica gel 200–400 mesh was used for flash column chromatography.
1H and 13C NMR spectra were recorded on a Bruker Avance III HD 250 MHz, a Bruker Avance III 400 MHz, or a Bruker Avance III 500 MHz and processed using MestreNova 12.0.4 software. The chemical shifts are reported in parts per million (ppm) on a delta (δ) scale. The residual solvent peaks were used as reference values; for 1H NMR, CHCl3 = 7.26 ppm; for 13C{1H} NMR, CDCl3 = 77.16 ppm. Signal multiplicity was reported as singlet (s), doublet (d), triplet (t), quartet (q), quintet (quint), and multiplet (m). High-resolution mass spectra (HRMS) were acquired on an Orbitrap Thermo QExactive mass spectrometer. Infrared (IR) spectra were recorded using a Thermo Scientific Nicolet IS5 spectrometer using a Thermo Scientific ID3 ATR, and the absorption frequencies are reported in cm−1.
4.1.2. Synthesis of the compounds
4-(3-Hydroxypropoxy)-3-(phenylsulfonyl)-furoxan (4)
To a solution 1,3-propanediol (509 mg, 6.48 mmol) and 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) (0.97 mL, 6.48 mmol) in anhydrous DCM (12.5 mL) was added furoxan 3 (500 mg, 1.30 mmol). The mixture was stirred at room temperature for two hours. The organic phase was washed with water and then with two portions of 1 M HCl (2 × 10 mL), followed by saturated sodium chloride solution (10 mL). The organic phase was dried with magnesium sulfate and concentrated on a rotary evaporator. Purification was carried out by liquid chromatography on silica gel (240–400 mesh) using a mixture of hexane and ethyl acetate as the eluent (gradient in a ratio of 2 : 1 to 1 : 1 v/v) to yield 0.164 g (0.55 mmol) of furoxan 4 as a white solid in 42% yield (m.p.: 105–106 °C; lit. 105–107,87 105–106).88 IR (cm−1, thin film): 680, 736, 1041, 1084, 1161, 183, 1254, 1355, 1375, 1448, 1553, 1617. 1H NMR (250 MHz, CDCl3): δ (ppm) 8.06 (d, J = 7.2 Hz, 2H), 7.74 (t, J = 7.4 Hz, 1H), 7.62 (t, J = 7.3 Hz, 2H), 4.60 (t, J = 6.0 Hz, 2H), 3.88 (t, J = 5.6 Hz, 2H), 2.13 (quint, J = 5.9 Hz, 2H). 13C NMR (62.5 MHz, CDCl3): δ (ppm) 158.9, 137.9, 135.7, 129.7, 128.5, 69.3, 59.3, 31.3. HRMS (ESI-TOF) m/z: [M + Na]+ calcd. for C11H12N2NaO6S: 323.03083; found: 323.03074.
4-((6-Hidroxyhexil)oxy)-3-(phenylsulfonyl)-furoxan (5)
To a solution of 1,6-hexanediol (782 mg, 6.48 mmol) and DBU (973 μL, 6.48 mmol) in anhydrous DCM (12.5 mL) was added furoxan 3 (500 mg, 1.30 mmol). The mixture was stirred at room temperature for two hours. The organic phase was washed with water and then with two portions (10 mL each) of 1 M HCl, followed by saturated sodium chloride solution (10 mL). The organic phase was dried with MgSO4 and concentrated on a rotary evaporator. Purification was carried out by column chromatography with flash silica using a mixture of hexane and ethyl acetate in a 1 : 2 ratio as the eluent. Furoxan 5 was obtained as a white solid (0.342 g, 1.00 mmol) in 77% yield (m.p.: 52–55 °C; lit.: 54–57 °C); IR (cm−1, film): 680, 701, 724, 999, 1167, 1181, 1255, 1367, 1383, 1449, 1554, 1681, 2856, 2936. 1H NMR (250 MHz, CDCl3): δ (ppm) 8.05 (d, J = 7.3 Hz, 2H), 7.74 (t, J = 7.5 Hz, 1H), 7.61 (t, J = 7.3 Hz, 2H), 4.42 (t, J = 6.5 Hz, 2H), 3.67 (t, J = 6.5 Hz, 2H), 1.88 (m, J = 6.5 Hz, 2H), 1.61 (quint, J = 6.5 Hz, 2H), 1.45–1.49 (m, 4H). 13C NMR (62.5 MHz, CDCl3): δ (ppm) 159.0. 138.1, 135.6, 129.6, 128.5, 110.5, 71.5, 62.7, 32.5, 28.3, 25.5, 25.3. HRMS (ESI+) m/z: [M + Na]+ calcd. for C14H18N2NaO6S: 365.07778; found: 365.07765.
4-((12-Hydroxydodecyl)oxy)-3-(phenylsulfonyl)-furoxan (6)
To a solution of 1,12-dodecanediol (828 mg, 4.09 mmol) and furoxan 3 (700 mg, 1.82 mmol) solubilized in anhydrous THF (8.2 mL) was added DBU (612 μL, 4.09 mmol). The mixture was stirred at reflux for 6 hours. Then, ethyl acetate was added (30 mL) and the organic layer was washed with water (20 mL), HCl 1 M solution and brine. The organic layer was dried over Na2SO4, filtered, and the solvent was removed under reduced pressure. The product was purified by column chromatography [(20 mm × 70 cm), H = 20 cm of SiO2, gradient elution from 20 to 35% ethyl acetate in hexane, 5% increases, 100 mL runs] to give 6 (159 mg, 46%) as a white solid. (m.p.: 91–93 °C). IR (cm−1, thin film): 680, 723, 746, 1166, 1380, 1455, 1558, 1628, 2852, 2923. 1H NMR (250 MHz, CDCl3) δ 8.06 (m, 2H), 7.75 (m, 1H), 7.61 (m, 2H), 4.41 (t, J = 6.6 Hz, 2H), 3.64 (t, J = 6.6 Hz, 2H), 1.87 (quint, J = 6.9 Hz, 2H), 1.57 (quint, J = 6.8 Hz, 2H), 1.49–1.13 (m, 16H). 13C NMR (125 MHz, CDCl3): δ (ppm) 159.2, 138.3, 135.7, 129.7, 128.7, 110.6, 71.8, 63.2, 32.9, 29.7, 29.7, 29.6, 29.6, 29.5, 29.2, 28.6, 25.9, 25.7. HRMS (ESI-TOF) m/z: [M + Na]+ calcd. for C20H30N2NaO6S: 449.17168; found: 449.17175.
(E)-1-(3-(4-(3-((5-(λ1-Oxidaneyl)-4-(phenylsulfonyl)-1,2,5λ4-oxadiazol-3-yl)oxy)propoxy)-3,5-dimethoxyphenyl)acryloyl)-5,6-dihydropyridin-2(1H)-one (7)
To a flask equipped with a stirring bar and under nitrogen atmosphere were added triphenylphosphine (78.7 mg, 0.30 mmol), alcohol 4 (97.0 mg, 0.22 mmol) and 4-hydroxypiplartine 2 (57.6 mg, 0.19 mmol), followed by anhydrous THF (1.0 mL). The solution was cooled to 0 °C and DIAD (64.7 mg, 0.32 mmol) was added dropwise, and the mixture was stirred for 10 min at the same temperature followed by heating at 30 °C for 2 h. The solvent was removed under reduced pressure, and the residue was purified by column chromatography [(20 mm × 50 mL SiO2) 5% MeCN in CHCl3], to give a yellow oil. A second purification [(20 mm × 50 mL SiO2) 50% ethyl acetate in hexane] gave pure hybrid furoxan 7 as a pale yellow solid (68.1 mg, 0.11 mmol) in 59% yield. IR (cm−1, thin film): 731, 826, 1038, 1126, 1169, 1182, 1215, 1245, 1278, 1318, 1352, 1385, 1419, 1450, 1503, 1553, 1558, 1613, 1685. 1H NMR (250 MHz, CDCl3) δ (ppm) 8.00 (m, 2H), 7.79–7.63 (m, 2H), 7.53 (t, J = 8.0 Hz, 2H), 7.43 (d, J = 15.6 Hz, 1H), 6.96 (dt, J = 9.5 and 4.1 Hz, 1H), 6.77 (s, 2H), 6.05 (dt, J = 9.7 and 1.8 Hz, 1H), 4.75 (t, J = 6.1 Hz, 2H), 4.19 (t, J = 5.7 Hz, 2H), 4.05 (t, J = 6.5 Hz, 2H), 3.79 (s, 6H), 2.49 (m, 2H), 2.27 (quint, J = 5.9 Hz, 2H). 13C NMR (62.5 MHz, CDCl3): δ (ppm) 168.8, 165.9, 159.1, 153.5, 145.6, 143.6, 138.7, 138.1, 135.9, 130.9, 129.6, 128.4, 125.8, 121.3, 105.4, 69.0, 68.5, 56.0, 41.7, 29.4, 24.8. HRMS (ESI-TOF) m/z: [M + Na]+ calcd. for C27H27N3NaO10S: 608.13094; found: 608.13078.
(E)-1-(3-(4-((6-((5-(λ1-Oxidaneyl)-4-(phenylsulfonyl)-1,2,5λ4-oxadiazol-3-yl)oxy)hexyl)oxy)-3,5-dimethoxyphenyl)acryloyl)-5,6-dihydropyridin-2(1H)-one (8)
To a flask equipped with a stirring bar and under nitrogen atmosphere were added triphenylphosphine (41.9 mg, 0.158 mmol), alcohol 5 (40.6 mg, 0.119 mmol) and 4-hydroxypiplartine 2 (30.0 mg, 0.0989 mmol), followed by anhydrous THF (1.0 mL). The solution was cooled to 0 °C and DIAD (33.7 mg, 0.17 mmol) was added dropwise, and the mixture was stirred for 10 min at the same temperature followed by heating at 30 °C for 2 h. The product was purified by column chromatography [(20 mm × 40 mL of SiO2) gradient elution from 2 to 5% MeCN in CHCl3] to give hybrid furoxan 8 (41 mg, 0.065 mmol, 66% yield) as a pale yellow solid. IR (cm−1, thin film): 684, 730, 825, 912, 997, 1053, 1127, 1169, 1182, 1244, 1277, 1317, 1354, 1382, 1419, 1415, 1502, 1552, 1579, 1614, 1684. 1H NMR (250 MHz, CDCl3) δ (ppm) 8.05 (m, 2H), 7.80–7.55 (m, 4H), 7.42 (d, J = 15.5 Hz, 1H), 6.95 (dt, J = 8.9 and 4.2 Hz, 1H), 6.80 (s, 2H), 6.05 (dt, J = 9.7 and 1.8 Hz, 1H), 4.43 (t, J = 6.5 Hz, 2H), 4.05 (t, J = 5.6 Hz, 2H), 4.02 (t, J = 5.9 Hz, 2H), 3.87 (s, 6H), 2.49 (m, 2H), 1.90 (quint, J = 6.7 Hz, 2H), 1.78 (quint, J = 6.8 Hz, 2H), 1.62–1.52 (m, 4H). 13C NMR (62.5 MHz, CDCl3): δ (ppm) 169.0, 166.0, 159.2, 153.7, 145.5, 143.8, 139.3, 138.1, 135.6, 130.5, 129.7, 128.6, 125.9, 121.1, 110.6, 105.7, 73.3, 71.7, 56.3, 41.8, 30.1, 29.8, 28.5, 25.5, 24.9. HRMS (ESI-TOF) m/z: [M + Na]+ calcd. for C30H33N3O10NaS: 650.17789; found: 650.17743.
(E)-1-(3-(4-((12-((5-(λ1-Oxidaneyl)-4-(phenylsulfonyl)-1,2,5λ4-oxadiazol-3-yl)oxy)dodecyl)oxy)-3,5-dimethoxyphenyl)acryloyl)-5,6-dihydropyridin-2(1H)-one (9)
To a flask equipped with a stirring bar and under nitrogen atmosphere were added triphenylphosphine (78.6 mg, 0.297 mmol), alcohol 6 (101.0 mg, 0.237 mmol) and 4-hydroxypiplartine 2 (60.0 mg, 0.198 mmol), followed by anhydrous THF (1.0 mL). The solution was cooled to 0 °C and DIAD (33.7 mg, 0.17 mmol) was added dropwise, and the mixture was stirred for 10 min at the same temperature followed by heating at 30 °C for 2 h. The product was purified by column chromatography [(20 mm × 50 mL SiO2) 2% MeCN in CHCl3 (1CV), then 5% MeCN in CHCl3], affording hybrid furoxan 9 (87 mg, 0.12 mmol, 62% yield) as a colorless gum. IR (cm−1, thin film): 684, 701, 730, 824, 998, 1072, 1127, 1170, 1215, 1277, 1317, 1381, 1450, 1502, 1551, 1579, 1614, 1686, 1725. 1H NMR (250 MHz, CDCl3) δ 8.06 (m, 2H), 7.82–7.55 (m, 4H), 7.42 (d, J = 15.5 Hz, 1H), 6.95 (dt, J = 9.0 and 4.2 Hz, 1H), 6.80 (s, 2H), 6.04 (dt, J = 9.7 and 1.8 Hz, 1H), 4.41 (t, J = 6.6 Hz, 2H), 4.04 (t, J = 5.6 Hz, 2H), 3.99 (t, J = 5.9 Hz, 2H), 3.87 (s, 6H), 2.49 (m, 2H), 1.86 (quint, J = 6.9 Hz, 2H), 1.73 (quint, J = 7.0 Hz, 2H), 1.49–1.22 (m, 16H). 13C NMR (125 MHz, CDCl3): δ (ppm) 168.9, 165.9, 159.1, 153.6, 145.6, 143.9, 139.4, 138.2, 135.6, 130.4, 129.6, 128.5, 125.8, 120.9, 110.5, 105.7, 73.6, 71.7, 56.2, 41.7, 30.1, 29.6, 29.6, 29.5, 29.5, 29.4, 29.1, 28.4, 25.8, 25.6, 24.8. HRMS (ESI-TOF) m/z: [M + H]+ calcd. for C36H46N3O10S: 712.28984; found: 712.28969.
4.2. Biology
4.2.1. Materials
The cell lines were purchased from Rio de Janeiro Cell Bank. Cell Counting Kit-8 (CCK-8), cisplatin, N-acetyl-l-cysteine (NAC), RNAse, and propidium iodide were purchased from Sigma-Aldrich (Brazil). RPMI-1640, Dulbecco's modified phosphate buffered saline (DMPBS, pH 7.4), 0.25% trypsin–EDTA solution (Gibco®), Alexa Fluor 488 Annexin V/Dead Cell Apoptosis kit and CellROX® green reagent were purchased from ThermoFisher Scientific (Itapevi, SP, Brazil). Fetal bovine serum (FBS) was supplied by Cultilab Materiais para Cultura de Células (Campinas, SP, Brazil).
4.2.2. Cell culture
The human MCF-7 (breast cancer), MCF-10A (normal breast), PC3 (prostate cancer), PNT2 (normal prostate) and OVCAR-3 (ovary cancer) cell lines were cultured in RPMI-1640 culture medium supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (penicillin at 100 U mL−1 and streptomycin at 100 μg mL−1). Cells were cultured at 37 °C under a 5% CO2 humidified atmosphere, and the culture medium was changed every 2–3 days. When the cell monolayer from each culture flask reached about 80% confluency, the adhered cell lines were detached with 0.25% trypsin–EDTA solution and homogenously distributed into the wells of 96-well culture plates.
4.2.3. In vitro cytotoxicity assay
Cell viability of compounds was evaluated using the CCK-8 assay according to the manufacturer's instructions. Briefly, cells were seeded in a 96-well plate at a density of 1 × 104 cells per well in 200 μL of cell culture medium and incubated at 37 °C in a humidified atmosphere with 5% CO2 for 24 h. Then the medium was replaced with a fresh one (200 μL per well) containing various concentrations of each compound, and the cells were incubated for further 48 h under the same cell culture conditions. The cytotoxicity of piplartine and cisplatin, used as positive controls, was evaluated in the same manner. Afterwards, 10 μL of the CCK-8 solution were added to each well, followed by incubation for another 3 h. The absorbance of the solution in each well was determined at a wavelength of 450 nm using a multi-well plate reader (FlashScan 530 Analitic Jena). Control cells were treated with vehicle alone, which exhibited cell viability of 100%. The half-maximal inhibitory concentration (IC50) values, expressed as mean ± SD, from at least two different experiments carried out in triplicate were obtained by nonlinear regression using GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA).
4.2.4. In vitro NO released amount measurement
The levels of NO generated by individual compounds in the cell supernatants were indirectly determined by the colorimetric Griess assay. Briefly, 85% phosphoric acid (0.75 mL) and sulfanilamide (0.25 g) were dissolved in ca. 20 mL of water and made up to 25 mL to obtain solution A. N-(1-Naphthyl)ethylenediamine dihydrochloride (0.025 g) was dissolved in 25 mL of water to make solution B. PC3 cells were seeded in a 96-well plate at a density of 1 × 104 cells per well in 200 μL of RPMI-1640 (plus 10% FBS and 1% penicillin/streptomycin) and cultured for 24 h. Then, the medium was replaced with RPMI only, and 100 μM concentration of each compound was added in 200 μL total volume. The cells were incubated for an additional 12, 24 and 48 h at 37 °C. Subsequently, 50 μL of the cell supernatant after treatment was transferred to a 96-well plate. Then, 50 μL of solution A (1% sulfanilamide in 2.5% phosphoric acid in water) was added, and the cells were incubated for 10 minutes in the dark. Finally, 50 μL of solution B (0.1% N-(1-naphthyl)ethylenediamine dihydrochloride) was added, and the cells were incubated at 37 °C for 10 minutes. The absorbance was measured at 540 nm using a multi-well plate reader (FlashScan 530 Analitic Jena). Sodium nitroprusside was used as the standard reference compound (100 μM). The cells treated with DMSO were used as negative controls for the background levels of nitrite production, while sodium nitrite solutions at different concentrations (0–100 μM in culture medium) were prepared as the positive control to construct the calibration curve, from which the concentration of nitrite (μM) released was calculated. Data were mean values ± SD of at least three independent experiments carried out in triplicate.
4.2.5. In vitro antiproliferative activity with the NO scavenger carboxy-PTIO
To determine the effect of NO on the antiproliferative activity of the studied compounds, we studied their effect on PC3 cancer cell growth in the absence and presence of the NO scavenger carboxy-PTIO (PTIO). Cells were seeded into a 96-well plate at 1 × 104 cells per well and were allowed to attach for 24 h. The cultures were pretreated with PTIO (0, 0.50, 3.13, 6.25, 12.50, 25 and 50 μM) for 1 h and then treated with 0.05 μM of compound 9 for an additional 48 h. Then, the cells were washed three times with PBS and the antiproliferative activity was assessed by the CCK-8 assay from two different experiments carried out in triplicate.
4.2.6. Morphological analysis
To monitor time-dependent morphological changes following treatment, PC3 cells were seeded in 6-well plates at a density of 1 × 105 cells per well. After 24 h, cultures were treated with compounds 7 and 9 at 0.24 and 0.05 μM respectively, with and without the addition of N-acetyl-l-cysteine (NAC) at 1 mM. The cell morphology was monitored, and images from the same observation field were captured at 1 h, 2 h, 3 h, 4 h, and 24 h after treatment using phase contrast microscopy (Carl Zeiss, Germany).
4.2.7. Cell viability analysis with N-acetyl-l-cysteine
For this in vitro cell viability assay, PC3 cells were seeded in a 96-well plate at a density of 1 × 104 cells per well. After 24 h, the cell culture was treated with the hybrid compound 7 or its precursor 4 at 0.24 μM, as well as with the hybrid compound 9 or its precursor 6 at 0.05 μM. All groups were treated in the presence or absence of N-acetyl-l-cysteine at 1 mM and incubated for 24 h (37 °C, 5% CO2). Then the cells were fixed with a 15% trichloroacetic acid (TCA) solution at 4 °C for 1 h, washed with Milli-Q water and left at room temperature overnight to dry. The cells were then stained with a 0.4% sulforhodamine B (SRB) solution, and the bound SRB was solubilized with Tris buffer (10 mM). The absorbance was read at 540 nm on a spectrophotometer.
4.2.8. Measurement of reactive oxygen species (ROS) levels
For the evaluation of ROS levels, cells were seeded in 6-well plates at a density of 1 × 105 cells per well. After 24 h, the cells were treated with compound 7 at 0.24 μM, or with compound 9 at 0.05 μM. For the positive control, PC3 cells under the same conditions received treatment with H2O2 at 1 mM. Simultaneously, cell cultures were incubated with CellROX® green reagent (C10444, Invitrogen) (at a final concentration of 5 μM) for 30 minutes at 37 °C. The cells were collected, and the analysis was performed by flow cytometry using GuavaSoft 2.7 software. Intracellular ROS production and functional mitochondria detection were assessed by confocal microscopy. Cells were loaded with 2.5 μM CellROX® green and 0.5 μM MitoTracker Red® (Molecular Probes, Eugene, USA) for 30 minutes at 37 °C. After three washings in PBS, the fluorescence images were immediately captured by a confocal microscope (Nikon, NY, USA).
4.2.9. Cell cycle progression analysis
Cell cycle progression of PC3 cells was assessed using flow cytometry. Cells were seeded into 6-well plates at a density of 1 × 105 cells per well and treated with the hybrid compound 7 at 0.24 μM, as well as with the hybrid compound 9 at 0.05 μM for 3 hours. All groups were pretreated in the presence or absence of N-acetyl-l-cysteine (NAC) at 1 mM. Cells were collected by enzymatic digestion (trypsin–EDTA solution), and fixed with 75% ethanol in PBS at 4 °C for 30 min. The samples were stained for 1 hour in a solution containing PBS, RNAse (1.5 mg mL−1), and propidium iodide (90 μg mL−1). DNA content was quantified by a flow cytometer, and the data were processed using GuavaSoft 2.7 software.
4.2.10. Apoptosis assessment using annexin V assay
Cells were seeded in 6-well plates at a density of 1 × 105 cells per well and treated with 7 (at 0.24 μM) or 9 (at 0.05 μM) for 6 h. Phosphatidylserine externalization was evaluated using the Alexa Fluor 488-Annexin V/Dead Cell Apoptosis kit following the manufacturer's instructions. Cells were collected by enzymatic digestion (trypsin–EDTA solution), centrifuged at 1000 rpm for 5 min at 4 °C, and washed with ice-cold PBS. Then, 2 × 104 cells were resuspended in 100 μL of a mixed solution containing Alexa Fluor 488-annexin V and propidium iodide (V13245, Molecular Probes). Samples were analyzed after a 20 min incubation at room temperature in a dark chamber. The analysis was performed by flow cytometry using GuavaSoft 2.7 software.
4.2.11. Gene expression analysis
The cells were seeded into 6-well plates at a density of 1 × 105 cells per well, and treated with compound 7 at 0.24 μM, or with compound 9 at 0.05 μM for 3 h. Total RNA was extracted using a RNeasy Mini kit (Qiagen, Mississauga, ON, Canada). Then, total RNA (1 μg) was treated with DNase I (1 U μg−1; Invitrogen, São Paulo, SP, Brazil) and subjected to reverse transcription using random primers and a high-capacity cDNA reverse transcription kit (Applied Biosystems, São Paulo, SP, Brazil). Expression of the target genes (BCL-2, BAX, CDKN1A, NRF2, CCND1 and c-MYC) (Table S1†) was investigated by real-time PCR with an ABI 7500 thermocycler using Power Sybr Green PCR Master Mix (Applied Biosystems). The genes were amplified using the following conditions: 95 °C for 10 s, and annealing and extension at 60 °C for 1 min (40 cycles). Although the reactions were run for 40 cycles, the detection and quantification of the amplified sequences were performed within the exponential phase. The Ct value, the PCR cycle at which the samples reach the threshold line, was used in relative quantity. For this, each target gene was normalized to the reference gene using the ΔΔCt method with efficiency correction and a control sample for calibration, according to Pfaffl, 2001.89 The average efficiency values for each gene were calculated through the amplification profile of each sample using the LinRegPCR program. The data are presented as mean ± standard error of the mean (SEM) from four independent experiments.
4.2.12. Protein expression analysis
The cells were seeded into 100 mm Petri dishes at a density of 1 × 107 cells per plate. The samples were treated with compound 7 at 0.24 μM, or with compound 9 at 0.05 μM for 3 h or 6 h. Cells were homogenized in RIPA lysis buffer containing both protease and phosphatase inhibitors (Sigma-Aldrich, Brazil). After total protein quantification, 50 μg of protein was separated by 12% sodium dodecyl–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Amersham Bioscience) using a transfer apparatus (100 V, 250 mA for 2 h). The membranes were blocked with 5% nonfat milk in Tris-buffered saline + 0.1% (v/v) Tween-20 for 1 h, then probed with β-actin antibody, HRP (1 : 25000, #A3854, Sigma-Aldrich) for 2 h at room temperature, or probed with the primary antibody for Cyclin D1 (1 : 1000, #55506, Cell Signaling) overnight at 4 °C. The primary antibody was detected using anti-Rabbit IgG (H + L) secondary antibody, HRP (1 : 500, #65-6120, Thermo-Fisher Scientific) for 2 h at room temperature. Immunoreactive bands were visualized using an ECL Western blotting detection kit (Amersham Pharmacia Biotech). A reprobing protocol was followed for detecting immunoreactive bands for different antibodies. Results were obtained from two independent experiments. The immunoreactive bands were quantified using Bio-Rad Image Lab software.
4.2.13. DNA damage evaluation through γ-H2A.X immunodetection
The immunocytochemistry for phopho-H2A.X (γ-H2A.X) was performed according to Noubissi et al.90 Briefly, the cells were seeded into 35 mm plates on coverslips at a density of 1 × 105 cells. After adherence, the cells were maintained in serum-free medium for 18 hours, and treated with compound 7 (0.24 μM) or compound 9 (0.05 μM) for 1 h. The samples were fixed with 3.7% formaldehyde for 15 minutes and treated with Triton X-100 (0.25%) for 10 minutes. After blocking (3% albumin for 2 hours), the samples were incubated with anti-phospho H2A.X (serine 139) (Cell Signaling, Danvers, MA, #9718, 1 : 500) at 4 °C for 2 hours at room temperature. Afterward, anti-rabbit secondary antibody-Alexa Fluor 488 conjugate (1 : 200, Abcam, #ab150073) was added to the samples (1 h at room temperature). The slides were mounted with Fluoroshield (Sigma, #F6057) and analyzed under a confocal laser scanning microscope (Nikon, NY, USA). The results refer to the mean of 3 independent experiments. 500 cells were quantified per slide. For the positive control, PC3 cells were irradiated with ultraviolet C light for 20 minutes.
4.2.14. Statistical analysis
All experimental data are expressed as the mean ± SD. One-way analysis of variance (ANOVA) followed by Tukey's post-test was performed to evaluate the differences in parameters across groups, with p < 0.05 as significantly different.
Data availability
The data supporting this article have been included as part of the ESI.†
Author contributions
Carolyne B. Braga: conceptualization, methodology, formal analysis, investigation, resources, writing – original draft, writing – review & editing, visualization. Julio C. Milan: conceptualization, methodology, visualization. Matheus A. Meirelles: methodology, investigation, resources, visualization. Bruno Zavan: investigation, writing – original draft, visualization. Ester S. Caixeta: investigation, visualization. Guilherme Álvaro Ferreira-Silva: investigation, methodology. Marisa Ionta: conceptualization, formal analysis, investigation, writing – original draft, writing – review & editing, supervision, resources, visualization. Ronaldo A. Pilli: conceptualization, methodology, investigation, writing – original draft, writing – review & editing, visualization, supervision, project administration, funding acquisition.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Sao Paulo Research Foundation (FAPESP) (fellowships #2017/06146-8 for C. B. B. and #2019/20735-1 for M. A. M. and research grants #2013/07607-8 and #2019/13104-5 for R. A. P.), National Council for Scientific and Technological Development (CNPq, 306747/2020-0 for R. A. P.) and Minas Gerais Research Foundation (FAPEMIG) for financial support to M. I. (#APQ02036-21). This study was financed in part by the Coordination for the Improvement of Higher Education Personnel (CAPES). The authors thank Arnaldo Gomes de Oliveira Junior for HPLC support.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4md00281d
Notes and references
- For a recent review on its traditional uses, phytochemistry, pharmacology, and health-promoting activities, see: ; Biswas P. Ghorai M. Mishra T. Gopalakrishnan A. V. Roy D. Mane A. B. Mundhra A. Das N. Mohture V. M. Patil M. T. Rahman M. H. Jha N. K. Batiha G. E. Saha S. C. Shekhawat M. S. Radha Kumar M. Pandey D. K. Dey A. Phytother. Res. 2022;36:4425–4476. doi: 10.1002/ptr.7649. doi: 10.1002/ptr.7649. [DOI] [PubMed] [Google Scholar]
- Rampogu S. Balasubramaniyam T. Lee J. H. Biomed. Pharmacother. 2022;155:113760. doi: 10.1016/j.biopha.2022.113760. doi: 10.1016/j.biopha.2022.113760. [DOI] [PubMed] [Google Scholar]
- Dodson C. D. Dyer L. A. Searcy J. Wright Z. Letourneau D. K. Phytochemistry. 2000;53:51–54. doi: 10.1016/S0031-9422(99)00446-X. doi: 10.1016/S0031-9422(99)00446-X. [DOI] [PubMed] [Google Scholar]
- Barcelos R. C. Pastre J. C. Vendramini-Costa D. B. Caixeta V. Longato G. B. Monteiro P. A. de Carvalho J. E. Pilli R. A. ChemMedChem. 2014;9:2725–2743. doi: 10.1002/cmdc.201402292. doi: 10.1002/cmdc.201402292. [DOI] [PubMed] [Google Scholar]
- Bharadwaj U. Eckols T. K. Kolosov M. Kasembeli M. M. Adam A. Torres D. Zhang X. Dobrolecki L. E. Wei W. Lewis M. T. Dave B. Chang J. C. Landis M. D. Creighton C. J. Mancini M. A. Tweardy D. J. Oncogene. 2015;34:1341–1353. doi: 10.1038/onc.2014.72. doi: 10.1038/onc.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. Li M. Yu X. Gao F. Li W. Int. J. Biol. Sci. 2019;15:826–837. doi: 10.7150/ijbs.31749. doi: 10.7150/ijbs.31749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Shi Z. Lin J. Zhao S. Hao M. Xu J. Li Y. Zhao Q. Tao L. Diao A. Biochem. Pharmacol. 2019;163:101–110. doi: 10.1016/j.bcp.2019.02.012. doi: 10.1016/j.bcp.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Zhang Q. Chen W. Lv X. Weng Q. Chen M. Cui R. Liang G. Ji J. Front. Pharmacol. 2019;10:1180. doi: 10.3389/fphar.2019.01180. doi: 10.3389/fphar.2019.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piska K. Gunia-Krzyżak A. Koczurkiewicz P. Wójcik-Pszczoła K. Pękala E. Eur. J. Med. Chem. 2018;156:13–20. doi: 10.1016/j.ejmech.2018.06.057. doi: 10.1016/j.ejmech.2018.06.057. [DOI] [PubMed] [Google Scholar]
- Zhu P. Qian J. Xu Z. Meng C. Zhu W. Ran F. Zhang W. Zhang Y. Ling Y. Eur. J. Med. Chem. 2021;220:113471. doi: 10.1016/j.ejmech.2021.113471. doi: 10.1016/j.ejmech.2021.113471. [DOI] [PubMed] [Google Scholar]
- Sun A. L. Mu W. W. Li Y. M. Sun Y. L. Li P. X. Liu R. M. Yang J. Liu G. Y. Molecules. 2021;26:3243. doi: 10.3390/molecules26113243. doi: 10.3390/molecules26113243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. J. Dai M. Pellegrino G. Wagner B. K. Stern A. M. Shamji A. F. Schreiber S. L. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15115–15120. doi: 10.1073/pnas.1212802109. doi: 10.1073/pnas.1212802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. B. Jin X. L. Zheng J. F. Wang F. Dai F. Zhou B. Eur. J. Med. Chem. 2017;126:517–525. doi: 10.1016/j.ejmech.2016.11.034. doi: 10.1016/j.ejmech.2016.11.034. [DOI] [PubMed] [Google Scholar]
- Ji L. Qu L. Wang C. Peng W. Li S. Yang H. Luo H. Yin F. Lu D. Liu X. Kong L. Wang X. Eur. J. Med. Chem. 2021;210:112965. doi: 10.1016/j.ejmech.2020.112965. doi: 10.1016/j.ejmech.2020.112965. [DOI] [PubMed] [Google Scholar]
- Wang Z. Mu W. Gong Z. Liu G. Yang J. Bioorg. Chem. 2021;117:105465. doi: 10.1016/j.bioorg.2021.105465. doi: 10.1016/j.bioorg.2021.105465. [DOI] [PubMed] [Google Scholar]
- Qin J. Li H. Wang X. Zhang Y. Duan Y. Yao Y. Yang H. Sun M. Eur. J. Med. Chem. 2022;243:114738. doi: 10.1016/j.ejmech.2022. doi: 10.1016/j.ejmech.2022. [DOI] [PubMed] [Google Scholar]
- Punganuru S. R. Madala H. R. Venugopal S. N. Samala R. Mikelis C. Srivenugopal K. S. Eur. J. Med. Chem. 2016;107:233–244. doi: 10.1016/j.ejmech.2015.10.052. doi: 10.1016/j.ejmech.2015.10.052. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Patel S. B. King M. R. Molecules. 2020;26:157. doi: 10.3390/molecules26010157. doi: 10.3390/molecules26010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong Hoang Q. Kim M. Kim B. C. Lee C. Y. Shim M. S. Colloids Surf., B. 2022;209:112189. doi: 10.1016/j.colsurfb.2021.112189. doi: 10.1016/j.colsurfb.2021.112189. [DOI] [PubMed] [Google Scholar]
- Truong Hoang Q. Lee D. Choi D. G. Kim Y.-C. Shim M. S. J. Ind. Eng. Chem. 2021;95:101–108. doi: 10.1016/j.jiec.2020.12.009. doi: 10.1016/j.jiec.2020.12.009. [DOI] [Google Scholar]
- Xu R. Yang J. Qian Y. Deng H. Wang Z. Ma S. Wei Y. Yang N. Shen Q. Nanoscale Horiz. 2021;6:348–356. doi: 10.1039/D0NH00674B. doi: 10.1039/D0NH00674B. [DOI] [PubMed] [Google Scholar]
- Braga C. B. Pilli R. A. Ornelas C. Weck M. Biomacromolecules. 2021;22:5290–5306. doi: 10.1021/acs.biomac.1c01196. doi: 10.1021/acs.biomac.1c01196. [DOI] [PubMed] [Google Scholar]
- Patel K. Jain P. Rajput P. K. Jangid A. K. Solanki R. Kulhari H. Patel S. Colloids Surf., A. 2022;652:129738. doi: 10.1016/j.colsurfa.2022.129738. doi: 10.1016/j.colsurfa.2022.129738. [DOI] [Google Scholar]
- Singh P. Sahoo S. K. Int. J. Pharm. 2022;616:121526. doi: 10.1016/j.ijpharm.2022.121526. doi: 10.1016/j.ijpharm.2022.121526. [DOI] [PubMed] [Google Scholar]
- Deng H. Zhang J. Yang Y. Yang J. Wei Y. Ma S. Shen Q. ACS Appl. Mater. Interfaces. 2022;14:24089–24101. doi: 10.1021/acsami.2c00574. doi: 10.1021/acsami.2c00574. [DOI] [PubMed] [Google Scholar]
- Qian J. Xu Z. Zhu P. Meng C. Liu Y. Shan W. He A. Gu Y. Ran F. Zhang Y. Ling Y. J. Nat. Prod. 2021;84:3161–3168. doi: 10.1021/acs.jnatprod.1c00618. doi: 10.1021/acs.jnatprod.1c00618. [DOI] [PubMed] [Google Scholar]
- Kulkarni M. R. Lad N. P. Khedkar V. M. Gaikwad N. D. J. Heterocyclic Chem. 2021;58:1359–1370. doi: 10.1002/jhet.4264. doi: 10.1002/jhet.4264. [DOI] [Google Scholar]
- Grigolo T. A. Braga C. B. Ornelas C. Russowsky D. Ferreira-Silva G. A. Ionta M. Pilli R. A. Bioorg. Chem. 2021;116:105292. doi: 10.1016/j.bioorg.2021.105292. doi: 10.1016/j.bioorg.2021.105292. [DOI] [PubMed] [Google Scholar]
- Cores Á. Carmona-Zafra N. Martín-Cámara O. Sánchez J. D. Duarte P. Villacampa M. Bermejo-Bescós P. Martín-Aragón S. León R. Menéndez J. C. Antioxidants. 2022;11:28. doi: 10.3390/antiox11010028. doi: 10.3390/antiox11010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y. Zhao D. Yan C. Ji Y. Liu J. Xu J. Lai Y. Tian J. Zhang Y. Huang Z. J. Med. Chem. 2018;61:1821–1832. doi: 10.1021/acs.jmedchem.7b01096. doi: 10.1021/acs.jmedchem.7b01096. [DOI] [PubMed] [Google Scholar]
- Lala P. K. Orucevic A. Cancer Metastasis Rev. 1998;17:91–106. doi: 10.1023/a:1005960822365. doi: 10.1023/a:1005960822365. [DOI] [PubMed] [Google Scholar]
- Fukumura D. Kashiwagi S. Jain R. K. Nat. Rev. Cancer. 2006;6:521–534. doi: 10.1038/nrc1910. doi: 10.1038/nrc1910. [DOI] [PubMed] [Google Scholar]
- Perrotta C. Bizzozero L. Falcone S. Rovere-Querini P. Prinetti A. Schuchman E. H. Sonnino S. Manfredi A. A. Clementi E. Cancer Res. 2007;67:7559–7564. doi: 10.1158/0008-5472.CAN-07-0309. doi: 10.1158/0008-5472.CAN-07-0309. [DOI] [PubMed] [Google Scholar]
- Wink D. A. Ridnour L. A. Hussain S. P. Harris C. C. Nitric Oxide. 2008;19:65–67. doi: 10.1016/j.niox.2008.05.003. doi: 10.1016/j.niox.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavida B. Baritaki S. Huerta-Yepez S. Vega M. I. Chatterjee D. Yeung K. Nitric Oxide. 2008;19:152–157. doi: 10.1016/j.niox.2008.04.018. doi: 10.1016/j.niox.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Bonavida B. Baritaki S. Nitric Oxide. 2011;24:1–7. doi: 10.1016/j.niox.2010.10.001. doi: 10.1016/j.niox.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Sugita H. Int. J. Oncol. 2012;40:807–815. doi: 10.3892/ijo.2011.1243. [DOI] [PubMed] [Google Scholar]
- Bian K. Murad F. Nitric Oxide. 2014;43:3–7. doi: 10.1016/j.niox.2014.08.006. doi: 10.1016/j.niox.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Vannini F. Kashfi K. Nath N. Redox Biol. 2015;6:334–343. doi: 10.1016/j.redox.2015.08.009. doi: 10.1016/j.redox.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. Fu J. Zhang Y. J. Med. Chem. 2017;60:7617–7635. doi: 10.1021/acs.jmedchem.6b01672. doi: 10.1021/acs.jmedchem.6b01672. [DOI] [PubMed] [Google Scholar]
- Khan F. H. Dervan E. Bhattacharyya D. D. McAuliffe J. D. Miranda K. M. Glynn S. A. Int. J. Mol. Sci. 2020;21:9393. doi: 10.3390/ijms21249393. doi: 10.3390/ijms21249393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasco A. Boulton A. J. Adv. Heterocycl. Chem. 1981;29:251–340. doi: 10.1016/S0065-2725(08)60789-8. doi: 10.1016/S0065-2725(08)60789-8. [DOI] [Google Scholar]
- Wang P. G. Xian M. Tang X. Wu X. Wen Z. Cai T. Janczuk A. J. Chem. Rev. 2002;102:1091–1134. doi: 10.1021/cr000040l. doi: 10.1021/cr000040l. [DOI] [PubMed] [Google Scholar]
- Cerecetto H. Porcal W. Mini-Rev. Med. Chem. 2005;5:57–71. doi: 10.2174/1389557053402864. doi: 10.2174/1389557053402864. [DOI] [PubMed] [Google Scholar]
- Miller M. R. Megson I. L. Br. J. Pharmacol. 2007;151:305–321. doi: 10.1038/sj.bjp.0707224. doi: 10.1038/sj.bjp.0707224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H. Nitric Oxide. 2008;19:205–216. doi: 10.1016/j.niox.2008.04.026. doi: 10.1016/j.niox.2008.04.026. [DOI] [PubMed] [Google Scholar]
- Medana C. Ermondi G. Fruttero R. Di Stilo A. Ferretti C. Gasco A. J. Med. Chem. 1994;37:4412–4416. doi: 10.1021/jm00051a020. doi: 10.1021/jm00051a020. [DOI] [PubMed] [Google Scholar]
- Burov O. N. Kletskii M. E. Fedik N. S. Lisovin A. V. Kurbatov S. V. Chem. Heterocycl. Compd. 2015;51:951–960. doi: 10.1007/s10593-016-1804-z. doi: 10.1007/s10593-016-1804-z. [DOI] [Google Scholar]
- For a recent review on the role of GSH in cancer, see: ; Kennedy L. Sandhu J. K. Harper M.-E. Cuperlovic-Culf M. Biomolecules. 2020;10:1429. doi: 10.3390/biom10101429. doi: 10.3390/biom10101429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay V. J. Townsend D. M. Morris T. E. Fraser J. P. He L. Tew K. D. Cancer Res. 2006;66:6800–6806. doi: 10.1158/0008-5472.CAN-06-0484. doi: 10.1158/0008-5472.CAN-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra D. Patel V. Banerjee D. Breast Cancer. 2020;14:117822341988268. doi: 10.1177/1178223419882688. doi: 10.1177/1178223419882688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina E. Novichkova M. Molecules. 2021;26:435. doi: 10.3390/molecules26020435. doi: 10.3390/molecules26020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B. K. Bortner C. D. Mason R. P. Cannon R. E. Biochim. Biophys. Acta, Gen. Subj. 2018;1862:2806–2814. doi: 10.1016/j.bbagen.2018.08.021. doi: 10.1016/j.bbagen.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovene C. Chugunova E. A. Goumont R. Mini-Rev. Med. Chem. 2013;13:1089–1136. doi: 10.2174/1389557511313080001. doi: 10.2174/1389557511313080001. [DOI] [PubMed] [Google Scholar]
- Fershtat L. L. Makhova N. N. ChemMedChem. 2017;12:622–638. doi: 10.1002/cmdc.201700113. doi: 10.1002/cmdc.201700113. [DOI] [PubMed] [Google Scholar]
- Chugunova E. Burilov A. Mini-Review. Curr. Top. Med. Chem. 2017;17:986–1005. doi: 10.2174/1568026616666160927145822. doi: 10.2174/1568026616666160927145822. [DOI] [PubMed] [Google Scholar]
- Seabra A. B. Durán N. Eur. J. Pharmacol. 2018;826:158–168. doi: 10.1016/j.ejphar.2018.02.040. doi: 10.1016/j.ejphar.2018.02.040. [DOI] [PubMed] [Google Scholar]
- Glynn S. A. Curr. Opin. Physiol. 2019;9:18–25. doi: 10.1016/j.cophys.2019.03.010. doi: 10.1016/j.cophys.2019.03.010. [DOI] [Google Scholar]
- Ingold M. Colella L. Hernández P. Batthyány C. Tejedor D. Puerta A. García-Tellado F. Padrón J. M. Porcal W. López G. V. ChemMedChem. 2019;14:1669–1683. doi: 10.1002/cmdc.201900385. doi: 10.1002/cmdc.201900385. [DOI] [PubMed] [Google Scholar]
- Ramazani A. Karimi M. Hosseinzadeh Z. Rezayati S. Hanifehpour Y. Joo S. W. Curr. Org. Chem. 2021;25:757–778. doi: 10.2174/1385272825666210208183751. doi: 10.2174/1385272825666210208183751. [DOI] [Google Scholar]
- Fershtat L. L. Zhilin E. S. Molecules. 2021;26:5705. doi: 10.3390/molecules26185705. doi: 10.3390/molecules26185705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola A., Bronte E., Crosetti M., Lazzarato L., Fruttero R. and Gasco A., EP Pat., EP2539328B1, 2010
- Wang T. Zhang Y. H. Kong X. W. Lai Y. S. Ji H. Chen Y. P. Peng S. X. Chem. Biodiversity. 2009;6:466–474. doi: 10.1002/cbdv.200800014. doi: 10.1002/cbdv.200800014. [DOI] [PubMed] [Google Scholar]
- Sorba G. Ermondi G. Fruttero R. Galli U. Gasco A. J. Heterocyclic Chem. 1996;33:327–334. doi: 10.1002/jhet.5570330220. doi: 10.1002/jhet.5570330220. [DOI] [Google Scholar]
- Beddoe R. H. Sneddon H. F. Denton R. M. Org. Biomol. Chem. 2018;16:7774–7781. doi: 10.1039/C8OB01929K. doi: 10.1039/C8OB01929K. [DOI] [PubMed] [Google Scholar]
- Munawar S. Zahoor A. F. Ali S. Javed S. Irfan M. Irfan A. Kotwica-Mojzych K. Mojzych M. Molecules. 2022;27:6953. doi: 10.3390/molecules27206953. doi: 10.3390/molecules27206953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasco A. M. Fruttero R. Sorba G. Gasco A. Liebigs Ann. Chem. 1991;11:1211–1213. doi: 10.1002/jlac.1991199101207. doi: 10.1002/jlac.1991199101207. [DOI] [Google Scholar]
- He F. Ru X. Wen T. Int. J. Mol. Sci. 2020;21:4777. doi: 10.3390/ijms21134777. doi: 10.3390/ijms21134777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlonka J. Rak B. Ambroziak U. Cancer Treat Res. Commun. 2021;27:100338. doi: 10.1016/j.ctarc.2021.100338. doi: 10.1016/j.ctarc.2021.100338. [DOI] [PubMed] [Google Scholar]
- Ala M. Cancer Biol. Ther. 2022;23:34–50. doi: 10.1080/15384047.2021.2017223. doi: 10.1080/15384047.2021.2017223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretones G. Delgado M. D. León J. Biochim. Biophys. Acta. 1849;2015:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Civenni G. Malek A. Albino D. Garcia-Escudero R. Napoli S. Marco S. D. Pinton S. Sarti M. Carbone G. M. Catapano C. V. Cancer Res. 2013;73:6816–6827. doi: 10.1158/0008-5472.CAN-13-0615. doi: 10.1158/0008-5472.CAN-13-0615. [DOI] [PubMed] [Google Scholar]
- Du S. Wang H. Cai J. Ren R. Zhang W. Wei W. Shen X. Biochem. Cell Biol. 2020;98:191–202. doi: 10.1139/bcb-2018-0230. doi: 10.1139/bcb-2018-0230. [DOI] [PubMed] [Google Scholar]
- Kale J. Osterlund E. J. Andrews D. W. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatok J. Racay P. Biomol. Concepts. 2016;7:259–270. doi: 10.1515/bmc-2016-0015. [DOI] [PubMed] [Google Scholar]
- Dadsena S. King L. E. García-Sáez A. J. Biochim. Biophys. Acta, Biomembr. 2021;1863:183716. doi: 10.1016/j.bbamem.2021.183716. doi: 10.1016/j.bbamem.2021.183716. [DOI] [PubMed] [Google Scholar]
- Liu Q. Y. Stein C. A. Clin. Cancer Res. 1997;3:2039–2046. [PubMed] [Google Scholar]
- Kojima H. Endo K. Moriyama H. Tanaka Y. Alnemri E. S. Slapak C. A. Teicher B. Kufe D. Datta R. J. Biol. Chem. 1998;237:16647–16650. doi: 10.1074/jbc.273.27.16647. doi: 10.1074/jbc.273.27.16647. [DOI] [PubMed] [Google Scholar]
- Li X. Marani M. Mannucci R. Kinsey B. Andriani F. Nicoletti I. Denner L. Marcelli M. Overexpression of BCL-XL underlies the molecular basis for resistance to staurosporine-induced apoptosis in PC-3 cells. Cancer Res. 2001;61(4):1699–1706. [PubMed] [Google Scholar]
- Castilla C. Congregado B. Chinchón D. Torrubia F. J. Japón M. A. Sáez C. Endocrinology. 2006;147:4960–4967. doi: 10.1210/en.2006-0502. doi: 10.1210/en.2006-0502. [DOI] [PubMed] [Google Scholar]
- Poudel S. Song J. Jin E. J. Song K. Biochem. Biophys. Res. Commun. 2013;431:572–578. doi: 10.1016/j.bbrc.2013.01.012. doi: 10.1016/j.bbrc.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Martin J. M. C. Balkenende A. Verschoor T. Lallemand F. Michalides R. Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res. 1999;59(5):1134–1140. [PubMed] [Google Scholar]
- Niu M. Y. Menard M. Reed J. C. Krajewski S. Pratt M. A. Oncogene. 2001;20:3506–3518. doi: 10.1038/sj.onc.1204453. doi: 10.1038/sj.onc.1204453. [DOI] [PubMed] [Google Scholar]
- Roué G. Pichereau V. Lincet H. Colomer D. Sola B. Oncogene. 2008;27:4909–4920. doi: 10.1038/onc.2008.126. doi: 10.1038/onc.2008.126. [DOI] [PubMed] [Google Scholar]
- Schieber M. Chandel N. S. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. Shi L. Zhang T. Hong L. He W. Cao P. Shen X. Zheng P. Xia Y. Zou P. Cell. Oncol. 2019;42:847–860. doi: 10.1007/s13402-019-00471-x. doi: 10.1007/s13402-019-00471-x. [DOI] [PubMed] [Google Scholar]
- Nortcliffe A. Ekstrom A. G. Black J. R. Ross J. A. Habib F. K. Botting N. P. O'Hagan D. Bioorg. Med. Chem. 2014;22:756–761. doi: 10.1016/j.bmc.2013.12.014. doi: 10.1016/j.bmc.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Wang T. Zhang Y. H. Kong X. W. Lai Y. S. Ji H. Chen Y. P. Peng S. X. Chem. Biodiversity. 2009;6:466–474. doi: 10.1002/cbdv.200800014. doi: 10.1002/cbdv.200800014. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi F. K. McBride A. A. Leppert H. G. Millet L. J. Wang X. Davern S. M. Sci. Rep. 2021;11:8945. doi: 10.1038/s41598-021-88296-3. doi: 10.1038/s41598-021-88296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article have been included as part of the ESI.†