Abstract
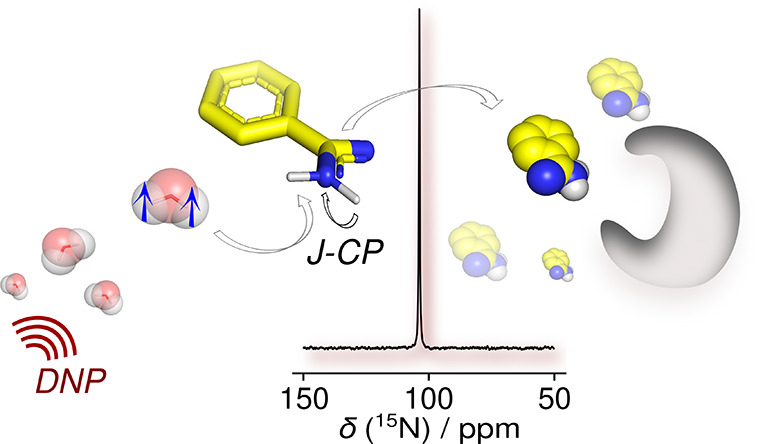
Hyperpolarization derived from water protons enhances the NMR signal of 15N nuclei in a small molecule, enabling the sensitive detection of a protein–ligand interaction. The water hyperpolarized by dissolution dynamic nuclear polarization (D-DNP) acts as a universal signal enhancement agent. The 15N signal of benzamidine was increased by 1480-fold through continuous polarization transfer by J-coupling-mediated cross-polarization (J-CP) via the exchangeable protons. The signal enhancement factor favorably compares to factors of 110- or 17-fold using non-CP-based polarization transfer mechanisms. The hyperpolarization enabled detection of the binding of benzamidine to the target protein trypsin with a single-scan measurement of 15N R2 relaxation. J-CP provides an efficient polarization mechanism for 15N or other low-frequency nuclei near an exchangeable proton. The hyperpolarization transfer sustained within the relaxation time limit of water protons additionally can be applied for the study of macromolecular structure and biological processes.
The amplification of signal from hyperpolarization enables NMR spectroscopy at physiologically relevant concentrations and improves the time resolution for the study of biochemical processes.1−3 Dissolution dynamic nuclear polarization (D-DNP)4 increases signals of low-frequency nuclei such as 13C and 15N by 3–4 orders of magnitude. Spectra measured in vitro or in cellulo reveal biosynthetic and metabolic pathways,5 reaction kinetics and mechanisms,6 protein folding,7 and other processes on the subsecond time scale.8 At equilibrium, hyperpolarization facilitates the measurement of macromolecular interactions, binding affinity,9 or the structures of binding interfaces and pockets.10
Although DNP readily polarizes the nuclear spins of most molecules in frozen solids, the decay of the hyperpolarization is accelerated after transfer to the liquid state. In aqueous solution, the available time and signal levels may be substantially extended by first hyperpolarizing water protons. Polarization from this reservoir can transfer repeatedly to the molecule of interest through proton exchange and dipolar interactions.11 Proton signals become enhanced for a period governed by the T1 relaxation time of water protons.12 This technique, also termed HyperW, removes the obstacle of fast relaxation for ex situ polarized bio-macromolecular spins, while retaining the physiological environment of the molecules to support native conformations.13,14 It holds promise to broaden the applications of NMR for structural biology, biomolecular dynamics,15 protein–ligand interactions, and others.16
We demonstrate the substantial potential that water hyperpolarization holds for the signal enhancement of a low-frequency nucleus, 15N. We then apply this technique for the measurement of the binding of benzamidine, a reversible inhibitor of trypsin and trypsin-like proteases. While the ability to hyperpolarize proton spins of a target molecule is improved by exchange with water protons, a short complex lifetime reduces the amount of polarization transferred to neighboring spins. This limitation is overcome by J-coupling-mediated cross-polarization (J-CP).
First, one of the nitrogen atoms in benzamidine was enriched with 15N (Scheme S1 and Figures S1–S5).17 Hyperpolarization of water was generated using DNP in a frozen solid at 1.4 K. Subsequent dissolution in D2O and rapid injection into a 400 MHz NMR spectrometer resulted in a water signal that was 40 ± 2-fold larger than the signal from a non-hyperpolarized pure water sample, despite a 1:26 dilution with deuterium.18 The signal enhancement of water protons from DNP was ∼2400-fold, resulting in an ∼1100-fold effective enhancement after introduction of residual protons of the dissolution solvent. J-CP transfer of this polarization under RF irradiation resulted in a 15N signal of benzamidine that was 757 ± 58-fold enhanced compared with non-hyperpolarized J-CP (Figure 1). Hyperpolarized J-CP also exhibited 1480 ± 110-fold 15N signal enhancement in contrast with non-hyperpolarized direct 15N measurements.
Figure 1.
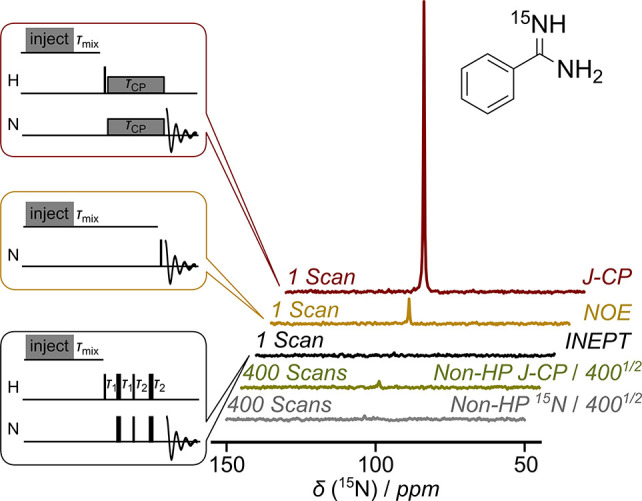
Polarization transfer from hyperpolarized water to 9.1 mM, 9.6 mM, and 9.1 mM benzamidine, employing J-CP, NOE, and INEPT, at pH 7.25. The injection of hyperpolarized water was followed by τmix = 500 ms, 15 s, and 500 ms, respectively. J-CP used τCP = 160 ms, γB1 = 3125 Hz, ωN = 105 ppm, and ωH = 8 ppm. Refocused INEPT included τ1 = 1/(4JHN), τ2 = 1/(6JHN), and JHN = 92 Hz. Non-hyperpolarized experiments were performed with a sample from a previous hyperpolarized experiment containing 9.08 mM benzamidine, in 400 scans. The vertical scale is reduced by 4001/2 to match noise levels.
Continuous transfer of water hyperpolarization can also occur through the NOE.12,19 A 15N signal enhancement of 110 ± 18-fold was observed after a contact time of 15 s. A refocused “insensitive nuclei enhanced by polarization transfer” (INEPT) scheme increased the 15N magnetization through the J-coupling merely by a factor of 17 ± 6 (Figure 1). As the NOE in small molecules is inefficient due to rapid motions and the otherwise preferred INEPT technique provided even less signal, neither of these options is ideal. The inefficiency of INEPT is explained by the fast-exchange kinetics of the water protons with the amidine protons. The exchange rate, kex, due to the hydroxide base catalyzed exchange mechanism at the physiological pH of 7.25 is estimated as 1700 s–1 (Figure S7).20,21 This rate is much higher than JHN = 92 Hz, which severely limits the INEPT polarization transfer. The limitation is overcome with J-CP, an effective method for sensitivity enhancement of low-frequency nuclei in solution.22,23 With proton exchange, the J-CP polarization transfer can occur continuously during the CP mixing time. The J-CP with Hartmann–Hahn matched spin-locking fields on 1H and 15N has provided improved polarization transfer under fast solvent exchange conditions in proteins without hyperpolarization.24,25 The data in Figure 1 demonstrate that HyperW with J-CP is an efficient approach for biomolecular polarization.
Because HyperW experiments generally dissolve hyperpolarized water in D2O, we explored the CP efficiency with variable water percentages (Figure S8). In these experiments performed without hyperpolarization, the maximum signal reached within 160 ms of CP buildup time is nearly independent of the water proton content. However, with higher deuterium enrichment, a shift in the maximum toward a longer CP time is observed. Concomitantly, upon lowering the water proton content from 90% to 10%, the initial slope of the buildup curve was reduced by 40%. This difference is explained by the fact that the polarization transfer originates primarily from the proton spins. Additionally, deuteration decreases the spin–lattice relaxation rates for water protons from 0.28 s–1 to 0.09 s–1 (Figure S9a–c). Under fast exchange, the 15N magnetization achieved in a given CP time depends on 15N and water proton T1 and not significantly on the T1 of the 15N-bound exchangeable proton.25 The reduced relaxation in the deuterated solutions, combined with the slower polarization transfer, explains the aforementioned shift of the signal maximum to a longer CP time.
The opposing effects on the relaxation and polarization transfer rates also equalize the maximum achievable 15N signal at different ratios of water and deuterium seen in Figure S8. The transferable signal in the J-CP experiment apparently depends primarily on the water proton polarization, even if the magnitude of the water signal is reduced with a lower proton concentration. In contrast, HyperW experiments that observe signals from exchangeable protons directly yield a signal proportional to the product of water proton concentration and polarization. By retaining its effectiveness at the lowered proton concentration, the J-CP experiment is ideally compatible with D-DNP. Due to the dilution of the hyperpolarized water with D2O during the dissolution, J-CP supports a higher polarization of the target nucleus than HyperW alone.
Figure 2 compares the signal buildup from hyperpolarized water to the 15N of benzamidine due to J-CP or NOE. In J-CP, the maximum buildup is beyond the longest mixing time of 160 ms. This result was expected, since hyperpolarized solutions contain only 3.9 ± 0.9% of water due to dilution in D2O. The water T1 was prolonged to 5.1 s, despite a relaxation contribution from 0.45 mM TEMPOL in the sample after dissolution (Figure S9e,f).
Figure 2.
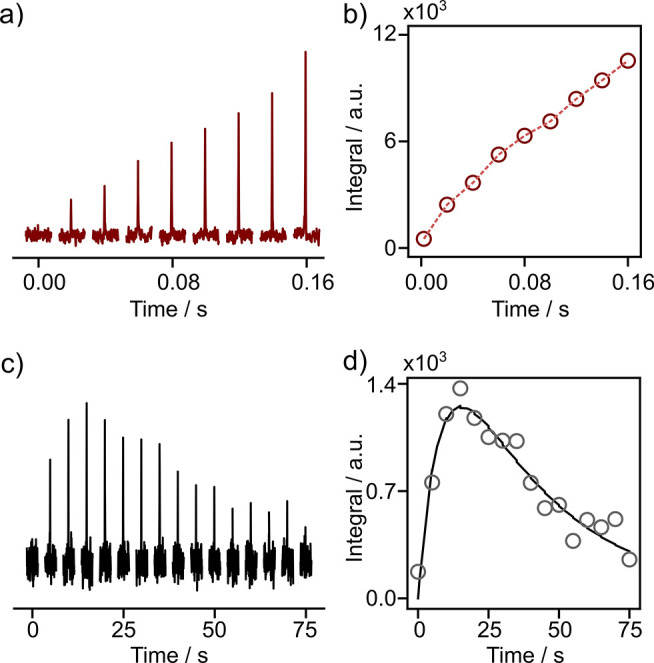
(a) 15N NMR signals of 1.23 ± 0.11 mM benzamidine transferred from hyperpolarized water using J-CP with different CP times. (b) Buildup curve of 15N magnetization achieved in (a). (c) Time-dependent 15N NMR signals measured from 14.6 mM benzamidine following NOE transfer of polarization from hyperpolarized water. Spectra were acquired with 9° pulses at intervals of 5 s. (d) Fitted curve of integrals from (c), resulting in R1N = 0.13 s–1, R1H = 0.03 s–1, and kH→N = 1.41 s–1. In (a) and (b), τmix = 500 ms. Integrals were normalized with the benzamidine concentration and for the flip-angle of the pulse.12,19
The J-CP efficiency leading to the observed buildup curve depends on the amidine proton exchange rate. The 15N signal integrals in a J-CP experiment with benzamidine decrease at high pH, where the rate is increased (Figure S10). A reduction of the J-CP efficiency with an exchange rate that is significantly larger than JHN has also been seen in ref (25). Nevertheless, the efficiency of J-CP sharply surpasses that of INEPT for a fast-exchanging proton.
The J-CP polarization transfer also depends on experimental parameters, including the RF field strength γB1, frequency offsets, and quality of radio frequency power matching (Figure S11). The DIPSI sequence is used in the experiment to improve the offset dependence.26,27 The γB1 = 3125 Hz of this sequence is sufficient to cover the line width of the amidine signal at pH 7.25 and at least partially spin-lock the signal of water, while preventing excessive RF power deposition.
The analysis of the NOE buildup and decay curve shows a maximum 15N polarization at only 15 s (Figure 2d). In addition, the starting slope of the CP buildup curve was 440 times larger than that of the NOE (Figure 2c). Different from the coherent magnetization transfer in J-CP, the NOE buildup depends on relaxation rates of amidine 15N and 1H, as well as hyperpolarized water.12 The curve reflects the longitudinal relaxation rate of 0.167 ± 0.061 s–1 and the overall polarization transfer rate from water to 15N due to exchange and NOE processes, kH→N = 1.57 ± 0.13 s–1. Since the amidine 1H is in fast exchange with hyperpolarized water at a pH of 7.25, the intramolecular polarization transfer rate is the rate-limiting step. As a result, the NOE enhanced 15N polarization is approximately 13 times less than that in the J-CP experiment.
The signal-to-noise ratio (SNR) in the J-CP experiment in Figure 1 is ≥550. The detection limit corresponding to an SNR of 3 would be reached with ∼50 μM substrate (SI Section 7). This substrate concentration enables the study of biological systems at or near physiological conditions, even while observing the otherwise insensitive 15N nucleus. It would further be possible to observe ∼12.5 mM of unenriched substrate at the 0.4% natural abundance of 15N with an SNR of 3.
In the following, enhanced 15N magnetization from J-CP polarization transfer was used to measure the interaction of benzamidine with the protein trypsin. The R2 relaxation rates of the polarized 15N spin in benzamidine were determined by a single-scan Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence incorporating J-CP transfer (Figure S6). The relaxation rate increased from R2 = 31.3 ± 2.5 s–1 for benzamidine alone to 41.1 ± 4.0 s–1 in the presence of trypsin protein (Figure 3 and Table S2; average of three measurements each with error ranges propagated from fit). The increased R2 relaxation rate indicates binding of benzamidine with trypsin.
Figure 3.
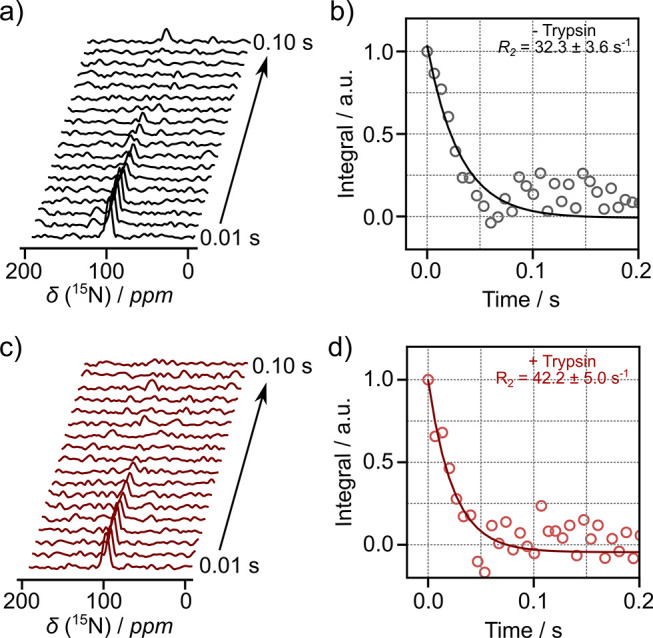
(a) 15N NMR spectra of 0.95 mM polarized benzamidine from the echoes in a single-scan CPMG experiment following J-CP. (b) Fitted exponential curve measuring R2 from (a). (c) 15N spectra of 0.88 mM polarized benzamidine with 50 μM trypsin. (d) Fitted curve from (c). All experiments were performed with hyperpolarized water at pH 7.25 ± 0.04, an echo time of 6.7 ms, and τmix = 500 ms. The integrated data points were normalized to the first point.
The R2 relaxation rate of the free ligand is relatively fast, likely due to exchange effects. These effects may among others include the influence of H/H and H/D exchange, which can result in the loss of antiphase coherences that appear between refocusing pulses.28 When the protein is bound, the exchange contribution may be reduced, but the increased rotational correlation time of the protein–ligand complex substantially increases R2. In Figure 3, the change in the relaxation rate upon binding to the protein was observable, despite the relatively fast relaxation rate of free benzamidine.
The data in Figure 3 emphasize the possibility of 15N NMR to prove a biological interaction in a single experiment. The binding affinity may further be determined by measuring R2 at different concentrations of the ligand or the protein, or using competitive binding experiments.29,30 Practically, a non-hyperpolarized 15N relaxation experiment will require ≥560,000 signal averages for gaining the same signal-to-noise ratio as in the hyperpolarized J-CP experiment with the signal enhancement of ≥750. In contrast, the single scan experiment using hyperpolarized water was performed in 40 min, including the water polarization time.
Hyperpolarization of 15N through HyperW opens new applications benefiting from well-dispersed and background-free 15N NMR signals. These include the screening of ligand interactions in drug development and the study of receptors and other targets that are important for cellular functions. Nitrogen nuclei with exchangeable protons are ubiquitous in biological macromolecules, including proteins and nucleic acids. Nitrogen is also part of small molecules such as adenosine diphosphate, adenosine triphosphate, cyclic guanosine monophosphate, biotin, dopamine, and gamma-aminobutyric acid, which are all involved in cell signaling. These molecules represent a large pool of substrates for NMR-based cell signaling studies. In proteins, polarized 15N relaxation experiments can provide insight into local backbone dynamics and transient protein–protein interactions. Hyperpolarized 15N relaxation dispersion may further identify binding epitopes of ligands with macromolecules.3115N polarization may also be adopted to perform in-cell NMR studies that provide structural and dynamic information about biological molecules and processes under physiological conditions.32 The 15N polarization may further be exploited to detect biomarkers for applications in metabolomic research.33,34
In summary, the 15N hyperpolarization of benzamidine using hyperpolarized water and J-CP was demonstrated. The transferred polarization outweighed both INEPT- and NOE-based pulse sequences in the molecule containing a fast-exchanging proton. The enhanced signals were used to measure the interaction of the molecule with trypsin as a target protein. The use of hyperpolarized water for the polarization of low-γ nuclei is applicable to various biological small molecules and macromolecules, for measuring protein–ligand binding, protein dynamics, protein–protein interactions, and others, in a native environment and short time frame.
Acknowledgments
Financial support from the National Institutes of Health (Grant R01GM132655) and the Welch Foundation (Grant A-1658) is gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.4c08241.
Synthesis of 15N-benzamidine, experimental methods, exchange rate calculation, J-CP buildup curve, water T1 relaxation experiments, measurement of J-CP efficiency, 15N signal-to-noise ratio, NOE data, and protein interaction data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Eills J.; Budker D.; Cavagnero S.; Chekmenev E. Y.; Elliott S. J.; Jannin S.; Lesage A.; Matysik J.; Meersmann T.; Prisner T.; Reimer J. A.; Yang H.; Koptyug I. V. Spin Hyperpolarization in Modern Magnetic Resonance. Chem. Rev. 2023, 123 (4), 1417–1551. 10.1021/acs.chemrev.2c00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen S.; Hilty C. Time-Resolved Dynamic Nuclear Polarization Enhanced NMR Spectroscopy. Angew. Chem., Int. Ed. 2008, 47 (28), 5235–5237. 10.1002/anie.200801492. [DOI] [PubMed] [Google Scholar]
- Lee Y.; Zeng H.; Ruedisser S.; Gossert A. D.; Hilty C. Nuclear Magnetic Resonance of Hyperpolarized Fluorine for Characterization of Protein-Ligand Interactions. J. Am. Chem. Soc. 2012, 134 (42), 17448–17451. 10.1021/ja308437h. [DOI] [PubMed] [Google Scholar]
- Ardenkjær-Larsen J. H.; Fridlund B.; Gram A.; Hansson G.; Hansson L.; Lerche M. H.; Servin R.; Thaning M.; Golman K. Increase in Signal-to-Noise Ratio of > 10,000 Times in Liquid-State NMR. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (18), 10158–10163. 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee S. S.; DiGialleonardo V.; Eskandari R.; Jeong S.; Granlund K. L.; Miloushev V.; Poot A. J.; Truong S.; Alvarez J. A.; Aldeborgh H. N.; Keshari K. R. Sampling Hyperpolarized Molecules Utilizing a 1 T Permanent Magnetic Field. Sci. Rep. 2016, 6 (1), 32846. 10.1038/srep32846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchak S.; Jagtap A. P.; Glöggler S. Signal-Enhanced Real-Time Magnetic Resonance of Enzymatic Reactions at Millitesla Fields. Chem. Sci. 2021, 12 (1), 314–319. 10.1039/D0SC04884D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragavan M.; Iconaru L. I.; Park C.-G.; Kriwacki R. W.; Hilty C. Real-Time Analysis of Folding upon Binding of a Disordered Protein by Using Dissolution DNP NMR Spectroscopy. Angew. Chem., Int. Ed. 2017, 56 (25), 7070–7073. 10.1002/anie.201700464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C.; Mankinen O.; Telkki V.-V.; Hilty C. Measuring Protein–Ligand Binding by Hyperpolarized Ultrafast NMR. J. Am. Chem. Soc. 2024, 146 (8), 5063–5066. 10.1021/jacs.3c14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratto R.; Bornet A.; Milani J.; Mammoli D.; Vuichoud B.; Salvi N.; Singh M.; Laguerre A.; Passemard S.; Gerber-Lemaire S.; Jannin S.; Bodenhausen G. Drug Screening Boosted by Hyperpolarized Long-Lived States in NMR. ChemMedChem. 2014, 9 (11), 2509–2515. 10.1002/cmdc.201402214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Kim J.; Hilty C. Determination of Protein–Ligand Binding Modes Using Fast Multi-Dimensional NMR with Hyperpolarization. Chem. Sci. 2020, 11 (23), 5935–5943. 10.1039/D0SC00266F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilty C.; Kurzbach D.; Frydman L. Hyperpolarized Water as Universal Sensitivity Booster in Biomolecular NMR. Nat. Protoc. 2022, 17 (7), 1621–1657. 10.1038/s41596-022-00693-8. [DOI] [PubMed] [Google Scholar]
- Harris T.; Szekely O.; Frydman L. On the Potential of Hyperpolarized Water in Biomolecular NMR Studies. J. Phys. Chem. B 2014, 118 (12), 3281–3290. 10.1021/jp4102916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epasto L. M.; Che K.; Kozak F.; Selimovic A.; Kadeřávek P.; Kurzbach D. Toward Protein NMR at Physiological Concentrations by Hyperpolarized Water—Finding and Mapping Uncharted Conformational Spaces. Sci. Adv. 2022, 8 (31), eabq5179 10.1126/sciadv.abq5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadet A.; Stavarache C.; Bacalum M.; Radu M.; Bodenhausen G.; Kurzbach D.; Vasos P. R. Hyperpolarized Water Enhances Two-Dimensional Proton NMR Correlations: A New Approach for Molecular Interactions. J. Am. Chem. Soc. 2019, 141 (32), 12448–12452. 10.1021/jacs.9b03651. [DOI] [PubMed] [Google Scholar]
- Kurzbach D.; Canet E.; Flamm A. G.; Jhajharia A.; Weber E. M. M.; Konrat R.; Bodenhausen G. Investigation of Intrinsically Disordered Proteins through Exchange with Hyperpolarized Water. Angew. Chem., Int. Ed. 2017, 56 (1), 389–392. 10.1002/anie.201608903. [DOI] [PubMed] [Google Scholar]
- Zhao E. W.; Maligal-Ganesh R.; Du Y.; Zhao T. Y.; Collins J.; Ma T.; Zhou L.; Goh T.-W.; Huang W.; Bowers C. R. Surface-Mediated Hyperpolarization of Liquid Water from Parahydrogen. Chem. 2018, 4 (6), 1387–1403. 10.1016/j.chempr.2018.03.004. [DOI] [Google Scholar]
- Creary X.; Sky A. F. Reaction of Arylbromodiazirines with Azide Ion. Evidence for N-Azidodiazirine Intermediates. J. Am. Chem. Soc. 1990, 112 (1), 368–374. 10.1021/ja00157a056. [DOI] [Google Scholar]
- Bowen S.; Hilty C. Rapid Sample Injection for Hyperpolarized NMR Spectroscopy. Phys. Chem. Chem. Phys. 2010, 12 (22), 5766–5770. 10.1039/c002316g. [DOI] [PubMed] [Google Scholar]
- Kim J.; Liu M.; Chen H.-Y.; Hilty C. Determination of Intermolecular Interactions Using Polarization Compensated Heteronuclear Overhauser Effect of Hyperpolarized Spins. Anal. Chem. 2015, 87 (21), 10982–10987. 10.1021/acs.analchem.5b02934. [DOI] [PubMed] [Google Scholar]
- Eigen M. Proton Transfer, Acid-Base Catalysis, and Enzymatic Hydrolysis. Part I: Elementary Processes. Angew. Chem., Int. Ed. 1964, 3 (1), 1–19. 10.1002/anie.196400011. [DOI] [Google Scholar]
- Perrin C. L.; Schiraldi D. A.; Lin G. M. L. Positional Selectivity in an Encounter-Controlled Reaction: Base-Catalyzed Proton Exchange in Amidinium Ions. J. Am. Chem. Soc. 1982, 104 (1), 196–201. 10.1021/ja00365a035. [DOI] [Google Scholar]
- Chingas G. C.; Garroway A. N.; Bertrand R. D.; Moniz W. B. Zero Quantum NMR in the Rotating Frame: J Cross Polarization in AX N Systems. J. Chem. Phys. 1981, 74 (1), 127–156. 10.1063/1.440866. [DOI] [Google Scholar]
- Bertrand R. D.; Moniz W. B.; Garroway A. N.; Chingas G. C. 13C-1H Cross-Polarization in Liquids. J. Am. Chem. Soc. 1978, 100 (16), 5227–5229. 10.1021/ja00484a063. [DOI] [Google Scholar]
- Kim J.; Grün J. T.; Novakovic M.; Kupce E.; Rosenzweig R.; Frydman L. Cross-Polarization Schemes for Improved Heteronuclear Transfers Involving Labile Protons in Biomolecular Solution NMR. Angew. Chem., Int. Ed. 2023, 62 (35), e202304900 10.1002/anie.202304900. [DOI] [PubMed] [Google Scholar]
- Novakovic M.; Jayanthi S.; Lupulescu A.; Concilio M. G.; Kim J.; Columbus D.; Kuprov I.; Frydman L. Heteronuclear Transfers from Labile Protons in Biomolecular NMR: Cross Polarization. Revisited. J. Magn. Reson. 2021, 333, 107083. 10.1016/j.jmr.2021.107083. [DOI] [PubMed] [Google Scholar]
- Rucker S. P.; Shaka A. J. Broadband Homonuclear Cross Polarization in 2D N.M.R. Using DIPSI-2. Mol. Phys. 1989, 68 (2), 509–517. 10.1080/00268978900102331. [DOI] [Google Scholar]
- Brown L. R.; Sanctuary B. C. Hetero-TOCSY Experiments with WALTZ and DIPSI Mixing Sequences. J. Magn. Reson. 1991, 91 (2), 413–421. 10.1016/0022-2364(91)90207-A. [DOI] [Google Scholar]
- Kim S.; Wu K.-P.; Baum J. Fast Hydrogen Exchange Affects 15N Relaxation Measurements in Intrinsically Disordered Proteins. J. Biomol. NMR 2013, 55 (3), 249–256. 10.1007/s10858-013-9706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossert A. D.; Jahnke W. NMR in Drug Discovery: A Practical Guide to Identification and Validation of Ligands Interacting with Biological Macromolecules. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 97, 82–125. 10.1016/j.pnmrs.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Hilty C. Affinity Screening Using Competitive Binding with Fluorine-19 Hyperpolarized Ligands. Angew. Chem., Int. Ed. 2015, 54 (16), 4941–4944. 10.1002/anie.201411424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C.; Wang Y.; Hilty C. Application of Relaxation Dispersion of Hyperpolarized 13C Spins to Protein–Ligand Binding. Angew. Chem., Int. Ed. 2021, 60 (45), 24018–24021. 10.1002/anie.202109430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchinat E.; Barbieri L.; Cremonini M.; Nocentini A.; Supuran C. T.; Banci L. Drug Screening in Human Cells by NMR Spectroscopy Allows the Early Assessment of Drug Potency. Angew. Chem., Int. Ed. 2020, 59 (16), 6535–6539. 10.1002/anie.201913436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. P.; Brahms A.; Janicaud V.; Anikeeva M.; Peschke E.; Ellermann F.; Ferrari A.; Hellmold D.; Held-Feindt J.; Kim N.; Meiser J.; Aden K.; Herges R.; Hövener J.-B.; Pravdivtsev A. N. Nitrogen-15 Dynamic Nuclear Polarization of Nicotinamide Derivatives in Biocompatible Solutions. Sci. Adv. 2023, 9 (34), eadd3643 10.1126/sciadv.add3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez B.; Durst M. A.; Lavie A.; Caffrey M. NMR-Based Metabolite Studies with 15N Amino Acids. Sci. Rep. 2019, 9 (1), 12798. 10.1038/s41598-019-49208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


