Abstract
Aedes aegypti densonucleosis virus (AeDNV) has two promoters that have been shown to be active by reporter gene expression analysis (B. N. Afanasiev, Y. V. Koslov, J. O. Carlson, and B. J. Beaty, Exp. Parasitol. 79:322–339, 1994). Northern blot analysis of cells infected with AeDNV revealed two transcripts 1,200 and 3,500 nucleotides in length that are assumed to express the structural protein (VP) gene and nonstructural protein genes, respectively. Primer extension was used to map the transcriptional start site of the structural protein gene. Surprisingly, the structural protein gene transcript began at an initiator consensus sequence, CAGT, 60 nucleotides upstream from the map unit 61 TATAA sequence previously thought to define the promoter. Constructs with the β-galactosidase gene fused to the structural protein gene were used to determine elements necessary for promoter function. Deletion or mutation of the initiator sequence, CAGT, reduced protein expression by 93%, whereas mutation of the TATAA sequence at map unit 61 had little effect. An additional open reading frame was observed upstream of the structural protein gene that can express β-galactosidase at a low level (20% of that of VP fusions). Expression of the AeDNV structural protein gene was shown to be stimulated by the major nonstructural protein NS1 (Afanasiev et al., Exp. parasitol., 1994). To determine the sequences required for transactivation, expression of structural protein gene–β-galactosidase gene fusion constructs differing in AeDNV genome content was measured with and without NS1. The presence of NS1 led to an 8- to 10-fold increase in expression when either genomic end was present, compared to a 2-fold increase with a construct lacking the genomic ends. An even higher (37-fold) increase in expression occurred with both genomic ends present; however, this was in part due to template replication as shown by Southern blot analysis. These data indicate the location and importance of various elements necessary for efficient protein expression and transactivation from the structural protein gene promoter of AeDNV.
Densoviruses are autonomous parvoviruses that infect arthropods. Aedes aegypti densonucleosis virus (AeDNV) is in the genus Brevidensovirus (4, 7). Its 4-kb negative-sense, single-stranded DNA genome can be divided into two parts, with the nonstructural protein genes at the left end and the structural protein gene at the right end (1). AeDNV has two nonstructural proteins, NS1 and NS2, that are encoded within the same DNA sequence in two different open reading frames (ORFs). NS1 is required for viral replication and has been implicated in the transactivation of viral promoters (2, 3). The structural proteins VP1 and VP2 are encoded within the same ORF (1). VP2 may be a proteolytic cleavage product of VP1 or the result of a different translation initiation codon.
Brevidensoviruses, like vertebrate parvoviruses, encode all of their proteins on the same strand (1, 10). Based on the location of TATAA boxes and ORFs, promoters were previously predicted to be at map units 7 and 61 for the nonstructural and structural genes of AeDNV, respectively (1, 2). These regions are conserved between AeDNV, Aedes albopictus parvovirus (AaPV), and a new isolate from mosquito cells (1, 10; B. N. Afanasiev, unpublished observations).
Extensive study of parvovirus promoter structure and transcriptional regulation has been mainly confined to the mammalian parvoviruses (10). These viruses use alternative splicing to yield different coterminal transcripts from the same promoter (for a review, see reference 8), which increases the number of protein species produced. Core promoter elements, which have been found to include a TATAA element and an upstream SP1 binding site (21), are sensitive to the presence of the viral NS1 protein (14, 17, 26, 30). However, the core promoter structure of densoviruses is not well defined. A parvovirus of cockroaches (Periplaneta fuliginosa DNV), of the genus Densovirus, is likely to utilize alternative splicing (31), but it is not clear whether members of the genus Brevidensovirus or Iteravirus do so as well. Some indirect evidence does suggest that AeDNV and members of the Densovirus genus can initiate translation at multiple AUG codons to produce multiple proteins from the same transcript (7, 17). Promoters of baculoviruses, another family of arthropod viruses, have been studied intensively. Early genes have been found to utilize TATAA sequences and an initiator sequence CAGT (9, 25). Late genes are expressed using a viral polymerase that initiates transcription at a TAAG sequence (9, 23, 25). The CAGT motif of the early genes can function without an accompanying TATAA sequence (9, 23, 25). This CAGT sequence has been shown to be important for expression from many arthropod and mammalian promoters whether or not they contain a TATAA sequence (9, 11, 27). This CAGT motif is observed downstream of TATAA sequences of both putative promoter regions of AeDNV.
This report presents analysis of expression from the structural protein gene promoter of AeDNV. We used Northern blot analysis to identify RNA species production and primer extension to map the transcription start site for the structural gene. Deletion analysis and site-directed mutagenesis were used to identify structural gene promoter sequence elements, and the sequences required for transactivation were also investigated.
MATERIALS AND METHODS
Cloning and mutagenesis.
All plasmid clones were grown in Escherichia coli DH5-α cells. pUCA, the infectious clone of the AeDNV genome, is described in detail elsewhere (2). pUCAINV is the transactivating construct used to supply NS1 without VPs (2). nsp61gal was derived from pUCA by inserting the lacZ gene in frame with the VPs at the SnaBI site at nucleotide 2674 as described elsewhere (2). pVPNco contains virus sequences from nucleotides 2043 to 2674 including the structural gene promoter driving expression of the VP–β-galactosidase fusion protein. This was accomplished by deleting the left end of the virus from nsp61gal by ligating Klenow enzyme-filled SstI and NcoI restriction digested nsp61gal DNA. This left 420 nucleotides of virus sequence upstream from the p61 TATA sequence. The right-hand terminal sequences were deleted by digestion with HindIII followed by religation.
pVPFsp is identical to pVPNco but with only 83 nucleotides (2381 to 2674) upstream of the p61 TATA sequence. pVPMsc is identical to pVPFsp but contains only 24 nucleotides (2440 to 2674) upstream of the p61 TATA sequence to the MscI site. This was created by digesting nsp61gal with MscI and religating to create a p7-p61 fusion. The p7 sequence was deleted by digestion with MscI and NarI, filling in with Klenow polymerase and ligation. ΔFsp/Msc is identical to pVPNco but with a deletion of nucleotides 2381 to 2440. This was created by digestion with MscI and partial FspI digestion to remove the region of interest, followed by Klenow polymerase repair of the FspI cohesive end and ligation. pATG.1 is a fusion of the lacZ gene to the first ATG in the structural gene transcript. It was made by partial BamHI digestion of pVPNco followed by MscI digestion, mung bean nuclease treatment, gel isolation of the 5,500-bp fragment, and ligation. This removes the region between the VP-LacZ fusion and the first ATG.
pVPNcoRLE is pVPNco with the right and left ends (5′ and 3′ nucleotides 1 to 268 and 3736 to 3999, respectively) of the virus left intact. It was made by digestion of nsp61gal with EcoNI and NcoI. These overhangs were filled in with Klenow enzyme and ligated. pVPNcoRE is pVPNco with the right end of the virus still intact. It was created by digestion of nsp61gal with NarI and NcoI; these ends were filled in with Klenow enzyme and ligated. pVPNcoLE is pVPNco with the left end of the virus intact. It was created by digestion of pVPNcoRLE with HindIII and religation.
For PCR mutagenesis, primers flanking the region of interest (structural gene region nucleotides 2045 to 2674) were synthesized (Gibco BRL, Gaithersburg, Md.). Primer Kasfwd (CAGATGCGTAAGGAGAAAATACCGC) binds to pUC sequences upstream of the region of interest. Primer βgalrev (GTTGTAAAACGACGGGATCC) binds to β-galactosidase sequences 120 nucleotides downstream from the VP-LacZ fusion.
To create mutations, two complementary primers were designed with a diagnostic restriction enzyme recognition site at the desired location. Two separate PCRs (one with Kasfwd and the mutation reverse primer and one with β-galrev and the mutation forward primer) were performed to yield two fragments with the mutation at either end. These fragments were purified from 1% agarose gel using a GeneClean kit (Bio 101, La Jolla, Calif.). Purified fragments were mixed together and denatured at 95°C for 10 min and allowed to anneal by cooling to 45°C for 10 min. Taq polymerase and deoxynucleoside triphosphates were then added, and the mixture was incubated for 10 min at 72°C. Finally, 10.5 pmol of the flanking primers was added, and the mixture was cycled 29 times at 95°C for 1 min, 45°C for 1 min, and 72°C for 2.5 min. To mutate the map unit 61 TATATAA sequence (designated p61), two complementary primers were made with a DraI restriction site (underlined) at nucleotide 2470 (CACAAAAATTTAAAATCTAATAGCAGAAGAAG [point mutations in bold]). For mutation of the map unit 60 TATAA sequence, complementary primers with an XhoI restriction sequence (GACAATATACCTCGAGTGCGCAAATAC) in the map unit 60 TATA sequence were used. For mutation of the transcription start site sequence, complementary primers with an EcoRV restriction sequence (CAAAATAAATTAGATATCCGTCCTCCAACTC) within the consensus start sequence were used. Fragments with these mutations were cut with NarI and BamHI and inserted into the pVPNcoΔgal subclone digested with the same enzymes. Successful insertions were then cut with BamHI, and the lacZ gene was added. The lacZ gene was obtained from nsp61gal digested with BamHI (3,072-bp fragment). All mutations were confirmed by sequencing using an automated sequencer at Colorado Sate University or the University of Colorado.
Cells and transfections.
For transfection, A. albopictus C6/36 cells were grown in L15 medium with 10% fetal bovine serum at 25°C in six-well plates at a density of 1.6 × 106 cells/well (17). Eighteen hours later, the cells were rinsed twice with phosphate-buffered saline (PBS) and 150 μl of the transfection mixture was added. The transfection mixture was made by combining 5 μg of plasmid DNA in 50 μl of L15 (3 μg of β-galactosidase expression construct, 1.5 μg of transactivating construct or pUC19, and 0.5 μg of pBSLuc) with 100 μl of L15–20% Lipofectin reagent (Gibco BRL). Cells were then incubated at 28°C for 6 h. The transfection mixture was removed, the cells were rinsed twice with PBS, and fresh complete L15 medium was added.
Protein expression assays.
At 48 h posttransfection, cells were rinsed twice with sterile PBS, lysed, and harvested using a Galactolight kit (Tropix, Bedford, Mass.). β-Galactosidase expression was quantified by incubating 5 μl of a 1:10 dilution of the cell lysates with Galacton reagent (Tropix) and measuring the resulting reaction with a TD-20e luminometer (Turner Designs, Sunnyvale, Calif.) as described previously (3, 17). Luciferase levels were determined using a luciferase assay system (Promega, Madison, Wis.) and TD-20e luminometer. Arbitrary light units from the β-galactosidase assays were normalized to the average light units of luciferase, which controls for transfection and lysis efficiency. Lysates from nontransfected C6/36 cells were included as a negative control.
Northern blot analysis.
Poly(A)+ RNA from both uninfected and infected C6/36 cells was isolated from total RNA (17). A 10-ml Poly Prep chromatography column (Bio-Rad, Richmond, Calif.) was washed with 10 ml of 5 M NaOH and then with 10 ml of diethyl pyrocarbonate-treated water. Dry oligo(dT)-cellulose powder (0.125 g) was suspended in 250 μl of 0.1 M NaOH, and the slurry was poured into the column. The oligo(dT) column was equilibrated with 10 ml of poly(A) loading buffer (0.5 M LiCl, 10 mM Tris-Cl [pH 7.5], 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]). Then 200 μg of total RNA dissolved in 0.5 M LiCl was heated to 70°C for 10 min and passed through the column. The column was washed, and the poly(A)+ RNA was eluted in 500 μl of 2 mM EDTA–0.1% SDS. The RNA was then precipitated overnight with 0.3 M sodium acetate and 2 volumes of ethanol at −20°C and centrifuged at 233,800 × g and 4°C for 1 h. The supernatant was decanted, and the pellet was allowed to air dry. RNA was dissolved in 100 μl of diethyl pyrocarbonate-treated water.
Two plasmids, pBluA and pBluA2, were constructed for transcribing hybridization riboprobes specific to structural and nonstructural genes of AeDNV, respectively. pBluA was made by cloning the 4,000-bp SstI/XhoI fragment from pUCA (2) into the like sites of pBluescript (Stratagene, La Jolla, Calif.). pBluA2 was made by removing the 2,800-bp BamHI fragment from pBluA. SnaBI and EcoRI were used to linearize pBluA and pBluA2, respectively, and were transcribed with T3 RNA polymerase (Promega). This yields a 1,186-nucleotide transcript that will hybridize to the structural gene transcripts (nucleotides 2792 to 3978) and a 900-nucleotide transcript that will hybridize to the nonstructural gene transcripts (nucleotides 307 to 707).
After gel electrophoresis using an garose-formaldehyde gel (17), the poly(A)+ RNA was transferred to a MagnaGraph nylon transfer membrane (Micron Separations Inc., Wesboro, Mass.). Probes were hybridized overnight at 45°C in hybridization buffer (50% formamide, 6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 5× Denhardt's solution, NS or VP probe [106 cpm]). Membranes were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.2% SDS at 50°C for 1 h. Images were captured by exposing the blots to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, Calif.) overnight. The captured images were digitized and imported into NIH Image 1.6 for densometric analysis.
Primer extension analysis.
A 10.5-pmol aliquot of oligonucleotide was labeled with [γ-32P]ATP using T4 polynucleotide kinase as recommended by the manufacturer (New England Biolabs, Beverly, Mass.). Oligonucleotide βgalrev (CCTAGGGCAGCAAAATGTTG) binds to the 5′ end of the lacZ gene just downstream from the BamHI site. p61rev108 (GGTACTGCCTCTTGTTGCT) binds to the viral sequence 108bp downstream from the p61 TATA at nucleotides 2583 to 2604. βgalrev and p61rev108 were used for primer extension on total RNA from cells transfected with nsp61gal and pUCA, respectively. RNA harvested from nontransfected C6/36 cells was used as a negative control. C6/36 cells were transfected as above except that 75-cm2 flasks seeded with 2.25 × 107 cells were incubated with 800 μl of transfection mixture (150 μl of Lipofectin and 30 μg of plasmid DNA in L15). Total RNA was collected from cells transfected as above by the guanidinium isothiocyanate method (17, 18) or by passage through an RNeasy spin column (Qiagen, Valencia, Calif.). RNA was aliquoted in 30-μg samples, treated with 100 U of DNase (Gibco BRL) for 30 min, precipitated with 2 volumes of ethanol and 0.1 volume of 3 M sodium acetate, and washed twice with 70% ethanol. The RNA was then resuspended in 12.5 μl of hybridization buffer (final concentration, 150 mM KCl, 10 mM Tris-HCl [pH 8.3], 1 mM EDTA); 0.8 pmol of labeled probe was added and allowed to anneal for 30 min at 65°C after being denatured for 5 min at 95°C. Primer extension buffer was added (final concentration, 50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol) with 10,000 U of Superscript AMVRT (Gibco BRL). The reaction mixture was incubated at 42°C for 50 min, and the reaction was stopped by heating to 70°C for 10 min. Then 4 μl of gel loading/stop buffer was added (New England Biolabs), and the samples were denatured at 95°C for 10 min and separated on a 5% acrylamide–8 M urea sequencing gel at 1,500 V for 2.5 h. To determine the precise transcriptional start site, sequencing was performed on nsp61gal or pUCA as a template with the same oligonucleotides as used for primer extension, using a Circumvent sequencing kit (New England Biolabs). These sequencing ladders were denatured as above and loaded next to the primer extension products for visualization of the transcriptional start site. Gels were then transferred to Whatman filter paper and dried. Gels were visualized by autoradiography using Fuji medical X-ray film (Fuji Medical Systems, Stamford, Conn.) for 5 to 72 h at −70°C.
Replication analysis.
C6/36 cells (7.5 × 106) were transfected with constructs containing the VP promoter and viral ends; 48 h posttransfection, low-molecular-weight DNA was extracted by the Hirt method (1). The DNA was precipitated with 10 M ammonium acetate and ethanol, washed twice with 70% ethanol, and resuspended in 50 μl of water. Each sample was digested with DpnI overnight at 37°C. The enzyme was heat killed; then the samples were loaded onto a 1% agarose gel and run for 6 h at 40 V. The DNA was transferred to a GeneScreen Plus membrane (DuPont), which was then prehybridized (50% formamide, 10% dextran sulfate, 2× SSC, 10% SDS) for 1 h at 45°C. Probe was prepared by random prime labeling (Boehringer Mannheim, Indianapolis, Ind.) a 3,072-bp lacZ gene fragment obtained by digesting nsp61gal with BamHI and purifying the 3,072-bp fragment by using agarose gel electrophoresis and a GeneClean kit (Tropix); 107 dpm of probe was hybridized to the membrane in 4.5 ml of hybridization buffer (50% formamide, 10% dextran sulfate, 2× SSC, 10% SDS) for 12 h at 45°C. The membrane was then washed twice with 2× SSC for 10 min and visualized by autoradiography using Fuji medical X-ray film (Fuji Medical Systems) for 5 to 72 hours at −70°C.
RESULTS
AeDNV mRNA species.
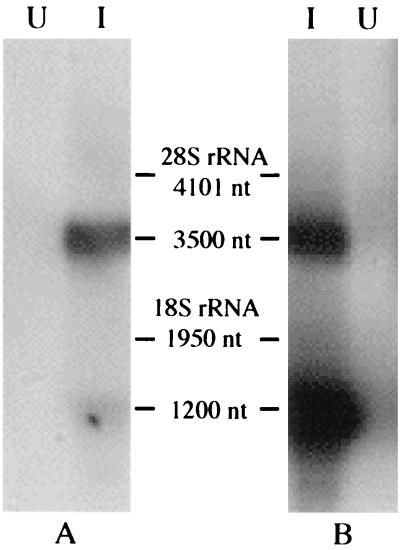
Northern blot analysis was performed to determine the size and relative abundance of viral RNA species present within AeDNV-infected cells. NS and VP probes that would hybridize to the NS1/NS2 transcript and the VP transcript, respectively, were generated. As shown in Fig. 1, two distinct transcripts of about 3,500 and 1,200 nucleotides were detected. The NS probe hybridized with the 3,500-nucleotide transcript. The VP probe bound to both transcripts, indicating that the longer transcript contains both NS and VP sequences. These transcripts correspond well to the expected sizes of the nonstructural and structural gene transcripts, respectively, assuming that both terminate at the same polyadenylation signal predicted near the right end of the viral genome (1). Quantification of the signal indicated that the VP transcript (1,200 nucleotides) is 2.2 times more abundant than the NS transcript.
FIG. 1.
Northern blot of poly(A) RNA from C6/36 cells infected with AeDNV (lanes I) or uninfected (lanes U). Membranes were probed with NS (A)- and VP (B)-specific probes. nt, nucleotides.
Structural gene transcript initiation site.
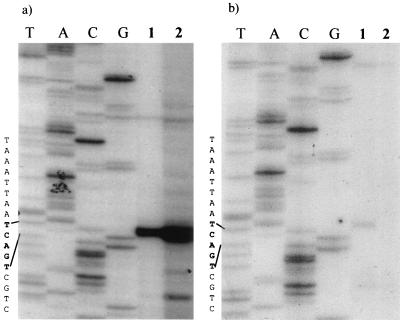
Primer extension analysis was used to precisely map the initiation site for the structural gene transcript. Primers that bound either to viral sequences (p61rev108) or to the 5′ end of the lacZ reporter gene (βgalrev) 108 or 200 nucleotides downstream from the map unit 61 TATAA sequence were used. LacZ fusions were included to confirm the identity of the transcription start site of reporter gene constructs, and the results were identical to those for the AeDNV-infected cells. As shown in Fig. 2a, when the 32P-labeled p61rev108 oligonucleotide was extended by reverse transcriptase, we observed a band approximately 200 nucleotides in length that corresponds to nucleotide 2402, which is 60 nucleotides upstream of the map unit 61 TATAA sequence. The putative start site is the first C within the sequence TCAGTC. Primer βgalrev (Fig. 2b) also mapped the transcript initiation to the C in the CAGT site, using nsp61gal-transfected cellular RNA.
FIG. 2.
Primer extension analysis of the structural gene transcript (sequence reads from top to bottom). (a) Primer extension using virus-specific primer p61rev108. Lanes T, A, G, and C, sequencing ladder produced using nsp61gal and primer p61rev108; lane 1, RNA from cells transfected with VP-LacZ fusion construct nsp61gal; lane 2, RNA from cells infected with AeDNV. (b) Primer extension using lacZ gene-specific primer βgalrev. Lanes T, A, G, and C, sequencing ladder produced using nsp61gal and primer βgalrev; lane 1, RNA from cells transfected with VP-LacZ fusion construct nsp61gal; lane 2, RNA from nontransfected cells.
Mutational analysis of the structural gene promoter.
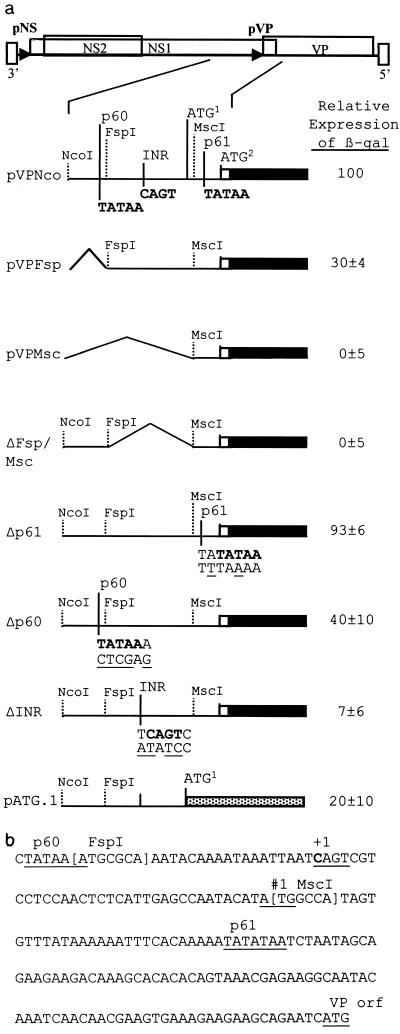
To determine the sequences critical for expression of the structural gene, different constructs containing the lacZ reporter gene fused to the VP reading frame at nucleotide 2674 (2) were compared for the efficiency of β-galactosidase expression. These constructs contained deletions of the viral sequences upstream of the VP gene. Plasmid constructs pVPMsc, pVPFsp, and pVPNco contain 24, 83, and 420 bp, respectively, upstream of the map unit 61 TATAA (Fig. 3). The level of expression from pVPNco was arbitrarily set to 100%. pVPFsp contains the initiation site defined by primer extension but lacks a TATAA sequence at map unit 60, 26 nucleotides upstream of the initiation site. This construct expressed at 30% of the level of pVPNco. pVPMsc lacks the initiation site and did not express above the background level. ΔFsp/Msc has a deletion of the 60 nucleotides between the FspI and MscI sites (nucleotides 2381 to 2440) including the initiator (Inr) site, no β-galactosidase expression was detected from this construct. This suggested that the region containing the Inr site is critical for expression. Fusion of the lacZ gene to the first ATG codon in the transcript, which is not in the VP reading frame (Fig. 3a, pATG.1), reduced expression by 80%.
FIG. 3.
(a) Effects of deletions and mutations on VP–β-galactosidase (β-gal) fusion protein expression. The VP–β-galactosidase fusion is indicated by black boxes with the relative upstream deletions or mutations indicated. The mutated sequences are shown below the wild-type viral sequence. Relative expression levels represent three experiments performed in duplicate and are normalized to the β-galactosidase expression of pVPNco. The dotted box (pATG.1) represents β-galactosidase fused to the first AUG of the transcript. AUG1, the first AUG in the VP transcript which is not in frame with the VP ORF; AUG2, AUG for the VP ORF. (b) Sequence of the VP promoter region (nucleotides 2372 to 2563).
To test their importance, the CAGT, map unit 60 TATAA, and map unit 61 TATAA sequences were modified via PCR-based mutagenesis. Changing the map unit 60 TATAA from TATAA to CTCGA reduced expression by 60% (Fig. 3, Δp60) compared to the wild-type construct pVPNco. Two nucleotide changes, T to A and A to T, in the map unit 61 TATATAA sequence were made yielding the sequence TTTAAAA. These changes were chosen because the region surrounding this sequence is very AT rich, with only five GC base pairs within 25 nucleotides. This mutation did not have a significant effect on expression of the β-galactosidase fusion protein (93% of that of pVPNco). The most dramatic reduction in expression was seen when the sequence surrounding the transcriptional start site was changed. A four-nucleotide change from TCAGTC to ATATCC (Fig. 3, ΔINR) reduced expression by 93% compared to pVPNco. This reduction is similar to the reduction observed with ΔFsp/Msc, in which the CAGT sequence and the surrounding 60 nucleotides are deleted.
Requirements for transactivation.
It has been reported that both of the AeDNV promoters can be transactivated by NS1 (2, 3, 17). The construct pVPNco was relatively insensitive to the presence of NS1 (provided by pUCAINV), it exhibited a 1.7-fold increase, compared to constructs studied previously that showed a 7-fold increase in gene expression (2; unpublished observations). Since NS1 is thought to interact with the terminal sequences of the viral genome, the 5′- and 3′-terminal sequences were added back to pVPNco. Constructs containing the right (5′) end (pVPNcoRE), left (3′) end (pVPNcoLE), or both ends (pVPNcoRLE) were transfected into C6/36 cells and analyzed for expression of β-galactosidase in the presence and absence of the transactivating construct pUCAINV. All four constructs had similar basal levels of gene expression without pUCAINV (Table 1). However, expression from the constructs containing viral terminal sequences was greatly enhanced by cotransfection with pUCAINV. pVPNcoRE and pVPNcoLE had increases of 9.7- and 7.9-fold, respectively, whereas pVPNcoRLE showed a 37-fold increase in β-galactosidase expression (Table 1). These results demonstrate that viral termini are necessary for increased expression from the structural gene promoter when NS1 is present.
TABLE 1.
Effect of NS1 on expression of β-galactosidase from constructs containing viral terminal sequencesa
| Construct | % Relative expression (mean ± SD)
|
||
|---|---|---|---|
| −NS1 | +NS1 | Fold increase (mean ± SD) | |
| pVPNco | 100 | 175 ± 22.8 | 1.7 ± 0.2 |
| pVPNcoRE | 140 ± 69.7 | 1,650 ± 563 | 9.7 ± 3.4 |
| pVPNcoLE | 93.3 ± 13.8 | 965 ± 297 | 7.9 ± 0.2 |
| pVPNcoRLE | 102 ± 41 | 2,750 ± 1,790 | 37.6 ± 8.3 |
| Controlb | 0.01 | 0.01 | |
Data are presented as relative light units and represents two to four experiments. The expression of β-galactosidase from pVPNco was set to 100% to allow standardization between experiments.
Lysates from nontransfected cells.
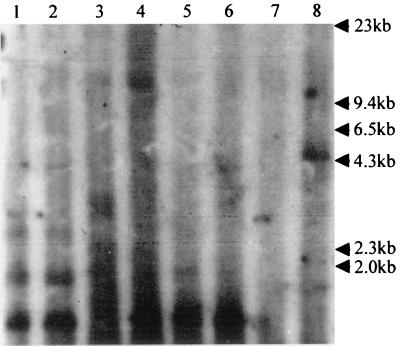
Since NS1 is involved in both the transactivation of parvovirus promoters and the replication of viral genomes (16, 30), we sought to differentiate between increased gene expression due to transactivation and replication of the template. Low-molecular-weight DNA was extracted from cells transfected as above with the construct pVPNco, pVPNcoRE, pVPNcoLE, or pVPNcoRLE with and without pUCA. This DNA was then digested with DpnI, which cleaves all Dam-methylated GATC sites in DNA of bacterial origin while leaving unmethylated viral replicative form DNA intact. These samples were analyzed by Southern blotting. The membrane was probed with a lacZ gene-specific probe to detect any replicated construct DNA. When both viral ends were present (pVPNcoRLE), a 4.6-kb, DpnI-resistant band was observed in the presence of NS1, indicating replication of the construct (Fig. 4, lane 8). Other constructs lacking either or both ends only show DpnI digestion fragments. Thus, the increase in protein expression observed when both viral ends and NS1 were present is due to both template replication and transactivation by NS1. This is consistent with previous observations with AeDNV and other parvoviruses (2, 16, 26).
FIG. 4.
Replication analysis of β-galactosidase expression constructs. Southern blot of Hirt extracts from C6/36 cells digested with DpnI after transfection as follows: with lane 1, pVPNco; lane 2, pVPNco plus NS1; lane 3, pVPNcoRE; lane 4, pVPNcoRE plus NS1; lane 5, pVPNcoLE; lane 6, pVPNcoLE plus NS1; lane 7, pVPNcoRLE; lane 8, pVPNcoRLE plus NS1. The blot was probed with a 32P-labeled lacZ gene-specific probe.
DISCUSSION
Previous studies have shown the existence of two functional promoters within the AeDNV genome, which were designated p7 and p61 according to the location of suspected TATAA boxes (1–3). In support of these observations, Northern blot analysis detected the presence of two RNA transcripts (Fig. 1). The smaller transcript exhibited a size (1,200 nucleotides) that corresponded well to the expected length of the structural protein gene transcript and hybridized only to a VP probe. The larger (3,500-nucleotide) transcript hybridizes to both VP and NS probes. This implies that it is expressed from the p7 promoter, bypasses the previously proposed polyadenylation site at the end of the NS1 gene (nucleotide 2730) (1), and uses the same polyadenylation site as the structural gene transcript (nucleotide 3679). This type of transcriptional organization in which all transcripts terminate at a polyadenylation signal near the right end of the genome is common among parvoviruses with the exception of B19 and members of the Densovirus genus (7, 8).
By using primer extension analysis, the transcriptional initiation site of the structural gene promoter was mapped to nucleotide 2402. It is located within a consensus Inr sequence TCAGTC, with the first C being +1. This Inr sequence fits the consensus Inr (TCA[G/T]T[T/C]) sequence of arthropods, including Drosophila, except the +1 is at the C and not A position. This motif is common in baculoviruses, arthropods, and mammalian systems (11, 23, 25, 27). The Inr is 60 nucleotides upstream of the TATAA sequence previously assumed to define the VP promoter (p61) (1, 2, 10). Deletions which encompass the Inr region (Fig. 3a, ΔFsp/Msc and pVPMsc), but retain the putative p61 TATAA sequence, severely crippled gene expression. Mutation of the putative p61 TATAA sequence (Δp61) had an insignificant effect on gene expression (Fig. 3a). Thus, the p61 TATAA sequence does not seem to be involved in gene expression. A different TATAA sequence located upstream of the transcriptional start site, at nucleotide 2373, and the Inr were mutated by PCR mutagenesis to confirm their function. A four-nucleotide change in the Inr sequence (ΔINR) resulted in a 93% reduction of gene expression (Fig. 3a). The TATAA sequence upstream of the Inr was found to be less important since constructs retained 30% (pVPFsp) to 40% (Δp60) of expression with this sequence deleted or mutated, respectively (Fig. 3a). These observations together place the dispensable TATA box of the structural gene promoter at nucleotide 2372 and demonstrate that the consensus Inr sequence, CAGT, is critical for efficient gene expression. This requirement of an Inr sequence for gene expression with a TATAA sequence only enhancing expression has been observed with a variety of Drosophila and mammalian genes, as well as baculovirus genes, containing the consensus sequence CAGT (9, 11, 23, 27, 28). Baculovirus early genes contain this Inr sequence (23), which is known to interact with cellular transcription factors such as TFIID (28) and would also be required for expression of densovirus genes which rely on cellular transcription machinery. In contrast, baculovirus late genes utilize a baculovirus-specific polymerase that recognizes the sequence TAAG and, with rare exceptions, lack a functional CAGT motif (23, 25).
Interestingly, with transcription beginning at nucleotide 2402, there is a short ORF starting 125 nucleotides upstream of the putative initiation codon of the VP gene (Fig. 3b). If expressed, this ORF would produce an 80-amino-acid protein corresponding to the carboxy terminus of the viral NS1 protein and could interfere with translation of the VP gene. A lacZ gene fusion to this upstream AUG (pATG.1) was expressed, though at a much relower level than VP fusions (Fig. 3a). The context of an AUG codon is important for the efficiency of translation initiation at that site, the optimal context being (A/G)CCAUGG (19). The context surrounding the AUG of the small ORF at nucleotide 2440 (CATAUGG) has a four-of-seven match to that of the optimal sequence (19). However, the first AUG in the VP reading frame (ATCAUGG) is more optimal, having matches of six of seven nucleotides. These observations may explain the reduced level of expression of the small ORF and the robust translation of the structural proteins from the downstream AUG (20). The sequences surrounding these AUG codons, the Inr, and the TATA box are completely conserved between AeDNV, AaPV, and a newly isolated mosquito DNV (1, 10; unpublished observations), suggesting that they are important for regulation of gene expression. It is interesting that the feline panleukopenia parvovirus and B19 virus were shown to contain one and many AUG codons, respectively, upstream from the structural gene AUG (13, 24). Deletion of these upstream AUG triplets resulted in increased gene expression, supporting the theory that a scanning ribosome was leaking past the first AUG to produce structural proteins. The presence of an upstream ORF may be another method of fine-tuning viral protein expression within infected cells, and may affect the pathogenesis of parvovirus diseases (12). It is interesting that expression of the NS2 protein of AeDNV would also require the ribosome to miss the NS1 start codon and scan further to translate this protein or perhaps to initiate via an internal ribosome entry site (17). Detailed examination of translation initiation will be required to elucidate the true function, if any, of the small ORF.
AeDNV promoters are known to be affected by the viral NS1 protein (2, 3). Expression from the base construct pVPNco, which lacks either of the viral ends, was found in this study to be relatively insensitive to stimulation by NS1 (1.7-fold [Table 1]). This is in contrast to previous work, which showed that the structural protein gene promoter can be transactivated by NS1 (2). However, the constructs used in the previous study contained the left end of the viral genome, which in many parvovirus systems is known to interact with NS1 (14, 15, 30). To determine the effect of the viral genome termini on transactivation of the structural protein gene promoter, the virus terminal sequences were added back to the pVPNco construct. Adding either the right or the left end had a dramatic effect on expression in the presence of NS1, increasing expression 9.7- or 7.9-fold with the right or left end, respectively (Table 1). This is in contrast to what has been found with minute virus of mice and feline panleukopenia virus, where sequences proximal to the viral promoter are fully functional in transactivation by NS1 without the viral ends (13, 21, 22). It remains possible that viral sequences other than the termini affect transactivation, since sequences of the VP gene and between the viral left end and the NcoI site were not tested. NS1 of other parvoviruses have been shown to bind sequences in the viral terminal regions (14–16, 30). This may indicate the presence of an enhancer-like sequence in the AeDNV viral ends similar to those observed under certain conditions for adeno-associated virus type 2 (6). The addition of both viral ends had a synergistic effect above that of either end alone, with a 37-fold increase in expression in the presence of NS1 (Table 1). Similar observations were made with the p4 promoter of the LUIII parvovirus (16). This is to be expected because the viral ends allow excision from the plasmid and subsequent replication of flanked sequences (2). Southern blot analysis (Fig. 4) confirms that replication of the viral DNA does indeed take place, provided that both viral ends and NS1 are present. Thus, the increase in template number was at least partially responsible for the greater increase in transactivated expression levels of pVPNcoRLE over constructs containing only one viral end. It is not clear whether both viral ends can produce a greater enhancer effect or if replication accounts for the entire increase in gene expression in the presence of NS1.
It is obvious from this study that although AeDNV has one of the smallest of DNA virus genomes, much can be learned from it that may apply to other mosquito densovirus promoters and add to a deeper understanding of gene expression and regulation in mosquitoes.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (NIAID AI25629, AI28781, and AI46793) and the John D. and Catherine T. MacArthur Foundation.
REFERENCES
- 1.Afanasiev B N, Gaylov E E, Buchatsky L P, Kozlov Y V. Nucleotide sequence and genomic organization of Aedes densonucleosis virus. Virology. 1991;185:323–336. doi: 10.1016/0042-6822(91)90780-f. [DOI] [PubMed] [Google Scholar]
- 2.Afanasiev B N, Kozlov Y V, Carlson J O, Beaty B J. Densovirus of Aedes aegypti as an expression vector in mosquito cells. Exp Parasitol. 1994;79:322–339. doi: 10.1006/expr.1994.1095. [DOI] [PubMed] [Google Scholar]
- 3.Afanasiev B N, Ward T W, Beaty B J, Carlson J O. Transduction of Aedes aegypti mosquitoes with vectors derived from Aedes densovirus. Virology. 1999;257:62–72. doi: 10.1006/viro.1999.9621. [DOI] [PubMed] [Google Scholar]
- 4.Afanasiev B N, Carlson J. Densovirinae as gene transfer vehicles. In: Faisst S, Rommelaere J, editors. Parvoviruses: from molecular biology to pathology and therapeutic uses. S. Basel, Switzerland: Karger; 2000. pp. 33–58 S. [DOI] [PubMed] [Google Scholar]
- 5.Bando H, Kusuda J, Gojobori T, Maruyama T, Kawase S. Organization and nucleotide sequence of a densovirus imply a host-dependent evolution of the parvoviruses. J Virol. 1987;61:553–560. doi: 10.1128/jvi.61.2.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergoin M, Tijssen P. Molecular biology of densoviruses. In: Faisst S, Rommelaere J, editors. Parvoviruses: from molecular biology to pathology and therapeutic uses. S. Basel, Switzerland: Karger; 2000. pp. 12–32. [Google Scholar]
- 8.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philidelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2173–2197. [Google Scholar]
- 9.Blissard G W, Kogan P H, Wei R, Rohrmann G F. A synthetic early promoter from a baculovirus: roles of the TATA box and conserved start site CAGT sequence in basal levels of transcription. Virology. 1992;190:783–793. doi: 10.1016/0042-6822(92)90916-d. [DOI] [PubMed] [Google Scholar]
- 10.Boublik Y, Jousset F, Bergoin M. Complete nucleotide sequence and genomic organization of the Aedes albopictus parvovirus (AaPV) pathogenic for Aedes aegypti larvae. Virology. 1994;200:752–763. doi: 10.1006/viro.1994.1239. [DOI] [PubMed] [Google Scholar]
- 11.Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- 12.Christensen J, Storgaard T, Viuff B, Aasted B, Alexandersen S. Comparison of promoter activity in Aleutian mink diseas parvovirus, minute virus of mice, and canine parvovirus: possible role of weak promoters in the pathogenesis of Aleutian mink disease parvovirus infection. J Virol. 1993;67:1877–1886. doi: 10.1128/jvi.67.4.1877-1886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemens D L, Carlson J O. Regulated expression of the feline panleukopenia virus P38 promoter on extrachromosomal FPV/EBV chimeric plasmids. J Virol. 1989;63:2737–2745. doi: 10.1128/jvi.63.6.2737-2745.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore S F, Christensen J, Nuesch J P F, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotmore S F, Tattersall P. The NS-1 polypeptide of minute virus of mice is covalently attached to the 5′ termini of duplex replicative-form DNA and progeny single strands. J Virol. 1988;62:851–860. doi: 10.1128/jvi.62.3.851-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson N D, Rhode S L., III Parvovirus NS1 stimulates P4 expression by interaction with the terminal repeats and through DNA amplification. J Virol. 1991;65:4325–4333. doi: 10.1128/jvi.65.8.4325-4333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimmick M W, Afanasiev B N, Beaty B J, Carlson J O. Gene expression from the p7 promoter of Aedes densonucleosis virus. J Virol. 1998;72:4364–4370. doi: 10.1128/jvi.72.5.4364-4370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingston R E, Chomczynski P, Sacchi N. Guanidinium methods for total RNA preparation, p 4.2.1–4.2.8. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Struhl K, editors. Current protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 19.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 21.Lorson C, Burger L R, Mouw M, Pintel D J. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J Virol. 1996;70:834–842. doi: 10.1128/jvi.70.2.834-842.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorson C, Pearson J, Burger L, Pintel D J. An SP-1 site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology. 1998;240:326–337. doi: 10.1006/viro.1997.8940. [DOI] [PubMed] [Google Scholar]
- 23.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. New York, N.Y: Oxford University Press, Inc.; 1995. [Google Scholar]
- 24.Ozawa K, Ayub J, Young N. Translational regulation of B19 parvovirus capsid protein production by multiple upstream AUG triplets. J Biol Chem. 1988;263:10922–10926. [PubMed] [Google Scholar]
- 25.Pullen S S, Freisen P D. The CAGT motif functions as an initiator element during early transcription of the baculovirus transregulator ie-1. J Virol. 1995;69:3573–3583. doi: 10.1128/jvi.69.6.3575-3583.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhode S L, III, Richard S M. Characterization of the trans-activation-responsive element of the parvovirus H-1 p38 promoter. J Virol. 1995;61:2807–2815. doi: 10.1128/jvi.61.9.2807-2815.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 28.Smale S T, Schmidt M C, Berk A J, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Biochemistry. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tal, J. 1994. Densonucleosis viruses, Encyclopedia of virology. p. 331–335. In R. G. Webster and A. Granoff (ed.), Academic Press, New York, N.Y.
- 30.Vanacker J-M, Rommelaere J. Non-structural proteins of autonomous parvoviruses from cellular effects to molecular mechanisms. Semin Virol. 1995;6:291–297. [Google Scholar]
- 31.Yamagishi J, Hu Y, Zheng J, Bando H. Genome organization and mRNA structure of Periplaneta fuliginosa densovirus imply alternative splicing involvement in viral gene expression. Arch Virol. 1999;144:2111–2124. doi: 10.1007/s007050050626. [DOI] [PubMed] [Google Scholar]