Abstract
Hepatitis C virus (HCV) infection causes acute and often also chronic liver disease. Worldwide, prevalence of infection is estimated to exceed that of human immunodeficiency virus infection fourfold. Because of the lack of appropriate animal models, knowledge of interactions between virus and host is still limited. Assumptions regarding pathogenesis or the activation status of innate antiviral host responses, for instance, derive mainly from clinical observations and from expression analyses of selected genes. To obtain a more objective insight into virus-host interrelationships, we used suppression-subtractive hybridization to compare gene expression in HCV-infected and non-HCV-infected liver tissues samples. Four differentially expressed genes were found: (i) the gamma interferon (IFN-γ)-inducible chemokine IP-10 gene; (ii) the IFN-α/β-inducible antiviral MxA gene; (iii) the gene encoding IFN-α/β-inducible p44, shown to be associated with ultrastructural cytoplasmic entities within hepatocytes of non-A, non-B hepatitis-infected chimpanzees; and (iv) the gene encoding IFN-α/β/γ-inducible IFI-56K, a protein recently shown to interact with the eukaryotic translation initiation factor eIF-3. Compared to hepatic gene expression in patients with liver diseases unrelated to viral infections, expression in patients with chronic HCV infection was up to 50-fold higher. While in patients with chronic HBV infection IP-10 was slightly activated as well, the IFN-α/β-regulated genes were not. Revealing a dominance of hepatic interferon-regulated processes in chronic HCV infection, data on the enhanced expression of the IFN-γ regulated IP-10 support earlier findings and may explain the composition of the hepatic cellular infiltrate. The data on enhanced expression of IFN-α/β inducible genes might be germane to therapeutic considerations.
Hepatitis C virus (HCV) infection causes acute and often chronic liver disease (8, 25). Worldwide, prevalence of infection is 0.3 to 4.0% (38). Since its molecular cloning, considerable advances have been made in the fields of diagnostics and molecular virology. However, knowledge of virus-host interactions is still limited because of the restricted species specificity of HCV and the resulting lack of an appropriate animal model. Clinical observations argue against a direct cytopathic effect of the virus and favor a destructive mechanism mediated by the host immune system (6, 14, 42).
Experimental approaches for understanding virus-host interactions addressed expression analyses of genes or proteins which are known to be involved in related pathological conditions. Proteins whose expression was reported to be elevated in chronic HCV infection are gamma interferon (IFN-γ) (3, 13, 32, 36), the inducible isoform of nitric oxide synthase (31, 44), and class I and class II major histocompatibility complex molecules (2, 4), which are known to be regulated by IFN-γ, as well as the accessory molecules CD80, CD40, and B7 (4, 34) and intercellular adhesion molecule 1 (2). Highly confirmatory of hepatic cytokine expression, phenotype and specificity analyses of infiltrating cells revealed Th1 lymphocytes as a predominant population. These cells were shown to be mostly antigen nonspecific and of a memory phenotype (3, 23, 33). Thus, it is assumed that in addition to CD8+ lymphocytes and monocytes, preferentially antigen-nonspecific activated Th1 helper lymphocytes are attracted to the liver. By secreting IFN-γ, the latter cells are thought to activate monocytes/macrophages, thereby initiating a delayed-type hypersensitivity reaction.
Regarding the activation of innate antiviral host responses, for instance, we have no conclusive evidence as to whether HCV is able to stimulate endogenous IFN-α production (5, 9, 22). This study aimed to obtain a more general view of the processes which might be involved in virus-host interrelationships, e.g., to identify genes of enhanced expression in the liver of HCV-infected patients by the generation of a subtracted library. Since HCV is assumed to infect only a minority of liver cells, at least transcripts which may be directly induced by the virus are expected to be of low abundance. We thus chose the method of suppression-subtractive hybridization (SSH) described by Diatchenko and colleagues (11) to compare gene expression in liver tissues from HCV-infected and non-HCV-infected patients. SSH was designed to generate a cDNA library which is enriched in differently expressed sequences and, more importantly, equalized for the number of individual cDNA species, thus allowing the detection of rare transcripts (19).
MATERIALS AND METHODS
Liver tissue.
Liver tissue that served as tester material for SSH was obtained directly after explantation from a 63-year-old female patient infected with HCV genotype 1b. Histologic evaluation revealed marked fibrosis, moderate infiltration, lymphoid aggregates, and steatosis with no signs of malignancy. The diagnosis indicated a moderate portal chronic-aggressive hepatitis C with transition to cirrhosis and mild to moderate active lobular hepatitis. For driver material, RNA preparations from three non-HCV-infected liver explants were pooled. Criteria for the selection of driver material were comparability in qualitative and quantitative histologic changes as far as possible, various etiologies of liver disease, and absence of hepatic viral infections. The liver disease of an 18-year-old male patient was diagnosed as cryptogenic cirrhosis with low activity. Histologic evaluation revealed moderate infiltration, marked fibrosis, and lymphoid aggregates without bile duct lesions or steatosis. Explanted liver tissue from a 53-year-old female patient showed marked fibrosis, moderate infiltration, lymphoid aggregates, bile duct lesions, marked intrahepatic cholestasis, and mild steatosis. The diagnosis indicated active primary biliary cirrhosis stage II, locally stage III, with marked intrahepatic cholestasis. A 48-year-old male patient suffered from nonactive alcoholic cirrhosis. Histologically, the tissue showed mild infiltration, locally lymphoid aggregates, and a low degree of siderosis of epithelial and Kupffer cells without steatosis.
Liver biopsy procedures from a total of 65 consecutive outpatients with chronic HCV infection (n = 38) (18 females and 20 males; mean age, 44.6 years; range, 18 to 67 years), chronic hepatitis B virus (HBV) infection (n = 12) (5 females and 7 males; mean age, 35 years; range, 17 to 54 years), or nonviral liver diseases (n = 15) (7 females and 8 males; mean age, 48 years; range, 27 to 63 years) were performed as part of a routine clinical evaluation. HCV infection was diagnosed by the presence of anti-HCV antibodies and HCV-specific RNA within the serum. Liver injury was proven histopathologically and categorized according to established criteria as described elsewhere (10, 30). Chronic HBV infection was diagnosed by the presence in sera of HBV surface and core antigen antibodies of the immunoglobulin G isotype. The diagnosis was confirmed histologically. Sera from all patients were found to be positive for HBV-specific DNA. Serum alanine transaminase activities ranged between 7 and 39 U/Iliter, with one exception of 167 U/liter. Histologic degree of inflammation was scored as absent to mild (n = 3), mild (n = 6), or moderate (n = 3). Histopathological findings for patients with no obvious hepatic viral infection comprised fibrosis (n = 2), primary biliary cirrhosis (n = 1), steatosis (n = 4), crytogenic hypodense liver foci (n = 1), hydropic swelling of hepatocytes (n = 1), cryptogenic cirrhosis (n = 2), primary sclerosing cholangitis (n = 1), Stauffer's syndrome (n = 1), and the absence of any pathological condition (n = 2). Patients with concomitant non-B or non-C viral infections and those with continued alcohol or drug abuse were excluded.
The study was approved by the local ethical committee of Georg August University, Göttingen, Germany.
Purification of poly(A)+ mRNA from liver tissue.
Total cellular RNA from liver tissue was isolated as described previously (32). Poly(A)+ mRNA was purified from the same source, using Oligotex spin columns (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Generation of a subtracted cDNA library (11, 19).
SSH was performed between the above-described tester and driver liver tissue RNA preparations using a PCR Select cDNA subtraction kit (Clontech, Heidelberg, Germany) essentially according to the manufacturer's manual. In brief, 2-μg aliquots each of poly(A)+ mRNA from the tester and the pooled driver were subjected to cDNA synthesis. Tester and driver cDNAs were digested with RsaI. The tester cDNA was subdivided into two portions, and each was ligated with a different cDNA adapter. In a first hybridization reaction, an excess of driver was added to each sample of tester. The samples were heat denaturated and allowed to anneal. Because of the second-order kinetics of hybridization, the concentration of high- and low-abundance sequences is equalized among the single-stranded tester molecules. At the same time single-stranded tester molecules are significantly enriched for differentially expressed sequences. During a second hybridization, the two primary hybridization samples are mixed together without denaturation. Only the remaining equalized and subtracted single-stranded tester cDNAs can reassociate forming double-stranded tester molecules with different ends. After filling in the ends with DNA polymerase, the entire population of molecules is subjected to nested PCR with two adapter-specific primer pairs.
Cloning the subtracted library into a TA vector.
Products of these amplified A overhangs containing a subtracted cDNA library (3 μl) were immediately ligated into a pT-Adv plasmid (advanTAge PCR cloning kit; Clontech). Subsequently, the material was desalted by ethanol precipitation. One-tenth of the purified plasmid was introduced into electrocompetent Escherichia coli DH5α (Clontech) by electroporation (1.8 kV, 150 Ω) (Electroporator II; Invitrogen). Bacteria were taken up in 600 μl of SOC medium (41a) and allowed to incubate for 60 min at 37°C and 225 rpm. Bacteria were plated onto agar plates containing ampicillin (50 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal; 20 μg/cm2), and isopropyl-β-d-thiogalactoside (IPTG; 12.1 μg/cm2) and incubated overnight at 37°C. Individual recombinant white colonies were picked and grown in Luria-Bertani medium containing ampicillin on 96-well microtiter plates.
Differential screening.
cDNA clones were subjected to a differential screening procedure (PCR-Select differential screening kit; Clontech) to identify those which hybridize to the subtracted library with preference. Briefly, cDNA inserts of the cloned cDNA library were amplified by subjecting an aliquot of the bacterial culture directly to PCR. The presence of one distinct PCR product was confirmed by agarose gel electrophoresis. The amplified material was then dot blotted onto nitrocellulose membranes. Four identical membranes with cDNA arrays in duplicate were prepared. Membranes were hybridized with a tester probe, a driver probe, a subtracted probe, and a reverse-subtracted probe as a control. Subtracted probes had previously been released from adapter sequences by restriction digestion. Probes were labeled with digoxigenin-11-UTP in a 3′ tailing reaction (DIG oligonucleotide tailing kit; Boehringer Mannheim, Mannheim, Germany). Hybridization and detection via a chemiluminescence reaction were carried out employing a DIG luminescent detection kit (Boehringer Mannheim), according to the supplier's standard protocol.
Bacterial clones harboring differentially hybridizing cDNA sequences were grown, and plasmids were purified using a Qiagen plasmid mini kit. Inserts were sequenced by the chain termination reaction using an automated sequencer (SEQLAB, Göttingen, Germany). Nucleic acid homology searches were performed using the BLAST program at the National Center of Biotechnology Information (National Institutes of Health, Bethesda, Md.).
Confirmation of differential screening results.
Differences in transcript expression within the original tester and driver RNA preparations were confirmed by a quantitative competitive reverse transcription-PCR (RT-PCR) technique (see below). In cases in which clones scored perfect or near-perfect matches with known human genes, primers were designed on the basis of the database entry unless indicated otherwise (Table 1).
TABLE 1.
Quantitative competitive RT-PCR specifications
| Gene (reference) | Primer | Annealing temp (°C) | No. of cycles | Size (bp) of product
|
|
|---|---|---|---|---|---|
| mRNA | Internal standard | ||||
| Albumin (12) | 5′ CTT GAA TGT GCT GAT GAC AGG 3′ | 58 | 28 | 157 | 223 |
| 5′ GCA AGT CAG CAG GCA TCT CAT C 3′ | |||||
| β-Actin (35) | 5′ GTG GGG CGC CCC AGG CAC CA 3′ | 62 | 28 | 538 | 370 |
| 5′ CTC CTT AAT GTC ACG CAC GAT 3′ | |||||
| G3PDH | 5′ ACC ACA GTC CAT GCC ATC AC 3′ | 59 | 30 | 452 | 260 |
| 5′ TCC ACC ACC CTG TTG CTG TA 3′ | |||||
| Cytochrome P450 reductase | 5′ GAC GTG GAT CTC TCT GGG GT 3′ | 61 | 30 | 508 | 343 |
| 5′ CAG AGT CGT TGG CTG GGT AC 3′ | |||||
| Glutaryl coenzyme A dehydrogenase | 5′ CTT GGG AGT TCT GCT TGC AC 3′ | 61 | 30 | 384 | 46 |
| 5′ GGA TTC CCG TGA TAG CTC TC 3′ | |||||
| IFI-56K | 5′ TAG CCA ACA TGT CCT CAC AGA C 3′ | 60 | 30 | 396 | 546 |
| 5′ TCT TCT ACC ACT GGT TTC ATG C 3′ | |||||
| IP-10 | 5′ GAT GGA CCA CAC AGA GGC TG 3′ | 62 | 30 | 408 | 546 |
| 5′ GGA GGA TGG CAG TGG AAG TC 3′ | |||||
| MxA | 5′ CTG TGG CCA TAC TGC CAG GA 3′ | 61 | 30 | 482 | 302 |
| 5′ ACT CCT GAC AGT GCC TCC AA 3′ | |||||
| p44 | 5′ ATT GGA GCA TAT GCA GAA GAG A 3′ | 59 | 30 | 339 | 443 |
| 5′ GGC AGA CAG TAA GCT CTT CCT G 3′ | |||||
| PA28α | 5′ AGA TTT CTG AGC TGG ATG CA 3′ | 60 | 30 | 566 | 443 |
| 5′ AGC ATA AGC ATT GCG GAT CT 3′ | |||||
| Pantophysin | 5′ CTG TAC TGC ATT GCT GCC CT 3′ | 61 | 30 | 373 | 549 |
| 5′ CTT GGC TAT GAG GGG CAG AT 3′ | |||||
| Ras-like protein | 5′ ACT CAT GAG CTA TGC CAA CG 3′ | 60 | 30 | 412 | 320 |
| 5′ TTC CAC ATA GCA GCA TGC TC 3′ | |||||
| Tumor-associated antigen L6 (43) | 5′ CTA GCA GAC CAC CAT GTG 3′ | 54 | 30 | 618 | 476 |
| 5′ TTA GCA GTC ATA TTG CTG 3′ | |||||
Analysis of transcript expression by quantitative competitive RT-PCR.
The expression of transcripts of interest and of housekeeping genes was quantified by a competitive RT-PCR technique using internal cDNA standards as described in detail previously (32). In brief, internal standards were constructed to be recognized by and to compete for oligonucleotide primers complementary to the target sequences. Target cDNA was amplified in the presence of 10- and 2-fold serial dilutions of the internal standard. The amount of target transcripts was then calculated on the basis of the known molecular quantity of the internal standard and related to the amount of housekeeping mRNA, which had been quantified in parallel analogously. Primer sequences, annealing temperatures, numbers of cycles, and sizes of the target and internal standard amplification products are given in Table 1. Specificity of amplification products was confirmed either by Southern hybridization using specific, digoxigenin-labeled probes or by restriction enzyme digestion analysis.
Statistical analyses.
Gaussian distributed data were analyzed by parametric t tests for independent samples. Correlation coefficients were calculated by linear regression analysis.
RESULTS
Identification of activated genes in HCV-infected liver tissue.
Using SSH, we generated a subtracted library using HCV-infected liver tissue sample as tester (minuend) and three pooled non-HCV-infected, histologically comparable liver tissue specimens as driver (subtrahend). In parallel, a reverse-subtracted library was prepared as a control using HCV-infected tissue as driver and non-HCV-infected tissue as tester. The subtracted library was cloned, and a total of 864 clones were obtained. The subsequent screening procedure (differential screening) revealed 55 clones hybridizing to the subtracted library but not to the reverse-subtracted control library. These 55 clones were sequenced, and 39 were found to correspond nearly entirely to database entries. Several of the clones represented fragments of same genes, thus decreasing the number of individual activated genes to 13 (Table 2). Increased expression within the original tester material was checked by quantitative competitive RT-PCR, a technique which had been established using primer pairs designed on the basis of the database entries (Table 1). Relative to the reference transcripts glyceraldehyde-3-phosphate dehydrogenase (G3PDH, β-actin, and albumin, and compared to the original driver RNA preparation, transcripts of four genes were found to be elevated severalfold in the tester material (Table 2): (i) the chemokine IP-10 gene, originally described as an IFN-γ-inducible early-response gene (26, 48); (ii) the IFN-α-regulated antiviral MxA gene (1, 40); (iii) the gene encoding the IFN-α/β-inducible 44-kDa microtubular polypeptide (p44) formerly shown to be associated with cytoplasmic ultrastructural entities in hepatocytes of chimpanzees inoculated with sera from patients suffering from non-A, non-B hepatitis (21, 24, 27, 46); and (iv) the gene encoding the IFN-α/β/γ-inducible 56-kDa protein (IFI-56K) recently described to interact with the eukaryotic translation initiation factor eIF-3 (7, 50). The differential screening hybridization showed that all clones represents low-abundance transcripts in the HCV-infected liver, since they were detectable with the labeled subtracted probe but not with the labeled tester probe (data not shown).
TABLE 2.
Relative increase of gene expression within the HCV-infected tester material
| Accession no. | Genea | Relative transcriptional expressionb
|
||
|---|---|---|---|---|
| G3PDH | Albumin | β-Actin | ||
| X02530 | IP-10 | 37 | 100 | 83 |
| M30817 | MxA | 30 | 15 | 53 |
| NM 006417.1 | p44 | 15 | 7.5 | 26 |
| X03557 | IFI-56K | 5 | 13 | 11 |
| U10360 | PA28α | 2 | 1 | 3.6 |
| S72481 | Pantophysin | 0.8 | 0.4 | 1.3 |
| M90657 | Tumor-associated antigen L6 | 1.5 | 0.8 | 0.3 |
| M31470 | Ras-like protein | 0.8 | 0.4 | 1.3 |
| S90469 | Cytochrome P450 reductase | 0.4 | 0.2 | 0.7 |
| U69141 | Glutaryl coenzyme A dehydrogenase | 0.04 | 0.4 | 0.4 |
| AF075587 | Myc-binding protein | NDc | ND | ND |
| Y00282 | Ribophorin II | ND | ND | ND |
| J02625 | Cytochrome P450j | ND | ND | ND |
Sequence identity based on comparison with GenBank/EMBL database.
Based on competitive RT-PCR analysis of tester and driver mRNAs. Values were normalized by relating them G3PDH, albumin, or β-actin transcript expression and are given as the factor by which tester exceeds driver expression.
ND, not determined.
Analysis of representative patient groups regarding hepatic activation of interferon-regulated genes.
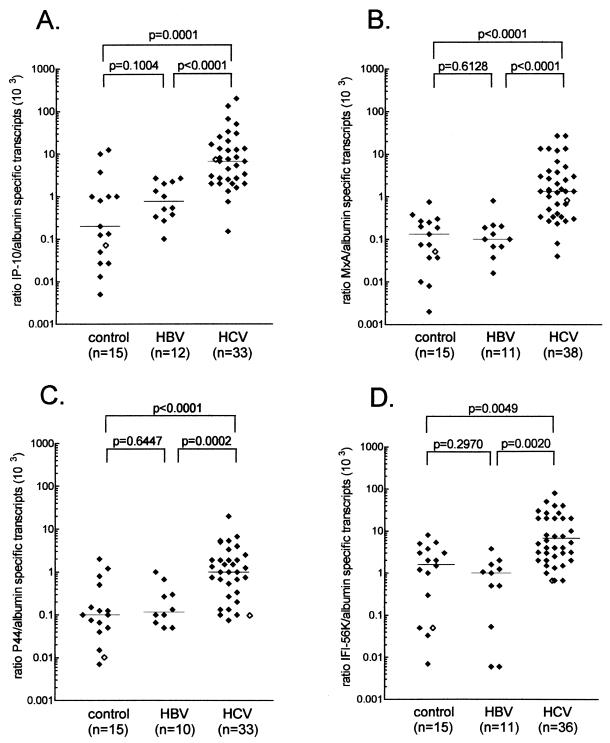
To confirm enhanced expression of the four interferon-regulated genes in chronic HCV infection in general, we compared a representative number of liver biopsy specimens from HCV-infected patients to a number of liver biopsy specimens derived from patients with liver diseases unrelated to any known viral infection (Fig. 1). Transcriptional expression of IP-10, IFI-56K, p44, and MxA was found to be significantly (4- to 50-fold) elevated in chronic HCV infection compared to nonviral liver diseases (Fig. 1).
FIG. 1.
Transcriptional expression of IP-10, MxA, p44, and IFI-56K in liver tissue from patients with chronic HCV or HBV infection and from patients with no known hepatic viral infection. RNA preparations from individuals with chronic HCV or HBV infection and from patients with liver disorders unrelated to any known virus infection were analyzed for IP-10 (A), MxA (B), p44 (C), and IFI-56K (D) transcript expression by competitive RT-PCR. Data were related to the amount of albumin-specific mRNA as a reference transcript. Expression of the four interferon-regulated genes was found to be higher in chronically HCV-infected patients than in both chronically HBV-infected patients and the control group (P values are indicated, t test for independent samples). Data on gene expression found within the original tester and driver material are included as open symbols. Comparable results were obtained when data were related to β-actin as a reference transcript.
As an additional control, liver biopsy specimens from patients with chronic HBV infection were included (Fig. 1). Although not reaching statistical significance, hepatic IP-10 expression was found to be on average sixfold higher than in the control group (Fig. 1A). In contrast, liver samples derived from patients with chronic HBV infection and from the control group showed comparable activation regarding the IFN-α/β-regulated genes (Fig. 1B to D). Whereas levels of transcriptional expression of MxA, p44, and IFI-56K were correlated to each other (MxA ∼ p44 [P = 0.0002, r = 0.58]; p44 ∼ IFI-56K [P = 0.0001, r = 0.62]; MxA ∼ IFI-56K [P = 0.0060, r = 0.42]) but not to IP-10 (MxA ∼ IP-10 [P = 0.14, r = 0.30]; p44 ∼ IP-10 [P = 0.35, r = 0.23]; IFI-56K ∼ IP-10 [P = 0.93, r = 0.01]) or IFN-γ (MxA ∼ IFN-γ [P = 0.18, r = 0.27]; p44 ∼ IFN-γ [P = 0.29, r = 0.26]; IFI-56K ∼ IFN-γ [P = 0.89, r = −0.03]), IP-10 mRNA expression was found to be related significantly although weakly to IFN-γ, as estimated by linear regression analysis (P = 0.0020, r = 0.5205) (data not shown).
DISCUSSION
The application of SSH to liver tissue from patients with chronic HCV infection as tester and non-HCV-infected patients as driver, the subsequent screening procedure, and the confirmatory controls revealed four genes which are expressed within the HCV-infected tissue to a higher degree than the noninfected control: (i) the CXC or alpha chemokine IP-10 gene (26, 48), (ii) the antiviral MxA gene (1, 40), (iii) a gene encoding a 44-kDa protein reported to be associated with ultrastructural changes in hepatocytes of chimpanzees infected with non-A, non-B hepatitis sera (21, 24, 27, 46), and (iv) a gene encoding 56-kDa interferon-inducible protein which was recently shown to interact with eIF-3 (7, 50). All of these genes are inducible by IFN-γ (IP-10), IFN-α/β (MxA and p44), or both (IFI-56K). Enhanced expression of these genes in chronic HCV infection was proven by analyzing a number of liver biopsy specimens from patients with chronic HCV infection, chronic HBV infection, and nonviral liver diseases (Fig. 1). Expression of IFN-α-inducible genes was found to be mutually correlated, whereas the expression of IP-10 was found to be related better, although weakly, to that of IFN-γ. The results support earlier findings on the role of IFN-γ as a mediator of the hepatic inflammatory process in chronic HCV infection (31, 32, 44). The findings might have implications for therapeutic considerations, since at least three IFN-α-regulated genes are activated at the transcriptional level in HCV-infected liver tissue.
In chronic HCV infection, the hepatic inflammatory infiltrate is composed mainly by antigen-nonspecific, IFN-γ-producing Th1 lymphocytes. IFN-γ is thought to activate monocytes/macrophages, thereby initiating an injurious delayed-type hypersensitivity reaction. This view is based (i) on observations of a positive correlation of peripheral as well as intrahepatic IFN-γ transcript expression and the inflammatory activity (32, 36), (ii) on the observation of a positive correlation between the number of CD4+ lymphocytes in the liver and inflammatory activity but not to the number of macrophages or CD8+ lymphocytes (23), (iii) on the finding that most of the T-cell clones isolated from liver tissue from chronically HCV-infected patients are able to produce IFN-γ after mitogenic stimulation in vitro (3), and (iv) on phenotype and specificity analyses of infiltrating cells (3, 23, 36).
The enhanced expression of the chemokine IP-10 may help to explain the cellular composition of the hepatic infiltrate. In addition to monocytes and NK cells, IP-10 recruits preferentially stimulated CD4+ memory T cells (48, 49). Nonstimulated, naive T cells, CD8+ lymphocytes, or neutrophils, on the other hand, are not attracted by IP-10 (48, 49). Data on the expression pattern of the recently cloned IP-10 receptor CXCR3 further confirmed a higher expression on Th1 and Th0 lymphocytes than on the Th2 subpopulation (41).
The results on IP-10 gene activation in chronic HCV infection are highly confirmatory of recent data obtained by Shields et al. (45) and earlier data published by Narumi et al. (37). Narumi and colleagues described higher serum concentrations of the IP-10 protein in patients with chronic HCV infection than in patients with rheumatoid arthritis or healthy volunteers; by applying in situ hybridization, the authors demonstrated IP-10 transcripts in hepatocytes surrounding infiltrated mononuclear cells (37). Shields and colleagues reported enhanced levels of the IP-10 receptor CXCR3 on infiltrating lymphocytes; on the other hand and in apparent contrast to data of Narumi et al., by immunohistochemistry they demonstrated the IP-10 protein on sinusoidal endothelial cells (45). Although the cellular source of IP-10 remains open, enhanced expression of IP-10 has now been demonstrated by three independent various experimental approaches.
IP-10 may represent the link between HCV-infected liver cells and recruitment of Th1 lymphocytes and monocytes. Since IP-10 is regulated by the product of the cells it attracts, namely, IFN-γ, and since conversely IP-10 has been described to be able to activate and enhance Th1 cytokine production in vitro (15), IP-10 might contribute to the maintenance of the Th1-dominated immune response in chronic HCV infection.
IFI-56K is one of the first interferon-inducible genes cloned (7). By a two-hybrid screen, it recently was described to interact with the cytoplasmic P48/Int6 subunit of eIF-3 (18). Binding to eIF-3 was shown to cause inhibition of translation (18).
MxA and p44 are induced preferentially by IFN-α/β. MxA belongs to the dynamin superfamily of large guanosine triphosphatases. It is active against pathogenic RNA viruses, such as members of the orthomyxovirus family, possibly by interfering with the viral polymerase protein (20). There is no evidence as to whether MxA interacts with HCV proteins during virus replication. However, MxA, like IFI-56K, is used as a surrogate marker for endogenous IFN-α induction and exogenous IFN-α application (22, 28).
In 1985, p44 was shown to be a component of the double-walled membranous tubules which appeared as a distinctive alteration in the cytoplasm of hepatocytes after intravenous administration of human non A, non B hepatitis inocula in chimpanzees (39, 46, 47). Neither the ultrastructural changes nor the expression of p44 was evident in chimpanzees experimentally infected with HBV.
The findings related to enhanced expression of IFN-α-regulated genes in chronic HCV infection indicate that HCV may be an IFN-α inducer. Moreover, comparison with material from chronically HBV-infected individuals suggests HCV to be stronger than HBV in that instance. However, direct measurements of IFN-α- and IFN-β-specific transcripts revealed no significant increase in either HBV- or HCV-infected individuals compared to the control group (unpublished data), possibly because the genes are activated by alternative pathways. At least for IFI-56K, MxA, and IP-10, there is substantial evidence on induction pathways by double-stranded RNA or viral infection that bypass interferons (16, 17, 29, 51). These findings might have implications for therapeutic considerations. If HCV is infact an inducer of IFN-α/β-inducible genes with antiviral effects, then its ability to persist would be due to resistance at least toward these cellular antiviral responses.
Regarding the experimental approach, it might be argued that many additional genes are expected to be differentially expressed in HCV-infected tissue and that the selected technique might be inadequate to identify them. We would agree, if the driver material chosen was simply an HCV-uninfected healthy control. In that case, mediators of inflammation, mediators of fibrosis, and enzymes of lipid metabolism, for instance, would have been expected. However, driver material was chosen extremely carefully (as outlined in Materials and Methods), aimed to exclude all transcripts which are generally involved in pathological processes. The result that no inflammatory mediator other than IP-10 was detected indicates that non-HCV-related mediators have been subtracted successfully.
ACKNOWLEDGMENTS
We thank B. Ringe and F. Braun, Division of Transplantation Surgery, for providing explanted liver tissue material, and we thank the physicians of the Division of Gastroenterology and Endocrinology who were involved in liver biopsy procedures for their kind cooperation. We also thank A. Fayyazi for histological evaluations, J. Blattner, F. Zschunke, and H. Siggelkow for theoretical and technical support regarding the cloning procedure, and W. Lendeckel and H. Dörler for expert technical assistance.
This work was supported by grants SFB 402/C1 and SFB 402/C6 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Aebi M, Fah J, Hurt N, Samuel C E, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol. 1989;9:5062–5072. doi: 10.1128/mcb.9.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballardini G, Groff P, Pontisso P, Giostra F, Francesconi R, Lenzi M, Zauli D, Alberti A, Bianchi F B. Hepatitis C virus (HCV) genotype, tissue HCV antigens, hepatocellular expression of HLA-A,B,C, and intercellular damage and response to interferon treatment in patients with chronic hepatitis C. J Clin Investig. 1995;95:2067–2075. doi: 10.1172/JCI117893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoletti A, D'Elios M M, Boni C, De Carli M, Zignego A L, Durazzo M, Missale G, Penna A, Fiaccadori F, Del Prete G, Ferrari C. Different cytokine profiles of intrahepatic T cells in chronic hepatitis B and hepatitis C virus infections. Gastroenterology. 1997;112:193–199. doi: 10.1016/s0016-5085(97)70235-x. [DOI] [PubMed] [Google Scholar]
- 4.Burgio V L, Ballardini G, Artini M, Caratozzolo M, Bianchi F B, Levrero M. Expression of co-stimulatory molecules by Kupffer cells in chronic hepatitis of hepatitis C virus etiology. Hepatology. 1998;27:1600–1606. doi: 10.1002/hep.510270620. [DOI] [PubMed] [Google Scholar]
- 5.Castelruiz Y, Larrea E, Boya P, Civeira M P, Prieto J. Interferon alfa subtypes and levels of type I interferons in the liver and peripheral mononuclear cells in patients with chronic hepatitis C and controls. Hepatology. 1999;29:1900–1904. doi: 10.1002/hep.510290625. [DOI] [PubMed] [Google Scholar]
- 6.Chazouilleres O, Kim M, Combs C, Ferrell L, Bacchetti P, Roberts J, Ascher N L, Neuwald P, Wilber J, Urdea M, Quan S, Sanchez-Pescador R, Wright T L. Quantitation of hepatitis C virus RNA in liver transplant recipients. Gastroenterology. 1994;106:994–999. doi: 10.1016/0016-5085(94)90759-5. [DOI] [PubMed] [Google Scholar]
- 7.Chebath J, Merlin G, Metz R, Benech P, Revel M. Interferon-induced 56,000 Mr protein and its mRNA in human cells: molecular cloning and partial sequence of the cDNA. Nucleic Acids Res. 1983;11:1213–1226. doi: 10.1093/nar/11.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 9.Cribier B, Schmitt C, Rey D, Lang J M, Kirn A, Stoll-Keller F. Role of endogenous interferon in hepatitis C virus (HCV) infection and in coinfection by HIV and HCV. Res Virol. 1996;147:263–266. doi: 10.1016/0923-2516(96)82284-9. [DOI] [PubMed] [Google Scholar]
- 10.Desmet V J, Gerber M, Hoofnagle J H, Manns M, Scheuer P J. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513–1520. [PubMed] [Google Scholar]
- 11.Diatchenko L, Lau Y-F C, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dugaiczyk A, Law S W, Dennison O E. Nucleotide sequence and the encoded amino acids of human serum albumin mRNA. Proc Natl Acad Sci USA. 1982;79:71–75. doi: 10.1073/pnas.79.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumoulin F L, Bach A, Leifeld L, El-Bakri M, Fischer H P, Sauerbruch T, Spengler U. Semiquantitative analysis of intrahepatic cytokine mRNAs in chronic hepatitis C. J Infect Dis. 1997;175:681–685. doi: 10.1093/infdis/175.3.681. [DOI] [PubMed] [Google Scholar]
- 14.Fong T L, Valinluck B, Govindarajan S, Charboneau F, Adkins R H, Redeker A G. Short-term prednisone therapy affects aminotransferase activity and hepatitis C virus RNA levels in chronic hepatitis C. Gastroenterology. 1994;107:196–199. doi: 10.1016/0016-5085(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 15.Gangur V, Simons F E, Hayglass K T. Human IP-10 selectively promotes dominance of polyclonally activated and environmental antigen-driven IFN-γ over IL-4 responses. FASEB J. 1998;12:705–713. doi: 10.1096/fasebj.12.9.705. [DOI] [PubMed] [Google Scholar]
- 16.Goetschy J F, Zeller H, Content J, Horisberger M A. Regulation of the interferon-inducible IFI-78K gene, the human equivalent of the murine Mx gene, by interferons, double-stranded RNA, certain cytokines, and viruses. J Virol. 1989;63:2616–2622. doi: 10.1128/jvi.63.6.2616-2622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J, Peters K L, Sen G C. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 18.Guo J, Sen G C. Characterization of the interaction between the interferon-induced protein P56 and the Int6 protein encoded by a locus of insertion of the mouse mammary tumor virus. J Virol. 2000;74:1892–1899. doi: 10.1128/jvi.74.4.1892-1899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurskaya N G, Diatchenko L, Chenchik A, Siebert P D, Khaspekov G L, Lukyanov K A, Vagner L L, Ermolaeva O D, Lukyanov S A, Sverdlov E D- Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemagglutinin and phorbol 12-myristate 13-acetate. Anal Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- 20.Haller O, Frese M, Kochs G. Mx proteins: mediators of innate resistance to RNA viruses. Rev Sci Technol. 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 21.Honda Y, Kondo J, Maeda T, Yoshiyama Y, Yamada E, Shimizu Y K, Shikata T, Ono Y. Isolation and purification of a non-A, non-B hepatitis-associated microtubular aggregates protein. J Gen Virol. 1990;71:1999–2004. doi: 10.1099/0022-1317-71-9-1999. [DOI] [PubMed] [Google Scholar]
- 22.Jakschies D, Zachoval R, Muller R, Manns M, Nolte K U, Hochkeppel H K, Horisberger M A, Deicher H, Von Wussow P. Strong transient expression of the type I interferon-induced MxA protein in hepatitis A but not in acute hepatitis B and C. Hepatology. 1994;19:857–865. [PubMed] [Google Scholar]
- 23.Khakoo S I, Soni P N, Savage K, Brown D, Dhillon A P, Poulter L W, Dusheiko G M. Lymphocyte and macrophage phenotypes in chronic hepatitis C infection. Am J Pathol. 1997;150:963–970. [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura A, Takahashi K, Okajima A, Kitamura N. Induction of the human gene for p44, a hepatitis-C-associated microtubular aggregate protein, by interferon-alpha/beta. Eur J Biochem. 1994;224:877–883. doi: 10.1111/j.1432-1033.1994.00877.x. [DOI] [PubMed] [Google Scholar]
- 25.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C virus by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 26.Luster A D, Unkeless J C, Ravetch J V. γ-Interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 27.Maeda T, Honda Y, Hanawa M, Yamada E, Ono Y, Shikata T, Shimizu Y K. Production of antibodies directed against microtubular aggregates in hepatocytes of chimpanzees with non-A, non-B hepatitis. J Gen Virol. 1989;70:1401–1407. doi: 10.1099/0022-1317-70-6-1401. [DOI] [PubMed] [Google Scholar]
- 28.Meier, V., S. Mihm, and G. Ramadori. MxA gene expression in peripheral blood mononuclear cells from interferon-α treated patients infected chronically with hepatitis C virus. J. Med. Virol., in press. [PubMed]
- 29.Memet S, Besancon F, Bourgeade M F, Thang M N. Direct induction of interferon-gamma-and interferon-alpha/beta-inducible genes by double-stranded RNA. J Interferon Res. 1991;11:131–141. doi: 10.1089/jir.1991.11.131. [DOI] [PubMed] [Google Scholar]
- 30.Mihm S, Fayyazi A, Hartmann H, Ramadori G. Analysis of histopathological manifestations of chronic hepatitis C virus infection with respect to virus genotype. Hepatology. 1997;25:735–739. doi: 10.1002/hep.510250340. [DOI] [PubMed] [Google Scholar]
- 31.Mihm S, Fayyazi A, Ramadori G. Hepatic expression of inducible nitric oxide synthase transcripts in chronic hepatitis C virus infection: relation to hepatic viral load and liver injury. Hepatology. 1997;26:451–458. doi: 10.1002/hep.510260228. [DOI] [PubMed] [Google Scholar]
- 32.Mihm S, Hutschenreiter A, Fayyazi A, Pingel S, Ramadori G. High inflammatory activity is associated with an increased amount of IFN-γ transcripts in peripheral blood cells of patients with chronic hepatitis C virus infection. Med Microbiol Immunol. 1996;185:95–102. doi: 10.1007/s004300050020. [DOI] [PubMed] [Google Scholar]
- 33.Minutello M A, Pileri P, Unutmaz D, Censini S, Kuo G, Houghton M, Brunetto M R, Bonino F, Abrignani S. Compartmentalization of T lymphocytes to the site of disease: intrahepatic CD4+ T cells specific for the protein NS4 of hepatitis C virus in patients with chronic hepatitis C. J Exp Med. 1993;178:17–25. doi: 10.1084/jem.178.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mochizuki K, Hayashi N, Katayama K, Hiramatsu N, Kanto T, Mita E, Tatsumi T, Kuzushita N, Kasahara A, Fusamoto H, Yokochi T, Kamada T. B7/BB-1 expression and hepatitis activity in liver tissues of patients with chronic hepatitis C. Hepatology. 1997;25:713–718. doi: 10.1002/hep.510250337. [DOI] [PubMed] [Google Scholar]
- 35.Nakajima-Iijima S, Hamada H, Reddy P, Kakunaga T. Molecular structure of the human cytoplasmic beta-actin gene: interspecies homology of sequences in the introns. Proc Natl Acad Sci USA. 1985;82:6133–6137. doi: 10.1073/pnas.82.18.6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Napoli J, Bishop G A, McGuinness P H, Painter D M, McCaughan G W. Progressive liver injury in chronic hepatitis C infection correlates with increased intrahepatic expression of Th1-associated cytokines. Hepatology. 1996;24:759–765. doi: 10.1002/hep.510240402. [DOI] [PubMed] [Google Scholar]
- 37.Narumi S, Tominaga Y, Tamaru M, Shimai S, Okamura H, Nishioji K, Itoh Y, Okanoue T. Expression of IFN-inducible protein-10 in chronic hepatitis. J Immunol. 1997;158:5536–5544. [PubMed] [Google Scholar]
- 38.Nishioka K. Epidemiological studies on hepatitis C virus infection: detection, prevalence, exposure and prevention. Intervirology. 1994;37:58–67. doi: 10.1159/000150359. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer U, Thomssen R, Legler K, Bottcher U, Gerlich W, Weinmann E, Klinge O. Experimental non-A, non-B hepatitis: four types of cytoplasmic alteration in hepatocytes of infected chimpanzees. Virchows Arch B. 1980;33:233–243. doi: 10.1007/BF02899184. [DOI] [PubMed] [Google Scholar]
- 40.Ronni T, Matikainen S, Lehtonen A, Palvimo J, Dellis J, Van Eylen F, Goetschy J F, Horisberger M, Content J, Julkunen I. The proximal interferon-stimulated response elements are essential for interferon responsiveness: a promoter analysis of the antiviral MxA gene. J Interferon Cytokine Res. 1998;18:773–781. doi: 10.1089/jir.1998.18.773. [DOI] [PubMed] [Google Scholar]
- 41.Sallusto F, Lenig D, Mackay C R, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sato K, Okamoto H, Aihara S, Hoshi Y, Tanaka T, Mishiro S. Demonstration of sugar moiety on the surface of hepatitis C virions recovered from the circulation of infected humans. Virology. 1993;196:354–357. doi: 10.1006/viro.1993.1488. [DOI] [PubMed] [Google Scholar]
- 43.Schiedeck T H, Christoph S, Duchrow M, Bruch H P. Detection of hL6-mRNA: new possibilities in serologic tumor diagnosis of colorectal carcinomas. Zentrabl Chir. 1998;123:159–162. [PubMed] [Google Scholar]
- 44.Schweyer S, Mihm S, Radzun H J, Hartmann H, Fayyazi A. Liver infiltrating T lymphocytes express interferon gamma and inducible nitric oxide synthase in chronic hepatitis C virus infection. Gut. 2000;46:255–259. doi: 10.1136/gut.46.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shields P L, Morland C M, Salmon M, Qin S, Hubscher S G, Adams D H. Chemokine and chemokine receptor interactions provide a mechanism for selective T cell recruitment to specific liver compartments within hepatitis C-infected liver. J Immunol. 1999;163:6236–6243. [PubMed] [Google Scholar]
- 46.Shimizu Y K, Feinstone S M, Purcell R H, Alter H J, London W T. Non-A, non-B hepatitis: ultrastructural evidence for two agents in experimentally infected chimpanzees. Science. 1979;205:197–200. doi: 10.1126/science.451589. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu Y K, Oomura M, Abe K, Uno M, Yamada E, Ono Y, Shikata T. Production of antibody associated with non-A, non-B hepatitis in a chimpanzee lymphoblastoid cell line established by in vitro transformation with Epstein-Barr virus. Proc Natl Acad Sci USA. 1985;82:2138–2142. doi: 10.1073/pnas.82.7.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taub D D, Lloyd A R, Conlon K, Wang J M, Ortaldo J R, Harada A, Matsushima K, Kelvin D J, Oppenheim J J. Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med. 1993;177:1809–1814. doi: 10.1084/jem.177.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taub D D, Sayers T J, Carter C R, Ortaldo J R. α and β chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol. 1995;155:3877–3888. [PubMed] [Google Scholar]
- 50.Wathelet M, Moutschen S, Defilippi P, Cravador A, Collet M, Huez G, Content J. Molecular cloning, full-length sequence and preliminary characterization of a 56-kDa protein induced by human interferons. Eur J Biochem. 1986;155:11–17. doi: 10.1111/j.1432-1033.1986.tb09452.x. [DOI] [PubMed] [Google Scholar]
- 51.Wu C, Ohmori Y, Bandyopadhyay S, Sen G, Hamilton T. Interferon-stimulated response element and NF kappa B sites cooperate to regulate double-stranded RNA-induced transcription of the IP-10 gene. J Interferon Res. 1994;14:357–363. doi: 10.1089/jir.1994.14.357. [DOI] [PubMed] [Google Scholar]