Abstract
Human immunodeficiency virus (HIV)-specific cytotoxic T lymphocytes (CTL) play a major role in control of viral replication. To understand the contribution of this antiviral response, an initial step is to fully define the specific epitopes targeted by CTL. These studies focused on CTL responses restricted by HLA-A∗3002, one of the HLA-A molecules most prominent in African populations. To avoid the time-consuming effort and expense involved in culturing CTL prior to defining epitopes and restricting alleles, we developed a method combining Elispot assays with intracellular gamma interferon staining of peripheral blood mononuclear cells to first map the optimal epitopes targeted and then define the HLA restriction of novel epitopes. In two A∗3002-positive subjects whose CTL responses were characterized in detail, the strongest response in both cases was to an epitope in p17 Gag, RSLYNTVATLY (residues 76 to 86). Using this method, CTL epitopes for which there were no motif predictions were optimized and the HLA restriction was established within 48 to 72 h of receipt of blood. This simple and convenient approach should prove useful especially in the characterization of CTL responses specific to HIV and other viruses, particularly in localities where performing cytotoxicity assays would be problematic.
Human immunodeficiency virus (HIV)-specific cytotoxic T lymphocytes (CTL) play a major role in controlling virus replication (1, 8, 9, 16, 21, 24). Development of vaccines designed to generate protective anti-HIV immune responses requires a fundamental knowledge of the CTL epitopes that are targeted by infected persons in populations most severely affected by the epidemic. So far, however, relatively little is known about the HIV-specific CTL epitopes that are presented by HLA class I molecules prevalent in sub-Saharan African populations, despite the estimate that 75% of HIV infection worldwide has occurred in this region. (UNAIDS website [http://www.unaids.org/epidemic_update/report/Epi_report.htm]). This study therefore focused on one allele prominent in southern Africa, HLA-A∗3002 (7, 15). HLA-A30 is in several countries the commonest HLA-A allele. For example, over 50% of Zimbabweans express HLA-A30 (14). In African Zulu, the phenotypic frequency of A30 is 44%, split approximately equally between A∗3001 and A∗3002 (13; M. Hammond, unpublished data). In contrast, in Caucasoids, A30 is uncommon (phenotypic frequency, 5%). No virus-specific A30-restricted epitopes have been optimally defined to date.
Initially one subject with A∗3002 was studied in detail; five novel A∗3002-restricted CTL epitopes were defined using a recently described method exploiting the sensitivity of the Elispot assay that allows CTL epitopes to be rapidly defined using peripheral blood mononuclear cells (PBMC), and the results were confirmed by the traditional approach via cytotoxicity assays (2). The HLA restriction, however, could not be defined by Elispot assays using PBMC incubated with a panel of peptide-pulsed Epstein-Barr virus (EBV)-transformed B-lymphoblastoid cell lines (BCL) because the background level became too high, possibly due to EBV-specific responses. The requirement to generate CTL clones or peptide-specific lines in order to define new epitopes is technically demanding and costly, which limits the number of laboratories that can be equipped to define new epitopes. To circumvent this need for culturing cells, a method for defining the HLA restriction of CTL responses by intracellular cytokine staining (ICS) assays was developed. Thus, using the Elispot assay for rapid epitope optimization and the ICS assay for rapid HLA restriction, novel CTL epitopes can now be defined from PBMC within 48 to 72 h of phlebotomy. In addition, this approach enables HIV-specific CTL responses to be characterized in laboratories that are not specialized in tissue culture techniques.
MATERIALS AND METHODS
Subjects studied.
Subject 199 (HLA A∗0201/∗3002 B∗4402/51 Cw2/5) is a Caucasian whose precise date of infection is unknown but who had had documented HIV infection for more than 6 years at the time of study. He is antiretroviral therapy naive. His viral load and CD4 count at the time of study were 3,700 HIV type 1 (HIV-1) RNA copies/ml of plasma and 811 cells/μl, respectively. Subject 6007 (HLA A∗3002/− B53/∗5801 Cw4/7) is an African-Caribbean who had been treated with highly active antiretroviral therapy for 1 year. Viral load at time of CTL analysis was below the level of detection (<50 copies of plasma HIV-1 RNA/ml), and the CD4+ T-cell count was 645 cells/μl.
HLA typing and subtyping.
HLA typing and subtyping were performed by sequence-specific primer PCR (6, 17).
Peptides.
Lymphocytes were tested for recognition of a panel of 290 overlapping peptides, 12 to 20 amino acids in length, spanning p17 Gag, p24 Gag, Nef, reverse transcriptase (RT), gp120, gp41, Rev, and Tat clade B SF2 sequence (35 Gag peptides provided by the NIBSC Centralized Facility for AIDS Reagents, supported by EU Program EVA and the United Kingdom Medical Research Council; the remainder synthesized either commercially [Research Genetics, Huntsville, Ala.] or at the Massachusetts General Hospital Peptide Synthesis Core). Peptides in each case overlapped by at least 10 amino acids.
Elispot assays.
Fresh (PBMC) were separated from whole blood by Ficoll-Hypaque (Sigma, St. Louis, Mo.) density gradient centrifugation and placed in 96-well polyvinylidene plates (Millipore, Bedford, Mass.) which had been precoated with (0.5 μg/ml; anti-gamma interferon (IFN-γ) monoclonal antibody (MAb) 1-DIK (Mabtech, Stockholm, Sweden). The peptides were added in a volume of 20 μl, and then PBMC were added at between 15,000 and 80,000 cells per well in a volume of 180 μl. The end concentration of the peptides was 10 μM. The plates were incubated overnight at 37°C and 5% CO2 and then washed with phosphate-buffered saline before addition of the second, biotinylated anti-IFN-γ MAb, 7-B6-1 biotin (Mabtech), at 0.5 μg/ml and incubation at room temperature for 100 min. Following washing, streptavidin-conjugated alkaline phosphatase (Mabtech) was added at room temperature for 40 min. Individual cytokine-producing cells were detected as dark spots after a 20-min reaction with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium using an alkaline phosphatase-conjugate substrate (Bio-Rad, Richmond, Calif.). The number of specific T cells was calculated by subtracting the negative control values. Background was <40/million PBMC (2 spots/well at 50,000 PBMC/well).
Generation of peptide-specific CTL lines.
Peptide-specific CTL lines were generated as previously described (19). Briefly, PBMC were incubated together with 200 μM peptide for 1 h and then resuspended in R10 medium (RPMI 1640 [Sigma], 10% fetal calf serum [Sigma], 10 mM HEPES buffer [Sigma]) with interleukin-7 (IL-7; R&D systems) added at 40 ng/ml. Following 1 week in culture at 37°C and 5% CO2 the medium was replaced twice weekly with R10 medium containing recombinant IL-2 (50-U/ml; kindly provided by M. Gately, Hoffmann-La Roche, Nutley, N.J.) instead of IL-7. Chromium release assays were performed following 2 to 4 weeks of culture.
Intracellular IFN-γ staining.
ICS assays were performed as described elsewhere (12, 22). Briefly, 0.2 × 106 to 1.0 × 106 PBMC were incubated with 4 μM peptide and anti-CD28 and anti-CD49d MAbs (each at 1 μg/ml; Becton Dickinson) at 37°C and 5% CO2 for 1 h before the addition of brefeldin A (10 μg/ml; Sigma). Following a further 6-h incubation at 37°C and 5% CO2, the cells were placed at 4°C overnight. PBMC were then washed and stained with surface antibodies, anti-CD8, and anti-CD3 (Becton Dickinson) at 4°C for 20 min. Following washing, the PBMC were fixed and permeabilized (Caltag, Burlingame, Calif.), and anti-IFN-γ MAb (Becton Dickinson) was added. Cells were then washed and analyzed. For assays using HLA-matched or mismatched BCL, BCL that were pulsed with 10 μM peptide for 1 h were washed thrice prior to incubation with PBMC or effectors at 105 BCL and 5 × 105 effectors in 1 ml of R10 medium. The anti-CD28 and anti-CD49d MAbs were then added, and the assay was carried out exactly as described above.
Generation of precursor frequency assays.
Precursor frequency assays were set up as previously described (26), using seven dilutions of PBMC from 8,000 down to 50 cells per well in 24 replicate wells. In brief, PBMC in 96-well plates were cultured with irradiated allogeneic feeder PBMC at 50,000 cells/well in a final volume per well of 200 μl of R10 medium with antibiotics (2 mM l-glutamine, 50 U of penicillin-streptomycin/ml). The anti-CD3 MAb 12F6 was added at 10 μg/ml. On day 5 and once weekly thereafter, the medium was replaced with R10 medium containing recombinant IL-2 (50 U/ml). Wells were screened for specific recognition of HLA-matched, peptide-pulsed, 51Cr (New England Nuclear, North Billerica, Mass.)-labelled EBV-transformed BCL target cells after 17 days in culture.
Bulk cultured lymphocyte assay.
As previously described (20), phytohemagglutinin-activated lymphoblasts were washed thrice and added to autologous PBMC in a ratio of 1:4 and incubated in R10 medium at 1.5 × 106/ml in 50-ml flasks (Costar). Cells were cultured in the same way as described above for the precursor frequency assays but with medium replacement with R10 medium containing recombinant IL-2 (50 U/ml) on day 7 and thereafter twice weekly. Expansions into additional 50-ml flasks were made as necessary to maintain cell numbers at 1 × 106 to 2 × 106/ml. The chromium release assay was performed on day 17.
RESULTS
Definition of a strong A∗3002-restricted response to epitope RSLYNTVATLY in p17 Gag.
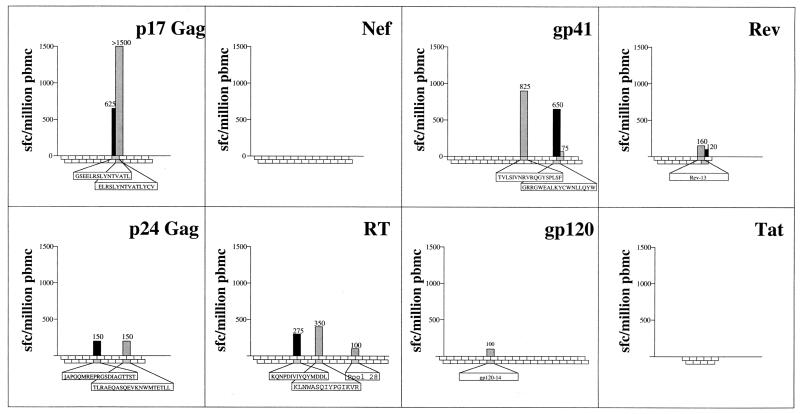
The approach adopted initially to screen for HIV-specific CTL responses is illustrated in Fig. 1, with 290 overlapping peptides spanning eight HIV-1 proteins used in the Elispot assay. As shown for subject 199, and as also observed in subject 6007 (not shown), strong responses were observed within the 15-mer peptide ELRSLYNTVATLYCV (p17 Gag residues 74 to 88). As previously described (2), optimization of CTL epitopes can be achieved with much greater rapidity by Elispot assays using PBMC, and the same optimal epitope RY11 was defined by this method (Fig. 2A). This was confirmed as the optimal epitope by generation of a CTL line specific for the longer 15-mer peptide ELRSLYNTVAT LYCV, followed by cytotoxicity assays using serial truncated peptides (Fig. 2B). This RY11-specific response was determined as HLA-A∗3002-restricted in standard chromium release assays using the peptide-specific CTL line (Fig. 2C), and the failure of A∗3001-positive targets to present this epitope to A∗3002-positive effectors (Fig. 2D and data not shown) demonstrated the need for accurate subtyping of A30-positive subjects.
FIG. 1.
Screening of PBMC from donor 199 for recognition of overlapping peptides spanning eight HIV-1 proteins as shown. Frequencies of responses >50 IFN-γ spot-forming cells (sfc) per million PBMC to individual peptides are indicated.
FIG. 2.
(A) Recognition in an Elispot assay using PBMC from donor 199 of peptides differing by one residue at the N and C termini of the optimal epitope peptide RSLYNTVATLY (RY11). (B) Recognition in a chromium release assay of truncations of the p17 Gag peptide ELRSLYNTVATLYCV by a peptide-specific CTL line from donor 199. Target BCL were from donor EBV-522 (HLA-A∗3002/0 B∗14/−Cw8/−). (C) HLA-A∗3002 restriction of the RY11-specific response using the same peptide-specific CTL line as in panel A and HLA-matched targets as shown. (D) HLA-A∗3001-positive targets do not present the RY11 peptide to A∗3002-positive effectors. A∗3002-positive BCL: EBV-522 (as in panels A and C) (diamonds); 009-BMC (A∗0201/∗3002 B14/27 Cw1/8) (squares); 016-TCH (A∗3001/− B42/− Cw17/−) (circles). Open symbols, BCL pulsed with no peptide; closed symbols; targets pulsed with 10 μM peptide.
Definition of HLA restriction by ICS.
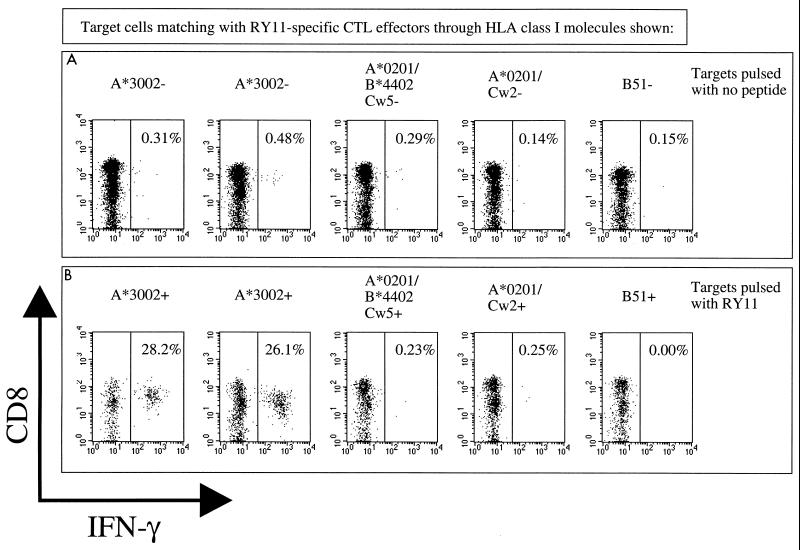
Previous studies had shown that although the optimal epitope sequence can be determined very rapidly and conveniently via Elispot assays using PBMC (2), this approach did not allow the HLA restriction of the response to be defined, since a high background of spot-forming cells was evident in Elispot plates following incubation of PBMC with HLA-matched BCL even when the BCL were not pulsed with any peptide. To circumvent this problem, an approach to defining HLA restriction via intracellular IFN-γ staining assays was adopted. Incubation of the RY11 peptide-specific effector cells with BCL matched through various of the individual HLA-A, -B, and -C alleles expressed by donor 199 enabled the HLA restriction of the response to be determined unequivocally as HLA-A∗3002 (Fig. 3).
FIG. 3.
HLA restriction of the A∗3002-RY11 response by intracellular IFN-γ staining assay using peptide-specific CTL as effectors. The BCL targets and CTL effectors used were the same as for Fig. 2C. (A) IFN-γ staining following incubation of effectors with BCL that had not been pulsed with RY11 peptide; (B) IFN-γ staining following incubation of effectors with BCL that had been pulsed with 10 μM RY11 peptide and washed thrice. Percentage of total lymphocytes gated is indicated for each plot.
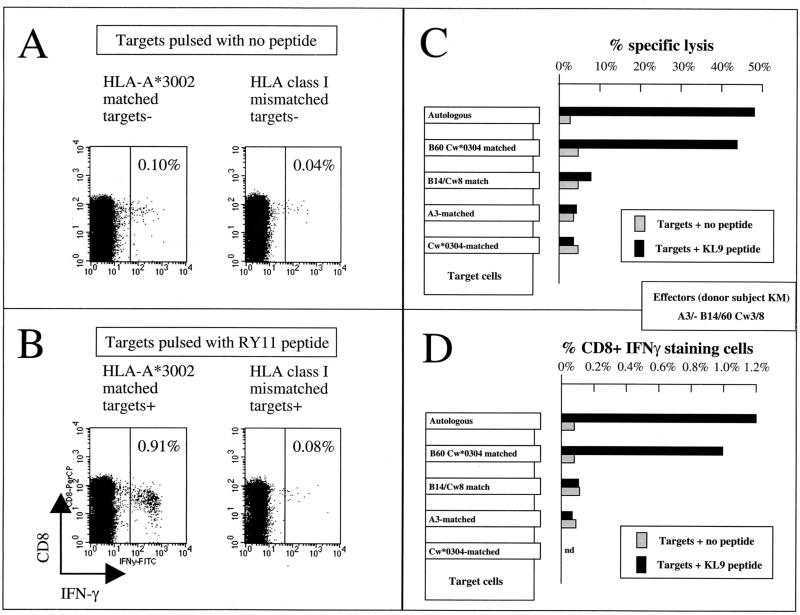
To determine whether this approach could also define the HLA restriction of a CTL response without the labor-intensive requirement for a peptide-specific CTL line, we repeated the assay using PBMC and the same panel of HLA-matched and mismatched BCL that had been employed previously. The same result was obtained by this method, as illustrated for the same A∗3002-restricted RY11-specific response in donor 6007 (Fig. 4A and B). A comparison of the methods of defining HLA restriction using peptide-specific CTL lines or clones in standard chromium release assays and using PBMC from a separate subject (described in reference 2) in intracellular IFN-γ staining assays is shown also for a B60-restricted Nef-specific response (Fig. 4C and D). These data show that this method of defining the HLA restriction of a CTL response by intracellular IFN-γ staining assay is equivalent to the standard method using CTL clones or peptide-specific CTL lines in terms of achieving the same result. However, this flow-based method represents a great saving in terms of the speed at which the result can be achieved.
FIG. 4.
HLA restriction of A∗3002- and B60-restricted CTL responses by intracellular IFN-γ staining assay using effectors within PBMC. (A and B) HLA-A∗3002 restriction of the RY11 response described above but using PBMC from donor 6007 (A∗3002/− B53/∗5801 Cw4/7) and A∗3002-matched BCL from donor 009-BMC (A∗0201/∗3002 B14/27 Cw1/8). HLA-mismatched BCL were from donor 027-BMC (A32/34 B51/71 Cw8/16). (C and D) Comparison of HLA restriction of a B60-restricted Nef-specific response using a CTL clone from donor KM (A3/− B14/60 Cw3/8) in a chromium release assay (C) and using PBMC from donor KM in an intracellular IFN-γ staining assay with the same BCL targets (D).
Definition of four further novel HLA-A∗3002-restricted CTL epitopes.
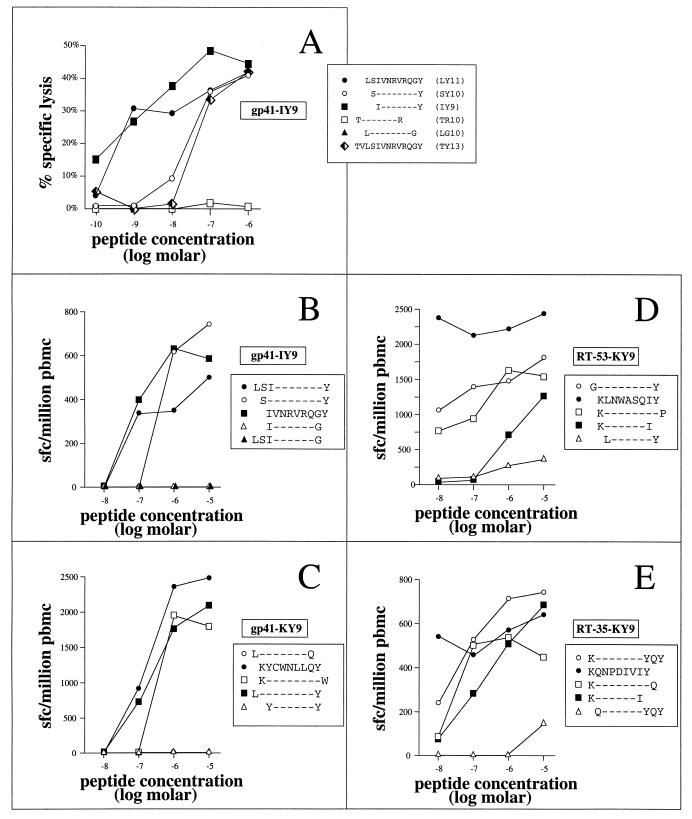
From the initial screening Elispot assays (Fig. 1), several other responses had been detected in PBMC from donor 199 using the sets of overlapping peptides, particularly in RT and gp41. None were in regions containing described epitopes for the class I alleles that this individual expressed (5). Optimization of one of the gp41-specific responses was determined by both chromium release assays using a peptide-specific line and Elispot assays using PBMC (Fig. 5A and B); optimization of three additional epitopes was determined by the latter approach alone (Fig. 5C to E). In each case, the restriction for these responses was again HLA-A∗3002 (Fig. 6 and data not shown), as determined in chromium release assays using peptide-specific lines.
FIG. 5.
Optimization of four additional novel A∗3002-restricted CTL epitopes. (A and B) Optimization of the gp41-specific epitope IVNRVRQGY (IY9) by chromium release assay using a peptide-specific line (A; BCL targets from donor EBV-522: A∗3002/− B14/− Cw8/−) and in an Elispot assay using PBMC from donor 199 (B). (C to E) Optimization of A∗3002-restricted epitopes KYCWNLLQY (KY9-gp41; C), KLNWASQIY (KY9-RT-35; D), and KQNPDIVIY (KY9-RT-53; E) by Elispot assay using PBMC incubated with peptide. The HLA restriction was confirmed in each case using peptide-specific CTL lines in standard chromium release assays (not shown). sfc, spleen-forming cells.
FIG. 6.
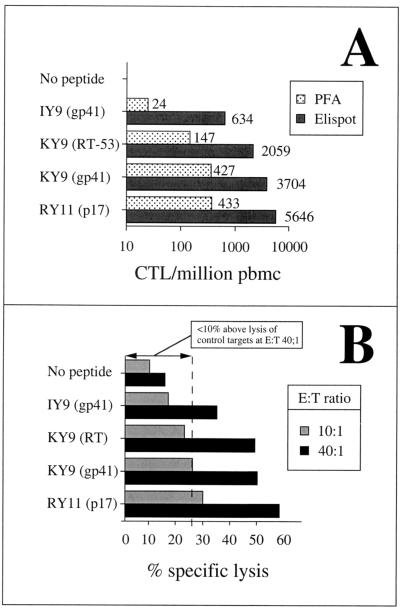
Comparison of the hierarchy of the A∗3002-restricted responses as determined by Elispot assays with the hierarchy determined from assays of cytolytic activity. (A) Frequencies of four of the A∗3002-restricted epitopes as shown in Elispot assays and in precursor frequency assays (PFAs). (B) Hierarchy of responses toward the same four epitope peptides in a chromium release assay using bulk cultured lymphocytes as effectors (E) and EBV-522 A∗3002-matched (A∗3002/− B14/−Cw8/−) BCL as targets (T).
Cytolytic function of A∗3002-restricted responses.
In assays using antigen-specific CD8+ T cells from HIV-infected subjects, we have observed a strong correlation between IFN-γ production and cytolytic function (12). However, recent data have described HIV-specific CD8+ T cells that can produce antiviral cytokines but are impaired in cytolytic function (3). We therefore compared cytolytic activities toward these newly described A∗3002-restricted epitopes to determine whether the hierarchy of response observed from measurements of IFN-γ production following peptide stimulation in Elispot or intracellular cytokine assays was equivalent to the hierarchy observed from two separate measurements of cytolytic function (Fig. 6). Comparisons of the CTL precursor frequencies in limiting dilution assays of responses toward four of the newly described epitopes with the Elispot assays supported other studies showing that the Elispot assay is more sensitive (12, 25), but the hierarchy of the responses was the same in the different assays. Similarly, the hierarchy was the same in cytotoxicity assays using bulk cultured lymphocytes as in Elispot assays. These data, albeit from study of a single subject showing successful control of viremia, are consistent with our previous studies (12) that show a strong correlation between levels of IFN-γ-producing cells in response to HIV epitope peptides and cytotolytic function.
DISCUSSION
These data show that strong HIV-specific responses toward HLA-A∗3002-restricted epitopes can be generated in infected persons. Five novel HLA-A∗3002-restricted epitopes are defined, of which the strongest in two subjects described was the epitope RSLYNTVATLY (RY11) within p17 Gag. All of the peptides have a tyrosine at the C terminus, indicating that this is an important anchor residue for F pocket binding in the A∗3002 binding groove. This epitope RY11 lies in a region of clustered epitopes that includes the dominant HLA-A∗0201-restricted epitope SLYNTVATL (5, 11). Characterization of A30-restricted CTL responses is important since A30 is so prevalent, found in up to 50% of populations such as those in southern Africa that are worst afflicted by the global HIV epidemic (UNAIDS website [http://unaids.org/epidemic_update/report/Epi_report.htm]).
The second principal result of the data described is that HLA restriction can be defined by use of PBMC incubated with peptide-pulsed BCL as antigen-presenting cells in ICS assays. This is a valuable advance in approach to characterizing CTL responses, since the combination of the Elispot assay as previously described (2) and the ICS assay described here would enable novel responses to be defined precisely within 2 to 3 days of receipt of the blood sample. This increases the speed with which novel epitopes can be defined and obviates the need for labor-intensive culture of peptide-specific CTL lines or CTL clones followed by chromium release assays. These factors together restrict CTL work to specialized laboratories. To make the dramatic progress that is urgently needed for a detailed understanding of the anti-HIV responses made by infected persons in developing countries, these assays ideally should be carried out on-site where access to the patients is least problematic. The radical simplifications to the process of defining CTL epitopes that are described here and recently (2) now enable the characterization of HIV-specific CTL responses to be undertaken in laboratories worldwide.
In common with many of the HLA class I molecules that are prevalent in areas most affected by the HIV epidemic, no HIV-specific HLA-A30-restricted CTL epitopes had previously been described. In recent studies of infected African Zulu and Xhosa in Durban, South Africa, and of African-Americans in Boston, Mass., it is noteworthy that strong responses to the RY11 epitope described here and detectable responses to the gp41- and RT-specific epitopes were observed in A∗3002-positive subjects (reference 11 and data not shown). However, despite detailed characterization of five A∗3001-positive subjects using methods similar to those described here, no A∗3001-restricted CTL epitopes have been defined to date (reference 11 and data not shown). The reasons for the infrequency of HIV-specific epitopes presented by prevalent HLA class I molecules such as HLA-A1 (5) and A∗3001 remain unknown.
A recently undertaken comparison of the peptide binding motifs of A∗3001 and A∗3002 reveals distinctive differences between the peptides bound by these two subtypes (18). As previously described for HLA-A2 subtypes (4), these binding differences correspond to functional differences, in keeping with the data above showing that A∗3001-positive targets pulsed with the A∗3002-defined epitopes were not recognized by A∗3002-positive effectors. These data underline the value of accurate HLA subtyping in order to define novel CTL responses.
A feature of the A∗3002-restricted epitopes described above that is somewhat unusual but that is consistent with the described motif for A∗3002 is the variability of residues that can be accommodated in position 2 (P2), normally a primary anchor position. To illustrate this point, the five HLA-A∗3002 epitope peptides defined above are aligned in Table 1 with the A∗3002-binding peptides that were sequenced (18). In contrast, there appears to be a greater restriction in the residues that occupy P1 than P2 for the A∗3002-binding peptides aligned in Table 1, with seven of nine peptides having either Arg or Lys in this position. However, pool sequencing of peptides eluted from A∗3002 did not identify a preference for positively charged residue at P1 (18). The significance of the variability of residues that can be accommodated at P2 is in the approach that can successfully be adopted to defining further A∗3002-restricted CTL epitopes. The method of reverse immunogenetics (13), which first predicts the sequence of peptides on the basis of the the peptide-binding motif, then tests candidate peptides fitting the motif for adequacy of binding, and finally tests the binding peptides for CTL recognition, is feasible only if the motif is relatively restricted. As can be seen from the first five A∗3002-restricted epitopes described here (Table 1), each peptide carries a different residue at P2. This suggests that an approach based on overlapping peptides as was used here is likely to be more successful for fully characterizing A∗3002-restricted virus-specific CTL responses.
TABLE 1.
Comparison of five newly defined A∗3002-restricted CTL epitopes with the A∗3002 peptide-binding motif and the sequences of four eluted A∗3002-binding peptides
| Sequence | Residue positiond
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9a | 10 | 11 | |
| A*3002 motifb | |||||||||||
| Anchors | VLY | Y | |||||||||
| Preferred | MF | HK | DEP | I | K | ||||||
| A∗3002-binding peptides | |||||||||||
| Ligandsb | A | Y | K | K | Q | F | S | Q | Y | ||
| R | I | S | G | V | D | R | Y | Y | |||
| R | L | A | W | E | V | G | W | K | Y | ||
| R | Y | L | P | P | M | W | K | Y | |||
| Epitopesc | R | S | L | Y | N | T | V | A | T | L | Y |
| I | V | R | N | V | R | Q | G | Y | |||
| K | Y | C | W | N | L | L | Q | Y | |||
| K | Q | N | P | D | I | V | I | Y | |||
| K | L | N | W | A | S | Q | I | Y | |||
C-terminal position.
Described in reference 18.
Described in this report.
Anchor positions P2 and P9, as identified by pool sequencing of eluted peptides (18), are underlined.
One important aspect of CTL epitope characterization that has not been addressed in these studies is the relevance to vaccine design of CTL epitopes that are detectable in chronically infected patients using the approach described above. The use of overlapping peptides whose sequence is based on a published consensus sequence is clearly limited by the fact that this sequence may not correspond to the autologous virus-specific responses being assayed. Previously this limitation was believed to be minor, since it was argued that conserved epitopes within which viral escape mutations would not occur early in infection would represent the most effective CTL responses that should be induced by vaccines (9, 20). However, recent data from the simian immunodeficiency virus macaque model (1) make the opposing argument, that epitopes in which escape occurs earliest may indeed represent the very responses that should be incorporated into vaccine design. If the epitopes that are most effective in containing virus are those that lie in variable regions, then the entire virus in each HIV-infected subject needs to be sequenced before initiation of CTL studies. This would clearly represent a huge investment of time and funding and thus would not be feasible for all patients investigated; however, studies should be undertaken in selected patients to determine the most effective CTL responses that should be induced by a future HIV vaccine. In the meantime, this critical issue is perhaps most likely to be resolved by future challenge experiments carried out in macaques previously given vaccines to induce different CTL specificities in different groups of animals.
In conclusion, strong HIV-specific HLA-A∗3002-restricted CTL responses in two infected subjects are described. These are the first A30-restricted CTL epitopes to be fully defined. These are of importance since A30 is one of the most prevalent HLA class I molecules expressed in populations such as those in southern Africa that have been most seriously affected by the global HIV epidemic. Alignment of the novel epitopes described illustrates a high degree of variability in the amino acid residues that can reside at P2 within A∗3002-binding epitope peptides. This finding is of significance, suggesting that an approach to defining further A∗3002-restricted epitopes specific to HIV-1 or indeed other viruses using reverse immunogenetics will be relatively unsuccessful. Finally, a rapid method of defining the HLA restriction of CTL responses using PBMC and intracellular IFN-γ staining assays is described. This will be of substantial value in reducing the time necessary to fully define novel CTL epitopes.
ACKNOWLEDGMENTS
This work was supported by grants to P.J.R.G. from the Elizabeth Glaser Pediatric AIDS Foundation, the United Kingdom Medical Research Foundation (G108/274), and the National Institutes of Health (NIH) (AI46995); to M.M.A. from the German Research Foundation; to M.A.A. from the German Academic Exchange Foundation; to E.S.R. from the Doris Duke Charitable Foundation and NIH (AI 01541); and to B.D.W. from the NIH (AI28568 and AI30914) and Doris Duke Charitable Foundation. P.J.R.G. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation. B.D.W. is a Doris Duke Distinguished Clinical Science Professor.
REFERENCES
- 1.Allen, T., D. O'Connor, P. Jing, J. Dzuris, B. Mothé, T. Vogel, E. Dunphy, M. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. Wolinsky, A. Sette, and D. I. Watkins. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386–390. [DOI] [PubMed]
- 2.Altfeld, Trocha M A A, Eldridge R L, Rosenberg E S, Addo M A, Phillips M, Sekaly R P, Kalams S A, Burchett S A, McIntosh K, Walker B D, Goulder P J R. Identification of dominant optimal HLA-B60- and B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J Virol. 2000;74:8541–8549. doi: 10.1128/jvi.74.18.8541-8549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appay V, Nixon D F, Donahue S M, Gillespie G A, Dong T, King A, Ogg G S, Spiegel H M L, Conlon C, Spina C A, Havlir D V, Richman D D, Waters A, Easterbrook P, McMichael A J, Rowland-Jones S. HIV-specific CD8+ T cells produce antiviral cytokines but are impaired in cytolytic function. J Exp Med. 2000;192:67–75. doi: 10.1084/jem.192.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch D, Friede T, Stevanovic S, et al. HLA-A2 subtypes are functionally distinct in peptide binding and presentation. J Exp Med. 1995;182:1847–1856. doi: 10.1084/jem.182.6.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brander C, Goulder P J R. Recent advances in the optimization of HIV-specific CTL epitopes. In: Korber B T M, Brander C, Walker B D, Koup R A, Moore J, Haynes B, Meyers G, editors. HIV molecular immunology database. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex. [Online.] http://www.hiv-lanl.gov. 1999. [Google Scholar]
- 6.Bunce M, O'Neill C M, Barnardo M C, Krausa P, Browning M J, Morris P J, Welsh K I. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence- specific primers (PCR-SSP) Tissue Antigens. 1995;46:355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 7.Clayton J, Lonjou C. Allele and haplotype frequencies for HLA loci in various ethnic groups. In: Charron D, editor. HLA: genetic diversity of HLA: functional and medical implication. 1997. pp. 665–820. Proceedings of the Twelfth International Histocompatibility Workshop and Conference. EDK, Paris, France. [Google Scholar]
- 8.Evans D T, O'Connor D H, Jing P, Dzuris J L, Sidney J, da Silva J, Allen T M, Horton H, Venham J E, Rudersdorf R A, Vogel T, Pauza C D, Bontrop R E, DeMars R, Sette A, Hughes A L, Watkins D I. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 9.Goulder P J R, Rowland-Jones S, McMichael A J, Walker B D. Anti-HIV cellular immunity: recent advances towards vaccine design. AIDS. 1999;13(Suppl. A):S121–S136. [PubMed] [Google Scholar]
- 10.Goulder P J R, Tang Y, Pelton S I, Walker B D. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal human immunodeficiency virus epitopes, one of which is entirely contained within the other. J Virol. 2000;74:5291–5299. doi: 10.1128/jvi.74.11.5291-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goulder P J R, Brander C, Annamalai K, Mngqundaniso N, Govender U, Tang Y, He S, Hartman K E, O'Callaghan C A, Ogg G S, Altfeld M, Rosenberg E S, Cao H, Kalams S A, Hammond M G, Bunce M, Pelton S I, Burchett S A, McIntosh K, Coovadia H M, Walker B D. Differential narrow focusing of immunodominant human immunodeficiency virus Gag-specific cytotoxic T-lymphocyte responses in infected African and Caucasoid adults and children. J Virol. 2000;74:5679–5690. doi: 10.1128/jvi.74.12.5679-5690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulder P J R, Tang Y, Brander C, Betts M, Altfeld M A, Annamalai K, Trocha A, He S, Rosenberg E S, Ogg G, O'Callaghan C A, Kalams S A, Mayer K, Koup R, Pelton S I, Burchett S K, McIntosh K, Walker B D. Functionally inert HIV-specific cytotoxic T lymphocyte do not play a major role in chronically infected adults and children. J Exp Med. 2000;192:1819–1831. doi: 10.1084/jem.192.12.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond M G, du Toit E D, Sanchez-Mazas A, Andrien M, Coluzzi M, de Pablo M R, de Stefano G, Kaplan C, Kennedy L J, Louie L, Migot F. HLA in sub-Saharan Africa: 12th International Histocompatibility Workshop SSAF report. In: Charron D, editor. Proceedings of the Twelfth International Histocompatibility Workshop and Conference. Paris, France: EDK; 1997. pp. 345–353. [Google Scholar]
- 14.Hill A V S, Allsopp C E, Kwiatkowski D, Anstey N M, Twumasi P, Rowe P A, Bennett S, Brewster D, McMichael A J, Greenwood B M. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 15.Imanishi T, Akaza T, Kimura A, Tokunaga K, Gojobori T. Allele and haplotype frequencies for HLA and complement loci in various ethnic groups. In: Tsuji K, Aizawa M, Sasaszuki T, editors. HLA 1991—Proceedings of the Xth International Histocompatibility Workshop and Conference. Vol. 1. Oxford, United Kingdom: Oxford University Press; 1992. pp. 1065–1220. [Google Scholar]
- 16.Jin X, Bauer D E, Tuttleton S E, Gettie A, Blanchard J, Irwin C E, Safrit J T, Lewin S, Mittler J, Weinberger L, Kostrikis L G, Zhang L, Perelson A S, Ho D D. Dramatic rise in plasma viremia after CD8+ T cell depletion in SIV-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krausa P, Carcassi C, Orru S, et al. Defining the allelic variants of HLA-A30 in the Sardinian population using amplification refractory mutation system-polymerase chain reaction. Hum Immunol. 1995;44:35–44. doi: 10.1016/0198-8859(95)00046-7. [DOI] [PubMed] [Google Scholar]
- 18.Krausa P, Munz C, Keilholz W, Stevanovic S, Jones E Y, Browning M, Bunce M, Rammensee H-G, McMichael A J. Definition of peptide binding motifs amongst the HLA-A∗30 allelic group. Tissue Antigens. 2000;56:8–10. doi: 10.1034/j.1399-0039.2000.560102.x. [DOI] [PubMed] [Google Scholar]
- 19.Lalvani A J, Dong T, Ogg G, Patham A A, Newell H, Hill A, McMichael A J, Rowland-Jones S. Optimization of a peptide-based protocol employing IL-7 for in vitro restimulation of human cytotoxic T lymphocyte precursors. J Immmunol Methods. 1977;210:65–77. doi: 10.1016/s0022-1759(97)00177-4. [DOI] [PubMed] [Google Scholar]
- 20.Nixon D F, Townsend A R, Elvin J G, Rizza C R, Gallwey J, McMichael A J. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature. 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 21.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon N F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 22.Pitcher C J, Quittner C, Peterson D M, Connors M, Koup R A, Maino V C, Picker L J. HIV-1 specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 23.Rammensee H G, Friede T, Stevanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 25.Tan L C, Gudgeon N, Annels N E, Hansasuta P, O'Callaghan C A, Rowland-Jones S, McMichael A J, Rickinson A J, Callan M F. A reevaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. J Immunol. 1999;162:1827–1835. [PubMed] [Google Scholar]
- 26.Walker B D. HIV-1-specific cytotoxic T lymphocytes. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. p. 201-233. [Google Scholar]
- 28.Wilkinson D, Connolly C, Rotchford K. Continued explosive rise in HIV prevalence among pregnant women in rural South Africa. AIDS. 1999;13:740. doi: 10.1097/00002030-199904160-00023. [DOI] [PubMed] [Google Scholar]