Summary
Angelman syndrome (AS) is a severe neurodevelopmental disorder caused by the loss of function of maternal UBE3A. The major cause of AS is a maternal deletion in 15q11.2-q13, and the minor causes are a UBE3A mutation, uniparental disomy (UPD), and imprinting defect (ID). Previous reports suggest that all patients with AS exhibit developmental delay, movement or balance disorders, behavioral characteristics, and speech impairment. In contrast, a substantial number of AS patients with a UBE3A mutation, UPD, or ID were reported not to show these consistent features and to show age-dependent changes in their features. In this study, we investigated 134 patients with AS, including 57 patients with a UBE3A mutation and 48 patients with UPD or ID. Although developmental delay was present in all patients, 20% of patients with AS caused by UPD or ID did not exhibit movement or balance disorders. Differences were also seen in hypopigmentation and seizures, depending on the causes. Moreover, patients with a UBE3A mutation, UPD, or ID tended to show fewer of the specific phenotypes depending on their age. In particular, in patients with UPD or ID, easily provoked laughter and hyperactivity tended to become more pronounced as they aged. Therefore, the clinical features of AS based on cause and age should be understood, and genetic testing should not be limited to patients with the typical clinical features of AS.
Keywords: uniparental disomy, imprinting defect, UBE3A, epigenetics
We investigated the clinical features of 134 patients with Angelman syndrome (AS). Our study revealed that patients with a UBE3A mutation or UPD/ID showed varying clinical features of AS depending on their age. Therefore, genetic tests should not be limited to patients with typical clinical features of AS.
Introduction
Angelman syndrome (AS; MIM: 105830) is a severe neurodevelopmental disorder caused by the loss of function of maternal ubiquitin protein ligase E3A (UBE3A; MIM: 601623).1,2,3 The UBE3A gene is located on the 15q11.2 region. This region is subject to genomic imprinting; therefore, gene expression is dependent on the parent of origin. Deletion of maternal 15q11.2-q13, a UBE3A mutation in the maternal allele, or paternal uniparental disomy (UPD)/maternal imprinting defect (ID) accounts for 70%, 5% and 5% of cases, respectively.4 The loss of function of the UBE3A gene affects the clinical features of AS, and leads to developmental delay and motor disorders in patients with AS.1,5,6 Furthermore, in the case of deletion, the other genes located in the deletion region also contribute to the clinical features.7,8 Clinical features of patients with AS are known to exhibit developmental delay, movement or balance disorders, seizures, speech impairment, microcephaly, behavioral characteristics, and sleep disorder.9 Developmental delay becomes apparent by the age of 6 months. Language expression is limited, and even with increasing age, it often remains just a few words.10 Abnormal electroencephalogram (EEG) is seen in almost all patients with AS.11 Seizures are observed in over 80% of patients, and the risk increases after age 5 years.12 Patients with AS also have behavioral characteristics, such as impulsivity, inattention, hyperactivity and easily provoked laughter,13 and a high prevalence of sleep problems in the form of disturbances in initiating and maintaining sleep.14 Furthermore, some clinical features are often observed by around age 3 years.15 Patients with AS are characterized by various distinctive features, which tend to become more apparent with age. Therefore, Williams et al.16 suggested that the clinical diagnosis in patients with AS becomes a more frequent diagnostic consideration between 1 and 4 years. Their report also suggested that all patients with AS exhibit developmental delay, movement or balance disorders, behavioral characteristics, and speech impairment. Additionally, over 80% of patients with AS display seizures, abnormal EEG, and microcephaly. Sleep disorders and hypopigmentation are observed in 20%–80% of patients with AS. Subsequent reports also suggested that movement disorders, developmental delay, and speech impairment were observed in almost all patients with AS, leading to similar results.17,18 Despite these reports, some previous reports of AS identified genotype-phenotype correlation.19,20,21,22,23,24,25 These reports suggested that patients with UBE3A mutations or UPD/ID were associated with higher rates of obesity and less frequently with seizures, hypopigmentation and microcephaly, milder symptoms with movement, or balance disorders. There are also differences in the age of symptom onset. Previous reports indicated that first onset of walking in AS with deletion occurred at an average age of 5.2 years, compared to 2.5 years in AS with non-deletion cases. Additionally, the age of first seizure in AS with deletion occurs before age 3 years, while in non-deletion cases, it is around 5.4 years.21 Therefore, we might encounter patients with AS who did not match these consistent features and miss the opportunity to undergo appropriate genetic testing, particularly in patients with a UBE3A mutation or UPD/ID. Since only a limited number of patients with a UBE3A mutation and UPD/ID have been reported, the clinical features and onset times of these features in patients with a UBE3A mutation and UPD/ID remain unclear.26 In cases of AS with deletion, the mean age at diagnosis was 5 years 8 months, whereas for AS with UPD, the mean age at diagnosis was 9 years.23 Here, we report on the clinical features of a large number of patients with AS. Furthermore, this study, involving a substantially large number of patients with a UBE3A mutation and UPD/ID, elucidated the clinical features of AS with these minor causes and clarified the differences in clinical features by their age.
Subjects and methods
We investigated the clinical features and genetic causes of patients with AS who received a genetic diagnosis in our laboratory in Japan by analyzing surveys and reviewing medical records. Our laboratory performed comprehensive genetic analysis of AS for patients who were suspected to have AS by their attending physicians. This patient information was provided by physicians treating these patients who requested genetic testing for AS. The survey inquired about the following clinical features: developmental delay, movement or balance disorders, easily provoked laughter, hand tremor, hyperactivity, absence of meaningful words, seizure, abnormal EEG, hypopigmentation, sleep disorder, microcephaly (2 SD smaller than average), age at onset of walking, and age at first seizure. Movement or balance disorders included ataxic gait and the inability to walk. This study was approved by the institutional review board of the Nagoya City University Graduate School of Medical Sciences.
We categorized the causes of AS into three groups: deletion, UBE3A mutation, or UPD/ID. Deletions were detected using fluorescence in situ hybridization tests. UBE3A mutations were identified through Sanger sequencing or next-generation sequencing. UPD/ID was identified when fluorescence in situ hybridization tests were negative and the methylation test in the 15q11.2 region was positive. One of these patients was previously reported on in a case report.27
In this study, we addressed missing data by using pairwise deletion. An analysis of the sex and frequency of various symptoms was conducted across the three patient groups as a qualitative parameter. To determine whether there were significant differences between the groups, the Fisher’s exact test was employed, and the Holm-Bonferroni correction was utilized for pairwise comparisons between groups. A p value of less than 0.05 was considered significant. To compare quantitative parameters (age and weight) among multiple groups, the Kruskal-Wallis test was used with the same p value threshold. Kaplan-Meier analyses were used to describe both the age at onset of walking and first seizure up to the age of 10 years. The log rank test was employed with the same p value threshold. All ages were rounded down to the whole number. All statistical analyses were performed with GraphPad Prism (version 10.2.0 for Windows, GraphPad Software, La Jolla, CA, www.graphpad.com), and EZR (version 1.64, Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user interface for R (version 4.3.1, R Foundation for Statistical Computing, Vienna, Austria).28
Results
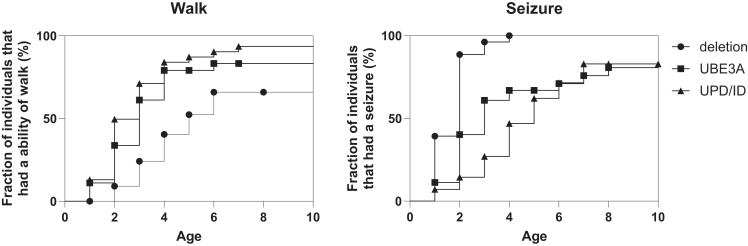
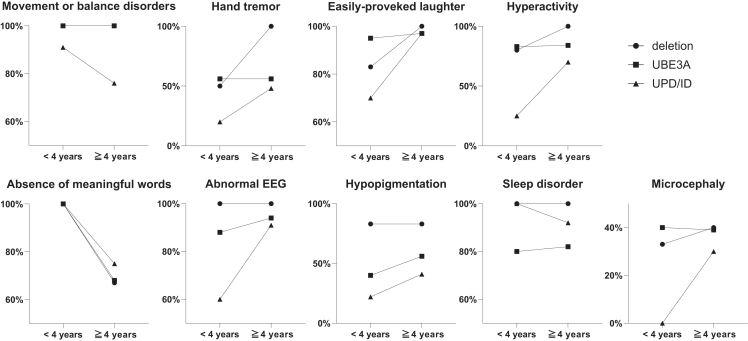
We investigated 134 patients with AS. Their ages ranged from 1 to 45 years of age, and the median was 6 years old, with 77 males and 57 females. A total of 29 cases were caused by a maternal deletion in the 15q11.2-q13 region, 57 cases by a UBE3A mutation, and 48 cases by UPD/ID (Table 1). There were no significant differences in sex, age, gestational week at birth, or birth weight among the groups. The median age of clinical diagnosis was 1 year in the deletion group, 3 years in the UBE3A mutation group, and 4 years in the UPD/ID group, showing significant differences (Table 1). Developmental delay, as detected by surveys or medical records, was present in all the patients (Table 2). Movement or balance disorders were observed in 92% of patients. Notably, in the UPD/ID group, some patients did not exhibit movement or balance disorders. Although the characteristic AS symptom of easily provoked laughter was observed in 94% of patients across all groups, interestingly, there were patients who did not have this symptom in all groups. Hand tremor was observed in 51% of patients and hyperactivity was observed in 72% of patients, with no significant difference among groups, although a lesser tendency was noted in the UPD/ID group. For language abilities, the absence of meaningful words was present in 79% of patients, with no significant differences among groups. Nearly all patients in the deletion group had a history of seizures, while patients in the UBE3A mutation and UPD/ID groups had significantly lower frequencies of seizures, 60%–70%. The nature of the seizures varied, including myoclonic, absence, and atonic seizures. Valproic acid was commonly used as an antiseizure medication. Differences in EEG abnormalities were not significant among groups and were observed in many patients with AS. These abnormalities often involved high-amplitude slow waves, predominantly in the frontal or occipital regions. Hypopigmentation was significantly more common in the deletion group, compared with the other groups. The prevalence of sleep disorders and microcephaly showed little variation across the groups. In the Kaplan-Meier analyses for the age at onset of walking, a significant difference was exhibited (p = 0.0012) among the groups. The onset of walking in AS with a UBE3A mutation (median 3 years) and UPD/ID (median 3 years) was earlier than AS with deletion (median 5 years) (Figure 1). In contrast, the age of first seizure in AS with deletion (median 2 years) was earlier than AS with a UBE3A mutation (median 3 years) and UPD/ID (median 5 years), with the difference being significant (p < 0.0001). To further investigate the differences in clinical features based on age, we separated the patients with AS into two age groups: those younger than 4 years and those 4 years or older (Figure 2). This division was based on the tendency for the diagnosis to be more frequent between children ages 1 to 4 years. Our analysis revealed that differences in movement or balance disorders tended to be fewer in patients with UPD/ID younger than 4 years of age. In the absence of meaningful words, similar results that tended to be fewer in patients younger than 4 years of age were observed across deletion, UBE3A mutation, and UPD/ID groups. In contrast, the clinical features in easily provoked laughter, hand tremor, hyperactivity, and abnormal EEG tended to become more prominent in patients with AS who were 4 years or older. In particular, the patients with UPD/ID showed a tendency in easily provoked laughter and hyperactivity to increase with age.
Table 1.
Clinical parameters across deletion, UBE3A mutation, and UPD/ID groups
| Deletion | UBE3A mutation | UPD/ID | p value | |
|---|---|---|---|---|
| Number of patients | 29 | 57 | 48 | |
| Males, n (%) | 14 (48) | 30 (53) | 33 (69) | 0.14 |
| Age, y (median, IQR1–IQR3) | 8 (3–11) | 6 (3–14) | 6 (4–11) | 0.96 |
| Gestational age, weeks (median, IQR1–IQR3) | 39 (37.25–40) | 38 (37–39) | 38 (37–39.25) | 0.44 |
| Birth weight, g (median, IQR1–IQR3) | 2,924 (2,570–3,304.25) | 2,800 (2,600–2,917.25) | 2,850 (2,664–3,178) | 0.19 |
| Clinically diagnosed age, y (median, IQR1–IQR3) | 1 (1–2) | 3 (2–5) | 4 (3–6) | <0.0001 |
IQR, interquartile range.
Table 2.
Clinical features of patients with AS by genetic cause
| Clinical feature | Deletion |
UBE3A mutation |
UPD/ID |
Fisher’s exact test |
|||
|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | p value | |
| Developmental delay | 15/15 | 100 | 46/46 | 100 | 41/41 | 100 | |
| Movement or balance disorders | 20/20 | 100 | 50/50 | 100 | 36/45 | 80a | 0.0004 |
| Hand tremor | 5/7 | 71 | 23/41 | 56 | 14/35 | 40 | 0.21 |
| Easily provoked laughter | 18/19 | 95 | 49/51 | 96 | 41/45 | 91 | 0.77 |
| Hyperactivity | 5/6 | 83 | 26/31 | 84 | 18/31 | 58 | 0.06 |
| Absence of meaningful words | 19/25 | 76 | 45/57 | 79 | 34/42 | 81 | 0.92 |
| Seizure | 28/29 | 97 | 37/54 | 69b | 26/43 | 60b | 0.0009 |
| Abnormal EEG | 15/15 | 100 | 46/50 | 92 | 32/37 | 86 | 0.35 |
| Hypopigmentation | 15/18 | 83 | 26/51 | 51b | 11/31 | 35b | 0.0049 |
| Sleep disorder | 16/16 | 100 | 18/22 | 82 | 14/15 | 93 | 0.17 |
| Microcephaly (2 SD smaller) | 8/21 | 38 | 18/46 | 39 | 6/26 | 23 | 0.38 |
Significantly less frequent with UPD/ID than with UBE3A mutation (p < 0.05).
Significantly (p < 0.05) less frequent with UBE3A mutation and UPD/ID than with deletion (p < 0.05).
Figure 1.
Kaplan-Meier analyses of cumulative walking and seizure events
Graph curves are shown up to age 10 years.
Figure 2.
Age-dependent changes of clinical features
Fraction of clinical features are demonstrated in children younger than 4 years and 4 years and older. In movement or balance disorders and absence of meaningful words, the symbols ● and ■ overlap.
Discussion
In this study, we observed that clinical features of AS varied depending on age and underlying cause. The age of diagnosis became earlier compared to the previous literature. This may be affected by increasing awareness and easier access to genetic testing for AS in Japan. Moreover, it was demonstrated that some typical features of AS, which were previously thought to always be present, might not be observed at certain ages. Developmental delay was observed in all patients with AS and should be considered a key feature of AS. Nevertheless, even in the absence of symptoms such as seizure, hypopigmentation, or movement or balance disorders, AS due to UBE3A mutation or UPD/ID should be considered. The incidence of seizures was significantly lower in the UBE3A mutation and UPD/ID groups compared with the deletion group. This may be attributed to the deletion including the GABAA receptor subunit genes.29 The abnormal EEG patterns and treatment with an antiseizure medication were similar to past reports.30 Hypopigmentation was more common in the deletion group and less common for the other groups. This is related to the fact that in deletion, not only UBE3A but also the neighboring OCA2 gene (MIM: 611409) is deleted.31,32 It is noteworthy that there were some patients with UPD/ID who did not exhibit movement or balance disorders. Despite this being one of the most characteristic symptoms of AS, it was surprising that 20% of patients with UPD/ID did not exhibit this symptom. Although some previous reports studied the difference in ataxic gait among deletion, UBE3A, and UPD/ID, the small number of patients with UBE3A mutation and UPD/ID made it difficult to clearly understand their symptoms.20,23 In this report, these differences became more clearly defined. Furthermore, while there was no difference among the three groups, easily provoked laughter was not observed in 5%–9% of patients with AS.
The clinical features of AS tend to become more consistent after the first year of life and increase the rate of clinical diagnosis.16 In this study, the acquisition of walking ability is earlier in UBE3A mutation and UPD/ID. Seizures were observed in less than half of the cases with a UBE3A mutation or UPD/ID even by 2 years of age, potentially leading to misdiagnosis. Within these age groups, it was suggested that the features of movement or balance disorders might disappear in older patients with UPD/ID. In contrast, it was found that the behavioral characteristics of AS with a UBE3A mutation and UPD/ID tend to be masked until the patients are older. This behavioral characteristic is a distinctive symptom in AS.33 Therefore, clinical features should be assessed considering the age of the patient. Given the recent development of novel therapeutics and suggestions for the effectiveness of earlier treatment initiation for AS,34,35,36 understanding the age of symptom onset can become even more important. In addition, patients with AS due to ID respond well to early behavioral interventions, and it is recommended that active intervention take place at an early age.37 It was also reported that patients with AS due to UPD/ID tended to become overweight within 44 months of age.38 Therefore, early diagnosis is crucial for the appropriate management of patients with AS. Additionally, the introduction of long-read sequencing for simultaneous testing of deletion, UBE3A mutation, and methylation analysis will enable earlier diagnosis.39
This study had several limitations. The first limitation is that our data contained missing information due to our data sources being based on surveys and medical records. The second limitation is the variability of evaluators. This variability might have influenced the stability of the evaluations conducted. The third limitation is the distribution of causes, which differs from the general population in AS. This is because this study was based on requests for genetic analysis of AS.
In conclusion, our study evaluated the clinical features of 134 patients with AS. Typical clinical features may not be present in some patients with AS, especially in patients with a UBE3A mutation or UPD/ID. Moreover, clinical features change depending on age in patients with AS. Therefore, the clinical features of AS based on cause and age should be taken into account, and genetic testing should not be limited to patients with the typical clinical features of AS.
Data and code availability
All data relevant to the study are included in the report. Please contact the corresponding author, S.S., for further information.
Acknowledgments
The authors thank the patients and guardians for participating in this study. This work was supported by the Japan Agency for Medical Research and Development (AMED) (JP23ek0109587).
Declaration of interests
The authors declare no competing interests.
Web resources
OMIM, https://www.omim.org/.
References
- 1.Jiang Y.H., Armstrong D., Albrecht U., Atkins C.M., Noebels J.L., Eichele G., Sweatt J.D., Beaudet A.L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 2.Matsuura T., Sutcliffe J.S., Fang P., Galjaard R.J., Jiang Y.H., Benton C.S., Rommens J.M., Beaudet A.L. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 3.Sutcliffe J.S., Jiang Y.H., Galijaard R.J., Matsuura T., Fang P., Kubota T., Christian S.L., Bressler J., Cattanach B., Ledbetter D.H., Beaudet A.L. The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Res. 1997;7:368–377. doi: 10.1101/gr.7.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton-Smith J., Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J. Med. Genet. 2003;40:87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heck D.H., Zhao Y., Roy S., LeDoux M.S., Reiter L.T. Analysis of cerebellar function in Ube3a-deficient mice reveals novel genotype-specific behaviors. Hum. Mol. Genet. 2008;17:2181–2189. doi: 10.1093/hmg/ddn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landers M., Calciano M.A., Colosi D., Glatt-Deeley H., Wagstaff J., Lalande M. Maternal disruption of Ube3a leads to increased expression of Ube3a-ATS in trans. Nucleic Acids Res. 2005;33:3976–3984. doi: 10.1093/nar/gki705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLorey T.M., Handforth A., Anagnostaras S.G., Homanics G.E., Minassian B.A., Asatourian A., Fanselow M.S., Delgado-Escueta A., Ellison G.D., Olsen R.W. Mice lacking the beta3 subunit of the GABAA receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome. J. Neurosci. 1998;18:8505–8514. doi: 10.1523/JNEUROSCI.18-20-08505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinkkonen S.T., Homanics G.E., Korpi E.R. Mouse models of Angelman syndrome, a neurodevelopmental disorder, display different brain regional GABA(A) receptor alterations. Neurosci. Lett. 2003;340:205–208. doi: 10.1016/s0304-3940(03)00123-x. [DOI] [PubMed] [Google Scholar]
- 9.Buiting K., Williams C., Horsthemke B. Angelman syndrome - insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 2016;12:584–593. doi: 10.1038/nrneurol.2016.133. [DOI] [PubMed] [Google Scholar]
- 10.Andersen W.H., Rasmussen R.K., Strømme P. Levels of cognitive and linguisticdevelopment in Angelman syndrome: a study of 20 children. Logoped Phoniatr. Vocol. 2001;26:2–9. doi: 10.1080/14015430117324. [DOI] [PubMed] [Google Scholar]
- 11.Boyd S.G., Harden A., Patton M.A. The EEG in early diagnosis of the Angelman (happy puppet) syndrome. Eur. J. Pediatr. 1988;147:508–513. doi: 10.1007/BF00441976. [DOI] [PubMed] [Google Scholar]
- 12.Dan B., Boyd S.G. Angelman syndrome reviewed from a neurophysiological perspective. The UBE3A-GABRB3 hypothesis. Neuropediatrics. 2003;34:169–176. doi: 10.1055/s-2003-42213. [DOI] [PubMed] [Google Scholar]
- 13.Pelc K., Cheron G., Dan B. Behavior and neuropsychiatric manifestations in Angelman syndrome. Neuropsychiatr. Dis. Treat. 2008;4:577–584. doi: 10.2147/ndt.s2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruni O., Ferri R., D’Agostino G., Miano S., Roccella M., Elia M. Sleep disturbances in Angelman syndrome: a questionnaire study. Brain Dev. 2004;26:233–240. doi: 10.1016/S0387-7604(03)00160-8. [DOI] [PubMed] [Google Scholar]
- 15.Margolis S.S., Sell G.L., Zbinden M.A., Bird L.M. Angelman Syndrome. Neurotherapeutics. 2015;12:641–650. doi: 10.1007/s13311-015-0361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams C.A., Beaudet A.L., Clayton-Smith J., Knoll J.H., Kyllerman M., Laan L.A., Magenis R.E., Moncla A., Schinzel A.A., Summers J.A., Wagstaff J. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am. J. Med. Genet. 2006;140:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- 17.Micheletti S., Palestra F., Martelli P., Accorsi P., Galli J., Giordano L., Trebeschi V., Fazzi E. Neurodevelopmental profile in Angelman syndrome: more than low intelligence quotient. Ital. J. Pediatr. 2016;42:91. doi: 10.1186/s13052-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler A.C., Sacco P., Cabo R. Unmet clinical needs and burden in Angelman syndrome: a review of the literature. Orphanet J. Rare Dis. 2017;12:164. doi: 10.1186/s13023-017-0716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lossie A.C., Whitney M.M., Amidon D., Dong H.J., Chen P., Theriaque D., Hutson A., Nicholls R.D., Zori R.T., Williams C.A., Driscoll D.J. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J. Med. Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan W.-H., Bacino C.A., Skinner S.A., Anselm I., Barbieri-Welge R., Bauer-Carlin A., Beaudet A.L., Bichell T.J., Gentile J.K., Glaze D.G., et al. Angelman syndrome: Mutations influence features in early childhood. Am. J. Med. Genet. A. 2011;155A:81–90. doi: 10.1002/ajmg.a.33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moncla A., Malzac P., Voelckel M.A., Auquier P., Girardot L., Mattei M.G., Philip N., Mattei J.F., Lalande M., Livet M.O. Phenotype-genotype correlation in 20 deletion and 20 non-deletion Angelman syndrome patients. Eur. J. Hum. Genet. 1999;7:131–139. doi: 10.1038/sj.ejhg.5200258. [DOI] [PubMed] [Google Scholar]
- 22.Gentile J.K., Tan W.-H., Horowitz L.T., Bacino C.A., Skinner S.A., Barbieri-Welge R., Bauer-Carlin A., Beaudet A.L., Bichell T.J., Lee H.-S., et al. A neurodevelopmental survey of Angelman syndrome with genotype-phenotype correlations. J. Dev. Behav. Pediatr. 2010;31:592–601. doi: 10.1097/DBP.0b013e3181ee408e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varela M.C., Kok F., Otto P.A., Koiffmann C.P. Phenotypic variability in Angelman syndrome: comparison among different deletion classes and between deletion and UPD subjects. Eur. J. Hum. Genet. 2004;12:987–992. doi: 10.1038/sj.ejhg.5201264. [DOI] [PubMed] [Google Scholar]
- 24.Smith A., Marks R., Haan E., Dixon J., Trent R.J. Clinical features in four patients with Angelman syndrome resulting from paternal uniparental disomy. J. Med. Genet. 1997;34:426–429. doi: 10.1136/jmg.34.5.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fridman C., Varela M.C., Kok F., Diament A., Koiffmann C.P. Paternal UPD15: further genetic and clinical studies in four Angelman syndrome patients. Am. J. Med. Genet. 2000;92:322–327. doi: 10.1002/1096-8628(20000619)92:5<322::aid-ajmg6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 26.Yang L., Shu X., Mao S., Wang Y., Du X., Zou C. Genotype-Phenotype Correlations in Angelman Syndrome. Genes. 2021;12 doi: 10.3390/genes12070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto M., Nakamura Y., Iwaki T., Sato E., Ieda D., Hattori A., Shiraki A., Mizuno S., Saitoh S. Angelman syndrome with mosaic paternal uniparental disomy suggestive of mitotic nondisjunction. J. Hum. Genet. 2023;68:87–90. doi: 10.1038/s10038-022-01088-z. [DOI] [PubMed] [Google Scholar]
- 28.Kanda Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cassater D., Bustamante M., Sach-Peltason L., Rotenberg A., Nespeca M., Tan W.-H., Bird L.M., Hipp J.F. Clinical Characterization of Epilepsy in Children With Angelman Syndrome. Pediatr. Neurol. 2021;124:42–50. doi: 10.1016/j.pediatrneurol.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thibert R.L., Conant K.D., Braun E.K., Bruno P., Said R.R., Nespeca M.P., Thiele E.A. Epilepsy in Angelman syndrome: a questionnaire-based assessment of the natural history and current treatment options. Epilepsia. 2009;50:2369–2376. doi: 10.1111/j.1528-1167.2009.02108.x. [DOI] [PubMed] [Google Scholar]
- 31.Low D., Chen K.-S. UBE3A regulates MC1R expression: a link to hypopigmentation in Angelman syndrome. Pigment Cell Melanoma Res. 2011;24:944–952. doi: 10.1111/j.1755-148X.2011.00884.x. [DOI] [PubMed] [Google Scholar]
- 32.Fridman C., Hosomi N., Varela M.C., Souza A.H., Fukai K., Koiffmann C.P. Angelman syndrome associated with oculocutaneous albinism due to an intragenic deletion of the P gene. Am. J. Med. Genet. A. 2003;119A:180–183. doi: 10.1002/ajmg.a.20105. [DOI] [PubMed] [Google Scholar]
- 33.Walz N.C., Benson B.A. Behavioral Phenotypes in Children with Down Syndrome, Prader-Willi Syndrome, or Angelman Syndrome. J. Dev. Phys. Disabil. 2002;14:307–321. doi: 10.1023/A:1020326701399. [DOI] [Google Scholar]
- 34.Silva-Santos S., van Woerden G.M., Bruinsma C.F., Mientjes E., Jolfaei M.A., Distel B., Kushner S.A., Elgersma Y. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Invest. 2015;125:2069–2076. doi: 10.1172/JCI80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bird L.M., Ochoa-Lubinoff C., Tan W.-H., Heimer G., Melmed R.D., Rakhit A., Visootsak J., During M.J., Holcroft C., Burdine R.D., et al. The STARS Phase 2 Study: A Randomized Controlled Trial of Gaboxadol in Angelman Syndrome. Neurology. 2021;96:e1024–e1035. doi: 10.1212/WNL.0000000000011409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dindot S.V., Christian S., Murphy W.J., Berent A., Panagoulias J., Schlafer A., Ballard J., Radeva K., Robinson R., Myers L., et al. An ASO therapy for Angelman syndrome that targets an evolutionarily conserved region at the start of the UBE3A-AS transcript. Sci. Transl. Med. 2023;15 doi: 10.1126/scitranslmed.abf4077. [DOI] [PubMed] [Google Scholar]
- 37.Heald M., Adams D., Walls E., Oliver C. Refining the Behavioral Phenotype of Angelman Syndrome: Examining Differences in Motivation for Social Contact Between Genetic Subgroups. Front. Behav. Neurosci. 2021;15 doi: 10.3389/fnbeh.2021.618271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan M.-L., Adam M.P., Seaver L.H., Myers A., Schelley S., Zadeh N., Hudgins L., Bernstein J.A. Increased body mass in infancy and early toddlerhood in Angelman syndrome patients with uniparental disomy and imprinting center defects. Am. J. Med. Genet. A. 2015;167A:142–146. doi: 10.1002/ajmg.a.36831. [DOI] [PubMed] [Google Scholar]
- 39.Yamada M., Okuno H., Okamoto N., Suzuki H., Miya F., Takenouchi T., Kosaki K. Diagnosis of Prader-Willi syndrome and Angelman syndrome by targeted nanopore long-read sequencing. Eur. J. Med. Genet. 2023;66 doi: 10.1016/j.ejmg.2022.104690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data relevant to the study are included in the report. Please contact the corresponding author, S.S., for further information.