Abstract
Actin networks are continuously reorganized in cells that rapidly change their shape. Applying total internal reflection fluorescence microscopy at acquisition rates of 10–20 Hz, we measured an average growth rate of 3 μm·sec–1 for filamentous actin structures throughout the entire substrate-attached cortex of Dictyostelium cells. New filaments often proceed along preexisting ones, resulting in bundle formation concurrent with filament growth. In cells that orientate in a gradient of chemoattractant, prominent assemblies of actin enriched in the Arp2/3 complex are inserted into the network, primarily at the base of filopods that point into the direction of the gradient. We propose that high turnover rates of actin filaments confer the plasticity to the cell cortex that is required for rapid accommodation to external stimuli.
Keywords: actin polymerization, Arp2/3 complex, Dictyostelium, latrunculin, total internal reflection fluorescence
The actin cytoskeleton undergoes rapid changes of its organization to fulfill its role in cell motility, phagocytosis, and cytokinesis. Thereby, interconnected and membrane-anchored actin filaments are assembled from soluble precursors in the cytoplasm and are subsequently depolymerized to replenish the precursor pool. In a moving cell, current view attributes most of the activity in actin filament nucleation, elongation, and branching to the leading edge of a lamellipodium (1, 2). Actin polymerized at the leading edge of fibroblasts, keratocytes, or neural growth cones is gradually depolymerized during its net transport toward the center of a cell. However, actin filaments do not only grow in contact with the membrane at the border of a cell (3, 4). Continuous reorganization of the entire actin system is particularly striking in fast migrating cells that rapidly change the direction of movement. Prominent examples are neutrophils and Dictyostelium cells, which adjust leading-edge formation to gradients of chemoattractant and readily engulf particles that adhere to their surface (5, 6). The question is how flexibility is conferred to the actin network that enables these cells to change their shape within seconds.
To visualize and measure actin dynamics in the cell cortex remote from a leading edge, we have applied total internal reflection fluorescence (TIRF) microscopy to Dictyostelium cells at acquisition rates of 10–20 Hz (7). TIRF microscopy is perfectly suited to visualize intricate network structures with a minimum of fluorescence emission from the cytoplasm. The evanescent field used for illumination extends typically 100–200 nm from the bottom surface into the cell, which corresponds to the thickness of the cortical actin network of Dictyostelium cells (8). Nevertheless, the background fluorescence turned out to be insufficiently reduced when GFP-actin was used as a marker, because half of the total actin exists in a nonfilamentous state within the cytoplasm (9). Only markers that selectively visualize filamentous actin structures provided the resolution necessary to study cortical actin network dynamics. Here, we apply fluorescent versions of a truncated or full-length Lim protein, LimEΔcoil-GFP or LimE-GFP (7), to measure increases of fluorescence intensity and to investigate actin dynamics in unstimulated cells or in cells responding to chemoattractant.
Methods
All strains, except HS1, were derived from Dictyostelium discoideum AX2-214. Full-length LimE-GFP was expressed in a LimE-null background (7), and LimEΔcoil-GFP was expressed either in a LimE-null or HS1 myosin II-null background (10). GFP-ABD120 was expressed in AX2-214 WT cells (7), and GFP-actin (11) was expressed in HS2205 myosin II-null cells (12). GFP-Arp3 (13) was combined with mRFPmars-LimEΔcoil (in which RFP indicates red fluorescent protein) (14).
Confocal scans were obtained by using the PerkinElmer Ultra View spinning-disk system. Through-the-objective TIRF microscopy of GFP labels was performed as described by Bretschneider et al. (7). For dual-emission TIRF imaging, both GFP and mRFPmars were excited at 488 nm, and emissions were split as described in ref. 15. Cells were treated with FM4-64 or 5 μM latrunculin A as in ref. 15. For chemotaxis, cells starved as a monolayer in 17 mM Na/K-phosphate buffer (pH 6) for 7–10 hr were stimulated through a micropipet filled with 10–4 M cAMP. Details are given in Supporting Text, which is published as supporting information on the PNAS web site.
Results
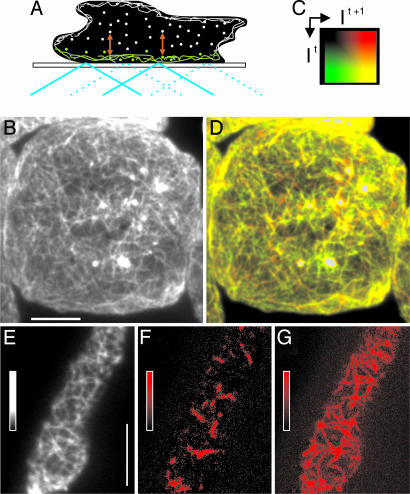
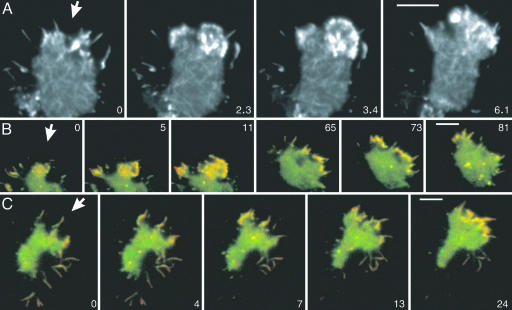
Rapid Reorganization of the Cortical Actin Network Independent of Leading-Edge Activity. The principle of TIRF microscopy in combination with an indirect label for the cortical actin network is outlined in Fig. 1A, and the structures visualized by this technique in a living Dictyostelium cell are shown in Fig. 1B. To suppress leading-edge activities, the cell shown was immobilized by compression with an agar sheet. The overall dynamics of actin network reorganization throughout the substrate-attached cell surface is illustrated by the overlay of two TIRF images acquired at an interval of 0.5 sec. These images are pseudocolored in a way such that new structures in the second image are shown in red, disappearing structures are in green, and persisting ones are in yellow (Fig. 1 C and D).
Fig. 1.
TIRF imaging of actin networks and their reorganization in the cortex of Dictyostelium cells. (A) Illumination at a high incidence angle (blue) generates an evanescent field that illuminates the cortical actin network of 100–200-nm thickness (green). Molecules diffusing in the cytoplasm are illuminated only for the fraction of time they persist within the evanescent field, a period that is prolonged by incorporation into the cytoskeleton (red double arrows). (B) TIRF image of the actin network visualized by LimEΔcoil-GFP in a cell immobilized by tight compression under an agar sheet, taken at an exposure time of 100 msec. (C) Two-dimensional color space where It indicates color coding of the gray scale for the first frame (green) and It + 1 indicates color coding of the gray scale for the second frame (red). Mixed colors (yellow tones) represent structures that persist from the first frame to the second frame with constant or slightly varying fluorescence intensity. (D) Superposition of two TIRF images of the cell shown in B, recorded at a 500-msec interval and color-coded as in C. Areas of very densely packed actin filaments with fluorescence intensities beyond the linear range of the 0–255 gray scale are left uncolored. (E–G) gainfi in a TIRF image series recorded at 100-msec intervals. (E) Snapshot at the beginning of the series. (F) gainfi during a period of 200 msec obtained by image subtraction, colored in red. (G) gainfi obtained by subtracting 20 consecutive frames from each other, presented as a maximum projection over the period of 2 sec. The gray and color bars indicate contrast enhancement. (Scale bars: 5 μm.)
To resolve details of actin network reorganization, we have taken images uninterruptedly at 50- or 100-msec exposure times. In the filamentous network of a freely moving cell (Fig. 1E), the gain of fluorescence intensity recorded at 100-msec intervals is mapped in red (Fig. 1 F and G). Integration of the gain over 200 msec reveals a pattern of short stripes (Fig. 1F), and integration over a period of 2 sec results in a distinct network of filamentous structures up to 4 μm in length (Fig. 1G), indicating that fluorescence is increasing along linear paths. In the following, we measure and analyze the gain in fluorescence intensity (gainfi) in comparison to the loss in fluorescence intensity (lossfi).
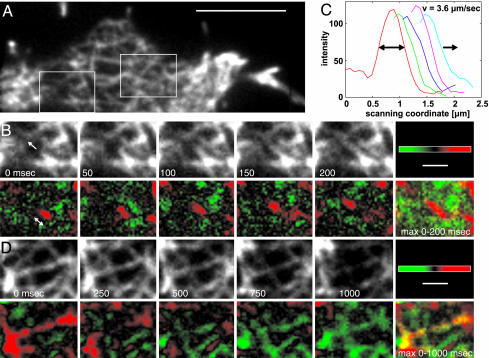
In Fig. 2, data recorded at 50-msec intervals are extracted from two subareas of a cell's network (framed in Fig. 2 A and also shown in Movie 1, which is published as supporting information on the PNAS web site). Linear extension of the tip of a fluorescent element is visualized in Fig. 2B Upper. The gainfi in consecutive frames and a maximum projection are shown in Lower. Profiles of the gain are plotted in Fig. 2C, indicating that the front progressed at a velocity of 3.6 μm·sec–1. No distinct pattern was found over the same time scale for lossfi (not shown). However, on a 5-fold extended time scale, it became evident that the fluorescence intensity does not recede in a linear manner (Fig. 2D). We will investigate the lossfi in detail later under conditions of blocked actin polymerization.
Fig. 2.
Linear progression of gainfi as opposed to irregular progression of lossfi.(A) TIRF image of an actin network. The framed areas are analyzed in B–D at an acquisition rate of 20 Hz. (Scale bar: 5 μm.) (B) gainfi within the left frame of A. The loose network in this region facilitates the separation of a single element from its texture. (Upper) Consecutive TIRF images. The arrow in the leftmost panel indicates the direction of tip progression. (Lower) Difference images color-coded for gainfi (red) and lossfi (green). Frames separated by 150 msec were subtracted from each other to make the tip region clearly visible. The double arrow indicates tip advancement within the first 150 msec. Maximum projection (max 0–200 msec) of the difference images results in a red stretch ≈1 μm in length. (C) Line-scan profiles of the advancing tip region at intervals of 50 msec, as shown in red in B Lower.(D) lossfi within the right frame of A, where the loss predominates gain. (Upper) Sequence of TIRF images. (Lower) Frames separated by 750 msec were subtracted from each other to make the decay of fluorescence intensity visible. Difference images are color-coded for lossfi (green) and gainfi (red). The maximum projection (max 0–1,000 msec) of the difference images is shown at far right. (Scale bars in B and D:1 μm.)
To confirm that the observed actin-filament dynamics is not artificially altered by the LimEΔcoil-GFP label used, full-length LimE-GFP was expressed in LimE-null cells at a level similar to endogenous LimE in WT cells (see Supporting Text). Apart from a higher cytoplasmic background, pattern dynamics was indistinguishable from that recorded by using LimEΔcoil-GFP. In addition, we have replaced the LimE label by the GFP-tagged actin-binding domain of the 120-kDa filamin-like protein (ABD120) from Dictyostelium (16). This marker also revealed a similar pattern, but, in accord with previous findings (7), the structures were less distinctly visualized (see Movie 2, which is published as supporting information on the PNAS web site).
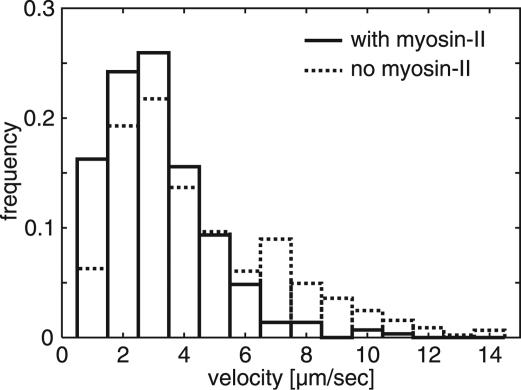
Linear Progression of Fluorescence Increases Measured in Cells With and Without Myosin II. The conventional, double-headed myosin II forms antiparallel filaments that are capable of cross-linking actin filaments. In cells lacking the myosin II heavy chains, contractility is substantially reduced (17). These mutant cells still form actin-filament networks visualized by TIRF microscopy (7), but the mechanical rigidity and force-generating power of the cell cortex is low (18). To check whether the progression in gainfi is a myosin II-dependent process, actin dynamics was visualized in the mutant cells in parallel to WT cells. Linear progression of the gainfi in the mutant was as clearly recognizable as in WT, and the reduced contractility in the mutant cells allowed the recording of longer time series (Movies 3 and 4, which are published as supporting information on the PNAS web site). Both velocity distributions peak at 3 μm·sec–1 (Fig. 3). However, in the myosin II-null background, the fraction of fast growing network elements is significantly higher, indicating that filamentous myosin slightly restricts the growth rate.
Fig. 3.
Velocity distribution of gainfi in cells having or lacking myosin II, tracked as in Fig. 2. For reproducible quantitation of tip velocities, the gain was thresholded. High velocities are significantly more abundant in myosin II-null cells (P < 0.1%, Mann–Whitney U test; n = 289 tracked positions in WT cells; n = 446 tracked positions in myosin II-null cells).
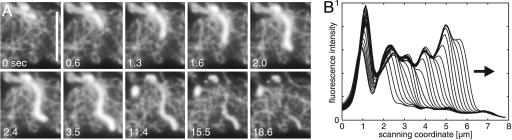
Growth of Actin Bundles Along Preexisting Filaments. A survey of our image series suggests that gainfi often advances along already existing elements of the actin network (see Movie 4). To establish that, we have made use of unusually thick actin bundles, the growth of which is easy to follow. The thickness of actin bundles varies in the cortex of Dictyostelium cells, and extremely thick ones are particularly abundant in cells lacking myosin II. Fig. 4A shows a network region with wide meshes and prominent actin structures, where the turning tip of a bundle follows an underlying filamentous element (in particular, see the 1.3- to 3.5-sec frames of Fig. 4A; see also Movie 5, which is published as supporting information on the PNAS web site).
Fig. 4.
Growth of a thick actin bundle in a myosin II-null cell. (A) The bundle labeled with LimEΔcoil-GFP and viewed by TIRF microscopy extends in line with the layout of the actin network. At 3.5 sec, the bundle reaches its maximal length of 6 μm. Thereafter, ≈15 sec are needed for disassembly. (Scale bar: 5 μm.) (B) Line scan of fluorescence intensities along the actin bundle shown in A, sampled at 100-msec intervals from the 0- to 2.4-sec frames in the direction of the arrow. The front of the bundle is characterized by a steep intensity gradient that advances at an average velocity of 1.7 μm·sec–1.
The Increases in Fluorescence Intensity Reflect Actin Polymerization. Fluorescence intensities along the axis of the actin bundle shown in Fig. 4A reveal peaks and valleys that stay in place while the tip progresses (Fig. 4B). The peaks are not only due to the crossing of other filaments, but also to differences in fluorescence intensity along the bundle. This stationary profile suggests that increases in fluorescence intensity are caused by the de novo polymerization of actin at the tip of a bundle rather than by the gliding of actin filaments along a track.
To clearly distinguish polymerization from gliding, we turned to spinning-disk microscopy, which allowed us to follow actin bundles over their entire length even if one of their ends was located too deep within the cell to be recognized by TIRF. To ensure that the entire bottom layer of a cell, including subcortical structures, is visualized, the cells were labeled with FM4-64. This red fluorescent styryl dye integrates into the plasma membrane and into membranes of endosomes and the contractile vacuole complex (Fig. 5). The bladders and tubules of this osmoregulatory organelle serve as markers for a layer between the actin network and the interior cytoplasmic space (19). We also took advantage of the fact that compact actin bundles contrast sufficiently against background to be directly monitored by the incorporation of GFP-actin.
Fig. 5.
Actin polymerization in thick bundles visualized by spinning-disk confocal microscopy. In myosin II-null cells, GFP-actin is expressed for incorporation into filamentous actin (green). The membranes of intracellular vesicles, primarily of the contractile vacuole complex, are labeled by FM4-64 (red). (A) A bundle that extends from its origin (open arrowhead) to the left and forms a kink when it hits the membrane (closed arrowhead). As in Fig. 4A, depolymerization starts at the rear of the filament (7-sec frame). (B) An actin bundle that gives rise to a filopod-like extension by applying force onto the cell border. (C) Velocities of tip progression in thick bundles labeled by the incorporation of GFP-actin. Data are from 85 measurements on 16 actin bundles. (Scale bars: 5 μm.)
Fig. 5A shows the origin of an actin bundle staying in place during progression of the tip (0- to 4-sec frames). This bundle sharply bends when it reaches the cell border and finally disassembles from behind (4- and 7-sec frames). In Fig. 5B, a growing bundle pushes the membrane forward, thus creating a long, filopod-like protrusion. In the bundles visualized by GFP-actin, the tip progressed with an average velocity of 1 μm·sec–1 and a maximal velocity of 2 μm·sec–1 (Fig. 5C).
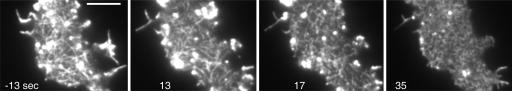
Speed and Mode of Actin Depolymerization. In a steady state, the rapid gain of polymerized actin must be balanced by efficient depolymerization. To view depolymerization separately, we have treated cells with latrunculin A. This drug entraps actin in a nonpolymerizable complex, thus allowing only depolymerization to proceed (20, 21). In WT cells, fragments of the actin network coalesce into large clusters, and the cells contract before actin structures disappear (Movie 6, which is published as supporting information on the PNAS web site). This process resembles the contraction of an actomyosin gel in vitro (22) and has prompted us to record the depolymerization in myosin II-null cells. In these cells, the network decays gradually into smaller fragments, while only dense clusters known to be associated with the Arp2/3 complex (7) are retained (Fig. 6).
Fig. 6.
Global depolymerization of actin in a myosin II-null cell. Numbers are seconds before and after the addition of 5 μM latrunculin A. Before addition of the drug, the cell shows typical actin network organization and a protrusion at its right side (–13-sec frame). The network is fragmented within the first 17 sec. Later on, rosette-like assemblies predominate (35 sec) before structures become diffuse (see Movie 7). Intensely labeled areas consist of dense actin clusters associated with the Arp2/3 complex (7). Notably, in the myosin II-null cells, filopods attached to the substrate are not retracted when they are depleted of their actin content. Exposure time = 100 msec. (Scale bar: 5 μm.)
Initially, the tips of filamentous actin structures are still progressing, and even small fragments are motile, suggesting that actin treadmilling is still superimposed on net depolymerization (Movies 7 and 8, which are published as supporting information on the PNAS web site). At an advanced stage of depolymerization, rosette-like structures become apparent (Fig. 6, 35-sec frame), probably produced by the annealing of actin bundles at their ends, similar to the rings produced in vitro by the actin bundling protein dynacortin (23).
Taking into account that latrunculin A has to pass the plasma membrane and sequester G-actin before it inhibits polymerization, our data indicate that actin filaments are visibly shortened within <20 sec. Importantly, the actin network disintegrates uniformly over the area of the cell surface that is visualized by TIRF, indicating that filamentous actin is rapidly and continuously renewed in the entire cortex of Dictyostelium cells.
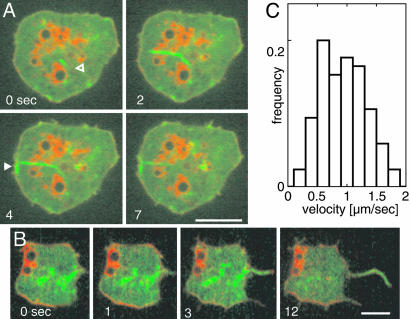
Actin Dynamics in Chemotaxis. The cells studied so far exhibited spontaneous actin dynamics in the absence of an external stimulus. To investigate how cells modulate their activities to orientate within a gradient of chemoattractant, we have stimulated cells with cAMP (Movie 9, which is published as supporting information on the PNAS web site). Three types of structures are distinguishable by TIRF microscopy in chemotaxing cells: the basal actin network, dense assemblies of actin, and filopods. As in unstimulated cells, increases in fluorescence intensity continuously progressed, with a mode of 3 μm·sec–1 throughout the entire actin network along the substrate-attached area of the cells. Directional activity was obvious in the dense actin assemblies integrated into the network, which extended predominantly into the direction of higher cyclic-AMP concentrations (Fig. 7A and Movie 10, which is published as supporting information on the PNAS web site).
Fig. 7.
Actin and Arp2/3 dynamics in chemotaxis. The cells shown respond to cAMP applied through a micropipet that is placed outside the frames shown. The direction of the diffusion gradients is indicated by arrows. (A) A cell labeled with full-length LimE-GFP recorded at a frame rate of 100 msec. (B and C) Cells labeled with GFP-Arp3 (green) and mRFP-LimEΔcoil (red). Yellow regions indicate merge of the two labels. The pattern of dense actin assemblies in B resembles the pattern in A, showing that these structures are enriched in the Arp2/3 complex. At subsequent stages, Arp2/3-rich actin structures are distributed along the region of the cell border that is exposed to higher cyclic-AMP concentrations. (C) Arp2/3-rich actin structures are preferentially accumulated at the base of filopods that point toward the source of the gradient. (In B and C, network structures are not visible, because the resolution of dual-emission fluorescence recordings is limited.) Time is indicated in seconds. (Scale bar: 5 μm.)
Dual-emission TIRF microscopy of cells double-labeled with GFP-Arp3 and mRFP-LimEΔcoil identified the dense actin assemblies as Arp2/3-enriched structures (Fig. 7B; see also Movie 11, which is published as supporting information on the PNAS web site). The GFP-Arp3 label distinguished these assemblies from filopods, which only occasionally contained patches of GFP-Arp3 along their length (Fig. 7C; see also Movie 12, which is published as supporting information on the PNAS web site), in accord with the immunolocalization of Arp3 (13). The Arp2/3 complex often accumulated at the base of filopods, from where clusters broadened and extended toward the source of the gradient.
Discussion
Rapid Actin Reorganization Analyzed by Time-Resolved TIRF Microscopy. Information on actin dynamics in live cells can be obtained by fluorescent probes in combination with speckle microscopy (24), selective photobleaching followed by ratio imaging (4), the use of photoactivatable GFP (25), or by TIRF microscopy (26). Single molecules of GFP-actin can be traced by speckle microscopy if incorporated into a filament (3), and the procession of a single motility unit of formin has been recorded (27). TIRF imaging as used here resolves cortical actin network structures in toto to study their dynamics in live cells on the scale of 50–100 msec. Owing to residual background, the detectability is currently limited to small bundles of two to five filaments (7).
As a parameter reflecting the speed of actin reorganization, we have measured progression of the tips of filamentous network elements (Fig. 2 and Movies 1–3). The tips advance over distances of several micrometers in a linear manner at velocities of 1–5 μm·sec–1, corresponding to the addition of up to 2 × 103 actin subunits per second to a filament (Fig. 3). This velocity means that, by uninterrupted progression, the diameter of a cell would be passed within ≈3 sec, which is more than 1 order of magnitude faster than the translocation of a whole cell. Because an indirect label was needed to visualize the actin filaments, its binding rate may become limiting so that we might miss velocities higher than the reported ones.
The principal question is whether the observed progression is due to the polymerization or to the gliding of actin filaments. If gliding were assumed, the end of a filamentous element should be seen as a lossfi that progresses at the same velocity as the gainfi at the tip. In our TIRF image series, which clearly revealed linear progression of the tips, no similar progression for the lossfi was found (Fig. 2 and Movies 1–3). To ensure that polymerization is monitored, we focused on actin bundles thick enough to be recognized in confocal scans by the incorporation of GFP-actin (Fig. 5). Such bundles are particularly abundant in cells lacking myosin II, probably because cross-bridges between actin filaments are efficiently broken in WT cells by the motor activity of myosin II (28). In the confocal images, the origin of a bundle can be seen even if polymerization starts outside the cortical actin network. The result was that bundles elongate while the origin stays in place, showing that polymerization, rather than gliding, is responsible for the tip progression measured. This notion does not rule out gliding of actin filaments in live cells, which might occur less frequently or on another time scale; in particular, myosin II-dependent actin movement remains to be analyzed in cells double-labeled for actin and the heads of myosin.
The maximal velocity obtained for the incorporation of GFP-actin into thick bundles is 2 μm·sec–1, which is about one-third of the value obtained for delicate actin structures (Fig. 3 and Fig 5C). A potential reason for the lower speed is local depletion of precursors, which, in thick bundles, might limit progression of the tip.
Rapid disintegration of the cortical network upon blockage of actin polymerization provides further evidence for its continuous reorganization. The network breaks down over the entire area of the cell surface visualized by TIRF microscopy, indicating that turnover is not linked to the activity of a leading edge (Fig. 6).
Mechanism of Actin Network Reorganization. The major nucleators of actin polymerization, the Arp2/3 complex and members of the formin family, give rise to different types of actin structures. The Arp2/3 complex nucleates clusters of branched filaments, whereas formins are processive motors that typically produce straight and often bundled filaments (for review see refs. 29 and 30). In a subclass of formins, the delivery of actin monomers to the plus end of actin filaments is gated by profilin, which binds to the formin homology (FH) 1 domain of formins, thus increasing the rate of polymerization (31, 32). The FH2 domain of yeast Bni1p forms a processive cap by continually walking to the plus end of the growing actin filament (33). Formins prevent the plus ends of actin filaments from being blocked by capping proteins, an activity that is required for the linear progression of polymerization over several micrometers, as shown in Fig. 2 and Movies 1–3. Such long runs have been observed in fibroblasts by using an FH1-FH2 construct of the formin mDia1 as an actin-driven motility machine (27). The speed of 2 μm·sec–1 measured for the FH1-FH2 motility in fibroblasts shows that the polymerization machinery of formins meets the rates of actin polymerization deduced from our data in Dictyostelium cells (Fig. 3). These data suggest that formins, which are represented by at least 10 members in the genome of Dictyostelium, are responsible for the linear growth of actin filaments in the cortical network (Fig. 1G). Consistent with this notion, the Arp2/3 complex is concentrated in Dictyostelium cells at the leading edge and in phagocytic cups, small foci, and traveling actin waves; not, however, along the filamentous elements of the actin network (7).
The observed bundling of actin filaments in statu nascendi suggests that the machinery promoting polymerization is capable of interacting with preexisting filaments that serve as tracks (Movies 4–5). A molecular basis for bundling concomitant with polymerization is provided by interaction of the yeast formin Bni1p with translation elongation factor 1A, an actin-filament bundling protein (34, 35).
Arp2/3 Accumulation in Chemotaxis. Orientation of cells in a gradient was reflected in the expansion of Arp2/3-rich actin assemblies toward the attractant. Often, the Arp2/3 complex started to accumulate at the base of filopods that were protruded into the direction of increasing cyclic-AMP concentrations (Fig. 7 B and C). This finding is consistent with the idea that while extending, filopods might sense temporal increases in attractant concentration and transmit information on the direction of the gradient to the actin system (36).
The reported dynamics of the actin system accounts for two opposite functions of the actin system: to stabilize cell shape and to enable changes of the shape. Actin polymerization along a template of actin filaments stabilizes the shape of a cell. Continuous renewal confers flexibility to the actin system that is needed for the immediate generation of new structures such as phagocytic cups, or leading edges in response to chemoattractant.
Supplementary Material
Acknowledgments
We thank our group members for expert assistance, Andreas Schröder for access to the spinning-disk microscope, and Dietmar Manstein (Hannover Medical School, Hannover, Germany) for HS1 cells. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to G.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TIRF, total internal reflection fluorescence; FH, formin homology; gainfi, gain in fluorescence intensity; lossfi, loss in fluorescence intensity.
References
- 1.Small, J. V., Stradal, T., Vignal, E. & Rottner, K. (2002) Trends Cell Biol. 12, 112–120. [DOI] [PubMed] [Google Scholar]
- 2.Pollard, T. D. & Borisy, G. G. (2003) Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe, N. & Mitchison, T. J. (2002) Science 295, 1083–1086. [DOI] [PubMed] [Google Scholar]
- 4.Zicha, D., Dobbie, I. M., Holt, M. R., Monypenny, J., Soong, D. Y. H., Gray, C. & Dunn, G. A. (2003) Science 300, 142–145. [DOI] [PubMed] [Google Scholar]
- 5.Gerisch, G. & Keller, H. U. (1981) J. Cell Sci. 52, 1–10. [DOI] [PubMed] [Google Scholar]
- 6.Gerisch, G., Albrecht, R., Heizer, C., Hodgkinson, S. & Maniak, M. (1995) Curr. Biol. 5, 1280–1285. [DOI] [PubMed] [Google Scholar]
- 7.Bretschneider, T., Diez, S., Anderson, K., Heuser, J., Clarke, M., Müller-Taubenberger, A., Köhler, J. & Gerisch, G. (2004) Curr. Biol. 14, 1–10. [DOI] [PubMed] [Google Scholar]
- 8.Hanakam, F., Albrecht, R., Eckerskorn, C., Matzner, M. & Gerisch, G. (1996) EMBO J. 15, 2935–2943. [PMC free article] [PubMed] [Google Scholar]
- 9.Haugwitz, M., Noegel, A. A., Karakesisoglou, J. & Schleicher, M. (1994) Cell 79, 303–314. [DOI] [PubMed] [Google Scholar]
- 10.Moores, S. L., Sabry, J. H. & Spudich, J. A. (1996) Proc. Natl. Acad. Sci. USA 93, 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westphal, M., Jungbluth, A., Heidecker, M., Mühlbauer, B., Heizer, C., Schwartz, J.-M., Marriott, G. & Gerisch, G. (1997) Curr. Biol. 7, 176–183. [DOI] [PubMed] [Google Scholar]
- 12.Manstein, D. J., Titus, M. A., DeLozanne, A. & Spudich, J. A. (1989) EMBO J. 8, 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Insall, R., Müller-Taubenberger, A., Machesky, L., Köhler, J., Simmeth, E., Atkinson, S. J., Weber, I. & Gerisch, G. (2001) Cell Motil. Cyotoskeleton 50, 115–128. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, M., Haase, I., Simmeth, E., Gerisch, G. & Müller-Taubenberger, A. (2004) FEBS Lett. 577, 227–232. [DOI] [PubMed] [Google Scholar]
- 15.Gerisch, G., Bretschneider, T., Müller-Taubenberger, A., Simmeth, E., Ecke, M., Diez, S. & Anderson, K. (2004) Biophys. J. 87, 3493–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang, K. M., Lee, E. & Knecht, D. A. (1998) Curr. Biol. 8, 405–408. [DOI] [PubMed] [Google Scholar]
- 17.Jay, P. Y., Pham, P. A., Wong, S. A. & Elson, E. L. (1995) J. Cell Sci. 108, 387–393. [DOI] [PubMed] [Google Scholar]
- 18.Laevsky, G. & Knecht, D. A. (2003) J. Cell Sci. 116, 3761–3770. [DOI] [PubMed] [Google Scholar]
- 19.Heuser, J., Zhu, Q. L. & Clarke, M. (1993) J. Cell Biol. 121, 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morton, W. M., Ayscough, K. R. & McLaughlin, P. J. (2000) Nat. Cell Biol. 2, 376–378. [DOI] [PubMed] [Google Scholar]
- 21.Yarmola, E. G., Somasundaram, T., Boring, T. A., Spector, I. & Bubb, M. R. (2000) J. Biol. Chem. 275, 28120–28127. [DOI] [PubMed] [Google Scholar]
- 22.Taylor, Lansing D. & Condeelis, J. S. (1979) Int. Rev. Cytol. 56, 57–144. [DOI] [PubMed] [Google Scholar]
- 23.Robinson, D. N., Ocon, S. S., Rock, R. S. & Spudich, J. A. (2002) J. Biol. Chem. 277, 9088–9095. [DOI] [PubMed] [Google Scholar]
- 24.Waterman-Storer, C. M. & Salmon, E. D. (1997) J. Cell Biol. 139, 417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson, G. H. & Lippincott-Schwartz, J. (2002) Science 297, 1873–1877. [DOI] [PubMed] [Google Scholar]
- 26.Sund, S. E. & Axelrod, D. (2000) Biophys. J. 79, 1655–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higashida, C., Miyoshi, T., Fujita, A., Oceguera-Yanez, F., Monypenny, J., Andou, Y., Narumiya, S. & Watanabe, N. (2004) Science 303, 2007–2010. [DOI] [PubMed] [Google Scholar]
- 28.Prassler, J., Stocker, S., Marriott, G., Heidecker, M., Kellermann, J. & Gerisch, G. (1997) Mol. Biol. Cell 8, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evangelista, M., Zigmond, S. & Boone, C. (2003) J. Cell Sci. 116, 2603–2611. [DOI] [PubMed] [Google Scholar]
- 30.Wallar, B. J. & Alberts, A. S. (2003) Trends Cell Biol. 13, 435–446. [DOI] [PubMed] [Google Scholar]
- 31.Kovar, D. R., Kuhn, J. R., Tichy, A. L. & Pollard, T. D. (2003) J. Cell Biol. 161, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krebs, A., Rothkegel, M., Klar, M. & Jockusch, B. M. (2001) J. Cell Sci. 114, 3663–3672. [DOI] [PubMed] [Google Scholar]
- 33.Zigmond, S. H., Evangelista, M., Boone, C., Yang, C., Dar, A. C., Sicheri, F., Forkey, J. & Pring, M. (2003) Curr. Biol. 13, 1820–1823. [DOI] [PubMed] [Google Scholar]
- 34.Umikawa, M., Tanaka, K., Kamei, T., Shimizu, K., Imamura, H., Sasaki, T. & Takai, Y. (1998) Oncogene 16, 2011–2016. [DOI] [PubMed] [Google Scholar]
- 35.Liu, G., Grant, W. M., Persky, D., Latham, V. M., Jr., Singer, R. H., & Condeelis, J. (2002) Mol. Biol. Cell 13, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerisch, G., Hülser, D., Malchow, D. & Wick, U. (1975) Philos. Trans. R. Soc. London B 272, 181–192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.