Abstract
Argania spinosa (L.) Skeels, an endemic Moroccan plant species from the Sapotaceae family, holds significant ecological, pharmaceutical, and socioeconomic value in the arid mid-western region. However, it is facing rapid degradation. Therefore, understanding its genetic diversity is critical for preserving this national heritage. We sequenced, assembled, and annotated the mitochondrial genome of A. spinosa and compared it to other plants in the Ericales order. Mitochondrial-like sequences from the A. spinosa genome were assembled using GetOrganelle, resulting in a 707,441 base pair mitochondrial genome with 45.75 % GC content. Annotation identified 32 protein-coding genes, 16 transfer RNAs, and 2 ribosomal RNA genes. Phylogenetic analysis of 15 Ericales species affirms that A. spinosa is closely related to the Theaceae family, which is in accordance with results from the chloroplast genome.
Keywords: Argan tree, Mitogenome, Assembly, Phylogeny, Sapotaceae
Specifications Table
| Subject | Omics: Genomics. |
| Specific subject area | Argan tree, Mitogenomics |
| Type of data | Table, Figure, GenBank, FASTA. |
| Data collection | A 9-year-old Argania spinosa specimen, "Amghar," was collected from the Souss Valley plain in Morocco (9°32′00″ N, 30° 24′ 00″ W, 126 m altitude). DNA was extracted from its leaves using the QIAgen Plant DNeasy Mini Kit, and a paired-end library was constructed using the Nextera DNA Library Prep Kit. Sequencing on the Illumina HiSeq XTen platform yielded 77,382,038 reads. Mitochondrial sequences were assembled using GetOrganelle v.1.7.1 with Camellia sinensis as a reference. Protein-coding genes (PCGs), rRNA, and tRNA were annotated using BlastX, BlastN, and tRNAscan-SE, respectively, and manually validated with Geneious Prime. The mitochondrial genome map was visualised with OGDraw 1.3.1. For phylogenetic analysis of Ericales, 7 common PCGs from 15 members were extracted, aligned with Mafft v.7.5.0.8, and analysed with MEGA v.11.0.10 under the best-fit model from Modeltest. |
| Data source location | Institution: Faculty of Sciences of Agadir Region: Souss Valley City: Agadir Country: Morocco Latitude: 9°32′00″ N Longitude: 30° 24′ 00″ W |
| Data accessibility | Repository name: NCBI Sequence Read Archive (SRA) Data identification number: SRR6062045 Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/SRR6062045\ Repository name: NCBI BioProject Data identification number: PRJNA294096 Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA294096 Repository name: NCBI BioSample Data identification number: SRX3207156 Direct URL to data: https://www.ncbi.nlm.nih.gov/biosample/SAMN04014715 Repository name: NCBI GenBank Data identification number: MZ151883.2 Direct URL to data: https://www.ncbi.nlm.nih.gov/nuccore/MZ151883 |
1. Value of the Data
-
•
The complete mitochondrial genome sequence of Argania spinosa, an endemic tree species from Morocco, serves as a valuable reference for accurate species identification and characterization, supporting conservation efforts and management strategies.
-
•
This comprehensive genetic data facilitates the resolution of taxonomic ambiguities and enhances our understanding of the evolutionary relationships of Argania spinosa within a broader botanical context.
-
•
Protein-coding sequences within the mitochondrial genome provide valuable markers for phylogenetic reconstruction, further elucidating the evolutionary history of Argania spinosa.
2. Background
The Argan tree (Argania spinosa (L.) Skeels), native to the mid-west of Morocco, is an endangered xerothermophilic species that uniquely represents the Sapotaceae family in subtropical regions [1]. Spanning approximately 8280 km², Argan tree forests thrive predominantly in the arid lowlands of the Souss Valley and the sunlit mountains of the Anti-Atlas. Renowned for yielding the worldʼs most expensive edible oil, extracted from its seeds, the Argan tree holds significant value for medicinal and cosmetic purposes. However, these vital ecosystems are experiencing alarming rates of degradation, underscoring the urgent need for management strategies to protect and conserve their genetic diversity.
In response to this imperative, the Argan genome has been sequenced, resulting in an initial draft assembly [2]. To further contribute to preserving this unique species, we have assembled and annotated the mitochondrial genome of the Argan tree. This endeavor aligns with the broader objective of understanding the genetic intricacies of Argania spinosa, providing valuable insights into its evolution and contributing to conservation efforts.
3. Data Description
In this study, a specimen of Argania spinosa named “Amghar,” a 9-year-old shrub collected from the Souss Valley plain, was utilised. This specimen is currently deposited at the Faculty of Sciences, Agadir, Morocco. The biosample is catalogued in the Biosample database under accession number SAMN04014715 (www.ncbi.nlm.nih.gov/biosample/SAMN04014715).
Genomic DNA was extracted from lyophilized leaf tissue of the “Amghar” specimen using the Plant DNeasy Mini Kit (Qiagen, Germantown, Maryland, USA). The extracted DNA was used to construct a paired-end library with an average insert size of 600 base pairs (bp) and sequenced using the Illumina HiSeq X Ten platform, resulting in raw whole genome sequence data comprising 77,382,038 reads with an average read length of 150 bp. This raw data is available in the SRA database under accession number SRR6062045 (www.ncbi.nlm.nih.gov/sra/SRX3207156).
All data pertain to the Argania spinosa whole genome sequence project, which is accessible in the BioProject database under accession number PRJNA294096 (www.ncbi.nlm.nih.gov/bioproject/PRJNA294096).
Mitochondrial reads were extracted from the whole genome sequence data and assembled to construct a circular genome representing the mitochondrial genome of A. spinosa. This process was carried out using the GetOrganelle software [3], with the database selected as embplant_mt and the mitochondrial genome of Camellia sinensis used as a seed reference. The mitochondrial genome assembly yielded a circular scaffold of 707,441 bp with a GC content of 45.8 %. The size of this mitochondrial genome falls within the known range for land plant mitochondrial genomes (100 kb to 2 Mb) and aligns with the range for Ericales mitochondrial genomes (400 kb to 1 Mb) [4].
For annotation, the assembled mitochondrial genome was mapped to gene features of Camellia sinensis using BlastX and BlastN [5] to identify protein-coding genes (PCGs), open reading frames (ORFs), and ribosomal RNAs (rRNAs), while tRNAscan [6] was employed to locate transfer RNA (tRNA) sequences. The annotated mitochondrial genome is available in the GenBank database under accession number MZ151883.2 (www.ncbi.nlm.nih.gov/nuccore/MZ151883).
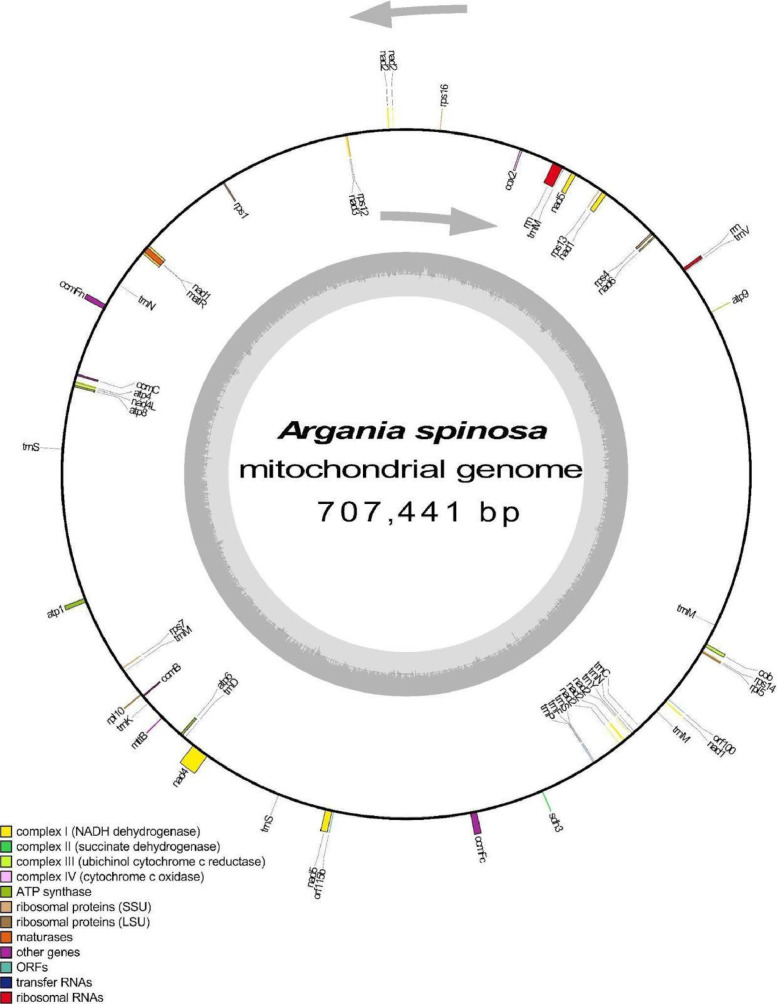
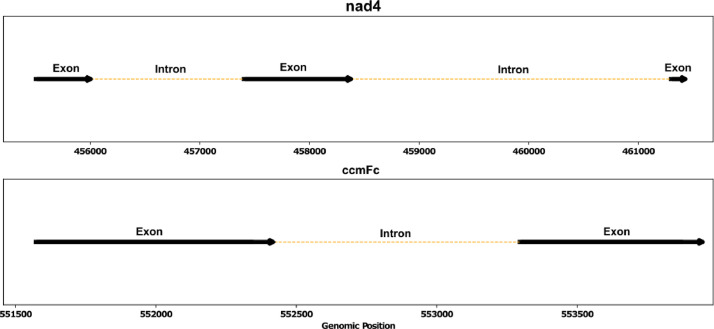
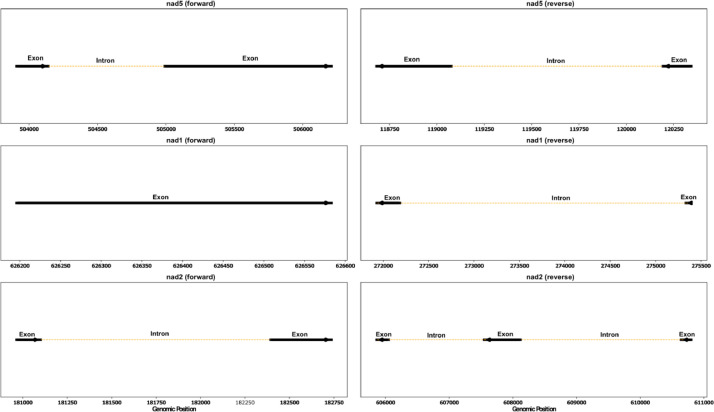
Annotation of the A. spinosa mitochondrial genome identified 50 coding sequences, including 2 rRNA sequences, 32 protein-coding genes, and 18 tRNA sequences (Fig. 1), collectively representing 9.6 % of the total mitochondrial genome (Table 1). Among the protein-coding genes, nad4 and ccmFc were found to be cis-splicing genes, with 3 and 1 introns, respectively. Additionally, three trans-splicing genes were detected, namely nad5, nad1, and nad2 (Figs. 2 and 3).
Fig. 1.
The mitochondrial genome of Argania spinosa was generated using OGDRAW software [7]. The transcription directions are indicated by outer and inner arrows.
Table 1.
Annotated genes in the mitochondrial genome of Argania spinosa. The number of sequences coding for each tRNA is indicated in brackets next to each gene.
| Gene group name | Genes | Number of genes |
|---|---|---|
| NADH dehydrogenase | nad2, nad6, nad3, nad5, nad1, nad4L, nad4 | 7 |
| Succinate dehydrogenase | sdh3 | 1 |
| Ubiquinol cytochrome c reductase | cob | 1 |
| Cytochrome c oxidase | cox2 | 1 |
| ATP synthase | atp8, atp1, atp6, atp4, atp9 | 5 |
| Cytochrome c biogenesis | ccmFc, ccmB, ccmFn, ccmC | 4 |
| Ribosomal proteins SSU | rps12, rps7, rps1, rps13, rps14, rps4, rps16 | 7 |
| Ribosomal proteins LSU | rpl5, rpl10 | 2 |
| Maturase | matR | 1 |
| ORFs | orf115b, orf100 | 2 |
| Transfer RNA | tRNA-Val (1), tRNA-Met (4), tRNA-Asn (2), tRNA-Ser (3), tRNA-Lys (1), tRNA-Asp (1), tRNA-Pro (1), tRNA-Phe (1), tRNA-Tyr (1), tRNA-Cys (1) | 16 |
| Ribosomal RNA | large subunit ribosomal RNA, small subunit ribosomal RNA | 2 |
| Other genes | mttB | 1 |
| Total number of genes | 50 | |
Fig. 2.
The diagram depicts the arrangement of cis-splicing genes within the mitochondrial genome of Argania spinosa. Exons are represented by black arrows, while introns are depicted as yellow dashed lines. The orientation of the genes in the mitogenome is indicated.
Fig. 3.
The diagram depicts the arrangement of trans-splicing genes within the mitochondrial genome of Argania spinosa. Exons are represented by black arrows, while introns are depicted as yellow dashed lines. The orientation of the genes in the mitogenome is indicated.
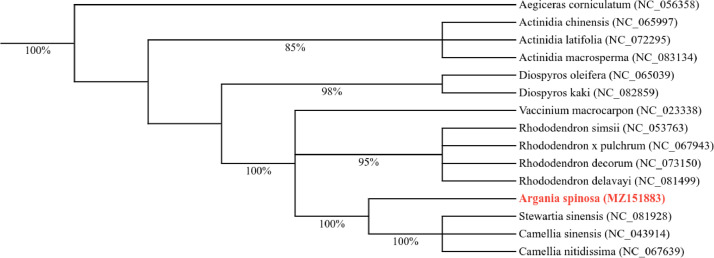
Sequences of seven common protein-coding genes (atp1, ccmB, cob, nad3, nad5, rpl10, rps13) from the mitochondrial genomes of 15 Ericales species, including Argania spinosa, were extracted and aligned using MAFFT [8]. The aligned sequences were concatenated and used as input for ModelTest-NG [9] to identify the optimal maximum likelihood (ML) model for constructing a phylogenetic tree of the Ericales. The model determined to be the best fit was the General Time Reversible model with Invariant Sites and Gamma Distribution (GTR+I + G), which was implemented using MEGA X [10] with 1000 bootstrap replicates.
The resulting phylogenetic tree demonstrates a close relationship between Argania spinosa and species of Camellia, supported by a bootstrap value of 100 % (Fig. 4). Additionally, our analysis corroborates the identification of a close relationship between the families Sapotaceae and Theaceae, consistent with previous phylogenetic studies based on chloroplast genomes [11].
Fig. 4.
A phylogenetic tree depicting the mitochondrial genomes of Argania spinosa and 14 other Ericales members is shown. The conserved protein-coding sequences from the mitochondrial genomes of these 15 species were aligned using MAFFT [6], and a phylogenetic tree was generated with MEGA [7]. Bootstrap values are provided on the branches, and the GenBank accession numbers for the mitochondrial genome sequences used in this tree are enclosed in parentheses.
4. Experimental Design, Materials and Methods
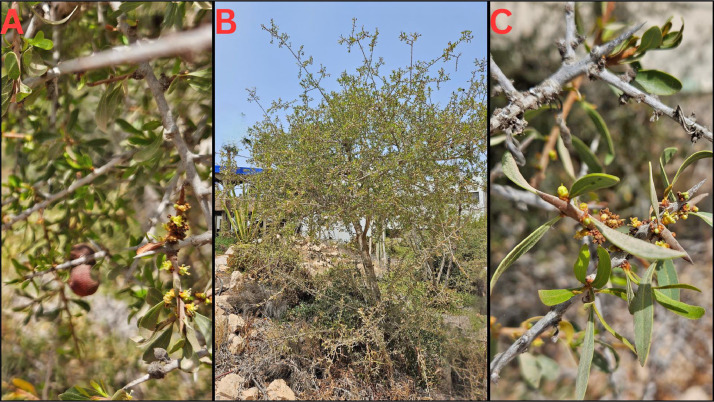
A specimen of A. spinosa named "Amghar" was deposited at the Faculty of Sciences of Agadir, Morocco (www.fsa.ac.ma, A. El Mousadik, a.elmousadik@uiz.ac.ma) under the voucher number Arg-Amr_2016. The specimen is a 9-year-old shrub collected from the Souss Valley plain, located at 9°32′00″ N, 30°24′00″ W, with an altitude of 126 m (Fig. 5).
Fig. 5.
Photo of the Argan shrub “Amghar” located in Agadir, Souss Valley, Morocco (Photograph taken by A. El Mousadik). A: shows branches with fruits, B : depicts the whole shrub, and C: highlights some branches with floral buds.
Genomic extracted from lyophilized leaf tissues of a single tree which is the "Amghar" specimen. DNA extraction was conducted from 1 g of leaf tissues using the Plant DNeasy Mini Kit manufactured by QIAGEN in Germantown, Maryland, USA. A paired-end library with an average insert size of 600 base pairs (bp) was then constructed using the Nextera DNA Library Prep Kit for Illumina, developed by New England Biolabs in New Brunswick, Massachusetts, USA. The library was sequenced on the Illumina HiSeq XTen platform in San Diego, California, USA, generating 77,382,038 reads with an average size of 150 bp.
Mitochondrial-like sequences from the Argania spinosa genome project were carefully isolated and assembled using GetOrganelle v.1.7.1 [8]. The assembly was performed with default parameters, utilizing the Camellia sinensis mitochondrial genome as a reference seed. Protein-coding genes (PCGs) were identified using BlastX [9], with PCGs from other Ericales species serving as reference queries. The annotation of rRNA and tRNA sequences employed BlastN [9] and tRNAscan-SE [10], respectively. Rigorous manual validation of the annotation was performed using the bioinformatics software Geneious Prime (www.geneious.com/prime/). Visualization of the meticulously assembled and annotated A. spinosa mitochondrial genome was achieved with OGDraw 1.3.1 [7], generating a comprehensive and informative genome map.
For the phylogenetic analysis of Ericales, seven common protein-coding genes from 15 members of the Ericales order were extracted using a GenBank Sequence Downloader script (www.pcggb.onrender.com/). These genes were individually aligned using MAFFT v.7.5.0.8 [6] and then concatenated into a single dataset. The "best-fit" maximum likelihood model was determined by ModelTest [11], and MEGA v.11.0.10 [7] was used for analysis with a bootstrap of 1000.
Limitations
Not applicable.
Ethics Statement
Your text is clear and effectively communicates your commitment to ethical research practices. Here is a revised version with minor adjustments for clarity and consistency:
The research involving the mitochondrial genome analysis of Argania spinosa was conducted in accordance with ethical principles. Although there is no specific local requirement for ethical committee approval for this type of study in Morocco, we ensured responsible conduct throughout the research process. The collection of plant material was non-invasive, and no harm was inflicted upon the ecosystem. The specimen was obtained with permission and in respect of local guidelines. We recognize the importance of ethical research practices and ecological preservation.
We, the authors, confirm that we have read and adhered to the ethical requirements for publication in Data in Brief. Furthermore, we confirm that this work does not involve human subjects, animal experiments, or data collected from social media platforms.
CRediT authorship contribution statement
Abdellah Idrissi Azami: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. Stacy Pirro: Data curation, Funding acquisition, Investigation, Resources, Writing – review & editing. Sofia Sehli: Data curation, Formal analysis, Visualization, Writing – original draft. Nihal Habib: Data curation, Formal analysis, Writing – original draft. Douae El Ghoubali: Data curation, Formal analysis, Visualization, Writing – review & editing. Najib Al Idrissi: Funding acquisition, Methodology, Supervision, Writing – review & editing. Bouchra Rahim: Data curation, Formal analysis, Resources, Visualization. Fatima Gaboun: Data curation, Formal analysis, Investigation, Visualization, Writing – review & editing. Fouad Msanda: Investigation, Methodology, Resources, Writing – review & editing. Abdelaziz Zahidi: Investigation, Methodology, Resources, Writing – review & editing. Aissam El Finti: Investigation, Methodology, Resources, Writing – review & editing. Abdelkhalek Legssyer: Conceptualization, Funding acquisition, Resources, Writing – review & editing. Tatiana Tatusova: Formal analysis, Investigation, Methodology, Writing – review & editing. Chakib Nejjari: Funding acquisition, Resources, Supervision, Writing – review & editing. Saaid Amzazi: Methodology, Resources, Supervision, Writing – review & editing. Lahcen Belyamani: Funding acquisition, Resources, Supervision, Writing – review & editing. Abdelhamid El Mousadik: Conceptualization, Funding acquisition, Investigation, Resources, Supervision, Writing – review & editing. Hassan Ghazal: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Writing – review & editing.
Acknowledgments
Acknowledgments
The computational resources HPC-MARWAN (www.marwan.ma/hpc) were provided by the National Center for Scientific and Technical Research (CNRST), Rabat, Morocco.
H. Ghazal is a recipient of an NIH grant through the h3abionet/h3africa consortium (grant number U24HG006941). Sequencing has been funded by Iridian Genomes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Abdellah Idrissi Azami, Email: idrissi.azami.abdellah@gmail.com, aidrissiazami@um6ss.ma.
Stacy Pirro, Email: stacy734@yahoo.com.
Sofia Sehli, Email: sofiasehli00@gmail.com, ssehl@um6ss.ma.
Nihal Habib, Email: nhlhabib98@gmail.com, nhabib@um6ss.ma.
Douae El Ghoubali, Email: douae1231@gmail.com, delghoubali@um6ss.ma.
Najib Al Idrissi, Email: nalidrissi@um6ss.ma.
Bouchra Rahim, Email: rahim.bouchra@gmail.com.
Fatima Gaboun, Email: fatima.gaboun@inra.ma, gabounf@gmail.com.
Fouad Msanda, Email: f.msanda@uiz.ac.ma.
Abdelaziz Zahidi, Email: a.zahidi@uiz.ac.ma.
Aissam El Finti, Email: a.elfinti@uiz.ac.ma.
Abdelkhalek Legssyer, Email: alegssyer@yahoo.fr.
Tatiana Tatusova, Email: tatusova@yahoo.com.
Abdelhamid El Mousadik, Email: a.elmousadik@uiz.ac.ma.
Hassan Ghazal, Email: h.ghazal@cnrst.ma, hassan.ghazal@fulbrightmail.org.
Data Availability
Argania spinosa Bioproject (Original data) (NCBI BioProject).
Argania spinosa BioSample (Original data) (NCBI BioSample).
Argania spinosa mitochondrion, complete genome (Original data) (NCBI GENEBANK).
Argania spinosa WGS Library (Original data) (NCBI SRA).
References
- 1.Ghazal H., et al. In: Oil Crop Genomics. Tombuloglu H., Unver T., Tombuloglu G., Hakeem K.R., editors. Springer International Publishing; Cham: 2021. Argane genetics and genomics; pp. 123–134. [DOI] [Google Scholar]
- 2.Khayi S., et al. First draft genome assembly of the Argane tree (Argania spinosa) F1000Res. 2020;7:1310. doi: 10.12688/f1000research.15719.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin J.-J., et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21(1):241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z.-Q., Liao X.-Z., Zhang X.-N., Tembrock L.R., Broz A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J. Syst. Evol. 2022;60(1):160–168. doi: 10.1111/jse.12655. [DOI] [Google Scholar]
- 5.Altschul S.F., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. Sep. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan P.P., Lowe T.M. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. Clifton NJ. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohse M., Drechsel O., Kahlau S., Bock R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. Jul. 2013;41(W1):W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Posada D., Crandall K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khayi S., et al. Complete chloroplast genome of argania spinosa: structural organization and phylogenetic relationships in Sapotaceae. Plants. 2020;9(10) doi: 10.3390/plants9101354. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Argania spinosa Bioproject (Original data) (NCBI BioProject).
Argania spinosa BioSample (Original data) (NCBI BioSample).
Argania spinosa mitochondrion, complete genome (Original data) (NCBI GENEBANK).
Argania spinosa WGS Library (Original data) (NCBI SRA).