Abstract
To identify proteins that can bind the 3′ untranslated region (UTR) of hepatitis C virus (HCV) we screened human cDNA libraries using the Saccharomyces cerevisiae three-hybrid system. Screening with an RNA sequence derived from the 3′-terminal 98 nucleotides (3′X region) of an infectious clone of HCV (H77c) yielded clones of human ribosomal proteins L22, L3, S3, and mL3, a mitochondrial homologue of L3. We performed preliminary characterization of the binding between the 3′X region and these proteins by a three-hybrid mating assay using mutant 3′X sequences. We have further characterized the interaction between 3′X and L22, since this protein is known to be associated with two small Epstein-Barr virus (EBV)-encoded RNA species (EBERs) which are abundantly produced in cells latently infected with EBV. The EBERs, which have similar predicted secondary structure to the HCV 3′X, assemble into ribonucleoprotein particles that include L22 and La protein. To confirm that L22 binds HCV 3′X we performed in vitro binding assays using recombinant L22 (expressed as a glutathione S-transferase [GST] fusion protein) together with a 3′X riboprobe. The 3′X region binds to the GST-L22 fusion protein (but not to GST alone), and this interaction is subject to competition with unlabeled 3′X RNA. To establish the functional role played by L22 in internal ribosome entry site (IRES)-mediated translation of HCV sequences we performed translational analysis in HuH-7 cells using monocistronic and bicistronic reporter constructs. The relative amount of core-chloramphenicol acetyltransferase reporter protein translated under the control of the HCV IRES was stimulated in the presence of L22 and La when these proteins were supplied in trans.
It is estimated that 170 million people worldwide are chronically infected with hepatitis C virus (HCV) (36). HCV infection is a leading cause of liver cirrhosis and hepatocellular carcinoma. As there is no vaccine or effective treatment available, HCV poses a significant threat to public health and there is thus an urgent need to understand the virus better and to develop vaccines and therapeutic agents (50).
HCV, a member of the Flaviviridae, is an enveloped virus containing a single stranded, approximately 9.6-kb genomic RNA molecule of positive polarity (10). The genome contains a single open reading frame flanked by 5′ untranslated regions (5′UTRs) and 3′UTRs. There are at least six genotypes of HCV whose sequences differ from each other by up to 30% over the complete genome, and the genotypes are grouped into subtypes according to sequence similarities (57). Recently, several infectious cDNA clones of HCV genome have been isolated (31, 68, 70). The genome encodes a polypeptide of approximately 3,010 amino acids which is cotranslationally processed by the host- and virus-encoded proteases to produce at least 10 mature proteins (11, 40, 50). In contrast to the recent progress made in understanding the genome organization of HCV, the proteolytic processing of the polyprotein and the biochemical characterization of the individual proteins, similar success in understanding the mechanisms of HCV replication has been hampered by the lack of an efficient in vivo replication system.
The HCV 5′UTR may play a role in viral replication and packaging. In addition, the structurally complex 5′UTR sequence contains an internal ribosome entry site (IRES), which drives expression of the HCV polyprotein in a cap-independent manner (22). The HCV 3′UTR consists of three distinct regions—a short nonconserved variable-length sequence, a variable-length polypyrimidine tract [poly(U-UC)], and a highly conserved stretch of 98 nucleotides (nt), termed the 3′X, which represents the authentic 3′ terminus of the HCV RNA genome (59). The HCV 3′X contains a considerable degree of secondary structure; three stem-loop structures have been demonstrated, (6, 26, 60), the most stable of which is formed by the 3′-terminal 46 nt (6, 26, 60). Recently, the 3′X region and the poly(U-UC) region of the HCV 3′UTR have been shown to be absolutely required for in vivo infectivity (69). In parallel with other members of the Flaviviridae family, whose 3′UTRs are highly conserved and contain stable secondary structures, it is possible that the 3′X region may serve as a cis-acting element in replication of HCV RNA. It is well established that host-encoded proteins play a role in the replication of many RNA viruses (5, 34) either by binding to the 3′UTR or as part of the replicase complex (17, 26, 38). Indeed, recent evidence has demonstrated that the HCV 3′UTR is capable of binding polypyrimidine tract binding protein (PTB) (19, 28, 65), hnRNP C (heterogeneous nuclear ribonucleoprotein C), (18), La (58), GAPDH (49), and other cellular proteins (p87 and p130) (24). While the roles played by PTB and La in modulation of IRES-mediated translation of HCV are being clarified (1, 25, 27, 28), the significance of the interaction between PTB, La, hnRNP C, GAPDH, and other unidentified 3′UTR binding proteins in HCV replication, or other aspects of HCV biology, is not yet understood.
The development of the Saccharomyces cerevisiae three-hybrid system allows the analysis and identification of RNA-binding proteins in vivo (56). Using this system, we identify human ribosomal proteins (RPs) L22, S3, L3, and mL3, the mitochondrial homologue of L3, as novel HCV 3′X RNA-binding proteins. These proteins may be involved in replication, translation, and/or packaging of HCV. We characterized the binding between these proteins and 3′X-derived sequences by three-hybrid mating analysis. The 3′X-L22 interaction was selected for further characterisation both in cell culture and in vitro. Translational analysis in cell culture using mono- and bicistronic reporter constructs showed that the 3′X-L22 interaction may modulate IRES-mediated translation of the HCV open reading frame.
MATERIALS AND METHODS
Plasmid constructs expressing HCV 3′UTR.
HCV 3′UTR sequences were amplified by PCR using an infectious HCV cDNA clone pCV-H77c (68) (kindly supplied by J. Bukh) as a template. Primers used for amplification of full-length (FL) 3′UTR were Upper, 5′ tctagaactagtggatccCCCGGGaggttggggtaaacactccggcctct 3′, and Lower, 5′ tctagaactagtggatccCCCGGGacatgatctgcagagaggccagtat 3′; primers used for amplification of the 3′X were Upper, 5′ tctagaactagtggatccCCCGGGaatggtggctcctcttagccc 3′, and Lower, 5′ tctagaactagtggatccCCCGGGacatgatctgcagagaggccagtat 3′ (where the Sma I site used for cloning is in uppercase and H77c DNA sequences are in boldface type). The resulting PCR products (268-bp FL 3′UTR; 148bp 3′X) were inserted by standard techniques into pBluescript SK vector (Stratagene) or the hybrid RNA expressing plasmids pIIIA/MS2-1 and pIIIA/MS2-2, which both carry the ADE2 and URA3 genes (56). Since it has been previously reported that the relative order of the RNA sequence of interest and the MS2 sites can affect signal strength in the three-hybrid assay (56), we cloned the 3′X sense (+) sequence into both pIIIA/MS2-1 and pIIIA/MS2-2 to generate 3′X-MS2 and MS2-3′X RNAs, respectively, in vivo. These constructs have been designated p3′X (+)-MS2 and pMS2-3′X (+), respectively. Additionally we fused the 3′X antisense (−) sequence to the 5′ end of the MS2 sequence into pIIIA/MS2-2 to generate plasmid p3′X (−)-MS2. The FL 3′UTR (sense) sequence was cloned into pIIIA/MS2-1 to generate pMS2-FL3′UTR. Plasmids encoding 3′X/MS2 mutants were prepared using standard PCR strategy, and the mutant 3′X sequences were cloned into pIIIA/MS2-2. The three mutants used in this study are designated p3′X SL1 (+)-MS2, p3′X SL2 (−)-MS2, and p3′X 23L1S (−)-MS2. p3′X SL1 (+)-MS2 corresponds to a 3′X sequence where CUCUGCAGA (nt 84 to 92) within the stem I has been replaced with ACAGCGCU. p3′X SL2 (−)-MS2 corresponds to a stem II mutation where the sequence UCACGGCU (nt 23 to 30) has been replaced with GCUAGCUC, in the antisense orientation. p3′X 23 L1S (−)-MS2 corresponds to a triple antisense mutant where the sequences GGCTG (stem III, nt 4 to 8), UCACGGCU (stem II, nt 23 to 30), and GCUCCUCUGCAGA (stem I, nt 80 to 92) have been replaced with CCGAG, GCUAGCUC, and GCUCCUGCAUCGG (stems III, II, and I, respectively). The sequence and orientation of all the constructs described in this study were determined using an ABI Prism 377 DNA sequencer (Perkin-Elmer).
Expression of L22 and La.
FL L22 (clone 66) was excised with appropriate restriction enzymes from the activation domain (AD) vector pGADGH and subcloned into the bacterial or mammalian expression vector pGEX-6P-3 (Pharmacia) or pcDNA3.1/Zeo(+) (Invitrogen), respectively. A DNA fragment encoding the FL La protein (a kind gift from Ger Pruijn) was cloned into pcDNA3.1/Zeo(+) to yield pcDNA-La. The glutathione S-transferase (GST)-L22 fusion protein was expressed in Escherichia coli BL21 cells following induction with IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM final), and purified by binding to glutathione agarose beads (Sigma). For in vitro RNA-binding studies, GST-L22 was treated with PreScission protease (Pharmacia) to remove the GST part of the fusion protein before use. For RNA gel mobility shift assays (RNA GMSA), the GST-L22 protein was purified further by gel filtration on a Superdex-75 column (Pharmacia) using fast-performance liquid chromatography equipment (Pharmacia). Proteins were eluted in RNA-binding buffer supplemented with 10% glycerol, and fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Construction of core-CAT reporter plasmids.
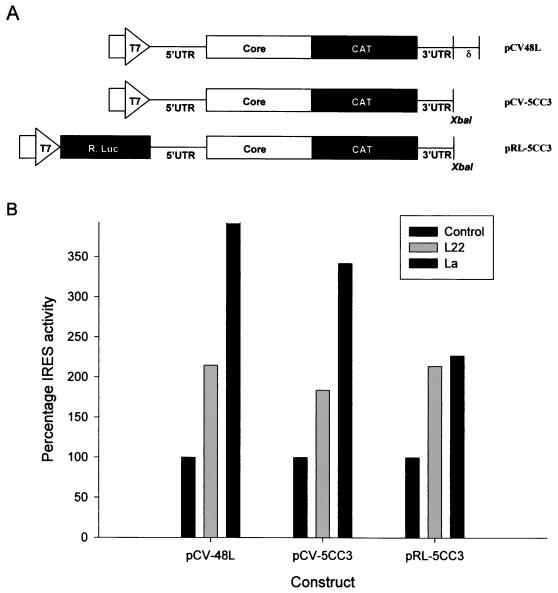
Using standard techniques, one bicistronic and two monocistronic constructs carrying sequences encoding HCV core and chloramphenicol acetyltransferase (CAT) were generated. The monocistronic constructs designated pCV-5CC3 and pCV-48L were generated in the pCV vector described by Yanagi et al. (68); they contain the sequences encoding the entire 5′UTR, the FL HCV core protein (nt 1 to 924 of HCV genome, [68]) fused in frame to the CAT gene, followed by a stop codon and the FL 3′UTR (nt 9378 to 9599 [68]). pCV-48L also contains the delta ribozyme sequence cloned 3′ of the HCV 3′UTR, which is designed to self-cleave and generate an authentic HCV 3′ terminus. As in pCV-H77c (68), the T7 promoter in both pCV-5CC3 and pCV-48L is expected to generate transcripts with the authentic 5′ end of HCV 5′UTR. The translation of these RNA transcripts should be mediated by HCV IRES and is expected to produce a core-CAT fusion protein. The core domain from this fusion protein is expected to be cotranslationally cleaved on account of the presence of the signal peptidase cleavage sites in these constructs. The bicistronic construct, pRL-5CC3 contains the Renilla luciferase gene cloned under the control of the T7 promoter in the vector pTZ-18i (Pharmacia). Following the luciferase sequence are the 5′UTR-core-CAT-3′UTR sequences as described above. The primary translation products from pRL-5CC3 are Renilla luciferase and the core-CAT fusion protein. These constructs are schematically represented below (see Fig. 4A).
FIG. 4.
L22 modulates HCV IRES-mediated translation. (A) Schematic diagram of the constructs used in this study. (B) Quantitation of the enzymatic activity of the translation products in cell lysates. For the monocistronic constructs pCV-5CC3 and pCV-48L, the effect of host proteins on HCV IRES activity was determined by comparing the CAT activity of 10 μg of lysate prepared from HuH-7 cells infected with a vaccinia virus expressing T7 RNA polymerase and transfected with either pcDNA3.1/Zeo(+) alone or pcDNA3.1/Zeo(+) expressing L22 or La protein. Results are expressed as a percentage, where the apparent IRES activity of extracts transfected with pcDNA3.1/Zeo(+) alone is arbitrarily set at 100%. Results shown represent means obtained in three separate experiments. For the bicistronic construct pRL-5CC3, the effect of host proteins on the relative IRES activity was calculated by determining the ratio of CAT activity (translated under the control of the HCV IRES) to Renilla luciferase (R. Luc) activity (translated from the upstream cistron of the same RNA molecule) in 10 μg of lysates prepared from HuH-7 cells infected with a vaccinia virus expressing T7 RNA polymerase and transfected with either pcDNA3.1/Zeo(+) alone or pcDNA3.1/Zeo(+) expressing L22 or La protein. Results are expressed as a percentage, where the IRES activity of extracts transfected with pcDNA3.1/Zeo(+) alone is arbitrarily set at 100%. Results shown represent means obtained in five separate experiments.
Northern blot analysis.
The hybrid RNA-expressing plasmids were used to transform yeast L40coat strain (which contains a gene expressing the LexA-MS2 coat protein fusion stably integrated into the yeast chromosome), and transformants were selected and maintained on plates lacking uracil. To confirm that the appropriate hybrid RNA molecules were produced in vivo, total RNA from each yeast strain was analyzed by Northern blotting, using an MS2 DNA probe or strand-specific 3′X riboprobes. Briefly, total RNA from the appropriate yeast strain was fractionated on denaturing formaldehyde agarose gels, capillary blotted onto nylon membrane, and UV cross-linked. For probe preparation, the MS2-encoding DNA fragment was excised from pIIIA/MS2-1, gel purified, and radiolabeled by random priming (Prime-It II kit; Stratagene) using [α-32P]dCTP. High-stringency Northern analysis was performed by hybridizing blots at 65°C for 1 h using Rapid Hyb buffer (Amersham) and washing three times at 65°C for 30 min in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS. The resulting blots were exposed to X-ray film for 3 days. Similar blots were prepared and hybridized under the same conditions of stringency using strand-specific riboprobes, i.e. 3′X sense and 3′X antisense to check for in vivo production of the appropriate HCV RNA.
Three-hybrid selection and screening.
Selection and screening were essentially performed as previously described (56, 72) with some minor modifications. A derivative of yeast strain L40coat, containing p3′X-MS2, was transformed with DNA from a HeLa, HeLa S3, or human liver cDNA library constructed in activation domain plasmids pACTII, pGADGH, and pACT, respectively (Clontech). Yeast transformants were plated on synthetic medium lacking leucine and histidine and supplemented with 2 to 5 mM 3-aminotriazole to select for higher activation of HIS3. RNA-independent positives were initially eliminated through red or white color selection, followed by 0.1% 5-fluoroorotic acid selection as described previously (71). To confirm that the RNA-dependent clones were true positives, hybrid RNA-expressing plasmids were introduced by mating with each of four derivatives of the yeast strain R40coat (opposite mating type to L40coat) carrying pMS2-3′X (+), p3′X (+)-MS2, p3′X (−)-MS2, or pMS2-FL 3′UTR (+). The mating mixtures were incubated overnight and plated on medium lacking uracil and Leu. After 3 days of growth, filters were lifted from each of the mating plates (lacking uracil and Leu) and the counterpart cured plates (without Leu but containing 0.1% 5′-fluoruorotic acid) and were tested for β-galactosidase (β-Gal) activity. For positive RNA-dependent clones, the mating should reintroduce the RNA-expressing plasmid and restore the previously observed positive phenotype in the β-Gal assay (blue). In contrast, filters lifted from the counterpart cured plate, where the yeast does not contain the RNA-expressing plasmid, should remain negative in the β-Gal assay (no blue color). DNA isolated from RNA-dependent positive yeast clones was used to transform E. coli TG1 cells. Plasmid DNA was sequenced using AD forward and reverse primers. For verification and determination of relative affinity of RNA-protein interactions a liquid o-nitrophenyl-β-d-galactopyranoside (ONPG) assay for β-Gal activity was used.
SDS-PAGE and immunoblotting.
Protein samples were mixed with SDS-PAGE denaturing buffer (50 mM Tris-HCl [pH 6.7] 2% SDS, 5% β-2-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue) and fractionated on SDS–12.5% polyacrylamide gels. For Western blotting, proteins were electrophoretically transferred to Hybond ECL membranes (Amersham) as previously described by Towbin et al. (64). Membranes were incubated with an appropriately diluted rabbit anti-L22 polyclonal antiserum (gift from Joan Steitz) or an anti-GST monoclonal antibody. The immunoreactive proteins were detected using protein A- or goat anti-mouse immunoglobulin G-horseradish peroxidase conjugate and enhanced chemiluminescence reagents (Amersham).
Riboprobe production.
Plasmids carrying 3′UTR, 3′X, and Epstein-Barr virus (EBV)-encoded RNA 1 (EBER1) sequences (kindly provided by M. Clemens [13]) were linearized by cleavage with an appropriate restriction enzyme immediately downstream from the respective inserts. The linearized DNA constructs were transcribed in vitro using T7 RNA polymerase (Promega) and [α-32P]CTP (NEN) according to the manufacturer's instructions. The resulting high-activity riboprobes were subjected to nondenaturing PAGE, visualized by autoradiography, excised, and eluted from the gel by incubation at 37°C in RNA elution buffer (0.5 M ammonium acetate, 0.1% SDS, 1 mM EDTA). The probes were then precipitated with ethanol and resuspended in nuclease-free water.
RNA GMSA.
Various amounts of protein were mixed with 20 μg of yeast tRNA (Sigma) and 40 U of RNasin (Promega) in a final volume of 25 μl of RNA-binding buffer [10 mM Tris-HCl (pH 7.2), 100 mM KC1, 3 mM magnesium acetate, 5% glycerol, 1 mM EDTA]. In competition assays, a fixed amount of protein in RNA-binding buffer was preincubated with different amounts of unlabeled competitor RNA. Following incubation at 30°C for 10 min, 32P-labeled RNA (50,000 cpm) was added and the reaction mixture was incubated for a further 20 min. After adding the tracking dye sample (1× RNA-binding buffer, 0.05% bromophenol blue, and 0.05% xylene-cyanol), the reaction mixtures were fractionated on a preelectrophoresed (200 V for 30 min) nondenaturing 6% polyacrylamide gel in 1× TBE (0.089 M Tris-borate, 0.089 M boric acid, 0.002 M EDTA). Electrophoresis was carried out at 200 V for 2 to 3 h. The gel was dried, exposed overnight to a phosphor screen, and visualized with a Bio-Rad Personal FX phosphorimager.
UV cross-linking analysis.
For UV cross-linking of RNA to proteins, various amounts of protein were mixed with 20 μg of yeast tRNA (Sigma) and 40 U of RNasin (Promega) in a final 25 μl volume of RNA-binding buffer. In competition assays, different amounts of unlabeled RNA were preincubated with a fixed amount of protein in the binding buffer. Following incubation at 30°C for 10 min, 32P-labeled RNA (50,000 cpm) was added and the reaction mixture was incubated at 30°C for a further 20 min. The reaction mixtures were then placed on ice and irradiated with UV light for 30 min at a wavelength of 254 nm (Stratalinker; Stratagene). After irradiation, 5 μl of RNase cocktail (containing RNase A and RNase T1) (Ambion) was added to each reaction mixture followed by incubation at 37°C for 15 min. For fractionation of samples by SDS-PAGE, 15 μl of 3× sample buffer was added to each reaction mixture, and the samples were boiled for 3 min, chilled on ice, and electrophoresed on a 12.5% polyacrylamide gel containing 0.1% SDS. The gels were dried, exposed to phosphor screens, and visualized with a Bio-Rad Personal FX phosphorimager.
Transfection of cultured cells.
HuH7 cells (45) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2% glutamine, 1% nonessential amino acids, penicillin (100 U/ml), and streptomycin (100 U/ml). Subconfluent monolayers of cells in six-well (35-mm-diameter well) plates were infected with vTF7-3, a recombinant vaccinia virus expressing T7 RNA polymerase (16), at a multiplicity of infection of 5, in 100 μl of medium for 1 h at 37°C. The cells were then transfected with plasmid DNA using liposomes as described previously (55). Following incubation at 37°C for 16 h, the cells were harvested and the cell lysates assayed for luciferase and CAT activity, as described below.
CAT and Renilla luciferase assays.
Transfected cells were washed with and harvested in 100 μl of 0.25 M Tris-HC1 (pH 7.4). Cell lysates were prepared by freeze-thaw followed by sonication and centrifugation at 13,000 rpm for 5 min in a Sanyo MSE Micro Centaur. The supernatant was removed, and total protein concentration was determined by the Bradford assay (8). CAT assays were carried out using 10 μg of protein and [14C]chloramphenicol (0.025 mCi/ml; NEN) essentially as described by Gorman et al. (20). The radioactivity in the substrate and the acetylated products was visualized with a Bio-Rad Personal FX phosphorimager and quantitated by using Quantity One volume analysis software (Bio-Rad). Approximately 10-μg protein samples were assayed for Renilla luciferase activity using a dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luciferase activities were measured using a Biotrace M3 benchtop luminometer.
RNA secondary structure analysis.
RNA secondary structures were predicted using GCG Mfold which uses the most recent energy minimization method of Jaeger et al. (30). The structures were displayed using GCG Plotfold and Squiggles output. The associated free energy of each structure is listed in kilocalories per mole. Plots were scanned using an AGFA Studioscan II flatbed scanner and manipulated in Abode Photoshop, version 4.01.
RESULTS
In vivo expression of hybrid RNAs.
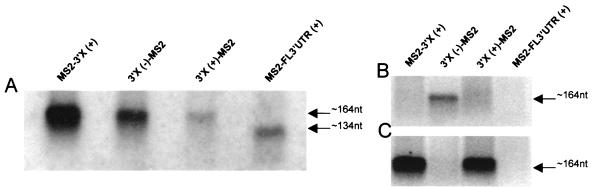
We generated plasmids expressing hybrid RNAs in which bacteriophage MS2 coat protein binding sites are fused to HCV 3′X or FL 3′UTR sequences. The 3′X sequence was inserted in either sense or antisense orientation, and the relative order of the MS2 and HCV sequences was varied. To verify that the appropriate RNA was transcribed by RNA polymerase III in the yeast strains expressing hybrid RNA sequences, Northern blotting of total RNA was performed. Results using the MS2-specific probe showed that the hybrid RNAs carrying 3′X sequences [MS2-3′X (+), 3′X (−)-MS2, and 3′X (+)-MS2] all contain MS2 sequence, as transcripts of ∼164 nt (Fig. 1A). The hybrid RNA carrying the FL 3′UTR sequence [MS2-FL 3′UTR(+)] also contained MS2 sequence, but the transcript size (∼134 nt) was smaller (Fig. 1A). Northern blot analysis using 3′X (sense) and 3′X (antisense) strand-specific riboprobes showed as expected that the yeast strain designated 3′X (−)-MS2 expressed the antisense 3′X sequence (Fig. 1B), and those designated MS2-3′X (+) and 3′X (+)-MS2 expressed the sense 3′X sequence (Fig. 1C). The sizes of the 3′X-MS2 hybrid RNAs are consistent with those obtained from the MS2 Northern blot (Fig. 1A). In contrast, the yeast strain designated MS2-FL 3′UTR (sense) expressed a shorter RNA transcript (∼134 nt), which contained the MS2 sequence (Fig. 1A) but no HCV 3′X sequence (Fig. 1C). The smaller size of the FL 3′UTR transcripts produced in vivo is probably a result of termination of RNA polymerase III within the poly(U-UC) region of the 3′UTR (preceding the 3′X region) (71). This hybrid RNA served as a useful negative control in mating experiments. Mfold (RNA folding program) analysis of the 3′X (+), 3′X (−), or FL 3′UTR (+) alone or when fused to the MS2 RNA sequence [MS2-3′X (+), 3′X (+)-MS2, 3′X (−)-MS2, and MS2-FL 3′UTR (+)] predicted that the secondary structures of the HCV RNA sequences were not altered in the context of the MS2 RNA (data not shown).
FIG. 1.
In vivo production of hybrid RNAs, as detected by Northern blotting. Total RNA (10 μg) from yeast strain L40coat carrying plasmids pMS2-3′X (+), p3′X (+)-MS2, p3′X (−)-MS2, or pMS2-FL 3′UTR (+) was fractionated by denaturing electrophoresis, blotted onto a nylon membrane and probed with an MS2 DNA probe (A), 3′X (+) riboprobe (B), or 3′X (−) riboprobe (C).
Yeast three-hybrid screen.
Having established that the 3′X (+)-MS2 hybrid RNA is correctly expressed in yeast (Fig. 1), we used this RNA in a three-hybrid assay to screen human liver and HeLa cell activation domain cDNA libraries prepared in GAL4 AD vectors (Clontech). Between 1.5 × 106 and 107 independent clones were screened in total. The screening on 3-aminotriazole plates lacking Leu and His yielded approximately 3,000 white colonies, which were subjected to further rounds of selection to eliminate false-positive clones.
The criteria imposed to identify genuine positives were that (i) they had to be RNA-dependent positives as verified by the loss of β-Gal activity in the absence of the RNA-expressing plasmid and (ii) these RNA-dependent positives had to be RNA-specific as shown by the restoration of β-Gal activity following mating with yeast strains expressing 3′X (+)-MS2 or MS2-3′X (+), but not MS2-FL 3′UTR (+), which does not express any 3′X sequence (Fig. 1). The cDNA inserts of library-derived plasmids from RNA-dependent, RNA-specific positive colonies were sequenced. A DNA database search identified these as independent cDNA clones encoding either human RP L22, S3, or L3 (each identified several times) or mL3, the mitochondrial homologue of L3 (identified once).
Characterization of 3′X-RP interactions.
To further characterize the interaction between the RPs and 3′X sequences, we selected clones of each protein containing minimum 5′UTR sequence and maximum coding sequences. For L22, we used clone 66, which carried the FL L22 cDNA with minimal 5′ noncoding sequence (a GCC triplet) between the EcoRI linker used to clone the library into the GAL4 AD and the initiating methionine codon. L40coat yeast transformants carrying constructs expressing AD-RP cDNAs fusion proteins were mated with R40coat transformants expressing wild-type or mutant 3′X/MS2 hybrid RNA sequences. The in vivo activity of each RNA-protein interaction (expressed as units of β-Gal activity) was determined in triplicate using quantitative ONPG assay (Table 1). Compared to the well-characterized iron-regulatory protein–iron-regulatory element (IRP-IRE) interaction (4, 21), the activity of the 3′X (+) and L22 or mRPL3 interaction was higher (Table 1). The binding of S3 and L3 to 3′X (+) produced lower β-Gal activity than that between IRP and IRE; S3 was approximately half as much, whereas L3 was approximately 1/10 as much. The use of sense and antisense 3′X RNA revealed that each protein had different sense specificity for the 3′X region. The binding of L22 to the 3′X (−) RNA produced nearly twofold more activity than binding to the 3′X (+) RNA. In contrast, S3 and L3 produced with lower activity, whereas mRPL3 failed to bind 3′X (−) (Table 1).
TABLE 1.
In vivo analysis of the interaction between 3′X sequences and RPsa
| Hybrid RNA | L22
|
mL3
|
S3
|
L3
|
IRP (β-Gal units) | ||||
|---|---|---|---|---|---|---|---|---|---|
| β-Gal units | % of WT(+) | β-Gal units | % of WT(+) | β-Gal units | % of WT(+) | β-Gal units | % of WT(+) | ||
| MS2 alone | 0.1 | 0.6 | 0.2 | 1.0 | 0.0 | 0.0 | 0.05 | 7.35 | 0.01 |
| MS2-IRE | 3.8 | 22.0 | 0.5 | 2.0 | 0.5 | 13.8 | 0.17 | 25.0 | 6.0 |
| MS2-3′X (+) | 17.0 | 100.0 | 23.6 | 100.0 | 3.9 | 100.0 | 0.68 | 100.0 | ND |
| MS2-3′X (−) | 32.5 | 190.8 | 0.7 | 2.8 | 3.2 | 82.0 | 0.3 | 42.0 | ND |
| MS2-3′X (stem I mutant) | 0.5 | 2.9 | 7.3 | 31.0 | 2.5 | 64.0 | 0.54 | 79.4 | ND |
| MS2-3′X (stem II mutant) (−) | 0.7 | 4.1 | 0.8 | 3.3 | 0.2 | 3.9 | 0.2 | 25.0 | ND |
| MS2-3′X (triple mutant) (−) | 0.16 | 1.0 | 8.0 | 34.0 | 0.5 | 12.8 | 0.23 | 33.0 | ND |
Assay of β-galactosidase reporter activity in extracts of yeast expressing AD-RP fusions, transformed with plasmids driving expression of the MS2-3′X element hybrid RNA. Assays were performed in triplicate, and the average results are listed as units of β-galactosidase and as a percentage of the 3′X sense binding, which is arbitrarily assigned as 100%. (+) and (−) refer to sense and antisense RNA, respectively. Abbreviations: WT, wild type.; ND, not determined.
The differences in β-Gal activities associated with each of the proteins and three RNA mutants revealed that the proteins are likely binding to different areas within the 3′X region. L22 did not bind to any of the three mutants, whereas mRPL3 was capable of binding to the stem-loop II mutant and the triple stem-loop mutant, although the β-Gal activity associated with binding to these two mutants was approximately 30% of the value associated with binding to 3′X (+). S3 and L3 were both able to bind to the stem-loop I mutant, although with reduced β-Gal activity (64 and 79.4%, respectively), whereas the binding to both the stem-loop II and the triple mutant was greatly reduced in each case.
Of the four RPs identified in the library screening procedure, mRPL3 and L22 exhibited the strongest activity with the 3′X sequence. Although mRPL3 expression has been shown to be upregulated in hepatocellular carcinoma (47) and may therefore be of interest with regard to HCV infection, little is known about the biochemical properties of this protein. In contrast, given the high number of independent L22 cDNA clones isolated in the screening procedure and the fact that L22 is well characterized (15, 17, 35, 66) and is known to bind to EBERs (63), we selected the interaction between 3′X and L22 for further investigation.
HCV 3′X interacts with human RPL22 in vitro.
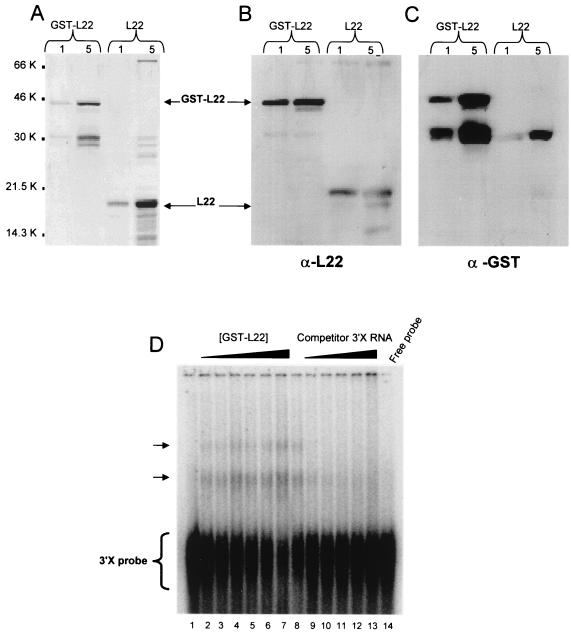
In order to confirm the binding of L22 to the 3′X sequence, we expressed FL L22 in E. coli as a GST fusion protein. As shown in Fig. 2A, purified GST-L22 fusion protein and L22 cleaved from the GST moiety both appeared as major bands with apparent molecular masses of 42 and 15 kDa, respectively, on SDS-PAGE. Western blot analysis using a rabbit anti-L22 polyclonal antibody and an anti-GST monoclonal antibody confirmed that the 42- and 15-kDa proteins are GST-L22 and L22, respectively (Fig. 2B and C). These protein preparations were further purified by gel filtration chromatography for use in in vitro RNA-binding assays. To confirm that the purified GST-L22 and L22 proteins interacted with the 3′X sequence, we performed RNA GMSA and UV cross-linking experiments using the 32P-labeled 3′X riboprobe. The results showed that two RNA-protein complexes of lower mobility were formed in which the intensity of these complexes was proportional to the GST-L22 protein concentration (Fig. 2D, lanes 2 to 7). Additionally, these RNA-protein complexes were subject to competition with increasing amounts of unlabeled competitor 3′X RNA, indicating that the interaction between L22 and 3′X RNA is specific (Fig. 2D, lanes 9 to 13). No RNA-protein complexes were formed when GST-L22 was replaced with GST alone or GST fused to HBV preS1 (GST-preS1) protein in the assay (data not shown).
FIG. 2.
Expression of human RPL22 in E. coli and electrophoretic mobility shift assay. Aliquots of L22 and GST-L22 (1 and 5 μg) were fractionated by SDS-PAGE and stained with Coomassie blue (A) or immunoblotted and probed with anti-L22 antibody (α-L22) (B) or anti-GST (α-GST) antibody (C) as described in Materials and Methods. The FL GST-L22 and L22 proteins are indicated. Molecular weights in thousands (K) are shown beside panel A. (D) GST-L22 protein was incubated with 50,000 cpm of 32P-labeled 3′X RNA and 20 μg of yeast tRNA in each binding reaction. Lanes 2 to 7 contain increasing amounts of GST-L22 protein (30, 40, 50, 60, 70, and 110 ng, respectively). Lanes 8 to 13 contain 50 ng of GST-L22 in each binding reaction, incubated with increasing amounts of unlabeled, competitor 3′X (sense) RNA (lanes 8 to 13, no competitor RNA, 10-, 20-, 30-, 40-, and 60-fold molar excess of 3′X [sense] competitor RNA, respectively). Lanes 1 and 14 contain 50,000 cpm of unbound 3′X probe only. Arrows indicate the RNA-protein complexes.
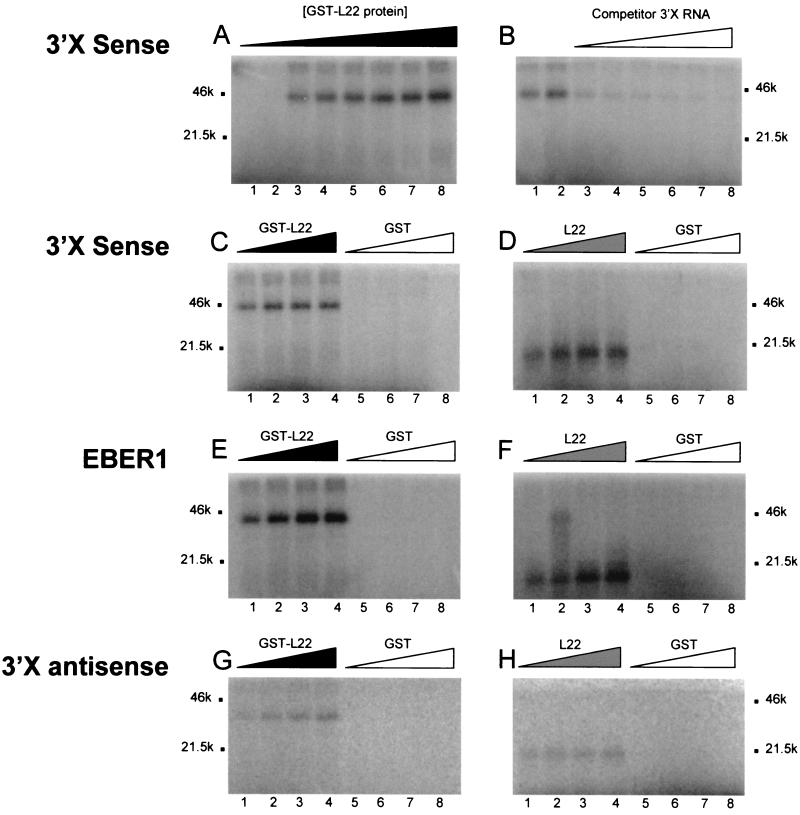
The interaction between L22 protein and 3′X was further confirmed in UV cross-linking experiments using purified GST-L22 or L22 alone and a 32P-labeled 3′X riboprobe. As shown in Fig. 3A, the 42-kDa GST-L22 cross-linked to the 3′X RNA in a dose-dependent manner. This binding was subject to competition by cold, unlabeled 3′X RNA, indicating that the L22-3′X interaction is specific (Fig. 3B). Similarly, the 15-kDa L22 bound the radiolabeled 3′X riboprobe, which could be competed for by cold probe (data not shown). To further confirm that the 3′X did not bind to protein in a nonspecific manner, we also performed UV cross-linking assays using GST protein alone. As shown in Fig. 3C and D the 3′X probe interacted with GST-L22 and L22, but not with GST.
FIG. 3.
UV cross-linking analysis of 3′X RNA. The UV cross-linking assay was as performed as described in Materials and Methods. (A) UV cross-linking analysis of GST-L22 protein with 3′X (sense) RNA. Lanes 1 to 8, 10, 20, 100, 200, 300, 400, 500, and 1,000 ng of GST-L22 protein preparation, respectively. (B) Competition assay with unlabeled 3′X RNA and GST-L22. Lane 1, 100 ng of GST-L22, no competitor RNA; lane 2, 200 ng of GST-L22, no competitor RNA; lanes 3 to 8, 200 ng of GST-L22 incubated with 10-, 20-, 30-, 40-, 50-, and 100-fold molar excess of unlabeled 3′X (sense) RNA. (C and D) UV cross-linking analysis of GST and GST-L22 or L22 protein with 3′X RNA. Lanes 1 to 4, 100, 200, 300, and 400 ng of GST-L22 protein (C) or L22 protein (D), respectively. Lanes 5 to 8, 100, 200, 300, and 400 ng of GST protein, respectively. (E and F) UV cross-linking analysis of GST and GST-L22 or L22 protein with EBER1. Lanes 1 to 4, 100, 200, 300, and 400 ng of GST-L22 protein (E) or L22 protein (F), respectively. Lanes 5 to 8, 100, 200, 300, and 400 ng of GST protein, respectively. (G and H) UV cross-linking analysis of GST and GST-L22 or L22 protein with 3′X (antisense) RNA. Lanes 1 to 4, 100, 200, 300, and 400 ng of GST-L22 protein (G) or L22 protein (H), respectively. Lanes 5 to 8, 100, 200, 300, and 400 ng of GST protein, respectively. Molecular weights (in thousands [k]) are shown beside each gel.
Since L22 was initially shown to bind EBER1, a small RNA molecule associated with latent EBV infection (63), we performed UV cross-linking experiments using the GST-L22, L22, and GST protein preparations together with an EBER1 riboprobe. As with HCV 3′X, the 32P-labeled EBER1 RNA interacted with the 42-kDa GST-L22 protein (Fig. 3E) and the 15-kDa L22 protein (Fig. 3F), but not with GST protein alone. The EBER1-L22 interaction was also verified in GMSA by an electrophoretic shift of the EBER1 probe (data not shown), and its specificity was confirmed in competition assays using unlabeled EBER1 RNA (data not shown).
Since our three-hybrid mating assays had demonstrated that L22 could interact with the 3′X antisense RNA, we performed UV cross-linking assays using GST-L22 or L22 protein and 3′X (antisense) riboprobe derived from HCV strain H77c cDNA. Similar to our results obtained with 3′X (sense) and EBER1, we observed the formation of RNA-protein complexes of ∼42 and ∼15 kDa between 3′X (antisense) and GST-L22 and L22, respectively; no complex formation was observed with GST alone (Fig. 3G and H).
Functional significance of L22-3′X interaction.
To establish the functional role of the 3′X-L22 interaction during viral infection we tested the possible effect of L22 on IRES-mediated translation of a panel of monocistronic and bicistronic constructs (Fig. 4A), both in vitro and in cell culture, as described previously (27, 28). Since La protein has also been shown to bind the HCV 5′UTR and 3′UTR (2, 58), we also tested the effect of this protein in our assays. HuH-7 cells were infected with a vaccinia virus expressing T7 RNA polymerase (vTF7.3), followed by transfection either of a plasmid vector expressing a reporter cDNA alone or cotransfection of a reporter cDNA with an L22 or La mammalian expression construct. Total cellular lysates were prepared, and equal amounts (in terms of total protein) of each extract were assayed for luciferase and CAT activity as described in Materials and Methods. The CAT activities were quantitated by directly comparing the amount of diacetylated chloramphenicol produced by extracts of cells cotransfected with each of the two monocistronic constructs pCV-48L or pCV-5CC3 and either pcDNA3.1/Zeo(+) alone, pcDNA-L22, or pcDNA-La (Fig. 4B). Since the amount of the diacetylated product reflects the CAT activity under the control of the HCV IRES, it should indicate any effect of the L22 or La proteins in trans. Results from three separate experiments revealed an effect of L22 and La on apparent IRES activity. For pCV-48L, L22 and La increased IRES activity to 2.1- and 3.8-fold, respectively. A slightly smaller effect was seen with pCV-5CC3, for which L22 and La increased apparent IRES activity by 1.8- and 3.4-fold, respectively. This enhancement of apparent IRES activity was consistent with results obtained from five separate experiments using the bicistronic construct pRL-5CC3 (Fig. 4B). L22 and La increased IRES activity to 2.3- and 2.2-fold, respectively.
We also tested the effect of L22 in reticulocyte lysates programmed with RNA transcribed from the linearized monocistronic construct pCV-5CC3. Consistent with the twofold increase in CAT activity seen with L22 in cell culture, we observed a twofold increase in CAT activity in reactions supplemented with either recombinant GST-L22 or His-L22 (data not shown) but not in reactions supplemented with GST alone (data not shown).
DISCUSSION
Using an in vivo library screening procedure we identified four human RPs (L22, L3, S3, and mL3) that can bind the HCV 3′X region. We further characterized the interaction between 3′X and L22 in vitro, demonstrating that RNA-protein complex formation is specific, concentration dependent, and subject to competition. Fulfillment of these three criteria suggests the validity of this RNA-protein interaction. L22 also stimulated HCV IRES-mediated expression of a bicistronic construct twofold in a human liver cell line.
Human L22 (35) was originally identified as a 15-kDa protein capable of binding two small, highly abundant EBERs (EBER1 and -2) in cells latently infected with EBV (63). L22 also binds HVP1, a small virally encoded RNA analogous to EBER1, detectable in baboon cells infected with herpesvirus papio (63). L22 is found in the 60S ribosomal subunit, and is located in both the cytoplasm and nucleolus. The cellular ligand of L22 is predicted to be a region of human 28S rRNA corresponding to the stem-loop between nt 302 and 317 (15).
Both EBERs are predicted to have stable secondary structures. L22 has been shown to bind EBER1 mainly in stem-loop III and less strongly in stem-loop IV (Fig. 5A) (62). Mutational analysis of stem-loop III has shown that L22 recognizes the majority of the nucleotides in this hairpin, interacting with both single-stranded and double-stranded regions in a sequence-specific manner (62). L22 binding to EBER1 stem-loop 3 has been suggested to be a critical aspect of EBER1 and HVP1 function, since this structure represents the region most highly conserved between EBER1 and HVP1 (62). The L22 binding site in EBER1 stem-loop III is largely palindromic (62). Interestingly, the double-stranded region of stem-loop I (46 nt) in the 3′X region also contains a sequence (CUGCAGA; nt 59 to 65 and 86 to 92 [Fig. 5B]) which is directly repeated on each side of the stem to form a palindromic region, similar to that observed for EBER1 (62). Our data show that L22 binding to a 3′X stem-loop I mutant, that does not contain a palindrome sequence is abrogated, implicating this region in binding.
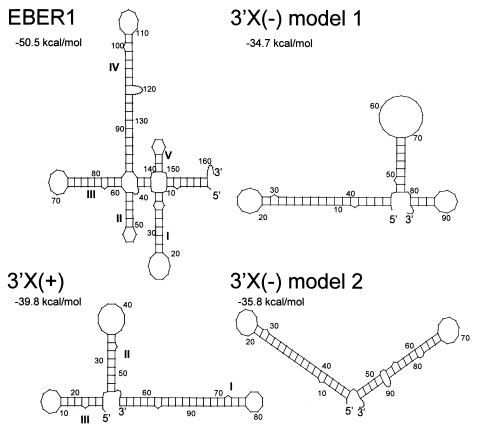
FIG. 5.
Secondary structure analysis of HCV 3′UTR and EBER1. Modeling was performed using the computer program Mfold with HCV H77c and EBER1 sequences. The structures were visualized using Plotfold with a Squiggles output. Established stem-loop structures are indicated with Roman numerals, and computed free-energy values are given. Two models for the 3′X(−) strand are shown since the exact structure of this region has yet to be experimentally determined.
The generalized RNA motif that mediates the interaction of RNA with L22, as determined by systematic evolution of ligands by exponential enrichment (SELEX) (15), consists of a stem with a GC base pair at the apical end and a loop of six to nine bases, the most 3′ of which is a U residue. An exact mirror image of this motif is present in stem-loop I of the 3′X sequence, further suggesting that this region is likely involved in binding to L22. It is possible that the 3′X region contains two L22 binding sites, as two RNA-protein complexes are observed in RNA GMSA (Fig. 3). This observation is consistent with evidence showing that EBER1 has two L22 binding sites and is capable of binding two L22 molecules simultaneously (15). Our data also demonstrate that a 3′X (antisense) riboprobe can bind L22 in UV cross-linking analysis. Secondary structure analysis of the antisense strand shows that the 3′-terminal 46 nt of the 3′X (domain I), which contains the palindromic region, is preserved in the two most stable predicted structures (Fig. 5C and D, nt 1 to 45), again suggesting that this region may be involved in binding to L22. Alternatively, strand-specific regions may be involved in 3′X binding to L22, as revealed by a comparison between EBER1 and H77c 3′UTR sequences, which showed distinct regions of similarity between the sense strand and EBER1, and the antisense strand and EBER1. Within the 3′X sense strand, nt 19 to 25 (GCCCUAG) are identical to nt 13 to 19 within EBER1, a region which lies in the first stem-loop structure (domain I, Fig. 5A). This region of EBER1 has not been shown to be involved in binding to L22 previously (61). In contrast, nt 92 to 98 (AGCCACC) within the antisense strand of 3′X are identical to a region encompassing nt 53 to 59 within EBER1, which lies in domains II and III (Fig. 5A), a region which has previously been shown to be involved in binding to L22 (61). Conceivably, these distinct regions of identity with EBER1 may be involved in binding of L22 to both 3′X sense and antisense strands. The requirement for multiple sequences or stem-loops within the 3′X region for binding to L22, particularly if more than one L22 molecule can bind, would be consistent with the EBER1-L22 interaction, which involves at least two stem-loop structures within the RNA molecule (61). Further mutational analysis is necessary to establish the exact regions within 3′X that are involved in binding to L22.
Analysis of the potential roles played by host cellular factors during HCV infection is difficult to establish since there is no efficient HCV propagation system and assays based upon HCV replication and packaging have not yet been developed. It is well-established that RNA viruses frequently subvert cellular proteins for replication and transcription of viral RNAs, and many of these factors are involved in host translation or RNA processing. To date, the only assays available to test the functional significance of HCV RNA-binding proteins are based upon translation, either in vitro or in cell culture, using monocistronic and bicistronic reporter constructs (14, 23, 28). The reporter constructs we used in our experiments contained the entire 5′UTR and the FL HCV core protein (191 amino acids). The 5′ end of core sequence is part of the IRES structure of HCV RNA (52, 53) and the 3′ end of the core coding sequence contains a PTB binding site, which together with the 3′UTR can modulate translation from the 5′ end (27). We initially tested the effect of host cellular factors in cell culture, since cell-free experiments using programmed reticulocyte lysates and recombinant protein are not always conclusive. Reticulocyte lysates translate RNAs very efficiently, and the effect of addition of exogenous protein on translation can be difficult to detect. The supplementation of recombinant protein produced in E. coli to reticulocyte lysates can be misleading if no apparent effect on translation is observed. This may be due to a lack of posttranslational modifications such as arginine methylation, or the presence of extra tag sequences which can alter the RNA-binding properties of a protein. Also, successful immunodepletion experiments, which can equivocally demonstrate the functional significance of a putative trans-acting factor, were complicated by the fact that L22 is a component of the ribosome. It would be difficult to physically deplete L22 from the extract, and depletion might have resulted in a decreased translational efficiency simply because the structure and function of the ribosome had been impaired. Additionally, depletion of the putative trans-acting factor often results in the removal of associated proteins whose loss may be responsible for the observed effect on translation (29). For translational analysis of L22 we used HuH-7 cells, which have recently been shown to support replication of a subgenomic HCV RNA (41). Experiments using three core-CAT reporter constructs (Fig. 4A) revealed that L22 and La, when supplied in trans, correlated with an upregulation of CAT activity by a factor of ∼2 (L22) or >2 (La) when compared to cells transfected with the same reporter construct and an empty vector. This upregulation of apparent IRES activity by L22 was confirmed in vitro, as a twofold increase was obtained using recombinant GST-L22 and histidine-tagged L22 (data not shown).
With regard to the interaction between L22 and the 3′X region, a number of roles for L22 are possible. L22 may be a factor directly involved in translation of HCV RNA, perhaps playing a role similar to the La protein which has been shown to bind both to the 5′UTR in the context of the initiating AUG codon (2) and also to the 3′UTR (58). L22 and La are both capable of binding EBERs in vivo, and both proteins are associated with ribosomes in the cytoplasm. La localizes with a subset of small ribosomal subunits, possibly by direct association with 18S rRNA. This is consistent with the putative role of this protein in translation regulation (48). Alternatively, L22 may be playing a regulatory role in translation (35), similar to that recently proposed for PTB (p57, hnRNP I), in which multiple binding sites within the HCV genome affect the efficiency of translation in vivo or ensure that only an FL genomic RNA is translated (27). Another interpretation of the apparent increase in HCV IRES activity in the presence of L22 and La could be an effect on RNA stability. If the half-life of the transcripts in vivo are increased in the presence of L22 or La, due to a stabilization by the bound protein within the 3′X region, greater amounts of the core-CAT fusion protein could be produced. An RNA stability effect of L22 would be consistent with the previously described role for La in stabilization of histone mRNAs (43) and for RPL3 in stabilization of human immunodeficiency virus long terminal repeat-directed RNA (51). However, our preliminary Northern blot analysis show that cells transfected with the reporter constructs used in this study produce, as expected, RNA transcripts of appropriate length both in the presence and absence of exogenously expressed L22 or La (data not shown). Furthermore, there was no evidence of instability of such transcripts in these cells, making it difficult to ascertain whether L22 (or La) plays a role in RNA stability.
Although L22 may have a role to play in translation of HCV, translation factors are not usually involved in regulation of the translation of RNA viruses since they do not bind to viral sequences directly involved in translation (34). More commonly, translation factors interact with either the RNA-dependent RNA polymerase (RdRp) or cis-acting replication signals, suggesting their roles in viral RNA synthesis. It is well-established that purified viral RNA polymerases lacking cellular factors are usually enzymatically inactive or lack template specificity (34). Cellular proteins have been shown to restore such inactivity or to provide template-specificity to the RdRp (reviewed by Lai [34]). The proteins can serve either directly as part of the replicase complex (7) or by association with RNA (3), or both (34), to direct the replicase complex to the template (3). Conceivably, any of the four RPs identified in this study as 3′X binding proteins could serve to direct the HCV replicase complex to the template or to govern template specificity of the RdRp.
Another intriguing possibility is that any of the 3′X-RP interactions may represent a mechanism by which HCV evades the effects of the interferon-inducible enzymes protein kinase R and 2′-5′ oligoadenylate synthetases. These enzymes are activated by double-stranded RNA and act in pathways that can inhibit host protein synthesis, thereby modulating cell growth and apoptosis (12, 42). Since both protein kinase R and 2′-5′ oligoadenylate synthetases play fundamental roles in the interferon-induced antiviral response, viruses have evolved multiple mechanisms to evade the shutoff of protein synthesis (reviewed by Mathews [42]). Recent evidence has demonstrated that individual RPs may also play a role in the prevention of shutoff of host protein synthesis (9, 33, 44, 54). It is clear that ribosome-associated proteins are involved in signaling pathways that affect rates of cellular protein synthesis and can thus govern processes such as regulation of cell proliferation and the cellular stress response. Although there is no direct evidence for the involvement of L22 in signaling pathways that affect cellular growth rate, the protein has recently emerged as a cellular factor that can bind both viral RNA (EBERs) (63) and viral proteins (herpes simplex virus type 1 ICP4) (39), although the significance of the interaction in each case is still not clear.
The diverse array of extraribosomal functions of individual RPs has recently become apparent (46). Of the four 3′X binding proteins identified in this study, three have proposed extraribosomal functions. S3 has been reported to play a role in processing of DNA damage and DNA repair (67); L3 binds and stabilizes human immunodeficiency virus RNA sequences (51); and L22, in addition to binding small viral RNA molecules (63), can also interact with human telomerase RNA (37) and poly(ADP-ribose) polymerase (in Drosophila melanogaster) (32).
Indeed, it has been recently suggested that the abundance of L22 in the nucleolus or associated with ribosomes and EBERs (in EBV-positive cells) places the protein in a variety of multiprotein complexes, consistent with a possible role in RNA processing and RNP assembly (37). The finding that HCV 3′X interacts with human L22, L3, S3, and mL3 adds HCV to the growing list of viruses that can interact with RPs. The significance of these interactions, however, may not be understood until efficient replication and packaging assays are developed or until the extraribosomal functions of the individual RPs within the cell are elucidated.
ACKNOWLEDGMENTS
We thank Jens Bukh for the H77c cDNA; Joan Steitz for the anti-L22 antibody; Ger Pruijn for the La cDNA; Michael Clemens for the EBER1 cDNA; and Nigel Stow, Chris Preston, and Duncan McGeoch for critical reading of the manuscript.
Stanley Fields is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′-noncoding region of the hepatitis-C virus-RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Nat Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrera I, Schuppli D, Sogo J M, Webster H. Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J Mol Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 4.Barton H A, Eisenstein R S, Bomford A, Munro H N. Determinants of the interaction between the iron-responsive element-binding protein and its binding site in rat L-ferritin mRNA. J Biol Chem. 1990;265:7000–7008. [PubMed] [Google Scholar]
- 5.Blackwell J L, Brinton M A. BHK cell proteins that bind to the 3′ stem loop structure of the West Nile Virus genome RNA. J Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight K J, Rice C M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumenthal T, Carmichael G G. RNA replication: function and structure of the Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Cassady, Gross K A M, Roizman B. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J Virol. 1998;72:8620–8626. doi: 10.1128/jvi.72.11.8620-8626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral-hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 11.Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397–2410. doi: 10.1099/0022-1317-78-10-2397. [DOI] [PubMed] [Google Scholar]
- 12.Clemens M J. PKR - a protein kinase regulated by double-stranded RNA. Int J Biochem Cell Biol. 1997;29:945–949. doi: 10.1016/s1357-2725(96)00169-0. [DOI] [PubMed] [Google Scholar]
- 13.Clemens M J. The small RNAs of Epstein-Barr-virus. Mol Biol Rep. 1993;17:81–92. doi: 10.1007/BF00996215. [DOI] [PubMed] [Google Scholar]
- 14.Collier A J, Tang S X, Elliott R M. Translation efficiencies of the 5′ untranslated region from representatives of the six major genotypes of hepatitis C virus using a novel bicistronic reporter assay system. J Gen Virol. 1998;79:2359–2366. doi: 10.1099/0022-1317-79-10-2359. [DOI] [PubMed] [Google Scholar]
- 15.Dobbelstein M, Shenk T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-Expressed RNA by using randomized and cDNA-derived RNA libraries. J Virol. 1995;69:8027–8034. doi: 10.1128/jvi.69.12.8027-8034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesises bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita Y, Okamoto T, Noshiro M, McKeehan W L, Crabb J W, Whitney R G, Kato Y, Sato J D, Takada K. A novel heparin-binding protein, Hbp15, is identified as mammalian ribosomal-protein L22. Biochem Biophys Res Commun. 1994;199:706–713. doi: 10.1006/bbrc.1994.1286. [DOI] [PubMed] [Google Scholar]
- 18.Furuya T, Lai M M C. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gontarek R R, Gutshall L L, Herold K M, Tsai J, Sathe G M, Mao J, Prescott C, DelVecchio A M. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457–1463. doi: 10.1093/nar/27.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haile D J, Hentze M W, Rouault T A, Harford J B, Klausner R D. Regulation of the interaction of the iron response element binding protein with iron-responsive elements. Mol Cell Biol. 1989;9:5055–5061. doi: 10.1128/mcb.9.11.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellen C U T, Pestova T V. Translation of hepatitis C virus RNA. J Viral Hepatitis. 1999;6:79–87. doi: 10.1046/j.1365-2893.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 23.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell G A, Lemon S M. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 24.Inoue Y, Miyazaki M, Ohashi R, Tsuji T, Fukaya K, Kouchi H, Uemura T, Mihara K, Namba M. Ubiquitous presence of cellular proteins that specifically bind to the 3′ terminal region of hepatitis C virus. Biochem Biophys Res Commun. 1998;245:198–203. doi: 10.1006/bbrc.1998.8315. [DOI] [PubMed] [Google Scholar]
- 25.Isoyama T, Kamoshita N, Yasui K, Iwai A, Shiroki K, Toyoda H, Yamada A, Takasaki Y, Nomoto A. Lower concentration of La protein required for internal ribosome entry on hepatitis C virus RNA than on poliovirus RNA. J Gen Virol. 1999;80:2319–2327. doi: 10.1099/0022-1317-80-9-2319. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito T, Lai M M C. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virol. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 28.Ito T, Tahara S M, Lai M M C. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson R, Hunt S, Reynolds J, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. In: Sarnow P, editor. Cap-independent translation. Berlin, Germany: Springer; 1995. pp. 1–25. [DOI] [PubMed] [Google Scholar]
- 30.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 32.Koyama Y, Katagiri S, Hanai S, Uchida K, Miwa M. Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal proteins, L22 and L23a, with unique histone-like amino-terminal extensions. Gene. 1999;226:339–345. doi: 10.1016/s0378-1119(98)00529-0. [DOI] [PubMed] [Google Scholar]
- 33.Kumar K U, Srivastava S P, Kaufman R J. Double-stranded RNA-activated protein kinase (PKR) is negatively regulated by 60S ribosomal subunit protein L18. Mol Cell Biol. 1999;19:1116–1125. doi: 10.1128/mcb.19.2.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 35.Laine R O, Laipis P J, Shay N F, Kilberg M S. Identification of an amino acid-regulated messenger-RNA from rat-liver as the mammalian equivalent of bacterial ribosomal protein-L22. FASEB J. 1992;6:A65. [PubMed] [Google Scholar]
- 36.Lavanchy D, Purcell R, Hollinger F B, Howard C, Alberti A, Kew M, Dusheiko G, Alter M, Ayoola E, Beutels P, Bloomer R, Ferret B, Decker R, Esteban R, Fay O, Fields H, Fuller E C, Grob P, Houghton M, Leung N, Locarnini S A, Margolis H, Meheus A, Miyamura T, Mohamed M K, Tandon B, Thomas D, Head H T, Toukan A U, VanDamme P, Zanetti A, Arthur R, Couper M, Damelio R, Emmanuel J C, Esteves K, Gavinio P, Griffiths E, Hallaj Z, Heuck C C, Heymann D L, Holck S E, Kane M, Martinez L J, Meslin F, Mochny I S, Ndikuyeze A, Padilla A M, Rodier G R M, Roure C, Savage F, Vercauteren G. Global surveillance and control of hepatitis C. J Viral Hepatitis. 1999;6:35–47. [Google Scholar]
- 37.Le S, Sternglanz R, Greider C W. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol Biol Cell. 2000;11:999–1010. doi: 10.1091/mbc.11.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leopardi R, Hukkanen V, Vainionpaa R, Salmi A A. Cell-proteins bind to Sites within the 3′ noncoding region and the positive-strand leader sequence of measles virus RNA. J Virol. 1993;67:785–790. doi: 10.1128/jvi.67.2.785-790.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leopardi R, Roizman B. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein ICP4 with EAP, a nucleolar-ribosomal protein. Proc Natl Acad Sci USA. 1996;93:4572–4576. doi: 10.1073/pnas.93.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmann V, Koch J O, Bartenschlager R. Processing pathways of the hepatitis C virus proteins. J Hepatol. 1996;24:11–19. [PubMed] [Google Scholar]
- 41.Lohmann V, Korner F, Koch J O, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 42.Mathews M B. Viruses and the protein synthesis machinery of the cell: offence, defence and dependence. In: McCrae M A, Saunders J R, Smyth C J, Stow N D, editors. Molecular aspects of host-pathogen interactions. Vol. 55. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 211–235. [Google Scholar]
- 43.McLaren R S, Caruccio N, Ross J. Human La protein: a stabilizer of histone mRNA. Mol Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulvey M, Poppers J, Ladd A, Mohr I. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J Virol. 1999;73:3375–3385. doi: 10.1128/jvi.73.4.3375-3385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakabayshi H, Taketa K, Miyano K, Yamane T, Sato J. Cloning of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 46.Naora H. Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol. 1999;77:197–205. doi: 10.1046/j.1440-1711.1999.00816.x. [DOI] [PubMed] [Google Scholar]
- 47.Ou J, Yen T, Wang Y, Kam W, Rutter W. Cloning and characterization of a human ribosomal protein gene with enhanced expression in fetal and neoplastic cells. Nucleic Acids Res. 1987;15:8919–8934. doi: 10.1093/nar/15.21.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peek R, Pruijn G J M, Van Venrooij W J. Interaction of the La (SSB) autoantigen with small ribosomal subunits. Eur J Biochem. 1996;236:649–655. doi: 10.1111/j.1432-1033.1996.0649d.x. [DOI] [PubMed] [Google Scholar]
- 49.Petrik J, Parker H, Alexander G J M. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly (U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J Gen Virol. 1999;80:3109–3113. doi: 10.1099/0022-1317-80-12-3109. [DOI] [PubMed] [Google Scholar]
- 50.Purcell R. The hepatitis C virus: overview. Hepatology. 1997;26:S11–S14. doi: 10.1002/hep.510260702. [DOI] [PubMed] [Google Scholar]
- 51.Reddy T R, Suhasini M, Rappaprt J, Looney D J, Kraus G, Wong-Staal F. Molecular cloning and characterization of a TAR-binding nuclear factor from T cells. AIDS Res Hum Retrovir. 1995;11:663–669. doi: 10.1089/aid.1995.11.663. [DOI] [PubMed] [Google Scholar]
- 52.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis-C virus-RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rijnbrand R, Bredenbeek P, Vanderstraaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis-c virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 54.Roller R J, Monk L L, Stuart D, Roizman B. Structure and function in the herpes simplex virus 1 RNA-binding protein US11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J Virol. 1996;70:2842–2851. doi: 10.1128/jvi.70.5.2842-2851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 56.Sengupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spangberg K, GoobarLarsson L, WahrenHerlenius M, Schwartz S. The La protein from human liver cells interacts specifically with the u-rich region in the hepatitis C virus 3′ untranslated region. J Hum Virol. 1999;2:296–307. [PubMed] [Google Scholar]
- 59.Tanaka T, Kato N, Cho M J, Shimotohno K. A novel sequence found at the 3′-terminus of hepatitis-C virus genome. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka T, Kato N, Cho M J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toczyski D P, Matera A G, Ward D C, Steitz J A. The Epstein-Barr-virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein-L22 in EBV-infected human B-lymphocytes. Proc Natl Acad Sci USA. 1994;91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toczyski D P, Steitz J A. The cellular RNA-binding protein EAP recognizes a conserved stem-loop in the Epstein-Barr-virus small RNA EBER-1. Mol Cell Biol. 1993;13:703–710. doi: 10.1128/mcb.13.1.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toczyski D P W, Steitz J A. EAP, a highly conserved cellular protein associated with Epstein-Barr-virus small RNAs (EBERs) EMBO J. 1991;10:459–466. doi: 10.1002/j.1460-2075.1991.tb07968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Unge J, Aberg A, AlKharadaghi S, Nikulin A, Nikonov S, Davydova N L, Nevskaya N, Garber M, Liljas A. The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance. Structure. 1998;6:1577–1586. doi: 10.1016/s0969-2126(98)00155-5. [DOI] [PubMed] [Google Scholar]
- 67.Wilson D M, 3rd, Deutsch W A, Kelley M R. Drosophila ribosomal protein S3 contains an activity that cleaves DNA at apurinic/apyrimidinic sites. J Biol Chem. 1994;269:25359–25364. [PubMed] [Google Scholar]
- 68.Yanagi M, Purcell R H, Emerson S U, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yanagi M, StClaire M, Emerson S U, Purcell R H, Bukh J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci USA. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanagi M, StClaire M, Shapiro M, Emerson S U, Purcell R H, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 71.Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M. A yeast three-hybrid system to detect and analyze interactions between RNA and protein. In: Bartel P L, Fields S, editors. The yeast two-hybrid system. New York, N.Y: Oxford University Press; 1997. [Google Scholar]
- 72.Zhang B L, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens M P. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]