Abstract
We have developed a sensitized screen to identify genes involved in gene silencing, using random N-ethyl-N-nitrosourea mutagenesis on mice carrying a variegating GFP transgene. The dominant screen has produced six mutant lines, including both suppressors and enhancers of variegation. All are semidominant and five of the six are homozygous embryonic lethal. In one case, the homozygous lethality depends on sex: homozygous females die at midgestation and display abnormal DNA methylation of the X chromosome, whereas homozygous males are viable. Linkage analysis reveals that the mutations map to unique chromosomal locations. We have studied the effect of five of the mutations on expression of an endogenous allele known to be sensitive to epigenetic state, agouti viable yellow. In all cases, there is an effect on penetrance, and in most cases, parent of origin and sex-specific effects are detected. This screen has identified genes that are involved in epigenetic reprogramming of the genome, and the behavior of the mutant lines suggests a common mechanism between X inactivation and transgene and retrotransposon silencing. Our findings raise the possibility that the presence or absence of the X chromosome in mammals affects the establishment of the epigenetic state at autosomal loci by acting as a sink for proteins involved in gene silencing. The study demonstrates the power of sensitized screens in the mouse not only for the discovery of novel genes involved in a particular process but also for the elucidation of the biology of that process.
Keywords: epigenetics, mutagenesis, X inactivation
Despite the fact that the epigenetic state of the eukaryotic genome has profound effects on ultimate phenotype, little is known about the mechanisms by which these states are established. In mammals, genomewide epigenetic reprogramming occurs during both gametogenesis and early embryonic development (1). For technical reasons, mainly related to the challenges of manipulating such small starting material, it has been difficult to study these events. We have designed a screen to detect genes involved in establishing and maintaining the epigenetic state of the genome in the mouse by screening for mutations that affect variegated gene expression in the adult. Similar screens have been undertaken in Drosophila melanogaster (2) and maize (3). Although two screens have been carried out in the mouse to identify genes involved specifically in X inactivation and parental imprinting (4, 5), our screen is designed to find genes that play a fundamental role in epigenetic modifications by assessing variegated gene expression.
Some genes in the adult animal appear to be more sensitive to epigenetic state than others, and at these loci the establishment of epigenetic state is to some degree stochastic. The Drosophila screens, described above, used flies that displayed variegated expression of the w locus to identify genes involved in position effect variegation (PEV), and these genes turn out, in many cases, to play a critical role in epigenetic gene silencing (6–8). HP1 and histone H3 lysine 9 methyltransferase [SU(VAR)3-9] are two notable examples (9). In the mouse, multicopy transgene arrays are particularly sensitive to gene silencing as evidenced by the fact that they often display variegation (10). We have used a murine transgenic line that expresses GFP in a variegated manner (11) in a random mutagenesis screen to identify unique genes involved in the establishment and maintenance of epigenetic state. The advantages of using this transgenic line are that changes in its expression will not alter the viability of the mouse, the measurement of the phenotype is carried out by flow cytometry, which is fast and extremely accurate (helpful when screening thousands of mice), and it is unlikely that we will pick up mutations allelic to the transgene itself, because point mutations in a multicopy transgene array would have a negligible effect on overall output.
In mice, stochastic gene silencing associated with variegation is not limited to transgenes. There are some endogenous alleles, metastable epialleles, that behave in this way. Their activity is sensitive to epigenetic state, and this state is to some degree unstable both within and between generations (12). The best characterized metastable epiallele is agouti viable yellow (Avy). The agouti gene encodes a protein that causes the production of yellow pigment in the hair. Expression of the WT agouti gene is under the control of promoters, which are active at only one stage in the hair growth cycle, resulting in a yellow band on a black hair (13). At Avy, a retrotransposon has integrated upstream of the agouti promoter and drives constitutive expression of Agouti, resulting in a completely yellow coat, along with late-onset obesity and diabetes (14). Expression from this retrotransposon can be epigenetically silenced in some cells, producing agouti-colored patches (mottled) or even a completely agouti-colored mouse (called pseudoagouti). So mice within a litter of isogenic individuals can have different phenotypes, termed variable expressivity. Given that a mottled mouse appears similar to a tabby mouse, where patches are produced because of random X inactivation (15), it is likely that the epigenetic silencing at Avy occurs at the same stage as X inactivation, around embryonic day 6.5. The silencing correlates with cytosine methylation at the Avy allele, but the mechanism by which the retrotransposon is stochastically silenced is unknown (16). At metastable epialleles, the range of phenotypes within a litter of isogenic animals is also influenced by the parental origin of the allele, e.g., maternal inheritance of Avy produces a higher proportion of yellow offspring than paternal inheritance (17, 18). In some genetic backgrounds the phenotype of the parent influences the phenotype of the offspring, termed epigenetic inheritance (16, 18). These phenomena are poorly understood but clearly involve epigenetic reprogramming during both gametogenesis and early development. The retrotransposon insertion alleles Avy and Aiapy both have been shown to be sensitive to haplo-insufficiency for an epigenetic modifier, DNA methyltransferase 1 (S. Chong, personal communication) (19), consistent with the fact that many modifiers of retrotransposon insertional alleles in Drosophila display dosage-dependent effects on PEV (20, 21).
We have identified six mutations in a dominant screen for transgene variegation. These genes must play a critical role in development because, in most cases, homozygosity is associated with embryonic lethality. We have introduced one copy of each of our mutations into a line carrying the Avy allele. In all cases tested, they alter penetrance at Avy. In addition, we have found that the mutations display interesting parent of origin and sex-specific effects on expression of the Avy allele.
Materials and Methods
Generation and Screening of Mutant Mice. The transgenic mouse line GFP1 carries a transgene array of ≈11 copies of a construct in which the human α-globin promoter and enhancer elements are linked to the GFP-coding sequence (11). The transgenic line was produced (and has been maintained) in the inbred FVB/NJ strain. The transgene is expressed in 55% of erythrocytes in 3-week-old offspring. This finding is highly reproducible among isogenic mice over many generations (11). Twelve homozygous GFP1 mice were treated with one dose of 100 mg/kg N-ethyl-N-nitrosourea (ENU) (22). The mice recovered fertility between 15 and 30 weeks later and were mated with homozygous GFP1 females. The offspring were screened at 3 weeks of age for changes in GFP expression by flow cytometry. Mice with alterations in expression were bred further to test for heritability. When heritability was confirmed, the mutant was named.
Flow Cytometry. Mice were analyzed by flow cytometry at 3 weeks of age. A drop of blood was collected in Osmosol buffer and analyzed on a FACSCalibur (Becton Dickinson), exciting at 488 and 550 nm. The 488-nm channel predominantly measures GFP fluorescence, and the 550-nm channel measures the autofluorescence of the cells. The data were analyzed by using cell quest software with a GFP-positive gate set to exclude 99.9% of WT erythrocytes. Mean fluorescence was calculated by using cells within the positive gate. Histograms shown depict only the GFP fluorescence channel.
Introduction of the Mutant Lines into Mice Carrying the Avy Allele. FVB/NJ mice heterozygous for the mutation (at least two generations down from the founder), and homozygous for GFP1, were mated with C57BL/6J mice heterozygous for the Avy allele. The coat color phenotype was classified at weaning by a trained observer as either yellow, mottled, or agouti. This observer had no knowledge of the genotype (with respect to the ENU mutation) of the individuals. Subsequently, GFP expression was determined by flow cytometry. FVB/NJ mice carry the A locus and C57BL/6J mice carry the a locus. Yellow and mottled mice carried the Avy allele. All agouti-colored mice were genotyped by PCR, to assess whether they were Avy/A and pseudoagouti, or A/a, as reported (18).
Linkage Analysis. Linkage analysis was performed on tail-tip DNA samples, prepared as described (16). Heterozygous mutant FVB/NJ mice at least two generations down from the founder were crossed into C57BL/6J. The mutant offspring from this mating were selected and crossed with C57BL/6J. We have mapped the transgene to chromosome 1 (data not shown). Initially, we used 40 microsatellite markers that differ in size between FVB/NJ and C57BL/6J, spaced evenly throughout the genome and excluding chromosome 1 (23). The PCR products were separated by gel electrophoresis, and the data were analyzed by interval haplotype analysis (24). When a linked chromosome was found, additional markers and mice were used to reduce the linked interval. WT littermates were used to confirm that there was no linkage to this region in these animals. A minimum of 80 mutant G2 progeny was used for mapping each dominant mutation.
Hprt Bisulfite Sequencing. Bisulfite conversion of DNA was performed as described (18), except that bisulfite treatment was for 40 h. The bisulfite oligonucleotides and cycling conditions were as described (25).
Sex Determination of Embryos. The presence of the Sry gene was detected by PCR, as described (26), with a modification to the PCR parameters: 94°C 2 min for 1 cycle; 94°C 30 s, 58°C 30 s, 72°C 30 s for 35 cycles; and 72°C 5 min for one cycle.
Results
Dominant Screen for Modifiers of GFP Transgene Variegation. We treated 12 FVB/NJ males, homozygous for a GFP transgene, with ENU (22). The GFP transgene in this inbred colony is under the control of an α-globin promoter and enhancer, which directs expression to erythrocytes (11). This transgene is expressed in a variegated manner; 55% of the erythrocytes express GFP when the mouse is homozygous for the transgene (11). The percentage of cells expressing GFP and the mean fluorescence of expressing cells is highly reproducible within this line. The line was chosen as it provided an opportunity to detect both suppressors and enhancers of variegation, and expression was known to be stable over many generations (11). The ENU-treated males were used in a dominant screen for mutations that altered either the percentage of GFP-expressing erythrocytes or the mean fluorescence of the expressing cells. We have named the mutations Modifiers of Murine Metastable Epialleles (Mommes). We screened 608 G1 offspring at 3 weeks of age and identified six dominant mutations, Momme D1–D6. The mutations arose in five different mutagenized males, and only Momme D1 and D6 were produced by the same male. These mutations produce distinct expression profiles (see Fig. 1 and Table 1). Each of these mutations have highly significantly different GFP expression profiles from their WT littermates, with respect to both percentage of GFP-expressing cells and mean fluorescence. The heritability of each mutation has been tested and confirmed over at least three generations by using at least 50 litters in each case (see Table 1 for quantitative analysis). Interestingly, Momme D6 has little effect on the percentage of expressing cells, but a large effect on mean fluorescence, possibly reflecting a role for the WT protein in stable, rather than variegating, position effects. Breeding studies confirmed that none of the mutations are allelic to the transgene itself (data not shown). The mutations include both suppressors and enhancers of variegation, because some shift the GFP fluorescence curve to the right (Momme D1, D2, D3, and D6), and some shift the curve to the left (Momme D4 and D5) (Fig. 1).
Fig. 1.
GFP expression profiles in Momme Ds. Erythrocytes from 3-week-old mice from six different mutant lines were analyzed by flow cytometry. In each case, the expression profile of phenotypically mutant offspring in a litter were averaged (red) and overlaid with the average of the WT littermates (black). The x axis represents the erythrocyte fluorescence on a logarithmic scale, and the y axis is the number of cells detected at each fluorescence level. The charts are representative of what occurs in at least 50 litters tested for each mutant line, but because of day-to-day variation in the flow cytometer readings, litters from different days cannot be pooled. For quantitative analysis and statistical significance see Table 1. The GFP-positive gate is indicated as GFP+.
Table 1. Quantitative analysis of Momme D expression in one representative litter.
| Mutant
|
WT
|
|||||
|---|---|---|---|---|---|---|
| Mutant | % Expressing cells | Mean fluorescence | n | % Expressing cells | Mean fluorescence | n |
| Momme D1 | 82 ± 3 | 464 ± 62 | 4 | 56 ± 4 | 325 ± 31 | 4 |
| Momme D2 | 76 ± 3 | 486 ± 44 | 4 | 53 ± 3 | 286 ± 18 | 4 |
| Momme D3 | 70 ± 2 | 507 ± 22 | 4 | 54 ± 2 | 302 ± 16 | 4 |
| Momme D4 | 43 ± 1 | 303 ± 8 | 4 | 60 ± 2 | 346 ± 17 | 4 |
| Momme D5 | 50 ± 1 | 223 ± 13 | 4 | 60 ± 4 | 298 ± 30 | 3 |
| Momme D6 | 66 ± 1 | 633 ± 24 | 4 | 57 ± 2 | 359 ± 25 | 4 |
Analysis of data presented in Fig. 1. The percentage of expressing cells was determined by using a GFP+ gate, which was set to exclude 99.9% of WT cells. In each case, data from one representative litter (of at least 50 analyzed) are shown, because day-to-day variation in the flow cytometer readings means litters from different days cannot be pooled. Each mutant line has a significantly different expression profile to WT littermates, reproducible over many generations.
Momme Ds Are Semidominant and Most Are Homozygous Lethal. Heterozygous intercrosses of each Momme D revealed that the proportion of expression types observed in the offspring was not that expected for dominant mutations (Table 2). For classic dominant mutations, three-quarters of the litter will have a mutant phenotype, and one-quarter will be WT. In most cases, i.e., Momme D2–D6, we found two phenotypic classes in a ratio consistent with semidominance combined with homozygous lethality; i.e., two-thirds of the litter display the mutant phenotype and one-third display the WT phenotype (Table 2). Momme D1 was the exception. It produced three distinct GFP expression profiles (see Fig. 2a), indicating that the mutation is semidominant and homozygous viable. There is a reduced proportion of homozygotes (Table 2), caused by lethality in females. Of the 13 homozygous offspring identified at weaning, all were male (P < 0.0005). The male homozygotes that survive to weaning are fertile and appear normal. Female homozygotes die in utero, between embryonic days 10.5 and 14.5 (data not shown).
Table 2. Phenotypes of offspring from heterozygous intercrosses.
| Observed | Expected if semidominant | Expected if dominant | |
|---|---|---|---|
| Momme D1 | |||
| Homozygous* | 13 | 43.75 | |
| Heterozygous* | 115 | 87.5 | |
| WT | 47 | 43.75 | |
| Momme D2 | |||
| Mutant* | 90 | 99.75 | |
| WT | 43 | 33.25 | |
| Momme D3 | |||
| Mutant* | 80 | 94.5 | |
| WT | 46 | 31.5 | |
| Momme D4 | |||
| Mutant | 119 | 138 | |
| WT | 65 | 46 | |
| Momme D5 | |||
| Mutant | 110 | 141 | |
| WT | 78 | 47 | |
| Momme D6 | |||
| Mutant | 55 | 66 | |
| WT | 33 | 22 |
P values, Momme D1, <0.005; Momme D2, 0.05; Momme D3, <0.005; Momme D4, <0.005; Momme D5, <0.0001; Momme D6, <0.01.
Progeny tested: 3 for Momme D1-/-, 18 for Momme D1-/+, 9 for Momme D2-/+, and 10 for Momme D3-/+.
Fig. 2.
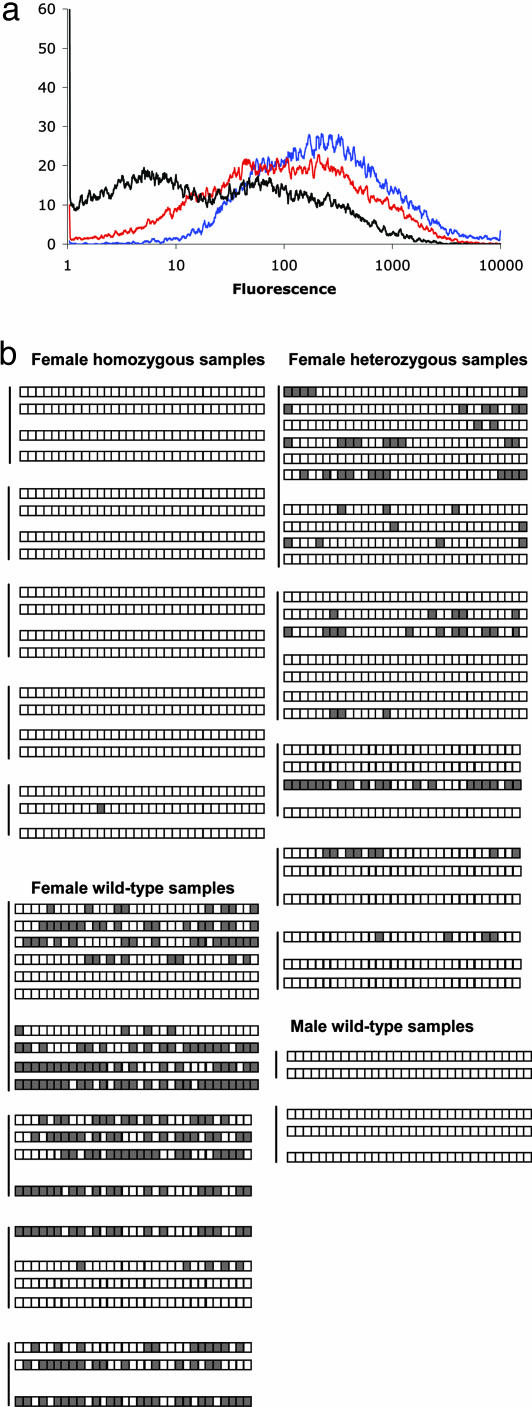
Momme D1 phenotypes. (a) Erythrocytes from 3-week-old mice from a Momme D1 heterozygous intercross were analyzed. The expression profile of two homozygote mutant offspring were averaged (blue) and overlaid with the average of the three heterozygote (red) and two WT littermates (black). The homozygotes are significantly different from the heterozygotes (P < 0.01). The x axis represents the erythrocyte fluorescence on a logarithmic scale, and the y axis is the number of cells detected at each fluorescence level. The charts are representative of the 29 litters tested, but because of day-to-day variation in the flow cytometer readings, litters from different days cannot be pooled. (b) Embryonic day 10.5 embryos derived from an FVB/C57 F1 heterozygous intercross were genotyped and sexed. Samples were bisulfite-converted, and the hprt promoter region was amplified. Clones from five homozygous, five heterozygous, and five WT females and from two WT males are shown. A gray box represents a methylated CpG, and a white box indicates unmethylated CpG. The homozygous females completely lack methylation at this locus. Where there is a separation between clones from the same embryo, clones were derived from different bisulfite conversions.
The classification of Momme D2–D6 as semidominant, homozygous lethal is supported by a significant reduction in litter size at weaning for each heterozygous intercross (see Fig. 4, which is published as supporting information on the PNAS web site). The fact that most of the dominant mutations are semidominant is consistent with the general finding in Drosophila that epigenetic processes are dose-dependent (2, 6, 7, 27).
MOMME D1 Plays a Role in Methylation of the X Chromosome. We performed embryonic dissections on FVB/C57 F1 Momme D1–/+ dams, mated to FVB/C57 F1 Momme D1–/+ sires, and genotyped the offspring by using microsatellite markers surrounding the linked interval on chromosome 17. Homozygous females appear macroscopically normal at embryonic day 10.5, yet show a complete lack of DNA methylation at the CpG island of the X-linked gene hprt (Fig. 2b). WT female embryos show both hypermethylated (presumably representing the inactive X chromosome) and hypomethylated (presumably representing the active X chromosome) clones. The locus in male embryos is unmethylated, as expected. Interestingly, whereas heterozygous females have both methylated and unmethylated clones, the methylated clones are less methylated than those seen in the WT samples. The WT samples show an average of 38% methylation, whereas heterozygous female embryos show an average of 11% methylation. Momme D1 appears to have a semidominant effect on cytosine methylation on the X chromosome, similar to its semidominant effect on transgene expression, and viability.
Momme Ds Affect the Coat Color Phenotype Associated with the Avy Allele. The Momme D1–D5 heterozygotes were mated with yellow or pseudoagouti Avy/a C57BL/6J mice, and the offspring were scored, at weaning, for the Avy coat color phenotype. Subsequently, the GFP expression profile was used to identify mutants from nonmutants (data not shown). Reciprocal matings were carried out because penetrance at metastable epialleles is affected by parent of origin (16, 18).
Momme D1 increases the proportion of yellow offspring when a Momme D1 sire is mated with a pseudoagouti dam (see Fig. 3a). This finding is consistent with Momme D1 increasing expression of the transgene (see Fig. 1). Furthermore, when the offspring from this type of mating are separated according to sex the shift toward yellow coat color (and thus the active allele) is limited to the female offspring. This finding is interesting in light of the fact that Momme D1 homozygosity in the inbred strain is associated with lethality of females. As expected, the shift in penetrance reflects a shift in methylation of the intracisternal A particle at the Avy allele (see Fig. 5, which is published as supporting information on the PNAS web site). These results suggest that the MOMME D1 protein is involved in X inactivation and in silencing of transgenes and retrotransposons. The inactive X chromosome may act as a sink for repressor proteins such as MOMME D1, and in situations where MOMME D1 is absent, disruption of X inactivation may result in female lethality. When there is a reduction in MOMME D1, as occurs in heterozygous females, presumably X inactivation does occur, but there is less MOMME D1 available for retrotransposon silencing (which we know occurs at approximately the same stage in development, see Introduction), leading to a shift to yellow at the Avy allele.
Fig. 3.
Momme D1, D2, and D4 shift penetrance at the Avy allele. Avy/a C57BL/6J mice of the indicated phenotype were mated with Momme D–/+ FVB/NJ mice. Data were produced from at least five different mating pairs. Offspring not carrying the Avy allele have been omitted from the pedigrees. (a) Paternal inheritance of Momme D1. Pedigrees from Avy/a dams mated with Momme D1–/+ FVB/NJ sires. The Momme D1–/+ mutants shift toward yellow when the Avy allele is inherited from a pseudoagouti dam (P < 0.0001). The data shown have been reanalyzed according to the sex of the offspring. There is no significant shift in the proportion of phenotypes observed between mutant and WT male littermates. In the female offspring, there is a significant shift toward yellow, removing the mottled class entirely (P < 0.0001). (b) Maternal inheritance of Momme D2. Pseudoagouti Avy/a C57BL/6J sires were mated with Momme D2–/+ FVB/NJ dams. When the data were analyzed according to the sex of the offspring, there was a significant shift in the mutant female offspring toward a yellow phenotype (P = 0.01). There is no shift in the proportion of phenotypes observed between mutant and WT male littermates. (c) Paternal inheritance of Momme D4. Yellow Avy/a C57BL/6J dams were mated with Momme D4–/+ FVB/NJ sires. There was a significant shift toward the mottled phenotype in both male and female offspring.
Momme D1 does not appear to affect coat color phenotypes after maternal transmission of the Momme D1 allele and paternal transmission of the Avy allele (data not shown). Momme D1 expression does not appear to be imprinted because its effect on transgene expression is independent of parent of origin (data not shown). So, this parent-of-origin effect may turn out to have more to do with the parent-of-origin effect always observed at the Avy allele (16, 18).
Momme D2 also affects expression at the Avy allele in complex ways. Maternal transmission of Momme D2 results in an increase in the proportion of yellow offspring and, again, only in females (see Fig. 3b). This finding is consistent with its role as a suppressor of variegation at the transgene. Although Momme D2 homozygosity is embryonic lethal in both males and females, approximately half the heterozygous females from a heterozygous intercross also do not survive to birth; of 90 heterozygous mutants, only 35 are female. Taken together, these results suggest that it may also be involved in X inactivation. Paternal transmission of Momme D2 has no effect on a maternally transmitted Avy allele (data not shown). As found for Momme D1, the effect of Momme D2 on transgene expression is independent of its parental origin (data not shown), and so it seems unlikely that Momme D2 is itself imprinted.
Momme D4 is an enhancer of variegation with respect to transgene expression, and we were interested to test its effect on the Avy allele. When a Momme D4 sire is mated to a yellow dam the offspring that inherit the Momme D4 mutant allele are more likely to be mottled. Momme D4 is shifting the penetrance toward the mottled, and therefore inactive state (Fig. 3c), consistent with it being an enhancer of variegation. In this case the effect is seen in both male and female offspring. Preliminary studies show that Momme D3 and D5 also influence penetrance at Avy in both parent-of-origin and sex-of-offspring specific ways (data not shown).
Linkage Analysis. We have mapped the location of each mutation, and for Momme D1-5 we have narrowed the interval to between 1.6 and 6 Mb (Table 3). During the breeding for the linkage analysis for each mutant, the expression profile remained constant, supporting the assumption that we are following a single mutation in each case. In most cases, the linked interval still contains too many genes to proceed to candidate gene analysis.
Table 3. Summary of Momme D screen for epigenetic modifiers.
| Mutant | Suppressor/enhancer | Effect on Avy expression? | Mapping data, Ensembl Build 33 base pair location |
|---|---|---|---|
| Momme D1 | Suppressor | Yes | Chr 17, 68,514,079 and 70,925,638 bp |
| Momme D2 | Suppressor | Yes | Chr 9, 26,951,956 and 29,544,890 bp |
| Momme D3 | Suppressor | Yes | Chr 11, 70,565,924 and 76,549,381 bp |
| Momme D4 | Enhancer | Yes | Chr 8, 79,018,677 and 81,467,429 bp |
| Momme D5 | Enhancer | Yes | Chr 4, 124,134,224 and 125,758,794 bp |
| Momme D6 | Suppressor | Not determined | Chr 14 |
Chr, chromosome.
Discussion
Using a sensitized screen for modifiers of transgene variegation in the mouse, we have identified a number of mutations that affect epigenetic reprogramming during gametogenesis and early development. Similar screens for modifiers of PEV in Drosophila identified ≈150 genes including both suppressor [Su(var)] and enhancer [E(var)] mutations (2). Of these, relatively few have been characterized. Although it is difficult to anticipate the frequency with which one will recover functionally important mutations from an ENU screen, previous reports estimate a frequency of ≈1 in 700 (28). Based on the fact that the Drosophila screens found 150 modifiers (2), we anticipated a mutation in ≈1 in 15 of the G1 offspring. Dominant mutations are known to occur far less frequently than recessive ones, so the frequency with which we found dominant mutations (1 in 100) seems reasonable. Interestingly, many of the mutations in the Drosophila screens were found to be haplo-dependent (2, 6, 7, 9, 27). Haplo-dependent effects indicate that the encoded gene product displays a concentration-dependent function. This result is similar to our finding that the Momme Ds are semidominant. The complexes that perform the processes of gene silencing are believed to be in a dynamic equilibrium that is sensitive to small changes in the amount of each protein partner (29).
The fact that the Momme Ds, identified because they play a role in transgene variegation, all affect penetrance at Avy suggests that many of the proteins involved in transgene silencing are the same as those involved in the silencing of retrotransposons. This finding is reminiscent of the fact that many dominant modifiers of retrotransposon insertional alleles in Drosophila display a dominant modifier effect on PEV (20, 21). Variegated expression at transgenes is generally associated with multicopy arrays. Although repeat-induced silencing of this type has been reported in a broad range of eukaryotic organisms, it remains poorly understood (30–32). Recent findings suggest RNA interference is likely to be involved (33).
We have uncovered female-specific effects that suggest that the X chromosome is acting as a sink for proteins involved in epigenetic silencing. It became apparent in the studies on PEV in Drosophila that the degree of variegation could be shifted by the presence of the Y chromosome (34). The Y chromosome results in suppression of variegation, which is thought to be because it acts as a sink for heterochromatin factors. In the case of Momme D1, homozygous female embryos die at midgestation and display abnormal patterns of cytosine methylation on the X chromosome. Therefore, at least in this case, it seems that the MOMME D1 protein is involved in X chromosome inactivation, and so it is reasonable to suggest that the inactive X chromosome may act as a sink for MOMME D1 in the heterozygous females.
The results reported here support the idea that at least some of the proteins involved in transgene and retrotransposon silencing are also involved in X inactivation, and they suggest that the presence of the X chromosome can influence transcriptional silencing at autosomal loci in mammals. It has been known for some time that early female embryos are developmentally retarded in mammals (including cows, humans, mice, and rats) compared with their male counterparts (35, 36). This difference appears before sexual differentiation of the gonads and so cannot be explained by hormonal differences relating to sex. Our studies raise the possibility that the entire X chromosome could be involved, by sequestering proteins that are involved in controlling gene expression at autosomal loci. An extension of this idea is that epigenetic differences may play a role in phenotypic differences between the sexes in mammals. We await the identification of the genes underlying the Momme D mutations with interest. These mutant lines should provide a valuable resource for those working in the field of epigenetics. The study demonstrates the power of sensitized screens not only for the discovery of novel genes involved in a particular process but also for the elucidation of the biology of that process.
Supplementary Material
Acknowledgments
We thank Michelle Holland for technical assistance and The National Health and Medical Research Council of Australia for Project Grant 301955. M.E.B., A.A., and T.J.B. were supported by Australian Postgraduate Awards. N.K.V. and J.I.P. were supported by International Postgraduate Awards.
Author contributions: M.E.B., N.K.V., and E.W. designed research; M.E.B., N.K.V., S.J.H., A.A., and T.J.B. performed research; J.I.P. and R.A. contributed new reagents/analytic tools; M.E.B., N.K.V., S.J.H., A.A., and T.J.B. analyzed data; and M.E.B. and E.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ENU, N-ethyl-N-nitrosourea; Avy, agouti viable yellow; Momme, Modifier of Murine Metastable Epiallele; PEV, position effect variegation.
References
- 1.Reik, W., Dean, W. & Walter, J. (2001) Science 293, 1089–1093. [DOI] [PubMed] [Google Scholar]
- 2.Schotta, G., Ebert, A., Dorn, R. & Reuter, G. (2003) Semin. Cell Dev. Biol. 14, 67–75. [DOI] [PubMed] [Google Scholar]
- 3.Hollick, J. B. & Chandler, V. L. (2001) Genetics 157, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Percec, I., Plenge, R. M., Nadeau, J. H., Bartolomei, M. S. & Willard, H. F. (2002) Science 296, 1136–1139. [DOI] [PubMed] [Google Scholar]
- 5.Tsai, T. F., Chen, K. S., Weber, J. S., Justice, M. J. & Beaudet, A. L. (2002) Hum. Mol. Genet. 11, 1659–1668. [DOI] [PubMed] [Google Scholar]
- 6.Spofford, J. B. (1961) Genetics 46, 1151–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henikoff, S. (1990) Trends Genet. 6, 422–426. [DOI] [PubMed] [Google Scholar]
- 8.Dorn, R., Krauss, V., Reuter, G. & Saumweber, H. (1993) Proc. Natl. Acad. Sci. USA 90, 11376–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuter, G. & Spierer, P. (1992) BioEssays 14, 605–612. [DOI] [PubMed] [Google Scholar]
- 10.Martin, D. I. & Whitelaw, E. (1996) BioEssays 18, 919–923. [DOI] [PubMed] [Google Scholar]
- 11.Preis, J. I., Downes, M., Oates, N. A., Rasko, J. E. & Whitelaw, E. (2003) Curr. Biol. 13, 955–959. [DOI] [PubMed] [Google Scholar]
- 12.Rakyan, V. K., Blewitt, M. E., Druker, R., Preis, J. I. & Whitelaw, E. (2002) Trends Genet. 18, 348–351. [DOI] [PubMed] [Google Scholar]
- 13.Bultman, S. J., Michaud, E. J. & Woychik, R. P. (1992) Cell 71, 1195–1204. [DOI] [PubMed] [Google Scholar]
- 14.Duhl, D. M., Vrieling, H., Miller, K. A., Wolff, G. L. & Barsh, G. S. (1994) Nat. Genet. 8, 59–65. [DOI] [PubMed] [Google Scholar]
- 15.Lyon, M. F., Rastan, S. & Brown, S. D. M. (1996) Genetic Variants and Strains of the Laboratory Mouse (Oxford Univ. Press, Oxford).
- 16.Morgan, H. D., Sutherland, H. G., Martin, D. I. & Whitelaw, E. (1999) Nat. Genet. 23, 314–318. [DOI] [PubMed] [Google Scholar]
- 17.Wolff, G. L. (1978) Genetics 88, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakyan, V. K., Chong, S., Champ, M. E., Cuthbert, P. C., Morgan, H. D., Luu, K. V. & Whitelaw, E. (2003) Proc. Natl. Acad. Sci. USA 100, 2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudet, F., Rideout, W. M., 3rd, Meissner, A., Dausman, J., Leonhardt, H. & Jaenisch, R. (2004) Mol. Cell. Biol. 24, 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birchler, J. A., Bhadra, U., Rabinow, L., Linsk, R. & Nguyen-Huynh, A. T. (1994) Genetics 137, 1057–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csink, A. K., Linsk, R. & Birchler, J. A. (1994) Genetics 138, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis, A. P., Woychik, R. P. & Justice, M. J. (1999) Mamm. Genome 10, 308–310. [DOI] [PubMed] [Google Scholar]
- 23.Neuhaus, I. M., Sommardahl, C. S., Johnson, D. K. & Beier, D. R. (1997) Mamm. Genome 8, 506–509. [DOI] [PubMed] [Google Scholar]
- 24.Neuhaus, I. M. & Beier, D. R. (1998) Mamm. Genome 9, 150–154. [DOI] [PubMed] [Google Scholar]
- 25.Park, J. G. & Chapman, V. M. (1994) Mol. Cell. Biol. 14, 7975–7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClive, P. J. & Sinclair, A. H. (2001) Mol. Reprod. Dev. 60, 225–226. [DOI] [PubMed] [Google Scholar]
- 27.Reuter, G., Giarre, M., Farah, J., Gausz, J., Spierer, A. & Spierer, P. (1990) Nature 344, 219–223. [DOI] [PubMed] [Google Scholar]
- 28.Hitotsumachi, S., Carpenter, D. A. & Russell, W. L. (1985) Proc. Natl. Acad. Sci. USA 82, 6619–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locke, J., Kotarski, M. A. & Tartof, K. D. (1988) Genetics 120, 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrick, D., Fiering, S., Martin, D. I. & Whitelaw, E. (1998) Nat. Genet. 18, 56–59. [DOI] [PubMed] [Google Scholar]
- 31.Linn, F., Heidmann, I., Saedler, H. & Meyer, P. (1990) Mol. Gen. Genet. 222, 329–336. [DOI] [PubMed] [Google Scholar]
- 32.Dorer, D. R. & Henikoff, S. (1994) Cell 77, 993–1002. [DOI] [PubMed] [Google Scholar]
- 33.Pal-Bhadra, M., Leibovitch, B. A., Gandhi, S. G., Rao, M., Bhadra, U., Birchler, J. A. & Elgin, S. C. (2004) Science 303, 669–672. [DOI] [PubMed] [Google Scholar]
- 34.Spofford, J. B. (1976) in The Genetics and Biology of Drosophila, ed. Ashburner, M. (Academic, London), Vol. 1c, pp. 955–1018. [Google Scholar]
- 35.Burgoyne, P. S., Thornhill, A. R., Boudrean, S. K., Darling, S. M., Bishop, C. E. & Evans, E. P. (1995) Philos. Trans. R. Soc. London B Sci. 350, 253–260 and discussion 260–261. [DOI] [PubMed] [Google Scholar]
- 36.Ray, P. F., Conaghan, J., Winston, R. M. & Handyside, A. H. (1995) J. Reprod. Fertil. 104, 165–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.