Abstract
Steroidogenic factor-1 (SF-1) and liver receptor homologue-1 (LRH-1) belong to the fushi tarazu factor 1 subfamily of nuclear receptors. SF-1 is an essential factor for sex determination during development and regulates adrenal and gonadal steroidogenesis in the adult, whereas LRH-1 is a critical factor for development of endodermal tissues and regulates cholesterol and bile acid homeostasis. Regulatory ligands are unknown for SF-1 and LRH-1. A reported mouse LRH-1 structure revealed an empty pocket in a region commonly occupied by ligands in the structures of other nuclear receptors, and pocket-filling mutations did not alter the constitutive activity observed. Here we report the crystal structures of the putative ligand-binding domains of human SF-1 at 2.1-Å resolution and human LRH-1 at 2.5-Å resolution. Both structures bind a coactivator-derived peptide at the canonical activation–function surface, thus adopting the transcriptionally activating conformation. In human LRH-1, coactivator peptide binding also occurs to a second site. We discovered in both structures a phospholipid molecule bound in a pocket of the putative ligand-binding domain. MS analysis of the protein samples used for crystallization indicated that the two proteins associate with a range of phospholipids. Mutations of the pocket-lining residues reduced the transcriptional activities of SF-1 and LRH-1 in mammalian cell transfection assays without affecting their expression levels. These results suggest that human SF-1 and LRH-1 may be ligand-binding receptors, although it remains to be seen if phospholipids or possibly other molecules regulate SF-1 or LRH-1 under physiological conditions.
Keywords: x-ray crystallography, phospholipid, nuclear receptor, steroid, bile
Steroidogenic factor-1 (SF-1; AD4BP/NR5A1) and liver receptor homologue-1 (LRH-1; CPF/FTF/NR5A2), expressed in man, are homologues of the fushi tarazu factor-1 of Drosophila (1) and FF1B of fish (2). Together, these factors constitute the NR5A subfamily of nuclear receptors (NRs) (3). SF-1 and LRH-1 function as monomers (4) to regulate genes by binding to similar response elements.
SF-1 is expressed in the adrenal, testes, ovary, pituitary, hypothalamus, spleen, and skin and regulates genes that direct biosynthesis of adrenal and gonadal steroids as well as Mullerian hormone and gonadotropins (5, 6). SF-1 is essential for normal adrenal and gonadal development given that SF-1 knockout in mice causes adrenal and gonadal agenesis and impaired gonadotropin expression, resulting in postnatal death due to severe adrenal insufficiency (7, 8). SF-1 knockout also causes abnormalities of the ventromedial hypothalamic nucleus, the control center for satiety and feeding, which suggests that SF-1 may have broader roles in the control of metabolism and obesity (9). In humans, partial loss-of-function mutations in SF-1 result in XY sex reversal and adrenal failure (10, 11). Although SF-1 is expressed in the ovary (12), a mutation of SF-1 was observed not to affect ovarian development; thus, SF-1 may not be crucial for female sexual development (13). Another NR, DAX-1, has been described to repress the SF-1 function (14).
LRH-1 is predominantly expressed in tissues of endodermal origin (15). LRH-1 is essential for normal hepatic and pancreatic development (16). In the adult, LRH-1 functions in the control of cholesterol and bile acid homeostasis in coordination with two other NRs, farnesol X receptor and short heterodimer partner (17). LRH-1 is also expressed in ovary (12), preadipocyte (18), placenta (19), and testis (20). LRH-1 in the ovarian granulosa cells induces progesterone biosynthesis after ovulation (21, 22).
Prototypical NRs, such as steroid receptors, thyroid hormone receptors, and retinoid receptors modulate gene transcription in response to small lipophilic molecules (23–25). Ligand binding induces the activation function-2 helix of the ligand-binding domain (LBD) to form a charge clamp for coactivator recruitment (26, 27). Several NRs previously referred to as orphan receptors have been shown to adopt the same regulatory paradigm, and physiologically relevant NR ligands now also include fatty acids (peroxisome proliferator-activated receptor), oxysterols (liver-X receptor), bile acids (farnesoid-X receptor), and xenobiotic compounds (pregnane-X receptor) (28, 29). For some NRs, ligands are reported to serve an essential structural role, not a regulatory one (30–35). The ligands for these receptors were first revealed by crystallographic studies highlighting a new structural genomics approach to the identification of ligands for orphan NRs (36). Structural analysis has also identified NRs lacking any pocket for a ligand, such as the estrogen-related receptors (37) and the NGFI-B/Nurr1 subfamily of NRs (38, 39), defining a mode of ligand-independent NR function.
The NR5A subfamily of NRs remain orphans with no identified bona fide ligands. Although an oxysterol was reported to activate SF-1 (40), this finding could not be confirmed (41). Alternative modes of regulation for SF-1, including phosphorylation (42, 43) and sumoylation (44), have been presented. A recent mouse LRH-1 (mLRH-1) structure revealed it to have a large empty pocket, but mutations made to fill this pocket did not reduce the transcriptional activity, which suggested that the activity of mLRH-1 may be ligand-independent (45). We have determined the crystal structures of human SF-1 (hSF-1) and human LRH-1 (hLRH-1). To our surprise, we find phospholipid molecules bound in both structures. Mass spectrometry and cell transfection experiments show that mutations of phospholipid-binding residues reduce the binding of phospholipids and the transcriptional activity of the receptors.
Materials and Methods
Protein Expression and Purification. The hSF-1 DNA encoding residues G219-T461 (National Center for Biotechnology Information accession no. NM_004959), and hLRH-1 DNA encoding residues S251-A495 (National Center for Biotechnology Information accession no. NM_003822) were PCR-amplified from cDNA (Clontech) and cloned into a modified pET vector (Novagen) encoding an N-terminal 6His tag and a tobacco etch virus protease cleavage site. Initial crystal diffraction of the wild-type hSF-1 was poor; therefore, based on structural modeling, a double mutation (C247S/C412S) was introduced by PCR mutagenesis (Stratagene) into the bacterial vector encoding the hSF-1 to avoid surface Cys-mediated protein aggregation.
hSF-1 and hLRH-1 LBDs were produced in 30-liter bioreactor cultures of Escherichia coli strain BL21(DE3) RIL (Stratagene) by using terrific broth with 15 h of induction at 20°C with 0.5 mM isopropyl β-d-thiogalactoside. Frozen cell pastes suspended in 40 ml of lysis buffer (50 mM Na/K phosphate, pH 8.0/250 mM NaCl/5% glycerol) per liter of cells were lysed by using a microfluidizer (model no. M-110H, Microfluidics) at 18,000 psi and clarified by centrifugation at 15,000 × g at 4°C for 2 h. Clarified lysates were fractionated by Ni-chelating sepharose (AP-Biotech), with elution using a gradient to 100% buffer B (20 mM Hepes, pH8.0/250 mM imidazole/250 mM NaCl/5% glycerol). Eluted LBDs were diluted 6-fold with buffer C (20 mM Tris, pH 8.0) and fractionated by using Source 30Q (AP-Biotech), with elution using a linear gradient from 2% to 25% buffer D (20 mM Tris, pH 8.0/1 M NaCl). Pooled fractions were incubated with tobacco etch virus protease at 50 μg/mg overnight at 4°C, and proteins (>95% pure) were concentrated to 20 mg/ml and stored at –80°C.
Protein Crystallization. Initial crystals of hSF-1 and hLRH-1 were obtained with sparse-matrix crystallization screen (46) kits (Hampton Research, Aliso Viejo, CA). The hSF-1 crystal diffraction quality was improved by introducing the C247S/C412S mutation. hSF-1 protein was diluted to 15 mg/ml in 20 mM Tris·HCl pH 8.0/100 mM NaCl/10 mM DTT with a 2× molar excess of the peptides NCOA1 (SRC-1) NID-2 (CPSSHSSLTERHKILHRLLQEGSPS) and/or NCOA-2 (TIF2, GRIP1) NID-3 (KENALLRYLLDKD). Crystals were grown by the sitting drop vapor diffusion method at 4°C, mixing equal volumes of protein/peptide sample with reservoir solution containing 18% polyethylene glycol 3350, 0.2M ammonium sulfate, 0.1M [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane, pH 5.5, and 2.5% sucrose. Crystals grew to a size of 0.6 mm × 0.3 mm × 0.3 mm in 5–8 days. For cryoprotection, sucrose was added to hSF-1 crystals before freezing.
hLRH-1 protein was diluted to 10 mg/ml in 20 mM Tris·HCl, pH 7.5/62 mM NaCl/100 mM ammonium acetate/2 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate with 2× molar excess of the peptide NCOA-2 NID-3 (KENALLRYLLDKD). Crystals were grown by the sitting drop vapor diffusion method at 20°C, mixing equal volumes of protein/peptide sample with reservoir solution containing 0.9 M NaH2PO4 and 0.1 M K2HPO4. Crystals grew to a size of 0.13 mm × 0.03 mm × 0.03 mm in 2 weeks. Glycerol was used for cryoprotection.
Data Collection and Structure Determination. The x-ray diffraction data of hSF-1 and hLRH-1 were collected at the Advanced Light Source beamline 8.3.1 under cryogenic temperature. The diffraction data were integrated and scaled by using mosflm and scala (47) (Table 1). To solve the hSF-1 structure, a homology model was generated based on the crystal structure of mLRH-1 [1PK5, (45)]. Molecular replacement with the data up to 3.5 Å was carried out by using epmr (48) obtaining a solution in space group P3121. Two molecules related by noncrystallographic symmetry were determined in each asymmetric unit. The initial model was then subject to refinement by using o (49), cnx (50), and refmac5 (51) against 2.1-Å data with least-squares refinement, individual B-factor refinement, and translation, liberation, and screw-rotation displacement refinement protocols. Well defined electron density indicated one NCOA2 NID-3 peptide bound to the surface and the unexpected phosphatidylethanolamine (PE) ligand bound inside the putative ligand-binding pocket. The structure of hLRH-1 was also determined through molecular replacement by using a homology model based on the mLRH-1 structure as the search model. The crystal is in space group P212121 with one molecule in each asymmetric unit. The structure refinement process is similar to that for hSF-1. During the refinement process, electron density appeared in the putative ligand-binding pocket, suggesting the structure of a phosphatidylglycerol-phopshpoglycerol. The NCOA-2 NID-3 peptide was found to bind at two different sites on the surface of hLRH-1.
Table 1. Statistics of crystallographic data and refinement.
| hSF-1 | hLRH-1 | |
|---|---|---|
| Crystal and data collection statistics | ||
| Unit cell dimensions, Å | a = b = 73.6 | a = 61.0, b = 67.0 |
| c = 195.7 | c = 78.2 | |
| Space group | P3121 | P212121 |
| Solvent content | 49% | 53% |
| Resolution range, Å | 50-2.1 | 50-2.5 |
| Unique reflections | 36,333 | 10,899 |
| Data redundancy | 4.2 | 4.6 |
| Completeness, % | 98.7 | 99.4 |
| 〈I/σ(I)〉 | 6.9 | 10.0 |
| Rsym, % | 11.2 | 4.9 |
| Refinement statistics | ||
| σ cut off | None | None |
| Total non-hydrogen atoms | 4,342 | 2,172 |
| Ave B factor, Å2 | 24.0 | 34.2 |
| Rcryst/Rfree, % | 21.6/26.5 | 23.9/28.1 |
| rms deviation bond lengths, Å | 0.012 | 0.008 |
| rms deviation bond angles, ° | 1.449 | 1.034 |
Rsym = Σ|Iavg — Ij|/ΣIj. Rcryst = Σ|Fo — Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factors, respectively, Rfree was calculated from a randomly chosen 5% of reflections excluded from the refinement, and Rcryst was calculated from the remaining 95% of reflections. rms deviation values are from ideal geometry.
Mass Spectrometry. Samples of hSF-1 and hLRH-1 (50 nmol, 1.5 mg in 100 μl of aqueous buffer) were spiked with 50 nmol of a standard, PE-12:0 (1,2-didocanoyl-sn-glycero-3-phosphoethanolamine, Sigma) before extraction, based on preliminary results showing the absence of this molecule in either of the protein preparations. Sample extraction was performed as described in ref. 52, ending in 200 μl of organic fraction, of which 20-μl samples were analyzed by using electrospray ionization MS in positive and negative modes (Micromass ZMD MS with masslynx software, Waters).
Biochemical Coactivator Recruitment Assay. The AlphaScreen histidine detection kit (PerkinElmer) was used to detect binding between bacterially expressed His-tagged hSF-1 LBD and coactivator NCOA1 (SRC-1/RIP160, residues M595 to Q780, NCBI NM_003743) cloned into a pGEX vector (53) but having a C-terminal biotinylation site (LNDIFEAQKIEWHR). Costar 384-well white polystyrene plates (Corning) were used. Reactions (15 μl per reaction), each with 50 nM His-tagged hSF-1 and a biotin-tagged NCOA1 fragment in 50 mM 1,3-bis[tris(hydroxymethyl)methylamino] propane, pH 7.5/50 mM KCl/0.05% Tween 20/1 mM DTT/0.1% BSA, were sealed and incubated at room temp for 2 h. A 5-μl mix of streptavidin donor beads (15 μg/ml) and Ni-chelate acceptor beads (15 μg/ml) was then added and incubated for a final 2 h. The Fusion Alpha reader was set to read for 1 s per well. Data analysis used prism (GraphPad, San Diego).
Mammalian Cell Transfection. The hSF-1 (G219-T461) and hLRH-1 (S251-A495) LBDs were engineered for expression as fusion proteins with the yeast GAL4 DNA-binding domain (DBD), using SG-GAL4 and CMV-GAL4 vectors. Mutations of the putative ligand cavity were introduced by PCR (Stratagene).
HEK293T cells were cultured at 37°C in DMEM with 100 units/ml penicillin, 100 units/ml streptomycin, and 10% heat-inactivated FCS (Invitrogen). For transient transfection, HEK293T cells were grown to 80% confluency in six-well plates, and medium was exchanged for 1.5 ml of serum-free medium before addition of 1-μg pSG-GAL4-hSF-1-LBD or pSG-GAL4-hLRH-1-LBD expression vectors, 400 ng of pFR-Luc reporter gene (Stratagene), and 120 ng of pRL-TK transfection control plasmids (Promega) mixed with 6 μl of Metafectene (Biontex, Munich). After 4 h, serum-containing medium was added. After 24 h, medium was removed and cells were lysed in 50 mM Tris, pH 7.5/150 mM NaCl/1% Nonidet P-40. Firefly luciferase was measured by using a Luciferase Reporter Gene Assay kit (Roche Diagnostics) and Renilla luciferase was measured by using the Renilla Luciferase Assay system (Promega).
Portions of the wild-type and mutated transiently expressed GAL-hSF-1-LBD and GAL-hLRH-1-LBD proteins were detected by Western blots using anti-GAL4 (DBD) SC510 horseradish peroxidase-conjugated antibody (Santa Cruz Biotechnology) with detection using ECL plus (Amersham Pharmacia Biosciences).
Results
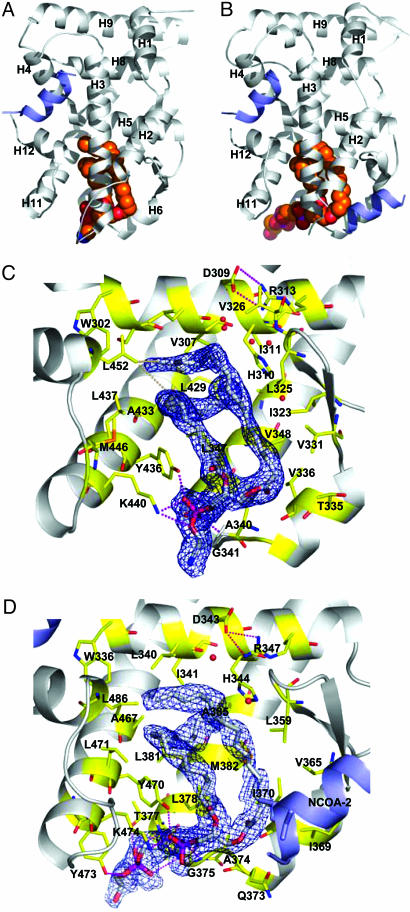
The hSF-1 and hLRH-1 putative LBD structures adopt an α-helical sandwich fold with 12 α-helices (H1–H12) and one β-hairpin between H5 and H6 (Fig. 1 A and B). Compared with most NR structures (36), hSF-1 and hLRH-1 each contain a unique fourth sandwich layer formed by H2, as described for the mLRH-1 structure (45). The hSF-1 and hLRH-1 LBDs show a high degree of structural similarity with an rms deviation of 1.25 Å (calculation based on the main chain Cα positions, excluding the loop between H1 and H2). The H2 of hSF-1 is shorter than that for hLRH-1 by one α-helical turn because of the presence of a helix-breaking residue (P251) in hSF-1. Consistent with reports that hSF-1 and hLRH-1 function as monomers, both appear to be isolated stable molecules in an asymmetric unit, with no crystallographic contacts forming through the canonical dimerization surface used by other NRs (54–56).
Fig. 1.
The hSF-1 and hLRH-1 LBD structures complexed with phospholipid and coactivator peptide. (A and B) The hSF-1 LBD (A) and the hLRH-1 LBD (B) (gray ribbon models) with phospholipid ligands (spherical model colored by atom type), and NCOA2 coactivator peptide (blue ribbon model). Note that two NCOA2 peptides bind to each LRH-1 molecule, one at the canonical activation function surface and the other at a site formed by H2, H3, and the β-sheet. (C) Residues of the hSF-1 ligand-binding pocket (stick models colored yellow and by atom type), showing salt bridge and hydrogen bonds (dotted lines) to the PE (stick models colored white and by atom type). The blue mesh indicates an unbiased 2Fo – Fc map covering the ligand. H2 and H3 are truncated to show the pocket features. (D) Residues of the hLRH-1 ligand-binding pocket depicted as in C showing interactions with the phosphatidylglycerol-phosphoglycerol.
For hSF-1 and hLRH-1, the difference Fourier map of the protein atom model exhibited a large, well defined electron density in and near the region occupied by ligands in other NRs (Fig. 1 C and D). These unaccounted electron densities extended to reach the solvent through a channel formed by the N-terminal end of H3, the C-terminal end of H11, and the loop between H6 and H7. Based on these electron densities, the molecule bound to hSF-1 was identified as a PE, and that bound to hLRH-1 was identified as a phosphatidylglycerol-phosphoglycerol (Fig. 1 A and B). In hLRH-1, the terminal phosphoglycerol is stabilized by a Thr residue (T377), whereas the corresponding residue in hSF-1 is Leu (L343). However, the headgroup selection seen in the crystal structures may also involve the purification and crystallization processes. The acyl chains in both cases are a palmitic ester (C16:0) linked to C1 and a palmitoleic ester (C16:1, Δ9) linked to C2 of the glycerol backbone. The Δ9-cis double bond causes a bend in the palmitoleic chain that allows it to pack around the palmitic chain, almost completely filling the interior cavity of the ligand-binding pocket (Fig. 1 C and D and Fig. 6, which is published as supporting information on the PNAS web site). Extensive van der Waals contacts with the pocket-lining hydrophobic residues are made (Fig. 1 C and D and Fig. 7, which is published as supporting information on the PNAS web site), including direct contacts with the activation function-2 helix (L452/hSF-1 and L486/hLRH-1) that support the active conformation.
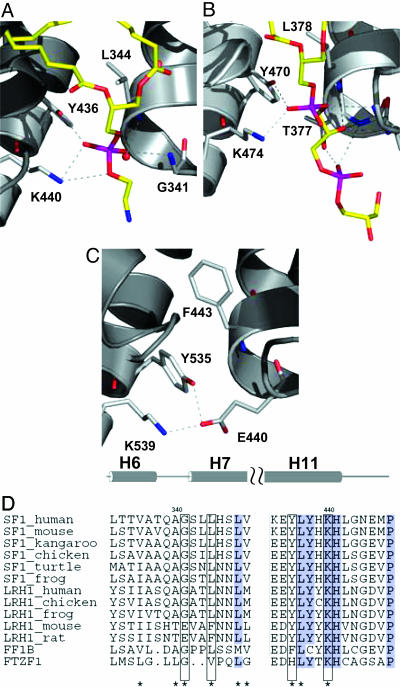
In hSF-1 and hLRH-1, the phospholipid phosphate interacts with three residues: a Lys from H11 (K440/hSF-1 or K474/hLRH-1), a Tyr from H11 (Y436/hSF-1 or Y470/hLRH-1), and a Gly from the H6–H7 loop (G341/hSF-1 or G375/hLRH-1) (Fig. 2). The Lys of this KYG triad makes hydrogen bonds with two phosphate oxygen atoms, whereas the side chain of the Tyr and the backbone NH of the Gly each forms a hydrogen bond with one phosphate oxygen. The KYG triad is highly conserved among most homologues of SF-1 and LRH-1 in the NR5 subfamily (Fig. 2D and Fig. 8, which is published as supporting information on the PNAS web site). But curiously, in mouse and rat LRH-1, a Glu (residue E440 in mouse) replaces the Gly of the KYG triad, and in the mLRH-1 structure (45), E440 forms a salt bridge with the Lys (residue K539 in mouse) of the KYG triad. In the mLRH-1 structure, a bulky Phe (F443 in mLRH-1) also replaces a Leu (L344 in hSF-1; L378 in hLRH-1) just inside the pockets of the human structures. The result of these differences is a change in the shape of the ligand-binding pocket that could reduce phosphate binding in mLRH-1.
Fig. 2.
Comparison of hSF-1 and hLRH-1 structures with mLRH-1. (A and B) A phosphate group in hSF-1 (A) and hLRH-1 (B) interacts with the Lys and Tyr of the KYG triad. (C) E440 in the APO mLRH-1 mimics the phosphate group interactions. Only the residues of the phosphate-binding triad (sticks colored yellow and by atom type) and the polar portions of the phospholipids (sticks colored by atom type) are shown. (D) Alignment of representative NR5A sequences. The two sequence segments that contain the KYG motif (helix H6–H7 and H11) are shown. A more complete alignment appears in Fig. 8.
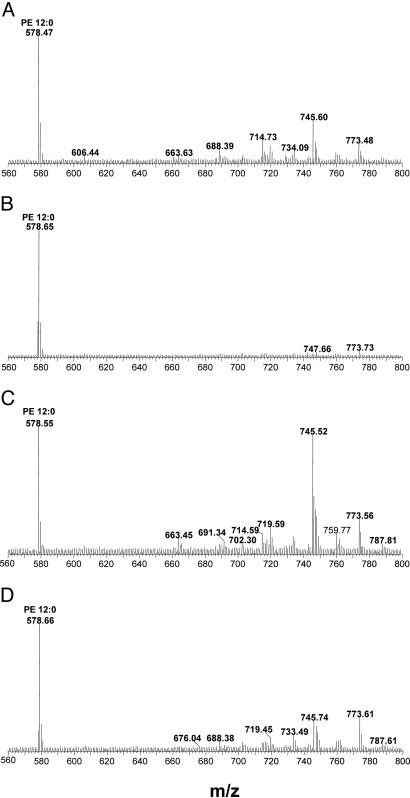
MS of lipids extracted from the bacterially expressed hSF-1 and hLRH-1 show several peaks that can be interpreted as PEs and phosphatidylglycerols having 14–18 carbons and containing various degrees of saturation. (Fig. 3), suggesting the cocrystallization with just one phospholipid species in each structure is fortuitous. After comparison to a standard PE-12:0 added in an amount equimolar to the LBDs before extraction, hLRH-1 appears fully occupied with the E. coli phospholipids, whereas hSF-1 appears only partially occupied. After double mutation of the phosphate-binding residues in hLRH-1 (Y470F/K474A) the phospholipid peaks were diminished by >50%, whereas after the equivalent double mutation in hSF-1 (Y436F/K440A), the phospholipid peaks were nearly completely lost (Fig. 3). These and other pocket mutants of hSF-1 and hLRH-1 were not observed to alter the soluble expression (≈20 mg per liter of culture) of these proteins in E. coli, indicating that the mutations do not have any deleterious effects that are obvious.
Fig. 3.
Mass spectral analysis of lipids bound to hSF-1 and hLRH-1 LBD proteins purified from E. coli: wild-type hSF-1 (A), hSF-1 Y436F-K440A (B), wild-type hLRH-1 (C), and hLRH-1 Y470F-K474A (D). The analyses were performed in negative mode. PE-12:0 (50 pmol) was mixed with 50 pmol of each LBD protein before extraction, giving the m/z = 578 standard peak.
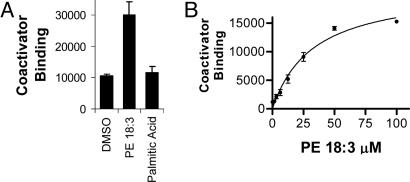
Coactivator recruitment to hLRH-1 in vitro was not affected by the addition of phospholipids (data not shown), which is consistent either with a ligand-independent mode of coactivator binding (45) or with the apparent preexisting full occupancy observed by MS. However addition of phospholipid, but not palmitic acid, did increase coactivator binding to the partially occupied hSF-1 (Fig. 4A). The PE-16:0/16:1 observed in the crystal structure is unavailable commercially, so it could not be tested. Several common PEs were tested (data not shown), and PE-18:3 showed the best stimulation, giving a dose-dependent increase in binding of NCOA1 (Fig. 4B), with an apparent half-maximal stimulation for this effect occurring at a concentration of 30 μM. This result shows that phospholipid promotes coactivator recruitment to hSF-1 in vitro.
Fig. 4.
PE dose-dependent increase in coactivator recruitment to hSF-1 in vitro. (A) PE-18:3 (50 μM 1,2-dilinolenoyl-sn-glycero-3-phosphoethanolamine) but not palmitic acid (50 μM) activates wild-type hSF-1 to bind NCOA1 by AlphaScreen. (B) Dose-dependent NCOA1 recruitment to hSF-1 by PE-18:3. Error bars indicate the standard deviations. The graphs shown are representative of three experiments.
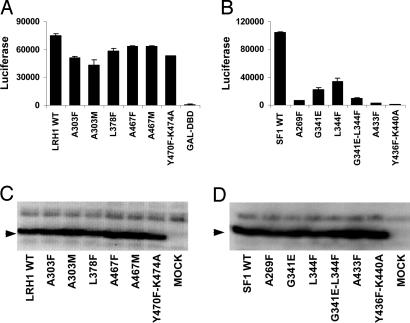
A selection of structure-guided mutations of the hLRH-1 and hSF-1 pockets were tested by using transfected mammalian cells. When the hLRH-1 or hSF-1 LBDs were fused to the DBD of GAL4, strong activation in transfected cells of a reporter gene containing GAL4-responsive elements was observed (Fig. 5 A and B). Six hLRH-1 pocket mutations, A303F, A303M, L378F, A467F, A467M, and Y470F/K474A, diminished activity 16–42% (Fig. 5A). Similar hSF-1 pocket mutations (A269F, G341E, L344F, G341E/L344F, A433F, and Y436F/K440A) diminished activity 68–99% (Fig. 5B). In neither the hLRH-1 nor the hSF-1 GAL fusions did the pocket mutations affect expression levels in the cells (Fig. 5 C and D). Similar mutations made in the mLRH-1 were previously reported to have no effect on the function, and this was cited as support for a ligand-independent mode of mLRH-1 function (45). The decreases in function observed here suggest that the activities of hSF-1 and hLRH-1 may be ligand-modulated in mammalian cells. However, whether phospholipids serve the role of ligand has yet to be examined.
Fig. 5.
Effects of pocket mutations on hLRH-1 and hSF-1 functions in HEK293T cells. (A) hLRH-1 LBD activity tested as GAL-DBD fusions acting at a GAL4-responsive LUC reporter gene. The mutations tested include residues A303, L378, A467, Y470, and K474. (B) hSF-1 LBD activity tested as GAL-DBD fusions. The mutations tested include residues A269, G341, L344, A433, Y436, and K440. (C) Western blot analysis of cells after transfection with vectors encoding GAL4-DBD–hLRH-1 LBD fusion proteins using anti-GAL4-DBD antibody. (D) Western blot analysis of GAL4-DBD hSF-1 LBD fusion proteins. Error bars indicate the standard deviations. The graphs shown are representative of three experiments.
In addition to the coactivator peptide bound to the canonical (26, 27, 53) coactivator-binding site of hLRH-1, we also found a second peptide molecule bound to a site (Fig. 1 B and D and Fig. 9, which is published as supporting information on the PNAS web site) comprised of residues that are conserved among LRH-1 homologues (Fig. 8). Interaction is also made to atoms of the C1-acyl chain of the phospholipid in coordination with the methyl group of T295. Unlike the canonical activation function-2 surface (26, 27, 53), there is no charge clamp to the coactivator peptide dipole in the second binding site. However, a Tyr of the NCOA2 peptide forms a hydrogen bond with D366 of the β-hairpin. The presence of the second peptide could be a fortuitous crystallographic effect. However, it has been shown that the negative coregulator PROX-1 interacts with LRH-1 through a site on H2 and H11 that may overlap with the site identified here (57). Further experiments are needed to address the biological role of this site.
Discussion
We have provided structural data to suggest that hSF-1 and hLRH-1 can bind phospholipids. Mutations of the residues that interact with the phospholipid were observed to diminish function in transient transfection experiments without affecting protein expression levels. In the case of hSF-1, addition of phospholipid enhanced the binding of coactivator peptide in a biochemical assay. However, a thorough analysis of whether phospholipids actually regulate hSF-1 and hLRH-1 will require cell biology and in vivo experiments that are out of the scope of the current structural analysis.
We have initiated experiments involving the addition of phospholipids to transiently transfected cells and have tested delipidated serum as used to investigate phospholipid regulation of SREBP (58), but these preliminary experiments failed to demonstrate a phospholipid-dependent regulation of activity (data not shown). A plethora of different phospholipids are present in cells and culture sera, making the task of identifying which phospholipids might be relevant very difficult. It is also known that phospholipids have low solubility in free solution and are associated inside cells with phospholipid transfer proteins (59) and scramblases (60), when they are not localized in membranes, or outside the cells in lipoprotein complexes (61, 62). Selective phospholipids are thought to be required for proper folding and function of membrane proteins (63). Soluble enzymes, such as CYP27A1, also bind phospholipids and are thought to have their activities regulated by such binding (52). At this point, although the phospholipid-bound crystal structures are intriguing, the most that can be said is that phospholipid cannot be ruled out as a possible ligand class.
With the current structures, hSF-1 and hLRH-1 join a few other orphan NRs found to cocrystallize with fortuitous ligands that are common cellular lipids (30–35). PE has also been observed in the structures of the insect RXR homologue ultraspiracle (64, 65). Evolutionary biologists have pondered how NRs arose and diversified (3). The fushi tarazu factor-1 subfamily of NRs has been considered one of the most evolutionarily conserved and, together with the COUP-TF and RXR/ultraspiracle subfamilies, perhaps closest to the ancestral NR (66). It will be interesting to see whether fushi tarazu factor-1, FF1B, and members of the COUP-TF subfamily also bind phospholipids and whether this has any evolutionary implications for the NR superfamily.
Supplementary Material
Acknowledgments
We thank Dr A. Kumar for data collection; Drs. P. Ibrahim, R. Bremer, J. Lin, B. England, K. Zhang, and A. Brunger for suggestions; Drs. R. Artis, G. Bollag, and K. P. Hirth for support; and Profs. J. Schlessinger and S.-H. Kim for stimulating discussions.
Author contributions: W.W., C.Z., A.M., H.I.K., M.T., R.S., U.M., K.E., H.N., B.P., M.V.M., and B.L.W. designed research; W.W., A.M., H.I.K., M.T., R.S., U.M., K.E., H.N., C.S., B.P., and B.L.W. performed research; A.M., C.S., and B.L.W. contributed new reagents/analytic tools; W.W., C.Z., A.M., H.I.K., M.T., R.S., U.M., K.E., H.N., C.S., B.P., and B.L.W. analyzed data; and W.W., C.Z., A.M., H.I.K., M.T., R.S., U.M., K.E., H.N., C.S., B.P., M.V.M., and B.L.W. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SF-1, steroidogenic factor-1; hSF-1, human steroidogenic factor-1; LRH-1, liver receptor homologue-1; hLRH-1, human liver receptor homologue-1; mLRH-1, mouse liver receptor homologue-1; NR, nuclear receptor; LBD, ligand-binding domain; DBD, DNA-binding domain; PE, phosphatidylethanolamine.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1ZDT and 1ZDU).
References
- 1.Lavorgna, G., Ueda, H., Clos, J. & Wu, C. (1991) Science 252, 848–851. [DOI] [PubMed] [Google Scholar]
- 2.Chai, C. & Chan, W. K. (2000) Mech. Dev. 91, 421–426. [DOI] [PubMed] [Google Scholar]
- 3.Laudet, V. (1997) J. Mol. Endocrinol. 19, 207–226. [DOI] [PubMed] [Google Scholar]
- 4.Ueda, H., Sun, G. C., Murata, T. & Hirose, S. (1992) Mol. Cell. Biol. 12, 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker, K. L. & Schimmer, B. P. (1997) Endocr. Rev. 18, 361–377. [DOI] [PubMed] [Google Scholar]
- 6.Val, P., Lefrancois-Martinez, A. M., Veyssiere, G. & Martinez, A. (2003) Nucl. Recept. 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo, X., Ikeda, Y. & Parker, K. L. (1994) Cell 77, 481–490. [DOI] [PubMed] [Google Scholar]
- 8.Parker, K. L. (2004) Endocr. Res. 30, 855. [DOI] [PubMed] [Google Scholar]
- 9.Majdic, G., Young, M., Gomez-Sanchez, E., Anderson, P., Szczepaniak, L. S., Dobbins, R. L., McGarry, J. D. & Parker, K. L. (2002) Endocrinology 143, 607–614. [DOI] [PubMed] [Google Scholar]
- 10.Achermann, J. C., Ito, M., Hindmarsh, P. C. & Jameson, J. L. (1999) Nat. Genet. 22, 125–126. [DOI] [PubMed] [Google Scholar]
- 11.Achermann, J. C., Ozisik, G., Ito, M., Orun, U. A., Harmanci, K., Gurakan, B. & Jameson, J. L. (2002) J. Clin. Endocrinol. Metab. 87, 1829–1833. [DOI] [PubMed] [Google Scholar]
- 12.Boerboom, D., Pilon, N., Behdjani, R., Silversides, D. W. & Sirois, J. (2000) Endocrinology 141, 4647–4656. [DOI] [PubMed] [Google Scholar]
- 13.Biason-Lauber, A. & Schoenle, E. J. (2000) Am. J. Hum. Genet. 67, 1563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Achermann, J. C., Meeks, J. J. & Jameson, J. L. (2001) Mol. Cell. Endocrinol. 185, 17–25. [DOI] [PubMed] [Google Scholar]
- 15.Fayard, E., Auwerx, J. & Schoonjans, K. (2004) Trends Cell Biol. 14, 250–260. [DOI] [PubMed] [Google Scholar]
- 16.Pare, J. F., Malenfant, D., Courtemanche, C., Jacob-Wagner, M., Roy, S., Allard, D. & Belanger, L. (2004) J. Biol. Chem. 279, 21206–21216. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin, B., Jones, S. A., Price, R. R., Watson, M. A., McKee, D. D., Moore, L. B., Galardi, C., Wilson, J. G., Lewis, M. C., Roth, M. E., et al. (2000) Mol. Cell 6, 517–526. [DOI] [PubMed] [Google Scholar]
- 18.Clyne, C. D., Speed, C. J., Zhou, J. & Simpson, E. R. (2002) J. Biol. Chem. 277, 20591–20597. [DOI] [PubMed] [Google Scholar]
- 19.Sirianni, R., Seely, J. B., Attia, G., Stocco, D. M., Carr, B. R., Pezzi, V. & Rainey, W. E. (2002) J. Endocrinol. 174, R13–7. [DOI] [PubMed] [Google Scholar]
- 20.Pezzi, V., Sirianni, R., Chimento, A., Maggiolini, M., Bourguiba, S., Delalande, C., Carreau, S., Ando, S., Simpson, E. R. & Clyne, C. D. (2004) Endocrinology 145, 2186–2196. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. W., Peng, N., Rainey, W. E., Carr, B. R. & Attia, G. R. (2004) J. Clin. Endocrinol. Metab. 89, 3042–3047. [DOI] [PubMed] [Google Scholar]
- 22.Saxena, D., Safi, R., Little-Ihrig, L. & Zeleznik, A. J. (2004) Endocrinology 145, 3821–3829. [DOI] [PubMed] [Google Scholar]
- 23.Renaud, J. P., Rochel, N., Ruff, M., Vivat, V., Chambon, P., Gronemeyer, H. & Moras, D. (1995) Nature 378, 681–689. [DOI] [PubMed] [Google Scholar]
- 24.Wagner, R. L., Apriletti, J. W., McGrath, M. E., West, B. L., Baxter, J. D. & Fletterick, R. J. (1995) Nature 378, 690–697. [DOI] [PubMed] [Google Scholar]
- 25.Schwabe, J. W. (1996) Curr. Biol. 6, 372–374. [DOI] [PubMed] [Google Scholar]
- 26.Feng, W., Ribeiro, R. C., Wagner, R. L., Nguyen, H., Apriletti, J. W., Fletterick, R. J., Baxter, J. D., Kushner, P. J. & West, B. L. (1998) Science 280, 1747–1749. [DOI] [PubMed] [Google Scholar]
- 27.Nolte, R. T., Wisely, G. B., Westin, S., Cobb, J. E., Lambert, M. H., Kurokawa, R., Rosenfeld, M. G., Willson, T. M., Glass, C. K. & Milburn, M. V. (1998) Nature 395, 137–143. [DOI] [PubMed] [Google Scholar]
- 28.Kliewer, S. A., Lehmann, J. M. & Willson, T. M. (1999) Science 284, 757–760. [DOI] [PubMed] [Google Scholar]
- 29.Chawla, A., Repa, J. J., Evans, R. M. & Mangelsdorf, D. J. (2001) Science 294, 1866–1870. [DOI] [PubMed] [Google Scholar]
- 30.Stehlin, C., Wurtz, J. M., Steinmetz, A., Greiner, E., Schule, R., Moras, D. & Renaud, J. P. (2001) EMBO J. 20, 5822–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhe-Paganon, S., Duda, K., Iwamoto, M., Chi, Y. I. & Shoelson, S. E. (2002) J. Biol. Chem. 277, 37973–37976. [DOI] [PubMed] [Google Scholar]
- 32.Wisely, G. B., Miller, A. B., Davis, R. G., Thornquest, A. D., Jr., Johnson, R., Spitzer, T., Sefler, A., Shearer, B., Moore, J. T., Willson, T. M. & Williams, S. P. (2002) Structure (Cambridge, Mass.) 10, 1225–1234. [DOI] [PubMed] [Google Scholar]
- 33.Kallen, J. A., Schlaeppi, J. M., Bitsch, F., Geisse, S., Geiser, M., Delhon, I. & Fournier, B. (2002) Structure (Cambridge, Mass.) 10, 1697–1707. [DOI] [PubMed] [Google Scholar]
- 34.Stehlin-Gaon, C., Willmann, D., Zeyer, D., Sanglier, S., Van Dorsselaer, A., Renaud, J. P., Moras, D. & Schule, R. (2003) Nat. Struct. Biol. 10, 820–825. [DOI] [PubMed] [Google Scholar]
- 35.Kallen, J., Schlaeppi, J. M., Bitsch, F., Delhon, I. & Fournier, B. (2004) J. Biol. Chem. 279, 14033–14038. [DOI] [PubMed] [Google Scholar]
- 36.Li, Y., Lambert, M. H. & Xu, H. E. (2003) Structure (Cambridge, Mass.) 11, 741–746. [Google Scholar]
- 37.Greschik, H., Wurtz, J. M., Sanglier, S., Bourguet, W., van Dorsselaer, A., Moras, D. & Renaud, J. P. (2002) Mol. Cell 9, 303–313. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Z., Benoit, G., Liu, J., Prasad, S., Aarnisalo, P., Liu, X., Xu, H., Walker, N. P. & Perlmann, T. (2003) Nature 423, 555–560. [DOI] [PubMed] [Google Scholar]
- 39.Baker, K. D., Shewchuk, L. M., Kozlova, T., Makishima, M., Hassell, A., Wisely, B., Caravella, J. A., Lambert, M. H., Reinking, J. L., Krause, H., et al. (2003) Cell 113, 731–742. [DOI] [PubMed] [Google Scholar]
- 40.Lala, D. S., Syka, P. M., Lazarchik, S. B., Mangelsdorf, D. J., Parker, K. L. & Heyman, R. A. (1997) Proc. Natl. Acad. Sci. USA 94, 4895–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellon, S. H. & Bair, S. R. (1998) Endocrinology 139, 3026–3029. [DOI] [PubMed] [Google Scholar]
- 42.Desclozeaux, M., Krylova, I. N., Horn, F., Fletterick, R. J. & Ingraham, H. A. (2002) Mol. Cell. Biol. 22, 7193–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammer, G. D., Krylova, I., Zhang, Y., Darimont, B. D., Simpson, K., Weigel, N. L. & Ingraham, H. A. (1999) Mol. Cell 3, 521–526. [DOI] [PubMed] [Google Scholar]
- 44.Chen, W. Y., Lee, W. C., Hsu, N. C., Huang, F. & Chung, B. C. (2004) J. Biol. Chem. 279, 38730–38735. [DOI] [PubMed] [Google Scholar]
- 45.Sablin, E. P., Krylova, I. N., Fletterick, R. J. & Ingraham, H. A. (2003) Mol. Cell 11, 1575–1585. [DOI] [PubMed] [Google Scholar]
- 46.Jancarik, J. & Kim, S.-H. (1991) J. Appl. Crystallogr. 24, 409–411. [Google Scholar]
- 47.Leslie, A. G. (1999) Acta Crystallogr. D 55, 1696–1702. [DOI] [PubMed] [Google Scholar]
- 48.Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999) Acta Crystallogr. D 55, 484–491. [DOI] [PubMed] [Google Scholar]
- 49.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 50.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 52.Murtazina, D. A., Andersson, U., Hahn, I. S., Bjorkhem, I., Ansari, G. A. & Pikuleva, I. A. (2004) J. Lipid Res. 45, 2345–2353. [DOI] [PubMed] [Google Scholar]
- 53.Marimuthu, A., Feng, W., Tagami, T., Nguyen, H., Jameson, J. L., Fletterick, R. J., Baxter, J. D. & West, B. L. (2002) Mol. Endocrinol. 16, 271–286. [DOI] [PubMed] [Google Scholar]
- 54.Bourguet, W., Vivat, V., Wurtz, J. M., Chambon, P., Gronemeyer, H. & Moras, D. (2000) Mol. Cell 5, 289–298. [DOI] [PubMed] [Google Scholar]
- 55.Gampe, R. T., Jr., Montana, V. G., Lambert, M. H., Miller, A. B., Bledsoe, R. K., Milburn, M. V., Kliewer, S. A., Willson, T. M. & Xu, H. E. (2000) Mol. Cell 5, 545–555. [DOI] [PubMed] [Google Scholar]
- 56.Bledsoe, R. K., Montana, V. G., Stanley, T. B., Delves, C. J., Apolito, C. J., McKee, D. D., Consler, T. G., Parks, D. J., Stewart, E. L., Willson, T. M., et al. (2002) Cell 110, 93–105. [DOI] [PubMed] [Google Scholar]
- 57.Qin, J., Gao, D. M., Jiang, Q. F., Zhou, Q., Kong, Y. Y., Wang, Y. & Xie, Y. H. (2004) Mol. Endocrinol. 18, 2424–2439. [DOI] [PubMed] [Google Scholar]
- 58.Dobrosotskaya, I. Y., Seegmiller, A. C., Brown, M. S., Goldstein, J. L. & Rawson, R. B. (2002) Science 296, 879–883. [DOI] [PubMed] [Google Scholar]
- 59.Schouten, A., Agianian, B., Westerman, J., Kroon, J., Wirtz, K. W. & Gros, P. (2002) EMBO J. 21, 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu, J., Dai, Q., Chen, J., Durrant, D., Freeman, A., Liu, T., Grossman, D. & Lee, R. M. (2003) Mol. Cancer Res. 1, 892–902. [PubMed] [Google Scholar]
- 61.Wang, N., Lan, D., Gerbod-Giannone, M., Linsel-Nitschke, P., Jehle, A. W., Chen, W., Martinez, L. O. & Tall, A. R. (2003) J. Biol. Chem. 278, 42906–42912. [DOI] [PubMed] [Google Scholar]
- 62.Vance, J. E. & Vance, D. E. (1986) J. Biol. Chem. 261, 4486–4491. [PubMed] [Google Scholar]
- 63.Bogdanov, M. & Dowhan, W. (1998) EMBO J. 17, 5255–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clayton, G. M., Peak-Chew, S. Y., Evans, R. M. & Schwabe, J. W. (2001) Proc. Natl. Acad. Sci. USA 98, 1549–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Billas, I. M., Iwema, T., Garnier, J. M., Mitschler, A., Rochel, N. & Moras, D. (2003) Nature 426, 91–96. [DOI] [PubMed] [Google Scholar]
- 66.Owen, G. I. & Zelent, A. (2000) Cell. Mol. Life Sci. 57, 809–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li, Y., Choi, M., Cavey, G., Daugherty, J., Suino, K., Kovach, A., Bingham, N. C., Kliewer, S. A. & Xu, H. E. (2005) Mol. Cell 17, 491–502. [DOI] [PubMed] [Google Scholar]
- 68.Krylova, I. N., Sablin, E. P., Moore, J., Xu, R. X., Waitt, G. M., MacKay, J. A., Juzumiene, D., Bynum, J. M., Madauss, K., Montana, V., et al. (2005) Cell 120, 343–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.