Abstract
Integration of retrovirus DNA is a specific process catalyzed by the integrase protein acting to join the viral substrate DNA (att) sequences of about 10 bases at the ends of the long terminal repeat (LTR) to various sites in the host target cell DNA. Although the interaction is sequence specific, the att sequences of different retroviruses are largely unrelated to one another and usually differ between the two ends of the viral DNA. To define substrate sequence specificity, we designed an “in vitro evolution” scheme to select an optimal substrate sequence by competitive integration in vitro from a large pool of partially randomized substrates. Integrated substrates are enriched by PCR amplification and then regenerated and subjected to subsequent cycles of selection and enrichment. Using this approach, we obtained the optimal substrate sequence of 5′-ACGACAACA-3′ for avian sarcoma-leukosis virus (ASLV) and 5′-AACA(A/C)AGCA-3′ for human immunodeficiency virus type 1, which differed from those found at both ends of the viral DNA. Clonal analysis of the integration products showed that ASLV integrase can use a wide variety of substrate sequences in vitro, although the consensus sequence was identical to the selected sequence. By a competition assay, the selected nucleotide at position 4 improved the in vitro integration efficiency over that of the wild-type sequence. Viral mutants bearing the optimal sequence replicated at wild-type levels, with the exception of some mutations disrupting the U5 RNA secondary structure important for reverse transcription, which were significantly impaired. Thus, maximizing the efficiency of integration may not be of major importance for efficient retrovirus replication.
Following retrovirus infection, the viral RNA genome is reverse transcribed into a linear blunt-ended DNA molecule, which has to be integrated into the host cell chromosome to complete the viral replication cycle (15). The integration step is catalyzed by the viral enzyme integrase (IN), whose recognition sequence (att) is located at the very ends of the viral DNA (12, 13, 16, 17, 36, 42). The att sequence is necessary for integrase to first catalyze the removal of a dinucleotide from the 3′ ends of viral termini in a 3′ end processing reaction and to then join the processed viral ends to the target DNA in a strand transfer reaction.
The most important feature of the viral att sequence for integration is the sequence 5′-CAXX-3′. The conserved CA is almost always located exactly 2 bases away from the end of the long terminal repeat LTR in unintegrated DNA. Substituting either one of the two bases substantially impairs 3′-end processing and strand transfer, although it does not completely abolish activity (8, 10, 11, 18, 29, 30, 38–40, 43). Alteration of both the U3 and U5 conserved CA to TG results in severe reduction in integration and thus in replication in vivo (6, 34).
The sequence internal to the conserved CA plays a significant but less important role in integration. In vitro mutational analysis shows that sequence specificity resides within 12 bases from the termini (8, 9, 16, 26, 29, 30, 35, 37, 38, 40, 43) and that most sequence specificity resides in the terminal 8 bases (8, 26, 29, 30, 35, 38, 40). However, extensive mutational analysis has not revealed a consensus sequence to account for the variations in activity that result from differences at these internal sites.
With the exception of the conserved CA dinucleotide, different retroviruses have largely unrelated att sequences (7), implying that integrases of different retroviruses have different substrate sequence specificities. In most viruses, the subterminal sequences on either end of the same virus are different, resulting in consistent differences in integration efficiency between oligonucleotide substrates corresponding to the U5 and U3 ends (8, 29, 30, 38, 45). For example, in avian sarcoma-leukosis virus (ASLV), the U3 end is a more efficient substrate than U5, while in human immunodeficiency virus type 1 (HIV-1), the U5 end is a more efficient substrate than U3 (8, 29, 30, 38, 45). The natural att sequences may not be the optimal substrate sequence for the integration since certain mutations of wild-type WT nucleotides of Rous sarcoma virus and Molorey murine leukemia virus substrate sequences result in a significant increase of integration efficiency in vitro (5, 44).
In an effort to define a consensus sequence for integrase, we designed a functional “in vitro evolution” system to competitively select an optimal substrate sequence from a large pool of substrate sequences. In this system, the nucleotide positions in the region of interest were randomized in a starting substrate pool. The selective force of “evolution” was conferred through a competitive integration reaction catalyzed by purified viral integrase. Integrated substrates were selectively enriched by PCR. The selected and enriched viral substrates were regenerated by digestion with a restriction enzyme that cuts at the substrate-target joining site. Regenerated substrates were subjected to subsequent selection and enrichment until an optimal sequence emerged. The sequences selected by ASLV and HIV integrases were distinct from one another and differed somewhat from those found in the viral DNA by a few bases. Insertion of the optimal sequence in place of the natural sequence in the ASLV genome did not enhance the rate of integration or overall replication, implying that integration may not be a rate-limiting step in replication. Rather, other constraints, such as the secondary structure of the genome-primer complex, may be more important than the att site sequence.
MATERIALS AND METHODS
Oligonucleotides.
Oligonucleotides were purchased from the Tufts University synthesis facility. All oligonucleotides used as starting substrates for the integration-selection procedure consisted of three parts (from 5′ to 3′): an 11-base sequence complementary to the 3′ end of U3 (underlined), some portion of which was randomized, the 5-base sequence for FokI (bold), and a 19-base sequence derived from the U3 region, which served as a site of primer binding for amplification and sequencing.
(i) Oligonucleotide sequences.
Oligonucleotides for ASLV were as follows: 5′-AATGNNNNNNNCATCCCTCCGTATCACATTGACTGG-3′ (AL-7NCA) and 5′-AANNNNNTCTTCATCCCTCCGTATCACATTGACTGG-3′ (AL-5N); oligonucleotides for HIV-1 were as follows: 5′-ACTGNNNNNNNCATCCGACAGCACGAAATACACCTTG-3′ (HVP-7NCA) and 5′-ACNNNNNGAGACATCCGACAGCACGAAATACACCTTG-3′ (HVP-5N); substrate oligonucleotides for the 3′-end processing assay were as follows: 5′-ACGAGCACAGGAGTATGGATGAAGACTACATT-3′ (AL-1) (U3 WT), 5′-ACGAGCACAGGAGTATGGATGACGACAACATT-3′ (AL-SA) (selected), 5′-ACGAGCACAGGAGTATGGATGAAGGATTAGTT-3′ (AL-M-1) (mutant); substrate oligonucleotides for the IN-PCR competition assay were as follows: 5′-CCAGTCAATGTGATACGGAGGGATGAAGACAACA-3′ (AL-31) (selected) and 5′-CCAGTCAATGTGATACGGAGGGATGAAGACTACA-3′ (AL-41) (U3 WT); complementary substrate oligonucleotides were as follows: 5′-AATGTTGTCGTCATCCATACTCCTGTGCTCGT-3′ (AL-SB) (complementary to AL-SA), 5′-AACTAATCCTCCATCCATACTCCTGTGCTCGT-3′ (AL-M-2) (complementary to AL-M-1), 5′-AATGTAGTCTTCATCCATACTCCTGTGCTCGT-3′ (AL-2) (complementary to AL-1), 5′-TTTGTTGTCTTCATCCCTCCGTATCACATTGACTGG-3′ (AL-32) (complementary to AL-31), and 5′-TTTGTAGTCTTCATCCCTCCGTATCACATTGACTGG-3′ (AL-42) (complementary to AL-41); primers (annealed to the randomized substrates for synthesis of double-strand oligonucleotides and amplification of integration products) were as follows: 5′-CCAGTCAATGTGATACGGAG-3′ (SP) (for AL-7NCA and AL-5N), 5′-TCAATGTGATACGGAGGGAT-3′ (SP-2) (for ASLV), 5′-CAAGGTGTATTTCGTGCTGTC-3′ (HVP) (for HIV-1), and 5′-CGAGCACAGGAGTATGGA-3′ (NBP-2); biotinylated primers for φX174 DNA were as follows: Bio-5′-AAACGTCGTTAGGCCAGT-3′ (B-NEW1) (1764–1781), Bio-5′-GAGCTTGAGTAAGCATTTGG-3′ (B-φX2) (1767–1748), and Bio-5′-TTTAGAGAACGAGAAGACGG-3′ (B-φX3) (4355–4374); biotinylated primers for pUC119 DNA were as follows: Bio-5′-GTAAAACGACGGCCAGT-3′ (B-20) (2872–2856), Bio-5′-GGGAGTCAGGCAACTATGG-3′ (B-UC1) (2148–2166), and Bio-5′-GAAACAGCTATGACCATGAT-3′ (B-OKT3) (208–277); oligonucleotides for site-directed mutagenesis were as follows (intended mutations are underlined) 5′-GGGAAATGTTGTCGTATGCAATAC-3′ (U3M1) (pAD1: 300–323), 5′-GTATTGCATACGACAACATTTCCC-3′ (U3M2) (pAD1: 323–300), 5′-GAATGAAGCAGACGACAACATTTGGTGACCC-3′ (U5M3) (pAD1: 617–647), 5′-GGGTCACCAAATGTTGTCGTCTGCTTCATTC-3′ (U5M4) (pAD1: 647–617), 5′-GACGACAACATTTGGTGAC-3′ (U5MC5) (pAD1: 627–645), and 5′-TACAACATTCAGGTGTTCG-3′ (U5MC6) (pAD1: 626–608).
(ii) Preparation of double-stranded oligonucleotides.
Substrate oligonucleotides (AL-SA/SB, AL-1/2, AL-M-1/2, AL-31/32, and AL-41/42) resembling the terminal sequences of the LTR were purified from 20% polyacrylamide gels. They were annealed to complementary oligonucleotides to form double-stranded substrates by boiling in 1× oligo buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and cooling overnight at room temperature. For substrates with random nucleotides, nonprocessing strand oligonucleotides with random nucleotides (AL and HVP) were annealed with three times the amount of the corresponding primer (SP and HVP) in 1× oligo buffer. Their 5′ overhangs were filled in to form blunt-end substrates by using T4 DNA polymerase in 50 mM NaCl–10 mM Tris-HCl–10 mM MgCl2–1 mM dithiothreitol (pH 7.9 25°C)–0.1 mM each deoxynucleoside triphosphate, (dNTP), 50 μg of bovine serum albumin per ml. The reactions were stopped by addition of 20 mM EDTA. The DNAs were phenol-chloroform extracted and precipitated with ethanol. The double-stranded oligonucleotides were then purified in 20% nondenaturing polyacrylamide gels.
Viral constructs and DNA probe.
All the viruses used in this study used the same gag, pol, and env DNA, a 6,610-bp SacI fragment from pNTRE-4B (19). The viruses were generated by cotransfecting cells with the gag-pol-env fragment and another SacI fragment containing an LTR. The LTR-containing Sac I fragment was from pAD1, pUC19 containing a 792-bp SacI fragment from pAS3 (4). Att mutants were generated by site-directed mutagenesis on the LTR fragment. DNA probes were prepared by digesting the gag, pol, and env DNA with PstI. The 900-bp fragment from the gag region, corresponding to bases 1775 to 2683 of pATV8, was gel purified and labeled using the random-prime labeling kit supplied by Life Technology, Inc.
Cell culture.
QT6 cells were maintained in modified Richter's medium (Tufts formula; Irvine Scientific, Irvine, Calif.) containing 5% fetal calf serum. The cells were incubated at 37°C in an atmosphere containing 5% CO2. Cultures were passaged by trypsinization every 2 days when the cells were confluent and were seeded at a density of about 107 cells per 100-mm plate. All transfections were done with Lipofectamine (Gibco-BRL) as recommended by the manufacturer.
In vitro integration and subsequent PCR.
Preparation of the MalE-ASLV integrase fusion protein has been described previously (27). HIV-1 integrase was a generous gift from A. Engelman. In vitro integration reactions were performed as described previously with minor modifications (27). Briefly, 1 pmol of oligonucleotide substrate, 6 pmol of recombinant MalE-IN fusion protein, and 0.1 pmol of target DNA (pUC119 and φX174) were incubated in 20 μl of 20 mM Tris (pH 8.0)–0.01% bovine serum albumin–1 mM dithiothreitol–10% dimethyl sulfoxide, 2 mM MnCl2 or 10 mM MgCl2 at 37°C for 30 min. The reactions were stopped by addition of 20 μg of proteinase K and 0.5 μmol of EDTA. DNA was extracted with phenol-chloroform and then precipitated with ethanol. PCR was used to amplify the plasmid-substrate junction of integration products. One primer (SP or HVP) annealed to the substrate; the other, biotinylated, primer (B-NEW1, B-UC1, B-OKT3, B-20, or B-φX) annealed to a fixed position on the plasmid target. One-tenth of each integration product was incubated in 100 μl of 10 mM Tris (pH 8.3)–50 mM KCl–3 mM MgCl2–200 μM each dNTP–1 μM primers with 2.5 U of AmpliTaq (Perkin-Elmer) and 1 U of PerfectMatch polymerase enhancer (Stratagene) for 35 cycles (94°C for 1 min, 60°C for 2 min, and 74°C for 2 min.). To reduce nonspecific amplification, the reaction mix was heated to 85°C before the AmpliTaq and dNTP were added.
Regeneration of selected substrates.
PCR products (600 to 1,000 μl) were first cleaned by passage through QIA (Qiagen) PCR purification columns and were then purified by gel electrophoresis on a 1% agarose gel to exclude nonspecific PCR products. Gel-extracted (Qiagen gel extraction kit) PCR products were then digested with 8 U of FokI (New England Biolabs) per 100 μl of PCR products. Digested substrates were separated by electrophoresis on a 15% polyacrylamide gel and extracted by the “crush-and-soak” method (33).
3′-end processing assay.
Oligonucleotides (AL-1, AL-SA, and AL-M-1) representing the processing strand of various substrates were 5′ phosphorylated with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) to a specific activity of approximately 3 × 106 cpm/pmol. The radiolabeled oligonucleotides were purified from a 20% denaturing polyacrylamide gel and then annealed to the complementary oligonucleotide to form blunt-ended substrate oligonucleotides. Reactions were performed at 37°C in 20 μl of 20 mM Tris (pH 8.0)–0.01% bovine serum albumin–1 mM dithiothreitol–8% glycerol–10% dimethyl sulfoxide–10 mM MgCl2 with 1 pmol of 32P-labeled oligonucleotide substrate and 6 pmol of recombinant MalE-IN fusion protein. The reactions were stopped at 10-min intervals by addition of 20 μg of proteinase K and 0.5 μmol of EDTA. After an equal volume of formamide was added, 1/10 of the reaction products were loaded onto a 20% denaturing polyacrylamide gel. After electrophoresis, the extent of 3′ processing was determined by phosphorimage analysis of the relative amounts of unprocessed and processed DNA.
Pool sequencing on magnetic beads.
Dynabeads M-280 streptavidin (Dynal Corp.) bound with single-stranded DNA were prepared using a magnetic particle concentrator (Dynal Corp.) as specified by the manufacturer. The sequence of the immobilized single-stranded DNA pool was determined by the dideoxynucleotide chain termination method (USB) with some modification of the protocol. Substrate primers were 5′-end labeled with [γ-P32]ATP to a specific activity of 1 × 106 to 3 × 106 cpm/pmol and purified on a 15% polyacrylamide gel. Labeled primers (106 cpm) were annealed to bound single-stranded PCR products in a 12-μl reaction volume that contained 2 μl of 5× Sequenase buffer (USB). The mixture was incubated at 65°C for 5 min and then at room temperature for 30 min. Extension and termination reactions were carried out by adding 1 μl of 0.1 M dithiothreitol and 2 μl of diluted T7 DNA polymerase (Sequenase 2.0) (1:8 in ice-cold Sequenase dilution buffer [USB]). A portion of this mixture (3.3 μl) was added to 2.5 μl of each of the four Sequenase dGTP termination mixes (USB) prewarmed at 45°C and incubated at 45°C for 5 min. Reactions were stopped by adding 4 μl of Sequenase stop solution (USB). To sequence the starting substrate pool, 1 pmol of starting substrate was mixed with 1 pmol of labeled primer (106 cpm) in a 12-μl reaction volume that contained 1 μl of Sequenase manganese buffer and 2 μl of 5× Sequenase buffer. This mixture was incubated at 95°C for 5 min and cooled on ice. The extension and termination reactions were same as above.
Cloning analysis.
PCR-amplified specific integration products were purified on a 1% agarose gel and cloned using the TA cloning kit (Invitrogen Corp.). Purified DNA was ligated into pCR2.1 and transformed into One Shot competent cells (Invitrogen) as suggested. Recombinant plasmids were analyzed by PCR for orientation and size.
Computer analysis of RNA structure.
RNA secondary-structure predictions were made by using a computer program (46). Nucleotides 1 to 270 of the U5 region and 8730 to 9180 of the U3 region were analyzed. Analysis of overlapping fragments of equal or smaller size did not produce different secondary-structure predictions in the region discussed in this study.
Site-directed mutagenesis.
Mutants with att substitution mutations at the U3 end (U3M) and the U5 end (U5M) were made using the QuikChange site-directed mutagenesis kit (Stratagene). Mutants with substitution mutants at the U5 end with correct secondary structure (U5MC) were made using the ExSite PCR-based site-directed mutagenesis kit (Stratagene).
RT assay.
Production of virus was assayed by determining the amount of reverse transcriptase (RT) activity in the culture medium. Filtered culture medium (12 μl) was incubated with 50 μl of assay buffer [50 mM Tris-Cl (pH 8.3), 6.5 mM NaCl, 10 mM MgCl2, 1% 2-mercaptoetanol, 100 μM ATP, 0.04 U of poly (A)-oligo(dT) (Sigma Chemical Co.), 2% NP-40, 1.0 μCi [α-32P]dTTP] at 37°C for 1 h. The assay made use of 96-well plates (MADENOB; Millipore Corp.) that have a DEAE paper at the bottom of every well. The plate was placed on a vacuum manifold. The reaction mixture was filtered through DEAE paper that was preequilibrated with 2× SSC (0.3 M NaCl, 30 mM Na3C6H5O7 [pH 7.0]). The wells were then washed five times with 150 μl of 2× SSC. The filters were punched out of the wells, dried for 5 min in a 70°C oven, added to 3 ml of scintillation fluid, and counted in a scintillation counter.
RNA isolation and RT-PCR sequencing.
The RNeasy minikit (Qiagen) was used to isolate viral RNA from virions for use in RT-PCR assays. Reverse transcription of an RNA sample and subsequent PCR amplification were carried out using an Access RT-PCR kit (Promega Corp.).
Isolation and Southern analysis of whole-cell DNA.
Whole-cell DNA was isolated as previously described (47). It was separated by electrophoresis through a 0.8% SeaKem LE agarose gel in 1× Tris-borate-EDTA (TBE) at 30 V overnight. The gel was soaked for 15 min in 0.25 N HCl, followed by 30 min in 0.5N NaOH–1.5 M NaCl, and then by 30 min in 1 M Tris-HCl (pH 8)–1.5 M NaCl. The gel was rinsed with distilled H2O between each step. The DNA was transferred to an Immobilon-Ny+ membrane (Millipore Corp.) in 20× SSC overnight, as specified by the manufacturer. Following transfer, the membrane was washed with 5× SSC. The membrane was then exposed to UV light for 30 s using a Stratalinker to cross-link the DNA to the membrane. The membrane was incubated in hybridization solution (5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7], 5× Denhardt's solution [0.1% bovine serum albumin, 0.1% polyvinylpyrrolidone, 0.1% Ficoll], 100 μg of sheared salmon sperm DNA per ml, 0.5% sodium dodecyl sulfate [SDS]) for 2 h at 68°C. It was then hybridized overnight at 65°C after the addition of fresh hybridization solution to which the probe (106 cpm/ml) had been added. The following day, it was washed twice for 15 min at room temperature in 2× SSC–0.1% SDS and then twice for 15 min at 68°C in 0.2× SSC–0.1% SDS. The membrane was air dried and then exposed to BIOMAX MS film (Kodak) and an intensifying screen at −70°C or exposed to a phosphorimager screen.
RESULTS
Strategy.
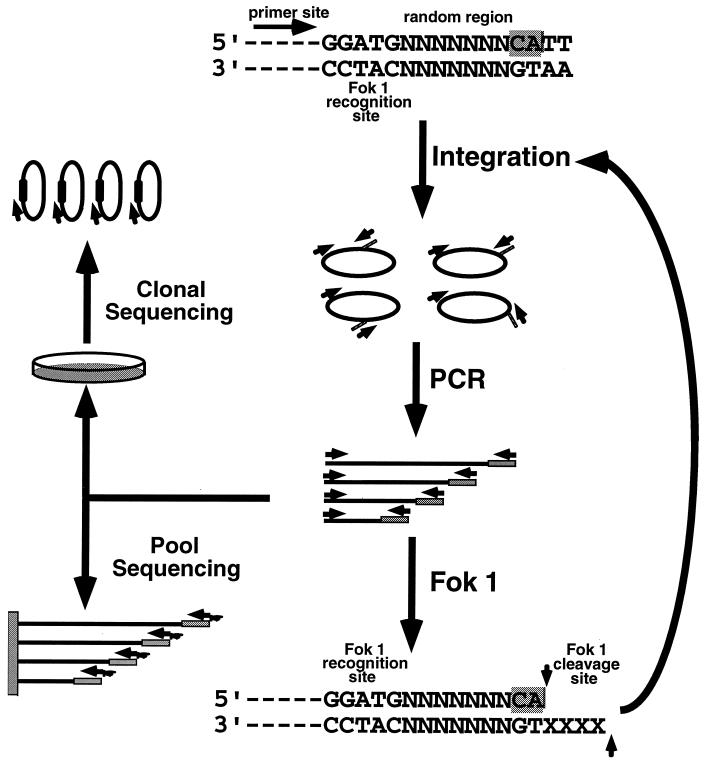
To define substrate sequence specificity, we designed an in vitro evolution scheme to select an optimal substrate from a large pool of oligonucleotides, in which the nucleotide sequence of the region important for integrase recognition was randomized. The principle of our approach is shown in Fig. 1. Substrate oligonucleotides consisted of three parts: a 20-base sequence complementary to the primer oligonucleotide used for amplification and sequencing, the 5-base recognition site for Fok 1, and a sequence based on the terminal 11 bases of U3, but with different numbers of bases (3 to 7) replaced by random sequence. Substrates were selected by competitive integration in vitro. The conditions of the reaction were such that every possible variant of sequence in the random region was present in sufficient amounts (1,000 to 15,000 molecules) in the starting pool, ensuring detection of the optimal substrate. Integrated substrates were enriched by PCR amplification using primers complementary to the end of the substrate oligonucleotide and a sequence in the target DNA and regenerated by digestion with FokI, which cuts 9 and 13 bases from its recognition site, at the junction of the substrate-target joining site. Regenerated substrates were subjected to subsequent rounds of integration selection and enrichment until an optimal sequence emerged. To monitor the selection process, a fraction of each amplified pool was sequenced either directly or (in some cases) after cloning.
FIG. 1.
In vitro evolution strategy. A pool of substrate oligonucleotides was synthesized with a sequence derived from the U3 end of the LTR, except for the presence of randomized nucleotides (designated as N) at the positions of interest, preceded by a primer binding site and a site for recognition by Fok 1 and followed by the pair of bases found in normal viral DNA. This pool was subjected to competitive integration into a circular DNA target in vitro, followed by PCR amplification and regeneration by digestion with Fok 1, a restriction endonuclease which cleaves downstream of its recognition site, at the junction of the substrate-target joining site. Regenerated substrate mixtures were subjected to subsequent cycles of integration and enrichment by amplification until an optimal sequence emerged. A fraction of each PCR pool was sequenced either directly after purification on streptavidin-coated magnetic beads or (in some cases) after cloning.
Selection of optimal substrates for ASLV integrase.
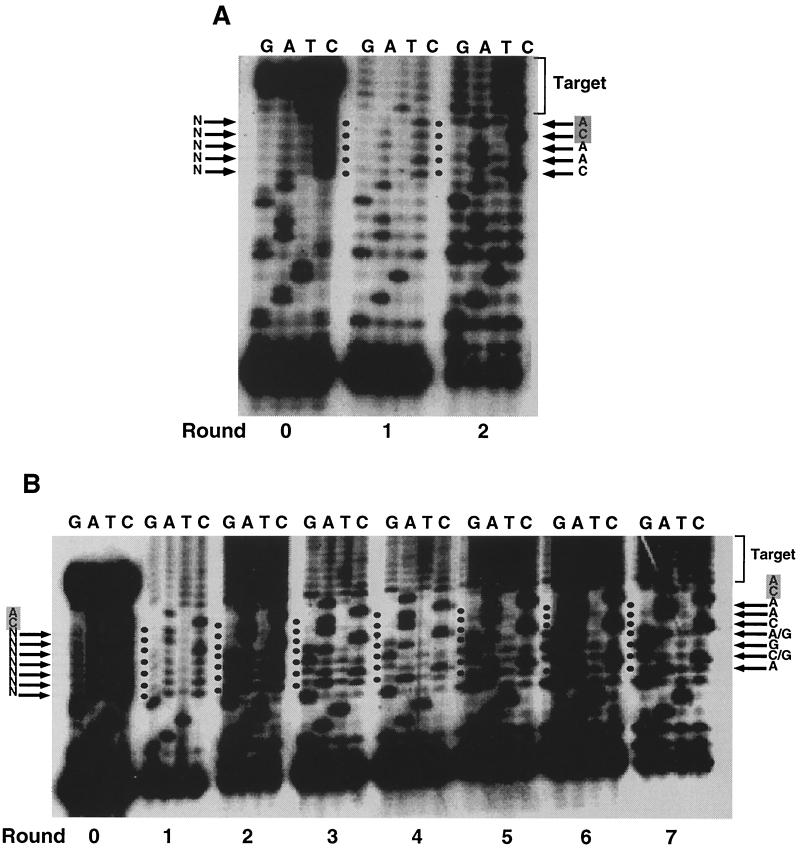
In preliminary experiments, an oligonucleotide pool containing all nine randomized nucleotides provided too few integration products to be amplified efficiently (data not shown). Therefore, two different substrates with overlapping random nucleotides were used in in vitro selection. First, we used a starting substrate pool with the terminal 5 nucleotides randomized (AL-5N). Figure 2A shows the pool sequencing of substrates after each round of integration selection and amplification. Round 0 was the initial substrate pool without any selection. The predominance of C in the randomized sequences did not seem to affect the outcome of the selection. After one round of selection, no nucleotide was obviously selected in the random region, but after another round of selection and amplification, 5′-CAACA-3′ was obviously the dominant sequence. This sequence was preceded by the fixed U3 substrate sequence 5′-GGATGAAGA-3′ and followed by the mixture of sequences in the target plasmid to which the substrate had been joined.
FIG. 2.
Selection of substrates for ASLV integrase. Multiple rounds of selection by integration, amplification, and regeneration were carried out using ASLV integrase and substrates with terminal 5 in which the nucleotides were randomized (AL-5N) (A) or the 7 nucleotides adjacent to the conserved CA were randomized (AL-7NCA) (B). A sample of the reaction mixture of each round was sequenced, and lanes corresponding to termination reaction mixtures are shown. Round 0 is the sequence of the starting pool of double-stranded oligonucleotide substrates without any selection. The starting sequence is shown to the left of the gel, while specific nucleotide sequences emerging in the final round are shown to the right of the gel. The cycling was terminated when the substrate pool showed stronger-than-WT integration efficiency, as judged by the intensity of the integration-PCR product.
Next, a starting pool in which 7 nucleotides adjacent to the conserved CA dinucleotide were randomized (AL-7NCA) was used (Fig. 2B). After two rounds of selection, 3 nucleotides (5′-CAA-3′) at positions adjacent to the conserved CA dinucleotide were visible. This is the identical sequence selected at the same position in the previous experiment (Fig. 2A). The preceding 4 bases emerged more slowly, taking seven rounds of selection to become visible.
Several conclusions can be made from the experiments in Fig. 2. First, the optimal sequence for the ASLV substrate was 5′-ACGACAACA-3′. This conclusion was supported by clonal analysis of the last pool sequence (see below). Second, the closer to the target-joining site of the substrate, the more rapidly the selected nucleotides emerged, implying that substrate nucleotides closer to the integration site played a more important role in integrase recognition and joining. Third, all bases were again approximately equally represented in the target sequence at each round of selection. This observation supports the idea that there is no strong preference for any specific base in the target sequence. Fourth, the optimal sequence selected resembled that found at the ends of the viral DNA but differed from that found at either end (5′-AAGACTACA-3′ at the U3 end and 5′-AAGGCTTCA-3′ at the U5 end of ASLV [conserved nucleotides are in boldface]). The selected sequence more closely resembled that at the U3 end, differing by only 2 bases (underlined), as compared to a 4-base difference from that at the U5 end. This result is consistent with previous reports that U3 ends are better substrates than U5 ends for in vitro integration (44).
Optimal conditions for integrase activity with purified enzyme differ from those for activity of preintegration complexes isolated from infected cells (27, 31). To test the sensitivity of the selected sequence to the reaction conditions, we repeated the selection with some conditions altered in the system. The differences included the divalent cation Mn2+ instead of Mg2+; the divalent ion concentration, from 4 to 20 mM; the enzyme-substrate ratio, from 6:1 to 2:1; and a different target DNA. In all cases, although the efficiency of the integration reaction varied, the same optimal sequence always emerged with approximately the same kinetics (data not shown).
Clonal analysis of selected sequences.
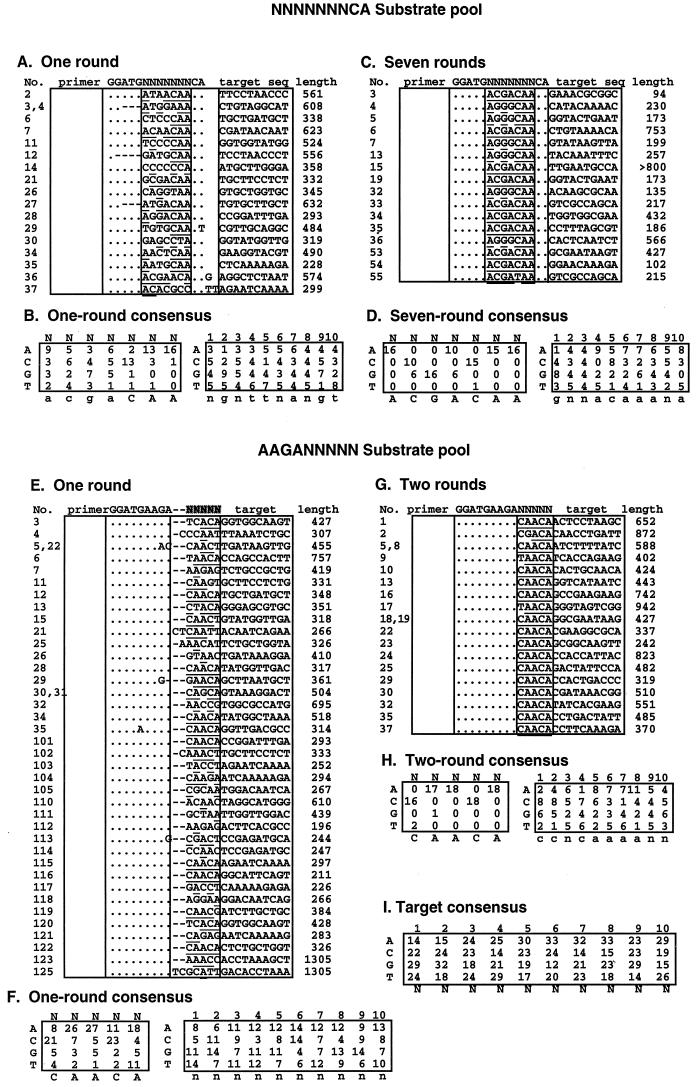
Additional bands can be seen in the sequential selection experiments in Fig. 2B, implying the presence of sequences other than the predominant one in the selected pools. To better understand the selection process and to ensure that the optimal sequence was indeed the predominant one, individual clones from the first and last pools were sequenced and aligned with the substrate sequence (Fig. 3A, C, E, and G). At each randomized nucleotide position, the nucleotides obtained from the sequence of each individual clone were counted and a consensus sequence was determined (Fig. 3B, D, F, and H). While clonal sequences from the last pool showed a very strong predominance of the optimal sequence, clonal analysis of the selected sequence after one round showed a diverse variety of sequences, indicating that ASLV integrase is able to use a wide variety of substrate sequences in vitro. Sequence variation was found at all positions, including the terminal CA, and no base was absolutely forbidden at any position. It is noteworthy that many different terminal dinucleotides were used by integrase and that A and T were approximately half as frequent as the conserved C and A after one round of selection (Fig. 3F). Despite the diversity of individual sequences of the first pool and the absence of optimal sequence in individual clones sequenced, a weak consensus sequence can be seen which is identical to the optimal sequence (Fig. 3A and B), an indication that each nucleotide of the optimal sequence contributes independently to integration. This conclusion was supported by a linkage analysis of every pair of nucleotides of the terminal 5 nucleotides of the 38 clonal sequences in the first-round pool (Fig. 3E and F). Using a χ2 test (results not shown), we determined that the probability that any apparent linkage of all possible pairs of nucleotides was due to chance was well above 5%.
FIG. 3.
Clonal analysis of integrase selected sequences. Amplified pools were cloned into pCR2.1, and the sequences of a number of independent clones were determined. (A, C, E, and G) The sequences of the Fok 1 recognition region, the randomized region, and the target at the joining site are shown, together with the original substrate sequences. The length of each amplified sequence (from the plasmid primer to the target site) is shown on the right side of each table. The nucleotides identical to the starting sequence are indicated as dots, and deleted nucleotides are indicated as dashes. Nucleotides matching the consensus are underlined. (B, D, F, and H) The selected nucleotides at each position from the randomized as well as the target sequence are summarized from the table above. Strong consensus bases are shown in capital letters, and weak consensus bases are shown in lowercase letters. (I) Summary of results for 89 target sequences.
A possible exception to the apparent lack of linkage between selected nucleotides was seen in the seventh-round pool. Of the 16 sequences cloned from this pool, 10 were identical to the optimal sequence, 5′-ACGACAA-3′. However, a second group of six clones, 5′-AGGGCAA-3′ was also present. This sequence was visibly present above random background in the sequencing reaction performed on the seventh pool (Fig. 2B). It did not diminish even after 10 rounds of selection (data not shown). It was presumably a highly competent substrate comparable in efficiency to the optimal sequence.
The first 10 nucleotides of the target sequences were also analyzed for consensus sequence (Fig. 3B, D, F, and H). Although weak preferences could be seen in each individual pool, they disappeared after all the target sequences from 89 clones were aligned (Fig. 3I). This result indicates that the preference for any specific base in the target sequence is weak, if it exists at all.
Selection of optimal substrates for HIV-1 integrase.
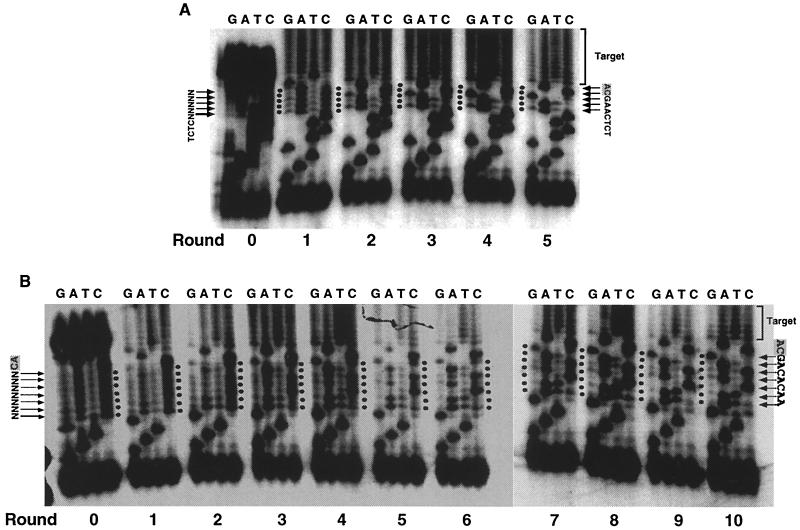
Retroviruses of different groups have quite different recognition sequences for integrase, conserving only the terminal CA dinucleotide (7, 32, 41). We took advantage of this fact to ensure that the selection we observed was on the basis of integration competence (not, for example, PCR amplification or Fok 1 cleavage), since HIV integrase should select a different sequence when used in the same system. Using HIV-1 integrase, we first used an oligonucleotide pool (HVP-5N) in which the terminal 5 nucleotides were randomized. After five rounds of selection, the predominant sequence was 5′-AAGCA-3′ (Fig. 4A). For a starting pool in which 7 nucleotides adjacent to the conserved CA dinucleotide were randomized (HVP-7NCA), the selected sequence that emerged after 10 rounds of selection was 5′-AACACAG-3′ (Fig. 4B). Unlike ASLV, the optimal nucleotide at position 5 depended on the initial substrate pool. Like ASLV, the optimal sequence of HIV-1 differed from either end of the viral DNA (5′-TCTCTAGCA-3′ at the U5 end and 5′-GCCCTTCCA-3′ at the U3 end). The resemblance of the optimal HIV sequence to either natural end was quite remote, with a slightly greater similarity to the sequence at the U5 end than the U3 end, consistent with the observation that the U5 end is a better substrate than the U3 end for HIV integrase (8, 29, 30). Thus, the substrate sequence selected was specific for the integrase used. This system should be useful for defining the optimal sequences of other integrases.
FIG. 4.
Selection of substrates for HIV-1 integrase. Multiple rounds of selection by integration, amplification, and regeneration were carried out using HIV-1 integrase and the substrates with the terminal 5 nucleotides randomized (HVP5N) (A) or the 7 nucleotides adjacent to the conserved CA randomized (HVP7NCA) (B). A sample of the reaction mixture of each round was sequenced, and lanes corresponding to termination reaction mixtures are shown. Round 0 is the sequence of the starting pool of double-stranded oligonucleotide substrates without any selection. The starting sequence is shown on the left of the gel, while specific nucleotide sequences emerging in the final round are shown on the right of the gel. The final round is determined by a stronger-than-WT integration efficiency, judging by the intensity of the integration-PCR product.
The selected nucleotide improves integration efficiency in vitro over the WT nucleotide.
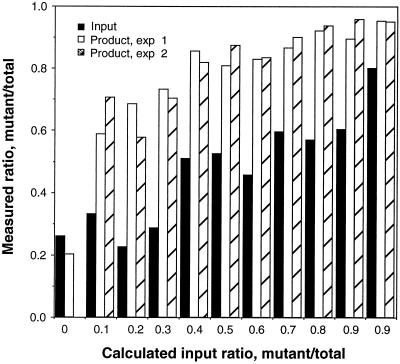
The optimal sequence for the ASLV substrate was different from that found at either end of the viral DNA. Also, the linkage test implied that each nucleotide contributes independently to the productive interaction between ASLV integrase and substrate DNA. To compare the integration efficiency of the selected sequence to the wild-type (WT) sequence, a single-nucleotide change from T to A was introduced at position 4 of the U3 WT substrate and used to compete with the U3 WT substrate in an integration-PCR assay. The two substrates were mixed at different ratios, and the mixture was used as a substrate for integration and subsequent PCR amplification. The PCR products were sequenced. The relative amounts of A and T at position 4, representing the two different substrate sequences, on the sequencing gel were measured. The density ratio of A/(A + T) of the output was plotted against the calculated input ratio, alongside the measured ratio for the starting mixture. The result (Fig. 5) showed a greater ratio of selected to WT substrate in the product than is the input for all ratios tested. This result shows that the nucleotide change increases the integration efficiency over and above that of the WT substrate and confirms that the original selection was on the basis of enhanced integration efficiency and not some other property of the system.
FIG. 5.
Competitive integration-PCR assay. AL-31/32, representing the ASLV U3 WT substrate, was mixed with AL-41/42, whose sequence is identical to AL-31/32 except for a single-nucleotide change from T to A at position 4. The mixture were subjected to a single round of integration and PCR amplification. The PCR products and initial substrate mixture pool were sequenced on a 20% sequencing gel. The relative amounts of A and T at position 4, representing the two integrated substrates, were determined by phosphorimage analysis. The x axis shows the ratio of the substrates added to the reaction mixture. The solid bars represent the value of A/(A+T) of the initial substrate mixture, determined by sequencing the pool prior to the integration-PCR assay. The first solid bar is a background value since there is no selected substrate. The shaded bars are values of A/(A+T) in the integration-PCR products from duplicate experiments.
The selected sequence is a better substrate for 3′-end processing than is the U3 WT substrate.
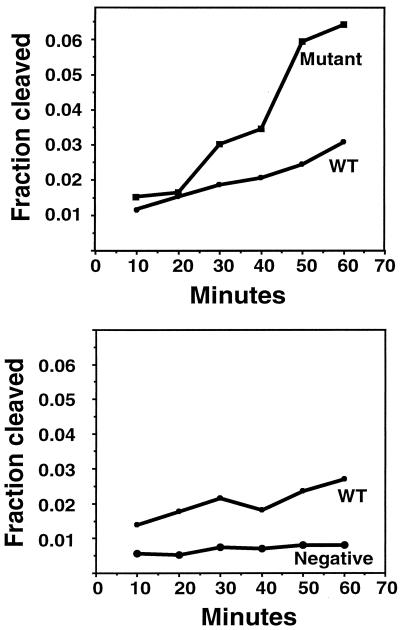
Integrase catalyzes two separate reactions: 3′-end processing and strand transfer. Although the two reactions are carried out by the same enzymatic active site and by a similar transesterification reaction (21), there are significant differences between these two reactions. In 3′-end processing, the viral substrate is attacked by a small nucleophile, usually—but not necessarily—water, while in the strand transfer reaction, the same viral substrate acts as a nucleophile to attack target DNA. How the viral DNA is arranged around the active site in these two reactions is not known, nor is the difference in substrate specificity in these two reactions. In our selection process, integrase carried out only the strand transfer reaction after the first round. It was possible that the sequence selected might be an optimal sequence only for strand transfer, not 3′-end processing. It was therefore of interest to determine whether a sequence selected for optimal strand transfer was also more efficiently used as a 3′-end-processing substrate. A standard 3′-end-processing reaction using ASLV integrase was performed on blunt-ended U3 WT substrate, the optimal substrate (AL-SA/SB), and a known inactive substrate (AL-M-1/2). As shown in Fig. 6, the rate of processing of the selected substrate was more than twice that of the WT. A similar result has been reported for a related mutant (44). Thus, selection for an efficient strand transfer substrate also led to a sequence with increased cleavage efficiency.
FIG. 6.
3′-end processing of selected sequences. A standard 3′-end-processing reaction using ASLV integrase was performed on a 5′ 32P-labeled, blunt-ended U3 WT substrate (AL-1/2), the selected substrate (AL-SA/SB), and a mutant substrate (AL-M-1/2). Reactions were stopped at different time points. 5′-end-labeled processed strands were separated from unprocessed strands on a 20% polyacrylamide gel and quantified by phosphorimager analysis. The percentages of substrate cleaved by IN were determined.
Growth of viral mutants bearing the optimal sequences.
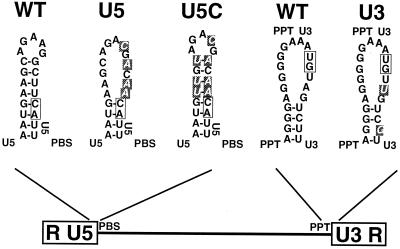
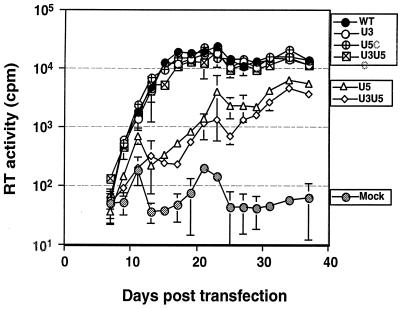
To evaluate the significance of the selected sequence to viral replication and integration, the optimal nucleotides were inserted into either one end or both ends of the WT virus, NTRE-4B (19). Because the base substitutions at the U5 end disrupt a secondary structure important for initiation of reverse transcription (1, 2, 12, 14), a second group of compensatory mutations that restore the secondary structure was also constructed (Fig. 7). Mutant and wild-type viral DNAs were introduced into QT6 cells by transfection, and the resultant virus production and spread were monitored by assaying for reverse RT activity in cell supernatants. We found that placing the mutation within U3 had no effect on the rate and extent of virus spread whereas mutants with base substitutions that disrupted the U5 secondary structure (mutants U5 and U3U5) spread much more slowly (Fig. 8). The introduction of compensatory mutations that restored the predicted secondary structure at the U5 end led to virus that spread at a rate similar to WT virus regardless of whether the U3 end was mutant or WT (Fig. 8). Similar results were noted when QT6 cells were infected with equal amounts (as estimated by RT units) of WT and mutant viruses (data not shown). Based on the studies of this U5 secondary structure by other groups, the slower growth of the U5 mutants is most probably related to a defect in the initiation of reverse transcription (1, 2, 12, 14). To detect possible compensatory mutations that might have arisen during repeated virus replication, sequences around the U3 and U5 att sites were monitored at the end of the passaging experiment using RT-PCR sequencing. We did not find any sequence changes in any of the passaged mutant viruses or in the WT virus (data not shown).
FIG. 7.
Substitutions with optimal nucleotides at the terminal sequences of LTR. Mutations substituted into the LTR are shown in white. Secondary structures were predicted by using M-fold (46).
FIG. 8.
Replication of viral mutants in QT6 cells. The results of RT assays performed on the culture medium of QT6 cells transfected with 10 μg of the indicated DNAs per ml are shown. Cells were passaged when confluent (every 2 days). Error bars indicate the standard deviation of values obtained from three separate transfections with the same DNA.
In vivo integration.
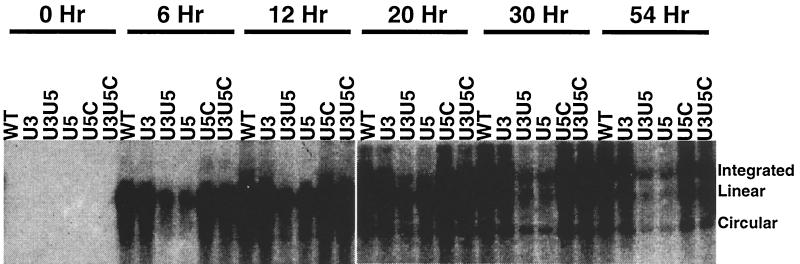
The relative integration efficiencies of WT and mutant viruses were examined by Southern analysis. QT6 cells were infected with equal amounts (as estimated by RT units) of the various viruses. At different time points, whole-cell DNA was extracted, analyzed (without digestion) by agarose gel electrophoresis, and probed with a 900-bp gag fragment. Three forms of DNA were detected by this method: high-molecular-weight DNA (integrated), unintegrated linear DNA, and circular forms (Fig. 9). The linear form of viral DNA, the precursor for integration, appeared earliest, and then its level gradually diminished after reaching a peak around 20 to 30 h after infection. The integrated proviral DNA appeared later, and its appearance was correlated with the decrease of the level of the linear DNA form. Circular DNA is a dead-end product (37, 48), and it appeared before the integrated form but persisted later after infection. The reduced yield of all DNA forms from viruses with U5 mutations that disrupted the RNA secondary structure is consistent with a defect in reverse transcription. This defect was reversed by the compensatory mutations. The integration efficiency, calculated as the density ratio of high-molecular-weight to linear-form DNA, was similar among the different mutants and WT virus (data not shown). In other words, no difference in integration efficiency between the viral mutants and the WT virus could be detected in this assay. The conclusion of the in vivo study is that substitution of U3 sequences by the optimal sequence did not affect viral replication while substitution of U5 sequences by the optimal sequence affected reverse transcription, due to an effect on secondary structure, but not integration.
FIG. 9.
In vivo integration of viral mutants. QT6 cells were infected with equal amounts of freshly harvested WT and mutant viruses (as estimated by RT units). At different time points, whole-cell DNA was extracted and loaded (undigested) onto agarose gels. The gels were blotted and probed with a 900-bp gag fragment, corresponding to bases 1775 to 2683 of pATV8.
DISCUSSION
Design of a functional in vitro evolution system.
In this paper, we describe a novel functional in vitro evolution system designed to obtain the optimal substrate sequence for strand transfer by retroviral integrase. The system comprised three steps repeated for multiple cycles: (i) selection of integration-competent substrates from a pool of substrate oligonucleotides with a portion of the substrate sequence replaced by random bases, (ii) amplification of the integrated sequences, and (iii) regeneration of suitable substrates from the amplified pool for the subsequent cycle of selection.
To the best of our knowledge, this report is the first use of an in vitro evolution strategy to study the viral substrate sequences based on the functional activity of the integrase. There are reports of the selection of high-affinity RNA ligands (3) and DNA substrate sequences for HIV-1 integrase (22). In the former study, selection was based on the RNA binding activity of HIV-1 integrase. The functional relevance of IN-RNA binding is unclear. In the latter study, selection was based only on the 3′-end-processing activity of HIV-1 integrase. Instead of cycled selection, only one round of selection was applied on the random oligonucleotides. In agreement with our result, Esposito and Craigie (22) identified the same first 4 nucleotides, 5′-AGCA-3′, after just one round of selection. Beyond these 4 nucleotides, from positions 5 to 9, as expected from our experience, they were not able to select a dominant nucleotide from the random. Our strategy allows continuous selection of substrates and thus is able to define an optimal sequence for the integrase and provide a dynamic view of the selection process.
Selection of the optimal sequences and their implications.
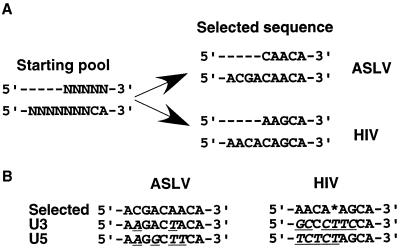
Starting from a pool of substrates with random bases, a consensus sequence emerged after 2 to 10 cycles, depending on the bases randomized. Where the randomized bases overlapped in separate runs, the same sequence was selected by ASLV integrase. The selected sequence resembled the sequence at the ends of the viral DNA, with bases nearer to the joined end being more quickly selected than distal sequences were. The type of integrase used in the integration reaction affected the optimal substrate sequence selected. We obtained the optimal substrate sequence of 5′-ACGACAACA-3′ for ASLV and 5′-AACA(A/C)AGCA-3′ for HIV-1. The HIV-1 sequences differed much more than the ASLV sequences from those naturally found at the two ends of the viral DNA (Fig. 10). With ASLV, the selected sequence differed by 2 and 4 bases from the U3 and U5 ends, respectively. With HIV-1, the equivalent differences were to 6 and 5 bases. This difference may reflect a reduced specificity of the integrase-DNA interaction for HIV-1 compared to ASLV, consistent with the greater difference between the terminal sequences, or greater constraints imposed on the natural sequence by other functions.
FIG. 10.
Summary of the in vitro evolution results. (A) Substrate sequence selection from two overlapping random regions using ASLV and HIV-1 integrases. (B) Comparison of the selected substrate sequences with the natural sequences. Differences are underlined. The nucleotide selected at the position marked by an asterisk is either A or C.
The consensus found with ASLV integrase was quite robust: the sequence obtained was independent of salt and divalent ion concentration, target sequence, and enzyme-substrate ratio in the integration reaction. The selected sequence provided a more active substrate for both strand transfer and 3′ cleavage than did the corresponding WT sequence. We conclude, therefore, that selection was based on integration efficiency and not on other aspects of the system employed. While it is remotely possible that the use of a MalE-IN fusion protein may have affected specificity, the robustness of the sequences obtained and their similarity to the natural substrates argue against this possibility. The optimal sequence we obtained gives a good prediction of the integration efficiency of natural viral substrates. The optimal sequence for ASLV is supported by the result of a mutagenesis analysis (44), in which a substrate of Rous sarcoma virus integrase containing a TT-to AA-mutation at positions 3 and 4 of the U5 LTR terminus showed a significant increase in strand transfer activity (as well as 3′ cleavage) relative to the WT U5 end.
The variety of bases found at each randomized site after the first round (Fig. 2A and 3E) implies that a very large number of different sequences provide usable substrates for strand transfer, in agreement with previous studies using more limited sets of variants (8, 26, 40, 43). The results of clonal analysis of selected mixtures from the first round imply that any base is possible at any site, although consensus sequences, identical to the final selected sequence, are clearly visible. The absence of significant linkage between any pair of bases in the terminal 5 bases implies that interaction between bases is not important for function as a substrate. Rather, it is likely that each base (or base pair) interacts with integrase independently. This conclusion from the observed behavior of the ASLV substrate may not be applicable to the HIV-1 substrate, however. In the HIV-1 system, the choice of optimal nucleotide at position 5 depended on the initial region of randomization (Fig. 4 and 10). A was selected when the terminal 5 nucleotides were randomized, and C was selected when the 7 nucleotides adjacent to CA were randomized. This difference may reflect interaction of this base with one in the upstream sequence.
The two reactions catalyzed by integrase, viral substrate 3′ cleavage and subsequent DNA strand transfer, are similar reactions but involve very different nucleophiles. In the 3′ cleavage step, the viral substrate is attacked by a small nucleophile, usually but not necessarily water. In the strand transfer step, the viral substrate serves as a nucleophile to attack target DNA (21). All evidence to date implies that both reactions occur at the same active site (20, 25, 28). Whether the viral substrate is bound to the same site for these two steps is an unanswered question. It is therefore interesting that the optimal substrate selected through the strand transfer reaction is also superior for 3′-end processing. This result suggests that the viral substrate does not change position on the integrase enzyme to accommodate the target DNA but that instead it is most probably bound in the same way for both reactions.
Limitation of our system.
Our in vitro evolution system has some potential limitations, but we do not believe that they detract seriously from our conclusions. First, after the first round of selection, the substrates we used have a 4-base overhang at the 5′ end of the unprocessed strands instead of the 2-base overhang found under natural conditions. A previous study concluded that a 5′ extension up to 6 bp did not affect the level of specific cleavage and strand transfer (43). The similarity of the consensus from the first round of selection, when the starting oligonucleotide is identical to the natural end, to the final selected sequence implies that the selection is independent of the structure of the end of the unprocessed strand. Furthermore, replacement of the nucleotide 3 bases from the target-joining site with the selected one improved the efficiency of a blunt-ended substrate relative to the WT.
Another potential limitation of the system is that it encompassed only a single integration event, while, in nature, both viral ends must be coordinated to integrate together at positions that are 4 to 6 bp apart on the target DNA (23, 24). The way in which the optimal substrate sequence affects concerted integration is not understood. It has been shown that mutations of the substrate affect both the half-site and full-site reactions in nearly a parallel quantitative fashion (44). Our in vivo study has shown that substitution of WT sequence by the optimal sequence at either end did not detectably affect integration.
In vivo analysis.
The fact that the selected sequences are different from those found in the virus suggests either that a different sequence is optimal in the in vivo context or that the sequences are under other constraints as well. For example, the U5 end sequence is believed to be involved in a complex secondary-structure interaction that is important in the initiation of reverse transcription (1, 2, 12, 14). Our in vivo study of viral mutants bearing the optimal nucleotide substitutions at the U3 end did not show any obvious defect (or improvement) in viral replication, while substitutions at the U5 end affected reverse transcription but not integration. A similar finding shows that many subterminal att mutants of HIV-1 did not affect viral growth and integration in vivo (6). These mutations obviously would affect in vitro integration efficiency. The reasons for the discrepancy between in vivo and in vitro integration are not known. Integration may not be the rate-limiting factor in viral growth. The integrase could naturally accommodate various substrate sequences without affecting the overall growth of the virus. In the virion, integrase is in large excess over the number of molecules required to join the two ends of the viral DNA to cellular DNA targets. If this ratio persists in the preintegration complex in the nucleus, where target DNA is also in large excess, then even relatively low-affinity substrates may be integrated quite rapidly. It is also possible that the conditions in an infected cell alter the recognition specificity, so that different sequences may be optimal in the two contexts. Further experimentation is required to distinguish these possibilities.
ACKNOWLEDGMENTS
We are grateful to Alan Engelman for the generous gift of HIV integrase.
This work was supported by grant R35 CA 44385 from the National Cancer Institute. J.M.C. is a Research Professor of the American Cancer Society.
REFERENCES
- 1.Aiyar A, Cobrinik D, Ge Z, Kung H J, Leis J. Interaction between retroviral U5 RNA and the T psi C loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J Virol. 1992;66:2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Ge Z, Leis J. A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J Virol. 1994;68:611–618. doi: 10.1128/jvi.68.2.611-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen P, Worland S, Gold L. Isolation of high-affinity RNA ligands to HIV-1 integrase from a random pool. Virology. 1995;209:327–336. doi: 10.1006/viro.1995.1264. [DOI] [PubMed] [Google Scholar]
- 4.Aschoff J M, Foster D, Coffin J M. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J Virol. 1999;73:7421–7429. doi: 10.1128/jvi.73.9.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balakrishnan M, Jonsson C B. Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J Virol. 1997;71:1025–1035. doi: 10.1128/jvi.71.2.1025-1035.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown H E, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 8.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman F D, Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow S A, Brown P O. Substrate features important for recognition and catalysis by human immunodeficiency virus type 1 integrase identified by using novel DNA substrates. J Virol. 1994;68:3896–3907. doi: 10.1128/jvi.68.6.3896-3907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 12.Cobrinik D, Aiyar A, Ge Z, Katzman M, Huang H, Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991;65:3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobrinik D, Katz R, Terry R, Skalka A M, Leis J. Avian sarcoma and leukosis virus pol-endonuclease recognition of the tandem long terminal repeat junction: minimum site required for cleavage is also required for viral growth. J Virol. 1987;61:1999–2008. doi: 10.1128/jvi.61.6.1999-2008.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobrinik D, Soskey L, Leis J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J Virol. 1988;62:3622–3630. doi: 10.1128/jvi.62.10.3622-3630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 16.Colicelli J, Goff S P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985;42:573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- 17.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 18.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 19.Dorner A J, Stoye J P, Coffin J M. Molecular basis of host range variation in avian retroviruses. J Virol. 1985;53:32–39. doi: 10.1128/jvi.53.1.32-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 22.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiwara T, Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci USA. 1989;86:3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz R A, Mack J P G, Merkel G, Kulkosky J, Ge Z, Leis J, Skalka A M. Requirement for a conserved serine in both processing and joining activities of retroviral integrase. Proc Natl Acad Sci USA. 1992;89:6741–6745. doi: 10.1073/pnas.89.15.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura Y, Lee Y M H, Coffin J M. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc Natl Acad Sci USA. 1992;89:5532–5536. doi: 10.1073/pnas.89.12.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequences transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaFemina R L, Callahan P L, Cordingley M G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leavitt A D, Rose R B, Varmus H E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y M H, Coffin J M. Efficient autointegration of avian retrovirus DNA in vitro. J Virol. 1990;64:5958–5965. doi: 10.1128/jvi.64.12.5958-5965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luciw P A, Leung N J. Mechanisms of retrovirus replication. In: Levy J A, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 34.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy J E, De Los Santos T, Goff S P. Mutation analysis of the sequences at the termini of the Moloney murine leukemia virus DNA required for integration. Virology. 1993;195:432–440. doi: 10.1006/viro.1993.1393. [DOI] [PubMed] [Google Scholar]
- 36.Panganiban A T, Temin H M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983;306:155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- 37.Roth M J, Schwartzberg P, Tanese N, Goff S P. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J Virol. 1990;64:4709–4717. doi: 10.1128/jvi.64.10.4709-4717.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman P A, Dickson M L, Fyfe J A. Human immunodeficiency virus type 1 integration protein: DNA sequence reuqirements for cleaving and joining reactions. J Virol. 1992;66:3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Den Ent F M I, Vink C, Plasterk R H A. DNA substrate requirements for different activities of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:7825–7832. doi: 10.1128/jvi.68.12.7825-7832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varmus H, Brown P. Retroviruses. In: Howe M M, Berg D E, editors. Mobile DNA. Washington, D.C.: ASM Press; 1989. pp. 53–108. [Google Scholar]
- 42.Vicenzi E, Dimitrov D S, Engelman A, Migone T-S, Purcell D F J, Leonard J, Englund G, Martin M A. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vink C, van Gent D C, Elgersma Y, Plasterk R H A. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vora A C, Chiu R, McCord M, Goodarzi G, Stahl S J, Mueser T C, Hyde C C, Grandgenett D P. Avian retrovirus U3 and U5 DNA inverted repeats. Role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 45.Vora A C, McCord M, Fitzgerald M L, Inman R B, Grandgenett D P. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter A E, Turner D H, Kim J, Lyttle M H, Muller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Withers-Ward E S, Kitamura Y, Barnes J P, Coffin J M. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 1994;8:1473–1487. doi: 10.1101/gad.8.12.1473. [DOI] [PubMed] [Google Scholar]
- 48.Yang W K, Kiggins J O, Yang D M, Ou C Y, Tennant R W, Brown A, Bassin R H. Synthesis and circularization of N- and B-tropic retroviral DNA in Fv-1 permissive and restrictive mouse cells. Proc Natl Acad Sci USA. 1980;77:2994–2998. doi: 10.1073/pnas.77.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]