Abstract
To study the physiological function of diacylglycerol (DAG) kinase ι (DGKι), which converts DAG to phosphatidic acid, we deleted this gene in mice. In contrast to previous studies showing that DGK isoforms decrease Ras activity, signaling downstream of Ras in embryonic fibroblasts was significantly reduced in cells lacking DGKι. DGKs regulate Ras signaling by attenuating the function of the DAG-dependent Ras guanyl nucleotide-releasing proteins (RasGRPs). We tested whether DGKι inhibited the four known RasGRPs and found that it inhibited only RasGRP3. In addition to activating Ras, RasGRP3 also activates Rap1, which in some cases can antagonize the function of Ras. We demonstrate that DGKι bound to RasGRP3 and inhibited its activation of Rap1 by metabolizing DAG. This inhibition consequently affected Ras signaling. We tested the physiological consequence of deleting DGKι by crossing wild-type or DGKι-deficient mice with mice carrying a v-Ha-Ras transgene, and then we assessed tumor formation. We observed significantly fewer tumors in DGKι-deficient mice. Because Rap1 can antagonize the function of Ras, our data are consistent with a model in which DGKι regulates RasGRP3 with a predominant effect on Rap1 activity. Additionally, we found that DGKζ, which is structurally similar to DGKι, inhibited RasGRPs 1, 3, and 4 and predominantly affected Ras signaling. Thus, type IV DGKs regulate RasGRPs, but the downstream effects differ depending on the DGK.
Diacylglycerol (DAG) is a potent activator of several signaling proteins, many of which, when abnormally active, can contribute to the initiation or promotion of cancer (1, 2). Several enzymes can metabolize signaling DAG, but the major route is thought to be by its phosphorylation, a reaction that produces phosphatidic acid and is catalyzed by the DAG kinases (DGKs) (reviewed in ref. 2).
DAG exerts its effects by activating classical and novel protein kinase C isoforms, as well as other proteins including the Ras guanyl nucleotide-releasing proteins (RasGRPs) (reviewed in refs. 3–5). Although the transforming effects of DAG have been attributed to activation of PKCs, identification of the RasGRP family suggested that DAG could also transform cells by directly activating RasGRPs, which in turn could lead to excess Ras signaling. The four known RasGRP proteins are RasGRP1 [calcium DAG guanine exchange factor II (CalDAG-GEFII)], RasGRP2 (CalDAG-GEFI), RasGRP3 (CalDAG-GEFIII), and RasGRP4 (reviewed in ref. 5). Each RasGRP activates either Ras or Rap1, except RasGRP3, which is unique because it facilitates exchange for both Ras and Rap1 (6–10).
Activating mutations in Ras are found in a number of tumors (reviewed in ref. 11). Using a mouse model, Chin et al. (12) showed that Ras expression was an absolute requirement for melanoma tumor maintenance, and other studies have demonstrated a role for Ras in metastasis (reviewed in ref. 13). Together, these data clearly show that abnormally active Ras contributes to the maintenance and progression of many types of cancer.
Rap1-dependent signaling is not as well understood. Rap1, which shares a high degree of homology with Ras (14), is also able to bind Raf-1 (15) and in doing so may antagonize the function of Ras. Indeed, overexpression of Rap1 can revert the transformation of cells caused by Ras (16). Additionally, Okada et al. (17) demonstrated that inhibiting Rap1 allowed Ras to bind Raf-1 and initiate downstream signaling (17). Conversely, other groups have shown that Rap1 fails to interfere with Ras signaling (18–20). One explanation for these conflicting observations is that competition between Ras and Rap1 depends on a number of factors, including the relative abundance of Ras and Rap1, expression of different Raf isoforms, and perhaps other regulatory proteins.
The antagonistic relationship between Ras and Rap1 places RasGRP3, which can activate both proteins, at an interesting crossroad, and the physiologic response elicited by RasGRP3 is not well characterized. Compared to activating only Ras, concurrently activating both Ras and Rap1 likely reduces downstream events dependent on Ras. For example, Yamashita et al. (6) compared the Ras-dependent responses, neurite outgrowth, and anchorageindependent cell growth in cells expressing RasGRP1, RasGRP2, or RasGRP3. Consistent with simultaneous activation of both Ras and Rap1, RasGRP3 caused responses intermediate between RasGRP1 (Ras activation) and RasGRP2 (Rap1 activation). However, these experiments were performed by overexpressing active RasGRP mutants, so they may not reflect the true physiologic response elicited by RasGRP3. In fact, one would predict that, rather than always activating both Ras and its antagonist, Rap1, a higher degree of regulation might exist in vivo that leads to predominant activation of one or the other.
We have demonstrated that DGKζ binds to and regulates the activity of RasGRP1 by metabolizing DAG (21). Additionally, Jones et al. (22) found that DGKα reduced Ras activation in T lymphocytes, presumably by reducing RasGRP1 activity. Together, these data demonstrate that specific DGK isotypes can regulate RasGRP1, and they suggest that DGKs may regulate other RasGRPs as well. While characterizing mice with targeted deletion of the gene encoding DGKι, we found that cells lacking this DAG kinase had reduced Ras activity. Further experiments demonstrated that DGKι could bind and regulate the activity of RasGRP3 and predominantly reduced Rap1 activation. We crossed transgenic mice expressing the v-Ha-Ras protooncogene with wild-type or DGKι knockout mice and found that mice lacking DGKι developed fewer tumors in response to a phorbol ester or after wounding. Together, our data demonstrate that DGKι regulates RasGRP3 but has a predominant effect on its activation of Rap1.
Experimental Procedures
Materials. Ras transgene (TG.AC) homozygous animals were provided by Judson Spalding (National Institute on Environmental Health Sciences via Taconic Farms. Gary Koretzky (University of Pennsylvania, Philadelphia) supplied dgkz–/+ animals. TG.AC mice were screened by using a Southern blotting probe (pASV) from Philip Leder (Harvard University, Boston) (23). The following supplies were used: anti-myc and anti-hemagglutinin (HA) antibodies (Calbiochem); normal IgG, protein A/G Sepharose, anti-phospho-ERK, anti-ERK1/2, and anti-Rap1 (Santa Cruz Biotechnology); anti-pan-Ras (Oncogene Science); glutathione Sepharose 4B (Amersham Pharmacia); cell culture reagents including Lipo-fectAMINE (Invitrogen); phorbol myristate acetate, platelet-derived growth factor, phosphatase inhibitors, and chemicals not listed (Sigma).
Expression Plasmids. Lawrence Quilliam (Indiana University, Indianapolis) provided RalGDS-GST. HA-DGKs, RasGRP1, Flag-DGKζ, and kinase-dead DGKζ were previously described (21). Kinase-dead DGKι was made by site-directed mutagenesis (Stratagene, 5′-GTGGTGGGGATGACACGGTGGGCTGG-3′). RasGRP3 was obtained from KIAA0351 (Kazusa DNA Research Institute, Kisarazu, Chiba, Japan) and cloned into pCDNA3.1myc/his. The Rap1 plasmid was from Johannes L. Bos (Utrecht University, Utrecht, The Netherlands). Rap1 mutants were made by using site-directed mutagenesis (Stratagene, V12: 5′-GTCCTTGGTTCAGTAGGCGTTGGGAAG-3′ and N17: 5′-AGGCGTTGGGAAGAATGCTCTGACAGTTCAG-3′).
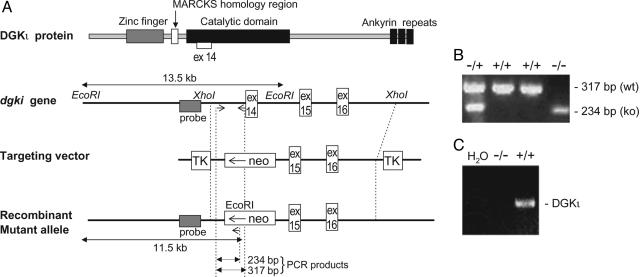
Interruption of the DGKι Gene in Mice. A DGKι gene (Fig. 1) in the phage-targeting vector MDASHII-2TK (Kirk Thomas, University of Utah) was electroporated into R1 ES cells, which were then isolated by positive-negative selection (24). To screen ES cell lines for homologous recombination, DNA was digested with EcoRI and probed by using a DGKι-specific probe external to the targeting vector sequence. Of the 31 selected clones, 8 contained the homologous recombined allele (26%). The homologous recombinant clones were injected into c57/BL6 blastocysts that were then implanted into uteri of pseudopregnant c57/BL6 females. Male chimeric mice were crossed with c57/BL6 females.
Fig. 1.
Generation of DGKι-deficient animals by homologous recombination. (A) DGKι knockout targeting vector with thymidine kinase (TK) and neomycin-resistance cassettes. EcoRI was used for Southern blotting, and the shaded box represents the probe. Also shown are PCR primers (arrows) used for genotyping and the size of the PCR products. (B) PCR to detect the genotype of the animals using genomic DNA from tail lysates. (C) RT-PCR analysis of mRNA from wild-type (+/+) or knockout (–/–) mouse livers to detect expression of DGKι mRNA.
Genotyping by PCR and Southern Blotting. Tail DNA (24) was either analyzed by Southern blotting or tested by PCR by using the primer 5′-AGGATGGTCCAGGAATGGCTTC-3′ and either 5′-AGGTGAGTGAGGCCAACTAGGC-3′ (wild type) or 5′-GAGGGAAGCGTCTACCTACTGG-3′ within the neomycin resistance cassette (knockout). The PCRs used 35 cycles, annealing at 65°C for 1.5 min and extending at 72°C for 1.5 min.
Tumor Induction in Mice. TG.AC mice (FVB/N) were crossed with dgkι–/– or wild-type mice. The Ras transgene was detected either by Southern blotting or by PCR by using primers in the SV40 promoter (5′-CCTCATCATCACTAGATGG-3′) and in H-Ras (5′-GCATGAGCTGCAAGTGTGT-3′). Within each experiment, animals of similar ages and colors were used for controls. For the phorbol 12-myristate 13-acetate (PMA)-induction experiment, shaved animals were treated three times per week for 8 weeks with 1.25 μg of PMA (in acetone), and tumor burden was assessed at 8 weeks. Two TG.AC+/dgki+/+ mice had >40 tumors, the maximum number of tumors counted on each animal in the PMA study (25). The wound-induction experiment was performed as described (26) by making a 1-cm full-thickness incision on the back. Tumor number was recorded at 20 weeks. Statistical analysis was performed by using one-tailed paired t tests.
Mouse Embryonic Fibroblasts/Transfections. HEK293 cells were maintained as described (21). Mouse embryonic fibroblasts from either DGKι or DGKζ mice were prepared by harvesting embryos at embryonic day 14.5. The head and liver were used for genotyping, and the remaining tissue was passed through an 18-gauge needle three times. Cells were cultured in DMEM with 10% FBS and 100 μg/ml penicillin/streptomycin. They were immortalized with SV40 T-antigen. All transfections were performed by using Lipo-fectAMINE, as described (27).
Assays to Detect Activity of H-Ras and Rap1. Activity of Elk-1 was monitored by using the Stratagene Elk-1 luciferase system, as described (21). In six-well plates, RasGRP3 (500 ng per well) was transfected with either a control vector or an HA-tagged DGK (500 ng/well). Elk-1 luciferase activity was normalized to total protein. Pull-down assays for GTP-bound H-Ras were performed as described (21, 28). The same protocol was used to precipitate GTP-bound Rap1, except the Rap1-binding domain of Ral-GDS was used.
Immunoprecipitations and Western Blotting. Transfected HEK293 cells were lysed in either magnesium lysis buffer or immunoprecipitation buffer (21). Cell lysates were precleared with IgG/Protein A/G beads, then mixed with anti-myc-conjugated beads (Santa Cruz Biotechnology) or protein A/G with Rap1 antibodies (Santa Cruz Biotechnology) for 2 h (4°C). Western blotting was performed as described (21). For phosphoERK experiments, cells were harvested into CS lysis buffer (20 mM Tris·HCl, pH 7.5/150 mM NaCl/1 mM EDTA/1 mM EGTA/1% Triton/2.5 mM sodium pyrophosphate/1 mM β-glycerophosphate/1 mM sodium orthovanadate/1 μg/ml leupeptin/1 mM phenylmethylsulfonyl fluoride). Cell lysates were separated by SDS/PAGE, and then phosphorylated ERK1/2 were detected by using antiphosphoERK antibodies and subsequently stripped and reprobed to detect total ERK1/2. In all cases, scanning densitometry was performed by using adobe photoshop to determine relative protein expression in both experimental and lysate/total protein controls.
Results
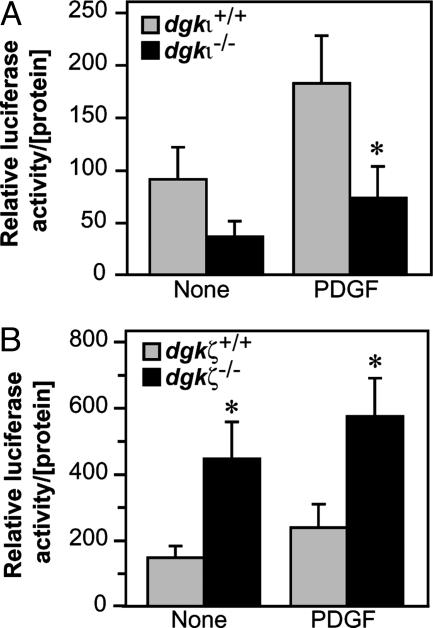
Deleting DGKι Attenuates Ras Signaling. By homologous recombination, we deleted part of the catalytic domain encoded by exon 14 of the DGKι gene in mice (Fig. 1). The knockout allele followed Mendelian distribution of inheritance, and mice lacking DGKι (dgki–/–) had a normal gender ratio, were grossly normal, and did not express truncated or full-length DGKι mRNA (Fig. 1 and data not shown). Because DGKζ, which is structurally similar to DGKι, regulates Ras signaling (21), we compared Ras signaling in primary embryonic fibroblasts derived from either wild-type or dgki–/– mice. Initial experiments demonstrated that wild-type embryonic fibroblasts expressed DGKι mRNA, whereas dgki–/– cells did not (data not shown). Using an assay that measures activity of the Ras-dependent transcription factor, Elk-1, we found that deleting DGKι reduced Elk-1 activity in serum-starved cells as well as cells exposed to platelet-derived growth factor (PDGF, Fig. 2A) or 10% serum (data not shown). Thus, cells lacking DGKι had lower Ras activity, suggesting that DGKι activated Ras signaling. This result was surprising, because we previously demonstrated that DGKζ inhibited Ras activation (21), whereas its deletion activated Ras (29). To directly compare the roles of these two closely related DGK isoforms, we assessed Ras activity in embryonic fibroblasts lacking DGKζ (dgkz–/–). Consistent with our prior observation that DGKζ inhibited Ras activation, cells lacking DGKζ had higher Elk-1 activity compared with wild-type cells (Fig. 2B). Thus, DGKζ/ι have opposing effects on Ras signaling: DGKζ inhibits Ras signaling, whereas DGKι activates it.
Fig. 2.
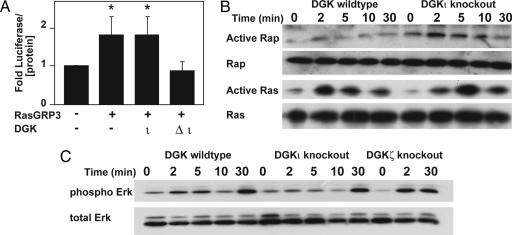
Type IV DGKs have different effects on Ras signaling. Immortalized fibroblasts from either wild-type, dgki–/– (A), or dgkz–/–(B) mouse embryos were transfected with the PathDetect plasmid constructs and starved overnight. They were then treated with platelet-derived growth factor (PDGF, 5.5 h, 40 ng/ml), and luciferase activity was detected. The data shown are the average + SE (n = 4–6). *, P < 0.05, compared with wild type.
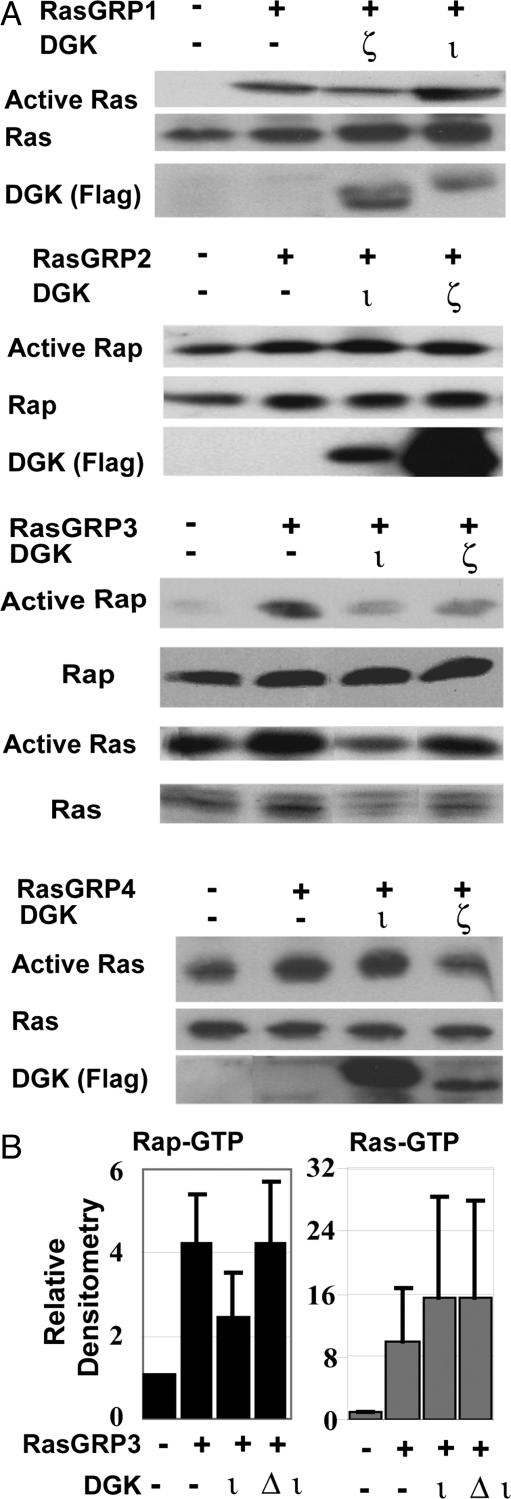
DGKι Inhibits RasGRP3. DGKs terminate DAG signaling and can regulate Ras activity by binding and inhibiting RasGRPs, which require DAG for activity (30). We considered the possibility that DGKι affected Ras signaling by altering RasGRP activity. Using RT-PCR, we found that mouse embryonic fibroblasts expressed RasGRP1–4 (data not shown), indicating that the effects on Ras signaling we observed may have been caused by regulation of any of these RasGRPs. Depending on the subtype, RasGRPs activate either Ras or Rap1. We tested whether DGKι could regulate RasGRP activity in HEK293 cells by coexpressing DGKι with one of the four RasGRP enzymes. Because it had a different effect on Ras signaling (Fig. 2), we also tested DGKζ in these experiments. To measure active Ras or Rap1, we used pull-down assays. As described (reviewed in ref. 30), RasGRP1/4 did not activate Rap1 (data not shown) and, consistent with our published work (21), we found that DGKι did not inhibit RasGRP1, whereas DGKζ inhibited it (Fig. 3A). RasGRP2 activates Rap1 (8), and we found that neither DGKι nor DGKζ affected Rap1 activation by RasGRP2 (Fig. 3A). DGKζ inhibited RasGRP4, whereas DGKι did not (Fig. 3A). Finally, we found that DGKι inhibited RasGRP3, consistently reduced Rap1 activation, and variably affected Ras activation (Fig. 3). DGKζ inhibited both Ras and Rap activation by RasGRP3 (Fig. 3A). When expressed with RasGRP3, a catalytically inactive DGKι mutant (Δι) did not inhibit Rap1 activity (Fig. 3B), consistent with our previous model where DGK activity is required to regulate RasGRP1 (21).
Fig. 3.
DGKι selectively inhibits RasGRP3. (A) HEK293 cells were transfected with RasGRPs along with the empty vector (–), Flag-DGKζ, or Flag-DGKι. After 24–48 h, active Ras (Ras-GTP) or Rap1 (Rap1-GTP) was detected by using GST-RBD pull-down assays. Total cell lysates were used to assess total Ras, Rap1, and DGK (n = 2–6 per experiment). (B) HEK293 cells were transfected with the indicated constructs. After 48 h, Rap1-GTP or Ras-GTP were affinity-precipitated and then detected by Western blotting. Data were analyzed by using scanning densitometry, and the results shown represent active Rap1 or active Ras normalized to total Rap1 or Ras in the lysates + SE (n = 3).
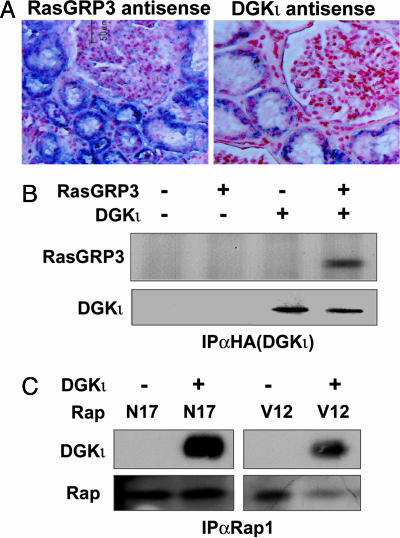
DGKι, RasGRP3, and Rap1 Associate with the Same Signaling Complex. RasGRP3 is expressed in kidney tissue (6). To test whether endogenous DGKι and RasGRP3 were expressed in the same regions within the kidney, we performed in situ hybridization using human kidney tissue. We found that both DGKι and RasGRP3 were expressed in the distal convoluted tubules but not in the glomeruli (Fig. 4A). Thus, the two proteins are expressed in the same cells. Additionally, we found that when both proteins were overexpressed in COS-7 cells/DGKι extensively colocalized with RasGRP3 in the perinuclear region and throughout the cytoplasm (data not shown). We were unable to examine colocalization of the endogenous proteins, because available antibodies do not recognize endogenous DGKι. We next tested whether DGKι could coimmunoprecipitate with RasGRP3 or Rap1 by expressing the proteins in HEK293 cells and then immunoprecipitating DGKι with anti-HA antibodies. Consistent with its inhibition of RasGRP3 and Rap1, we found that DGKι coimmunoprecipitated with both proteins (Fig. 4 B and C). It associated with both inactive (N17) and active (V12) mutants of Rap1. Δι also coprecipitated with both RasGRP3 and Rap1 (data not shown). Furthermore, when we immunoprecipitated Rap1, we detected coprecipitation of endogenous DGK activity (data not shown). We were unable to detect direct in vitro association of purified DGKι and Rap1, suggesting they do not bind directly to each other. Combined, these data suggest that DGKι, RasGRP3, and Rap1 associate in the same signaling complex, allowing the DGK to regulate the activity of RasGRP3 and consequently Rap1.
Fig. 4.
DGKι colocalizes and coimmunoprecipitates with both RasGRP3 and Rap1. (A) Human kidney sections were incubated with either sense or antisense RNA probes specific for DGKι or RasGRP3. The blue signal indicated mRNA for both proteins in the distal convoluted tubules. Little or no expression was observed in the glomeruli. There was minimal signal in the sense (negative) control (data not shown). (B) Cell lysates of HEK293 cells transfected with the indicated plasmids were precipitated with anti-HA antibody. The lysates or precipitated proteins were separated by SDS/PAGE, and then DGKι and RasGRP3 were detected by Western blotting (n = 2–4). (C) HEK293 cells were transfected with the indicated plasmids. Anti-Rap1 antibody was used for immunoprecipitation, and then DGKι and Rap1 were detected by Western blotting (n = 3).
DGKι Predominantly Affects Rap1 Signaling. Active Rap1 is known to interfere with, and thus inhibit, Ras signaling. By predominantly affecting Rap1 activity through RasGRP3, deletion of DGKι would lead to activation of Rap1, which in turn could reduce Ras signaling. This model is consistent with the reduced Ras-dependent luciferase activity we observed in dgki–/– embryonic fibroblasts (Fig. 2A). To further pursue this, we assessed the overall effect that expression of Δι had on Ras-dependent signaling. In HEK293 cells, we found that expression of RasGRP3 increased Ras signaling measured by Elk-1-dependent luciferase activity. Coexpression of wild-type DGKι did not affect Elk-1 activity, but simultaneous expression of Δι and RasGRP3 reduced Elk-1 activity to basal levels (Fig. 5A). This result was consistent with Δι having a dominant-negative effect, causing increased Rap1 activation that interfered with signaling downstream of Ras. However, we found (Fig. 3B) that Δι did not consistently increase Rap1 activation. The inconsistent dominant-negative effects may have been due to the relative expression levels of RasGRP3 and kinase-dead DGKι.
Fig. 5.
DGKι regulates Rap1 activity through RasGRP3. (A) HEK293 cells were transfected with the PathDetect plasmids, RasGRP3, and either DGKι or kinase-dead DGKι. After 48 h, luciferase activity was determined and then normalized to total protein. Data shown are the average + SE (n = 5; *, P < 0.05 compared to control). (B) Immortalized embryonic fibroblasts were starved overnight and then treated with 2 units/ml thrombin. Active Rap1 and Ras were detected by using affinity precipitation followed by Western blotting (n = 2–3 per condition). (C) Immortalized wild-type DGKζ knockout or DGKι knockout embryonic fibroblasts were treated as above, and then phospho-ERK and total ERK levels were measured by Western blotting (n = 3).
The previous results were obtained by overexpressing DGKι and RasGRP3, which may have caused artifacts by altering the normal stoichiometry of these proteins. For example, differing relative expression levels of Δι and RasGRP3 may have caused inconsistent dominant-negative effects by kinase-dead DGKι and could have masked the predicted increase in luciferase activity when wild-type DGKι was overexpressed with RasGRP3. To more accurately examine the effects of DGKι on RasGRP3 and the consequent changes in Ras and Rap signaling, we examined the effects of deleting endogenous DGKι by treating wild-type or dgki–/– embryonic fibroblasts with thrombin and then measuring active Rap1, Ras, and ERK. Based on its selective inhibition of Rap1 (Fig. 3B), we predicted that deletion of DGKι would augment Rap1 activity. Indeed, we found elevated levels of active Rap1 (Rap1-GTP) in embryonic fibroblasts isolated from dgki–/– mice (Fig. 5B). Using densitometry of Western blots at the 2-min time point (n = 3), we found an average of 9.7-fold more active Rap1 in dgki–/– cells. Rap1 can interfere with Ras signaling by forming nonproductive complexes with Raf-1 (reviewed in ref. 31). In doing so, it should not interfere with the levels of Ras-GTP, which we found in Fig. 3B, but should reduce signaling downstream of Ras. We thus predicted that deletion of DGKι, which increased Rap1-GTP (Fig. 5B), should have the same effect. Consistent with this, we found similar levels of Ras-GTP in wild-type and dgki–/– embryonic fibroblasts (Fig. 5B, no significant change by densitometry in two experiments). Although there was no change in active Ras, we found reduced phosphorylated ERK in dgki–/– cells compared with wild-type cells (Fig. 5C). Using densitometry of Western blots at the 2-min time point (n = 3), there was a 30% reduction in ERK phosphorylation in dgki–/– cells compared with wild-type cells. ERK phosphorylation was not reduced in dgki–/– cells at the latest time point (30 min), presumably due to signaling by RasGRP1 at the Golgi apparatus (32). These data are consistent with the reduced Ras-dependent luciferase activity in dgki–/– fibroblasts (Fig. 2A). In contrast, there were higher levels of phosphorylated ERK in dgkz–/– fibroblasts (Fig. 5C), consistent with its effects on RasGRP1 (21). However, because the fibroblasts also express RasGRP3/4, the augmented ERK activity in dgkz–/– cells may have been due to an effect on one of these RasGRPs as well. Together, our data suggest a model where DGKι inhibits RasGRP3 and has a predominant effect on Rap1.
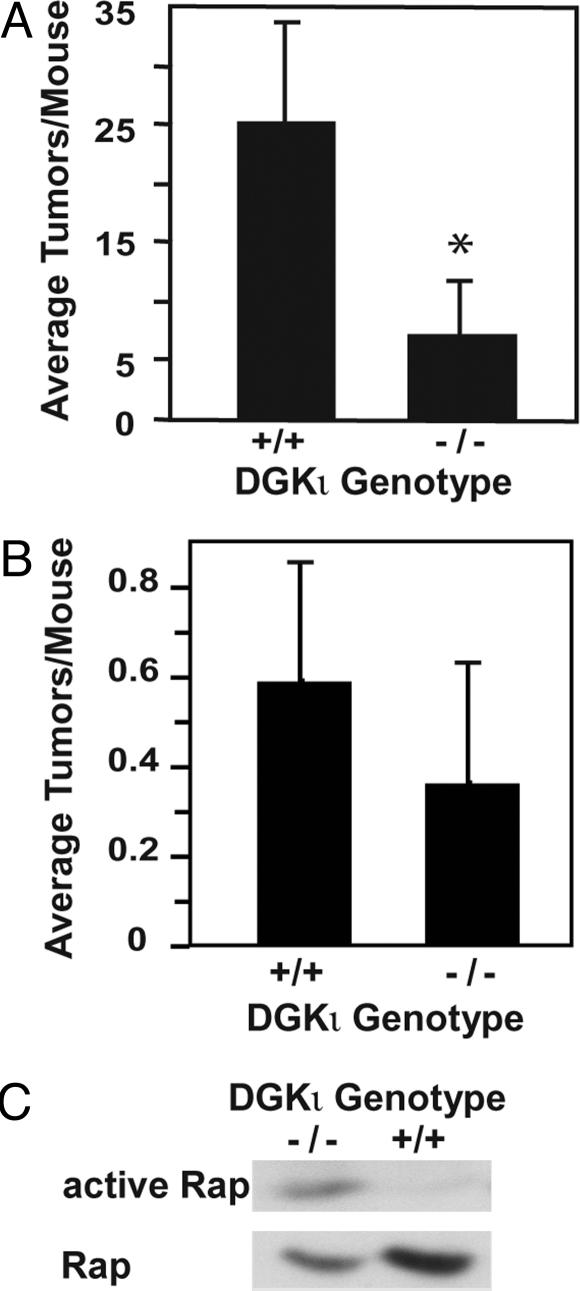
Deletion of DGKι Reduces Ras-Induced Tumor Formation. The reduced Ras activity in embryonic fibroblasts lacking DGKι suggested that deleting DGKι may affect Ras-dependent tumor formation. To investigate this, we used TG.AC mice, which carry multiple copies of a v-Ha-Ras transgene (23, 25), which encodes an active form of Ras that makes them prone to form tumors. For example, skin tumors can be induced by topical application of various carcinogens or by simply wounding the skin (26). In initial experiments, we confirmed using RT-PCR that DGKι mRNA was expressed in skin homogenates from wild-type mice (data not shown). By crossbreeding dgki–/– mice with TG.AC mice, we generated four groups of animals: dgkι+/+/TG.AC– (DGKι wild-type without Ras transgene), dgkι–/–/TG.AC– (DGKι knockout without Ras transgene), dgkι+/+/TG.AC+ (DGK wild-type with Ras transgene), and dgkι–/–/TG.AC+ (DGK knockout with Ras transgene). We first induced skin tumors using PMA. Control mice receiving vehicle alone (acetone) did not develop skin tumors, but application of PMA caused tumor formation. Likewise, full-thickness skin wounding also caused skin tumors. Mice lacking the Ras transgene (TG.AC–) did not develop tumors when wounded or when exposed to PMA (data not shown), indicating that tumor formation in the TG.AC mice was caused by expression of v-Ha-Ras. We found, using either wounding or PMA to induce tumors, that the dgkι–/–/TG.AC+ animals had fewer tumors than the dgkι+/+/TG.AC+ animals (Fig. 6 A and B). The v-Ha-Ras transgene encodes an active form of Ras, indicating that the reduced tumor load in DGKι knockout mice was caused by inhibition downstream of Ras. This was consistent with Fig. 5, where deletion of DGKι activated Rap1, which consequently inhibited ERK phosphorylation but did not affect Ras activation. We were unable to measure active Ras or Rap in the skin tumors but, in spontaneous oral tumors that developed in one dgkι+/+/TG.AC+ and one dgkι–/–/TG.AC+ mouse, we detected higher levels of active Rap1 in the DGKι knockout mouse (Fig. 6C). Thus, deleting DGKι in mice reduced Ras-dependent tumor formation, which was consistent with the attenuated Ras signaling we observed in dgki–/– embryonic fibroblasts. Taken together, our data indicate that DGKι regulates RasGRP3 and Rap1, which consequently modulates Ras signaling.
Fig. 6.
Deletion of DGKι enhances Ras-dependent tumor formation. TG.AC+/dgki+/+ or TG.AC+/dgki–/– animals were either treated with PMA three times per week for 8 weeks (A, n = 8 per condition) or subjected to full-thickness wounding (B, n = 22–25 per condition, tumors assessed at 20 weeks). *, P < 0.05 as compared with vehicle control animals. (C) Using affinity precipitation, levels of active Rap1 were determined in spontaneous mouth tumors from one TG.AC+/dgki+/+ or one TG.AC+/dgki–/– animal.
Discussion
We have demonstrated that DGKι regulates RasGRP3 by associating with it and metabolizing DAG. Inhibition of RasGRP3 by DGKι appears to primarily affect activation of Rap1, consequently altering Ras-dependent signaling events. Indeed, in dgki–/– embryonic fibroblasts, we found higher levels of active Rap1 and reduced ERK phosphorylation. Further, we noted reduced tumor formation in TG.AC mice lacking DGKι. It is important to note that TG.AC mice express an active v-Ha-Ras transgene (23, 25), indicating that the effect of deleting DGKι occurs downstream of Ras. Rap1 can activate B-Raf or act as an antagonist of Raf-1 (reviewed in ref. 31). Consistent with a predominant effect on Raf-1, we found that mouse embryonic fibroblasts expressed high levels of Raf-1 and little, if any, B-Raf (data not shown). Previous studies testing the antitumor effects of Rap1 have demonstrated interesting similarities to our studies. Wani et al. (33) found that expressing Rap1 could suppress H-Ras-induced genomic instability in transformed mouse fibroblasts. Damak et al. (34) found that expressing Rap1 reduced the incidence of lung tumors in a transgenic mouse model. Finally, expression of Rap1 reduced growth rates of the human prostate cancer cell lines PC-3 and TSU-Pr1 and attenuated tumor growth (35). These studies using overexpression of Rap1 (16, 33–35) support our data where changes in the activity of RasGRP3 similarly alter Ras signaling and consequent tumor growth.
Regulation of RasGRP3 by DGKι is consistent with previous models where enzymes that modify lipids associate in signaling complexes with proteins activated by the lipids. This allows the enzymes to cause discrete, rather than global, changes in signaling lipids that may comprise specificity. Combined with previous work demonstrating that both DGKζ and -α can regulate RasGRP1 (22, 36) and the inhibition of RasGRP3/4 by DGKζ (Fig. 3A), these data demonstrate that regulation of RasGRPs by DGKs is common.
Because each DGK isoform appears to be regulated uniquely, individual DGKs may regulate RasGRP proteins in specific tissues, cells, or even compartments within a cell. We also found that DGKζ coimmunoprecipitated with RasGRP3 (data not shown). And like DGKι, DGKζ was also coexpressed with RasGRP3 in kidney tissue. Consistent with its structural similarities to DGKι, DGKζ also reduced activation of both Ras and Rap1 (Fig. 3A). We tested the effects of DGKζ on signaling downstream of Ras by measuring Elk-1 luciferase activity induced by RasGRP3. In contrast to DGKι, wild-type DGKζ, when expressed with RasGRP3, reduced Elk-1 activity, whereas kinase-dead DGKζ augmented its activity (data not shown), suggesting that DGKζ inhibits Ras signaling. Thus, both type IV DGKs inhibit RasGRP3, but DGKι predominantly affects Rap1 signaling, whereas DGKζ appears to primarily affect Ras signaling. In fact, DGKζ appears to broadly affect Ras signaling, because it also inhibited the Ras-specific RasGRP1/4 (Fig. 3 and ref. 21), and its deletion enhanced phosphorylation of ERK in embryonic fibroblasts treated with thrombin for 2 min (Fig. 5C). Finally, we crossed DGKζ knockout mice with TG.AC mice and found that dgkz–/–/TG.AC+ mice had more tumors than dgkz+/+/TG.AC+ mice (2.8 +/–1.2 vs. 4.5 ± 3.7, n = 8) when treated with phorbol ester. Thus, DGKζ appears to inhibit the RasGRPs that activate Ras, whereas DGKι appears to selectively affect Rap1 activation.
The Ras- or Rap-selective effects that DGKζ/ι have on RasGRP3 may be due to the ability of the DGKs to bind either Ras or Rap1 in addition to RasGRP3. For example, we found that overexpressed DGKζ could associate with H-Ras (21), which is consistent with its inhibition of Ras signaling, whereas DGKι associated with Rap1 (Fig. 4B). Selectivity may also be caused by the ability of a DGK to access subcellular compartments along with RasGRP3, Ras, or Rap1. Because there are no antibodies sensitive enough to recognize endogenous DGKι, we have not been able to compare its affinity for endogenous Ras or Rap1 or its true subcellular localization. However, our data clearly demonstrate that DGKι inhibits RasGRP3 and Rap1. More broadly, we have shown that type IV DGKs regulate RasGRP3, but the downstream events affected by these DGKs differ.
Acknowledgments
Diana Stafforini, Thao Doan, John Gannon, Shannon Tew, Rebecca Abbey, Tracy Crotty, and Bai Luo provided technical and editorial advice. This work was supported by the Huntsman Cancer Foundation and National Institutes of Health Grant CA95463 (to M.K.T.). D.S.R. is supported by a National Institutes of Health, National Research Service Award (1F32CA93048) from the National Cancer Institute.
Author contributions: D.S.R., F.S., and M.K.T. designed research; D.S.R., J.H., K.M.L., and M.K.T. performed research; D.S.R., S.M.P., and M.K.T. analyzed data; and D.S.R. and M.K.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RasGRP, Ras guanyl-releasing protein; DAG, diacylglycerol; TG.AC, Ras transgene; PMA, phorbol 12-myristate 13-acetate; HA, hemagglutinin.
References
- 1.Chang, J.-S., Noh, D. Y., Park, I. A., Kim, M. J., Song, H., Ryu, S. H. & Suh, P.-G. (1997) Cancer Res. 57, 5465–5468. [PubMed] [Google Scholar]
- 2.Topham, M. K. & Prescott, S. M. (1999) J. Biol. Chem. 274, 11447–11450. [DOI] [PubMed] [Google Scholar]
- 3.Ron, D. & Kazanietz, M. G. (1999) FASEB J. 13, 1658–1676. [PubMed] [Google Scholar]
- 4.Kazanietz, M. G. (2000) Mol. Carcinog. 28, 5–11. [DOI] [PubMed] [Google Scholar]
- 5.Quilliam, L. A., Rebhun, J. F. & Castro, A. F. (2002) Prog. Nucleic Acid Res. Mol. Biol. 71, 391–444. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita, S., Mochizuki, N., Ohba, Y., Tobiume, M., Okada, Y., Sawa, H., Nagashima, K. & Matsuda, M. (2000) J. Biol. Chem. 275, 25488–25493. [DOI] [PubMed] [Google Scholar]
- 7.Ebinu, J. O., Bottorff, D. A., Chan, E. Y. W., Stang, S. L., Dunn, R. J. & Stone, J. C. (1998) Science 280, 1082–1086. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki, H., Springett, G. M., Shinichiro, T., Canales, J. J., Harlan, P., Blumenstiel, J. P., Chen, E. J., Bany, A., Mochizuki, N., Ashbacher, A., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 13278–13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuther, G. W., Lambert, Q. T., Rebhun, J. F., Caligiuri, M. A., Quilliam, L. A. & Der, C. J. (2002) J. Biol. Chem. 277, 30508–30514. [DOI] [PubMed] [Google Scholar]
- 10.Yang, Y., Li, L., Wong, G. W., Krilis, S. A., Madhusudhan, M. S., Sali, A. & Stevens, R. L. (2002) J. Biol. Chem. 277, 25756–25774. [DOI] [PubMed] [Google Scholar]
- 11.Bos, J. L. (1989) Cancer Res. 49, 4682–4689. [PubMed] [Google Scholar]
- 12.Chin, L., Tam, A., Pomerantz, M. W., Wong, M., Holash, J., Bardeesy, N., Shen, Q., O'Hagan, R., Pantginis, J., Zhou, H., et al. (1999) Nature 400, 468–472. [DOI] [PubMed] [Google Scholar]
- 13.Scharovsky, O. G., Rozados, V. R., Gervasoni, S. I. & Matar, P. (1999) J. Biomed. Sci. 7, 292–298. [DOI] [PubMed] [Google Scholar]
- 14.Kawata, M., Matsui, Y., Kondo, J., Hishida, T., Teranishi, Y. & Takai, Y. (1988) J. Biol. Chem. 263, 18965–18971. [PubMed] [Google Scholar]
- 15.Nassar, N., Horn, G., Herrmann, C., Scherer, A., McCormick, F. & Wittinghofer, A. (1995) Nature 375, 554–560. [DOI] [PubMed] [Google Scholar]
- 16.Kitayama, H., Sugimoto, Y., Matsuzaki, T., Ikawa, Y. & Noda, M. (1989) Cell 56, 77–84. [DOI] [PubMed] [Google Scholar]
- 17.Okada, S., Matsuda, M., Anafi, M., Pawson, T. & Pessin, J. E. (1998) EMBO J. 17, 2554–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwartkruis, F. J. T., Wolthuis, R. M. F., Nabben, N. M. J. M., Franke, B. & Bos, J. L. (1998) EMBO J. 17, 5905–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.York, R. D., Yao, H., Dillon, T., Ellig, C. L., Eckert, S. P., McCleskey, E. W. & Stork, P. J. (1998) Nature 392, 622–626. [DOI] [PubMed] [Google Scholar]
- 20.Vossler, M. R., Yao, H., York, R. D., Pan, M. G., Rim, C. S. & Stork, P. J. (1997) Cell 89, 73–82. [DOI] [PubMed] [Google Scholar]
- 21.Topham, M. K. & Prescott, S. M. (2001) J. Cell Biol. 152, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, D. R., Sanjuan, M. A., Stone, J. C. & Merida, I. (2002) FASEB J. 16, 595–597. [DOI] [PubMed] [Google Scholar]
- 23.Leder, A., Kuo, A., Cardiff, R. D., Sinn, E. & Leder, P. (1990) Proc. Natl. Acad. Sci. USA 87, 9178–9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez de Turco, E. B., Tang, W., Topham, M. K., Sakane, F., Marcheselli, V. L., Chen, C., Taketomi, A., Prescott, S. M. & Bazan, N. G. (2001) Proc. Natl. Acad. Sci. USA 98, 4740–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spalding, J. W., Momma, J., Elwell, M. R. & Tennant, R. W. (1993) Carcinogenesis 14, 1335–1341. [DOI] [PubMed] [Google Scholar]
- 26.Battalora, M. S., Spalding, J. W., Szczesniak, C. J., Cape, J. E., Morris, R. J., Trempus, C. S., Bortner, C. D., Lee, B. M. & Tennant, R. W. (2001) Carcinogenesis 22, 651–659. [DOI] [PubMed] [Google Scholar]
- 27.Bunting, M., Tang, W., Zimmerman, G. A., McIntyre, T. A. & Prescott, S. M. (1996) J. Biol. Chem. 271, 10230–10236. [PubMed] [Google Scholar]
- 28.Franke, B., Akkerman, J.-W. N. & Bos, J. L. (1997) EMBO J. 16, 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong, X.-P., Hainey, E. A., Olenchock, B. A., Jordan, M. S., Maltzman, J. S., Nichols, K. E., Shen, H. & Koretzky, G. A. (2003) Nat. Immunol. 4, 882–890. [DOI] [PubMed] [Google Scholar]
- 30.Springett, G., Kawasaki, H. & Spriggs, D. (2004) BioEssays 26, 6311–6323. [DOI] [PubMed] [Google Scholar]
- 31.Reuther, G. W. & Der, C. J. (2000) Curr. Opin. Cell Biol. 12, 157–165. [DOI] [PubMed] [Google Scholar]
- 32.Caloca, M. J., Zugaza, J. L. & Bustelo, X. R. (2003) J. Biol. Chem. 278, 33465–33473. [DOI] [PubMed] [Google Scholar]
- 33.Wani, M. A., Denko, N. C. & Stambrook, P. J. (1997) Somat. Cell. Mol. Genet 23, 123–133. [DOI] [PubMed] [Google Scholar]
- 34.Damak, S., Harnboonsong, Y., George, P. M. & Bullock, D. W. (1996) Mol. Carcinog. 17, 84–91. [DOI] [PubMed] [Google Scholar]
- 35.Burney, T. L., Rockove, S., Eiseman, J. L., Jacobs, S. C. & Kyprianou, N. (1994) Prostate 25, 177–188. [DOI] [PubMed] [Google Scholar]
- 36.Topham, M. K., Bunting, M., Zimmerman, G. A., McIntyre, T. M., Blackshear, P. J. & Prescott, S. M. (1998) Nature 394, 697–700. [DOI] [PubMed] [Google Scholar]