Abstract
Homologous proteins occurring through gene duplication may give rise to novel functions through mutations affecting protein sequence or expression. Comparison of such homologues allows insight into how morphological traits evolve. However, it is often unclear which changes are key to determining new functions. To address these ideas, we have studied a system where two homologues have evolved clear and opposite functions in controlling a major developmental switch. In plants, flowering is a major developmental transition that is critical to reproductive success. Arabidopsis phosphatidylethanolamine-binding protein homologues TERMINAL FLOWER 1 (TFL1) and FLOWERING LOCUS T (FT) are key controllers of flowering, determining when and where flowers are made, but as opposing functions: TFL1 is a repressor, FT is an activator. We have uncovered a striking molecular basis for how these homologous proteins have diverged. Although <60% identical, we have shown that swapping a single amino acid is sufficient to convert TFL1 to FT function and vice versa. Therefore, these key residues may have strongly contributed to the selection of these important functions over plant evolution. Further, our results suggest that TFL1 and FT are highly conserved in biochemical function and that they act as repressors or activators of flowering through discrimination of structurally related interactors by a single residue.
Keywords: FLOWERING LOCUS T, phosphatidylethanolamine-binding protein, Raf-kinase inhibitor protein, TERMINAL FLOWER 1
Novel morphologies arise through the evolution of new protein functions. Duplicated genes are a key source of new functions, acquiring mutations that affect expression and/or protein sequence (1, 2). Studies of large gene families show that different members diverge and participate in different developmental pathways, for example, homeobox and MADS box genes in various species (3–5). However, it is often unclear and difficult to determine which changes in homologues are critical to establish a novel function.
The Arabidopsis homologues TERMINAL FLOWER 1 (TFL1) and FLOWERING LOCUS T (FT) provide an excellent model to address this question (6–9). Flowering plant species arose > 100 million years ago, and FT and TFL1 have been conserved in diverse species, including monocots and eudicots (10–15). Both TFL1 and FT are key controllers of flowering and plant architecture but act in an opposite manner. TFL1 is a repressor, and FT is an activator. Further, gain-of-function studies gave clear and opposite phenotypes in vivo, showing that protein sequence, rather than expression pattern, largely determines the different functions of TFL1 and FT (8, 9, 16).
TFL1 is expressed in the shoot apical meristem (SAM) and represses the transition to flowering; tfl1 mutants flower early (6, 7, 17, 18). TFL1 also maintains indeterminate growth of the SAM by repressing floral meristem identity genes; tfl1 mutants have their SAMs converted into terminal flowers. TFL1 therefore controls plant architecture by determining where flowers are made and controls when flowers are made by delaying the switch from the vegetative phase to flowering.
The switch to flowering is a critical developmental change in the life cycle of a plant, giving rise to seed production for the next generation. The importance of this flowering transition is reflected in the many genetic pathways that have evolved to respond to diverse external signals, such as day length and temperature, and internal signals such as hormones and developmental controls (19, 20). Integration of these various signals leads to flowering. FT is a key target and integrator of many flowering pathways, and induction of FT expression leads to activation of flowering (8, 9, 21). In contrast, induction of TFL1 results in a suppression of flowering (6, 7, 17, 18). Genetic analyses show that TFL1 and FT act independently in flowering control but, so far, studies in different plant species have not revealed the biochemical mechanism of this family of proteins (8, 9, 11, 13–15, 22–24).
TFL1 and FT are homologous to phosphatidylethanolamine-binding proteins (PEBPs), a wider group of proteins that have diverse roles in animals, yeast, and bacteria. The PEBP family regulates signaling pathways to control growth and differentiation. PEBPs are neural peptide precursors, protease and kinase inhibitors, and Ras-signaling modulators (25–29). Studies on the mammalian homologue Raf-kinase inhibitor protein (RKIP) show that it modulates Raf action, G-protein signaling, and NF-κB activity (30–33). RKIP has been shown to act as a suppressor of cancer metastasis (29). Some PEBPs appear to act biochemically as stoichiometric inhibitors, binding signaling components to modulate the flux through their pathways. However, the biochemical modes of TFL1 and FT action, their interactions, and the molecular nature of their antagonistic effects are unclear.
Here, we reveal a molecular basis for how TFL1 and FT act as opposing functions. We have used crystal structures of PEBPs to identify potentially important residues for their activity. By swapping these residues between TFL1 and FT, we tested whether any residue was not only important but sufficient for determining activator or repressor functions. Our unexpected findings showed that one residue was critical in both proteins and leads us to previously uncharacterized models for how this family of proteins evolved and controls flowering and plant development.
Materials and Methods
Plant Materials, Growth Conditions, and Scoring Phenotypes. All Arabidopsis plants were of the Columbia (Col) ecotype. The tfl1;ft double mutant was obtained from crosses of tfl1-1 [1] and ft-10 (GABI-Kat line 290E08). This ft-10 allele is a T-DNA mutant insertion generated in the context of the GABI-Kat program and was provided by Bernd Weisshaar (Max Planck Institute for Plant Breeding Research, Cologne, Germany) (34). This null allele has a T-DNA insertion in the first intron and was named ft-10. Plants were grown in soil as described in ref. 16, under long days in the greenhouse, with supplementary light as required to maintain long-day conditions (16 h light/8 h dark) at ≈20–24°C. Numbers of nodes generated in each phase of growth were recorded. Batches of at least 10 plants were recorded, and the mean was determined with standard error calculated.
Plasmid Constructions and Transgenic Plants. The TFL1 cDNA (Arabidopsis Stock Center clone 129D7T7, recloned as pD71) and FT cDNA (GenBank accession no. AB027504, recloned as pD298) were used as templates.
For amino acid changes, we made complementary oligos that had appropriate mismatches in their middles, corresponding to the required base pairs needing to be changed to alter the sequence (Table 1, which is published as supporting information on the PNAS web site). These mismatch oligos were used for PCR with appropriate end oligos and templates to generate each version of FT and TFL1 by using the overlap extension PCR method (35). The 5′ and 3′ end oligos carried sites suitable for cloning behind the 35S promoter (via HindIII and BamHI sites) in the pGreen II basic binary vectors 0229 or 0029 used for plant transformations (36). For FT, the 5′ oligo carried a HindIII site and was s163 (cctgaagcttccatggctataaatataagag), whereas the 3′ oligo carried a BclI site (BamHI compatible) and was s164 (cttgatcatctagactataggcatcatcaccg). For TFL1, the 5′ oligo carried a HindIII site and was s165 (cctaagcttacatgtatggagaatatgggaac), whereas the 3′ oligo carried a BamHI site and was s166 (ttcggatcctctagacggattcaactcatctttg). All PCRs used PFU DNA polymerase according to the manufacturer's directions (Promega) and were sequenced by using the Applied Biosystems system, and bigdye 3.1 (PerkinElmer), and samples were processed by the John Innes Centre Genome Laboratory.
Wild-type plants were transformed with constructs in pGreen0229 (which confers basta resistance in plants), whereas tfl1;ft double mutants were transformed with pGreen0029 (which confers kanamycin resistance in plants) (37).
Results and Discussion
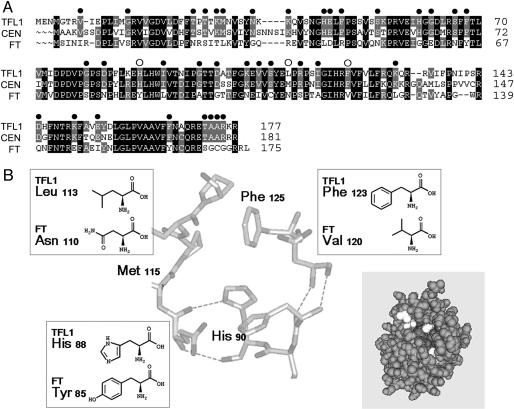
Loss of TFL1 function causes early flowering and conversion of the shoot to a flower. Constitutive TFL1 expression causes late flowering, and all phases are greatly extended (16). Conversely, FT activates flowering with constitutive FT causing early flowering and conversion of the shoot meristem to a flower (8, 9). TFL1 and FT share ≈59% identity. Sequence comparison of FT with TFL1 and CENTRORADIALIS (CEN) (the functional TFL1 homologue from Antirrhinum; refs. 11, 12, and 22) identified 38 amino acid differences, but these differences did not cluster into any clear domain that might confer functional specificity (Fig. 1A).
Fig. 1.
Comparison of TFL1 and FT amino acid sequences. (A) Sequence comparison of TFL1 (and its functional homologue CEN) with FT. Identical amino acids are in black; conserved in gray. Filled circles highlight residues conserved in TFL1 and CEN, but different from FT. Open circles highlight the three residues at the potential binding pocket that were tested for function. (B) Structure of the potential binding pocket of CEN, highlighting the positions and structures of the three residues that differ between TFL1 and FT. [Reprinted with permission from ref. 40 (Copyright 2000, Elsevier).] The insert (gray) view of the complete CEN structure is shown with the three residues highlighted in white. Structure is from Protein Data Bank ID code 1QOU and viewed by cn3d 4.1 (40, 41).
Protein crystal structures have identified a potential phosphate ligand-binding pocket in the PEBP family (38–40). The crystal structure of CEN shows that it is highly conserved with animal PEBPs, including the potential binding pocket. Although the sequence of the pocket is highly conserved between CEN/TFL1 and FT, we focused on three amino acid positions corresponding to residues 90, 115, and 125 in CEN that surround the pocket and which show structural and charge differences (Fig. 1B).
Amino acid variation at the pocket could affect molecular interactions and function. In particular, we asked whether TFL1 and FT specificity depend on the appropriate residue. To test this hypothesis, each amino acid at the corresponding positions in TFL1 (88, 113, and 123) were changed (singly or in combinations) to the corresponding amino acids of FT (Fig. 1B). Similarly, the amino acids of FT were changed to those of TFL1/CEN. The resulting proteins were expressed through the constitutive CaMV 35S promoter in wild-type Arabidopsis plants, and their functions were examined.
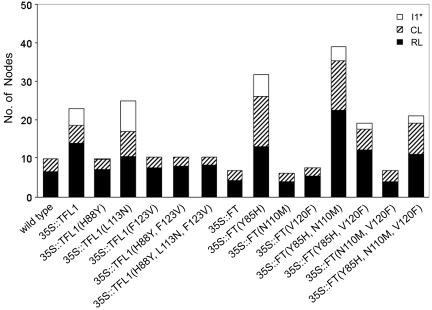
Changing one corresponding residue altered both TFL1 and FT action. Plants overexpressing TFL1 showed late-flowering phenotypes with an extended first inflorescence phase (I1) of cauline leaves subtending shoots1 phase, followed by a characteristic inflorescence phase I1*, where shoots or shoot-like flowers are produced with no subtending cauline leaf (Fig. 2; see also Table 2, which is published as supporting information on the PNAS web site; ref. 16). In contrast, 35S::TFL1 (H88Y or F123V) plants showed a clear shift to early flowering (Fig. 2 and Table 2). Plants overexpressing FT flowered early and made terminal flowers as described in refs. 8 and 9, whereas 35S::FT (Y85H) showed greatly extended phases closely resembling the phenotype of 35S::TFL1 plants.
Fig. 2.
Numbers of nodes on the main shoot of wild-type Arabidopsis plants overexpressing different versions of TFL1 or FT. RL, rosette leaves; CL, cauline leaves subtending secondary shoots; I1*, secondary shoots/shoot-like flowers not subtended by a leaf. The data are from Table 2.
The phenotypes of wild-type plants expressing altered forms of TFL1 and FT suggested that the corresponding first residue (His-88 in TFL1 and Tyr-85 in FT) might be critical in determining both TFL1 and FT functions. However, an alternative possibility was that the phenotypes of the plants overexpressing modified TFL1 or FT were due to gene silencing or dominant-negative effects on endogenous TFL1 or FT function. Therefore, to clarify their actions, we overexpressed wild-type TFL1 or FT and the key proteins, TFL1 (H88Y) and FT (Y85H) in tfl1;ft double-mutant plants. The tfl1;ft plants show an additive phenotype of late flowering and conversion of the shoot meristem into a terminal flower. Therefore, if a protein functioned as FT, it would give early flowering; if a protein functioned as TFL1, it would prevent terminal flower formation in this background.
First, we examined how TFL1 and FT act in tfl1;ft double-mutant plants. Overexpression of TFL1 caused later flowering and prevented formation of terminal flowers (Fig. 3). This finding was consistent with the phenotypes seen in wild-type plants. Overexpression of FT in tfl1;ft mutants caused very early flowering and very early production of a terminal flower. These observations showed that TFL1 and FT acted as predicted and that they did not depend on internal TFL1 and/or FT.
Fig. 3.
Phenotypes of tfl1;ft mutants expressing different versions of TFL1 or FT. (A) Photographs of Arabidopsis plants after 3 weeks of growth. From left to right are tfl1;ft, 35S::FT, 35S::FT (Y85H), 35S::TFL1, and 35S::TFL1 (H88Y). (Scale bar: 1 cm.) (B Upper) Schematic diagrams of mature, flowering plants corresponding to (A). Leaves (ellipses), stems/shoots (lines), flowers (circles) and shoot meristems (arrows). (B Lower) Number of leaves and shoots on the main shoot. RL, rosette leaves; CL, secondary shoots subtended by cauline leaves; I1*, secondary shoots/shoot-like flowers not subtended by a leaf; TF, conversion of the shoot meristem into a terminal flower. Values are given with their standard error of the mean. More than 26 individual T1 plants for each construct were scored. (C) The shoot apex of tfl1;ft (Upper) and 35S::FT (Y85H) (Lower) plants. The terminal flower was absent in 35S::FT (Y85H) expressing plants, showing that this version of FT acted as TFL1 and complemented the tfl1 phenotype. (Scale bar: 0.5 cm.)
The identity of the first residue was sufficient to confer specific TFL1 or FT function. In the tfl1;ft double mutant, 35S::TFL1 (H88Y) flowered early and produced a terminal flower, like plants overexpressing FT (Fig. 3). In contrast, 35S::FT (Y85H) prevented formation of the terminal flower as seen in plants overexpressing TFL1 (Fig. 3C). Thus, the residues were sufficient to change TFL1 and FT functions reciprocally. This result suggests that the biochemical actions of TFL1 and FT are highly conserved despite their diverged protein sequences. Because only one residue was changed in each protein, it indicates that the rest of the TFL1 and FT proteins are sufficiently similar to then act in each other's pathways. Interestingly, to convert His to Tyr and vice versa, only a single nucleotide mutation was required. Such a mutation could therefore arise naturally to change either of the two proteins function.
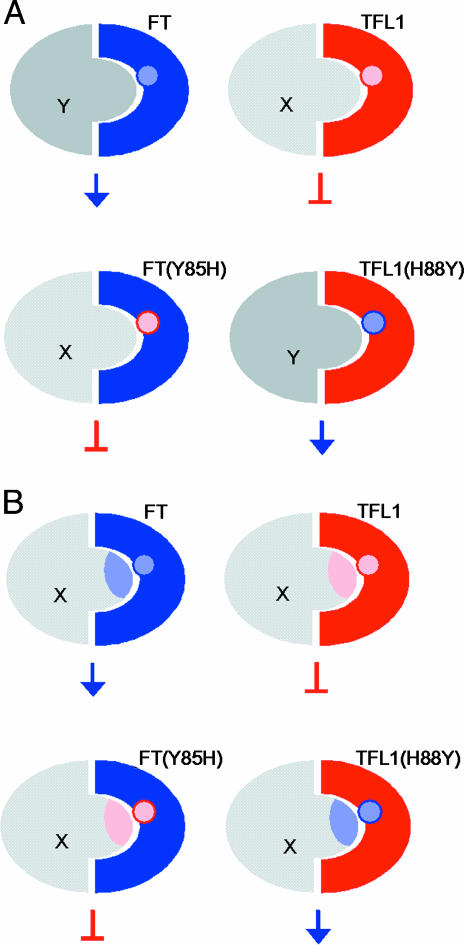
Our data suggests two possible models to explain how one key residue can determine overall protein function as either a repressor or an activator. In one model, TFL1 and FT interact with related components X and Y that carry structurally similar domains (Fig. 4A). The specificity of their interactions depends on the identity of the key residue and, by swapping this residue, FT and TFL1 can swap interacting partners. This change is sufficient to swap pathways and thus changes a repressor to an activator of flowering and vice versa. In the second model, both TFL1 and FT interact with the same component (Fig. 4B). The key residue must confer some change to X or to the complex, structurally or in its activity and, thus, lead to repression or activation.
Fig. 4.
Molecular models for how TFL1 and FT act through the key amino acid. (A) TFL1 and FT recruit structurally related components X and Y depending on the identity of the key amino acid. Activated X complex then leads to repression of flowering, whereas activated Y complex promotes flowering. TFL1 and FT are structurally similar, as is X and Y. Therefore, swapping the key residue is sufficient for TFL1 and FT to swap partners and, thus, their effects on shoot meristem identity. (B) TFL1 and FT interact with the same component X. The key residue specifies whether the component functions as a repressor or an activator. Binding of FT to X induces some change (represented by blue shading of X) that leads to activation of flowering. TFL1 binds X and induces repression (pink). These changes depend on the key residue.
There are many examples of the second model. For instance, a single transcription factor may be an activator or repressor, depending on which protein it interacts with (42, 43). However, there are no clear examples showing that such interactions are specified by a single amino acid. For the first model, there is a clear example. Proteins carrying SH2 domains bind specific phosphotyrosine peptides present in interactors (44). Altering the SH2 domain of Src by a single residue changed its binding specificity and, when this modified SH2 domain was placed into Sem-5 protein, it rescued the sem-5 mutant in Caenorhabditis elegans. However, in the case of FT and TFL1, a single residue is sufficient to switch signaling pathways for a whole protein, not just a domain.
In control of flowering time, conversion between FT and TFL1 was incomplete (Fig. 3). Although constitutive expression of TFL1 affected both flowering time and prevented terminal flower formation, FT (Y85H) only acted like TFL1 in controlling terminal flower formation: FT (Y85H) acted like weak FT in controlling flowering time. This finding suggests that the His residue is sufficient to control shoot meristem identity, but that the rest of the protein is required to give full TFL1 function and control flowering time. Interestingly, the early flowering effect of TFL1 (H88Y) was not as strong as FT. This result indicates that the rest of the FT protein is also important to control flowering time. Loss-of-function alleles have also highlighted the importance of other regions of TFL1 and FT for their action (6–9, 40). These results suggest that repression or activation of floral genes in the shoot meristem may be different from their regulation in leaf primordia or floral meristems as measured by flowering time. This observation may reflect a difference in threshold for TFL1 and FT signaling. Alternatively, the distribution of interacting components may be different between the shoot meristem and leaf primordia or floral meristems.
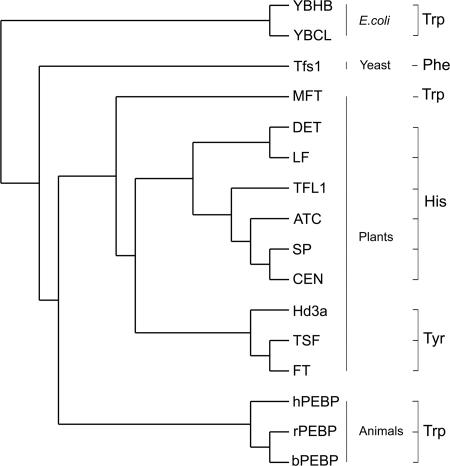
Although the whole protein is required for full function, our results reveal the importance of a key residue in the plant family of PEBPs. Our work shows that to evolve new functions from homologous genes, certain changes may be critical. It is likely that potential binding sites for protein interaction may be strong targets for selection. In diverse plant species, the key residue is always His in those members shown to have TFL1-like function and Tyr in those with FT-like function (Fig. 5). One exception is MFT in Arabidopsis, whose residue is Trp. MFT has weak FT activity: overexpression caused slightly early flowering but no conversion of shoots to flowers (24). This finding suggests that Trp is not sufficient to affect meristem identity, unlike His and Tyr. Further phylogenetic and functional analyses of PEBPs in various plant species will help to reveal how the potential binding pocket functions.
Fig. 5.
A cladogram of PEBP proteins from different organisms. Databases reveal a much greater number of PEBPs, but only those shown to have function or crystal structure are compared. The key residue that swaps function in FT and TFL1 is shown. All plant proteins with His have TFL1-like effects, those with Tyr have FT-like effects, and other organisms have Trp or other residues such as Phe. MFT has Trp and has a weak FT-like effect when overexpressed in Arabidopsis. The tree was generated by the neighbor-joining method. Gen-Bank accession nos.: YBHB (NP_415294), YBCL (NP_415077), Tfs1 (CAA44015), human hPEBP (AAB32876), rat rPEBP (S18358), bovine bPEBP (P13696), Arabidopsis MFT (AF147721), rice Hd3a (BAB61029), Arabidopsis TSF (AB027506), Arabidopsis FT (AB027504), Arabidopsis TFL1 (U77674), Arabidopsis ATC (AB024715), Antirrhinum CEN (S81193), tomato SP (O82088), pea DET (AY340579), and pea LF (AY343326).
Overall sequence comparison places FT-like and TFL1-like proteins into two separate clades. Further, this comparison shows that only about five residues are conserved between members of each clade that distinguishes FT from TFL1. One of these residues is the key residue, but how the others contribute to function awaits further study. Interestingly, the CEN crystal structure reveals that one of these residues (D148 in CEN, equivalent to D144 in TFL1 and Q140 in FT) maps very close to the key residue at the binding pocket.
Our work shows that a single base change could cause conversion of FT to TFL1 and vice versa. If this conversion happened in nature, plant architecture and flowering time could be dramatically altered, with phenotypes similar to those above or completely novel. It may therefore be interesting to look at the phenotypes of plants expressing such novel proteins under native TFL1 or FT promoters. This analysis may also reveal whether any spatially restricted interactions have evolved to contribute to TFL1 or FT function.
Supplementary Material
Acknowledgments
We thank Enrcio Coen, Kim Baumann, and Lucio Conti (John Innes Centre) for useful discussion and comments on the manuscript and Detlef Weigel (Max Planck Institute, Tubingen, Germany) and Leo Brady (University of Bristol, Bristol, U.K.) for helpful discussions. This work was supported by a Biotechnology and Biological Sciences Research Council (U.K.) Core grant to the John Innes Centre, Human Frontier Science Program Organization Grant Ref. RGP0235/2001-M, and a Japan Society for the Promotion of Science grant (to Y.H.).
Author contributions: Y.H. and D.B. designed research; Y.H., T.M., and D.B. performed research; Y.H., T.M., and D.B. analyzed data; and Y.H. and D.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CEN, CENTRORADIALIS; FT, FLOWERING LOCUS T; PEBP, phosphatidylethanolamine-binding protein; TFL1, TERMINAL FLOWER 1.
References
- 1.Lynch, M. & Conely, J. S. (2000) Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- 2.Taylor, J. S. & Raes, J. (2004) Annu. Rev. Genet. 38, 615–643. [DOI] [PubMed] [Google Scholar]
- 3.Gehring, W. J., Affolter, M. & Burglin, T. (1994) Annu. Rev. Biochem. 63, 483–526. [DOI] [PubMed] [Google Scholar]
- 4.Messenguy, F. & Dubois, E. (2003) Gene 316, 1–21. [DOI] [PubMed] [Google Scholar]
- 5.De Bodt, S., Raes, J., Van de Peer, Y. & Theiβen, G. (2003) Trends Plant Sci. 8, 475–483. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R. & Coen, E. (1997) Science 275, 80–83. [DOI] [PubMed] [Google Scholar]
- 7.Ohshima, S., Murata, M., Sakamoto, W., Ogura, Y. & Motoyoshi, F. (1997) J. Mol. Gen. Genet. 254, 186–194. [DOI] [PubMed] [Google Scholar]
- 8.Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., Chory, J., Harrison, M. J. & Weigel, D. (1999) Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M. & Araki, T. (1999) Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- 10.Davies, T. J., Barraclough, T. G., Chase, M. W., Soltis, P. S., Soltis, D. E. & Savolainen, V. (2004) Proc. Natl. Acad. Sci. USA 101, 1904–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S. & Coen, E. (1996) Nature 379, 791–797. [DOI] [PubMed] [Google Scholar]
- 12.Cremer, F., Lönnig, W.-E., Saedler, H. & Huijser, P. (2001) Plant Physiol. 126, 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T. & Yano, M. (2002) Plant Cell. Physiol. 43, 1096–1105. [DOI] [PubMed] [Google Scholar]
- 14.Pnueli, L., Carmel-Goren, L., Hareven, D., Gutfinger, T., Alvarez, J., Ganal, M., Zamir, D. & Lifschitz, E. (1998) Development (Cambridge, U.K.) 125, 1979–1989. [DOI] [PubMed] [Google Scholar]
- 15.Foucher, F., Morin, J., Courtiade, J., Cadioux, S., Ellis, N., Banfield, M. J. & Rameau, C. (2003) Plant Cell 15, 2742–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratcliffe, O. J., Amaya, I., Vincent, C. A., Rothstein, S., Carpenter, R., Coen, E. S. & Bradley, D. J. (1998) Development (Cambridge, U.K.) 125, 1609–1615. [DOI] [PubMed] [Google Scholar]
- 17.Shannon, S. & Meeks-Wagner, D. R. (1991) Plant Cell 3, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez, J., Guli, C. L., Yu, X.-H. & Smyth, D. R. (1992) Plant J. 2, 103–116. [Google Scholar]
- 19.Putterill, J., Laurie, R. & Macknight, R. (2004) BioEssays 26, 363–373. [DOI] [PubMed] [Google Scholar]
- 20.Boss, P. K., Bastow, R. M., Mylne, J. S. & Dean, C. (2004) Plant Cell 16, S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C. & Coupland, G. (2004) Development (Cambridge, U.K.) 131, 3615–3626. [DOI] [PubMed] [Google Scholar]
- 22.Mimida, N., Goto, K., Kobayashi, Y., Araki, T., Ahn, J. H., Weigel, D., Murata, M., Motoyoshi, F. & Sakamoto, W. (2001) Genes Cells 6, 327–336. [DOI] [PubMed] [Google Scholar]
- 23.Pnueli, L., Gutfinger, T., Hareven, D., Ben-Naim, O., Ron, N., Adir, N. & Lifschitz, E. (2001) Plant Cell 13, 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo, S. Y., Kardailsky, I., Lee, J. S., Weigel, D. & Ahn, J. H. (2004) Mol. Cells 17, 95–101. [PubMed] [Google Scholar]
- 25.Tohdoh, N., Tojo, S., Agui, H. & Ojika, K. (1995) Brain Res. Mol. Brain Res. 30, 381–384. [DOI] [PubMed] [Google Scholar]
- 26.Hengst, U., Albrecht, H., Hess, D. & Monard, D. (2001) J. Biol. Chem. 276, 535–540. [DOI] [PubMed] [Google Scholar]
- 27.Bruun, A. W., Svendsen, I., Sorensen, S. O., Kielland-Brandt, M. C. & Winther, J. R. (1998) Biochemistry 37, 3351–3357. [DOI] [PubMed] [Google Scholar]
- 28.Chautard, H., Jacquet, M., Schoentgen, F., Bureaud, N. & Benedetti, H. (2004) Eukaryotic Cell 3, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu, Z., Smith, P. C., Zhang, L., Rubin, M. A., Dunn, R. L., Yao, Z. & Keller, E. T. (2003) J. Natl. Cancer Inst. 95, 878–889. [DOI] [PubMed] [Google Scholar]
- 30.Yeung, K., Seitz, T., Li, S., Janosch, P., McFerran, B., Kaiser, C., Fee, F., Katsanakis, K. D., Rose, D. W., Mischak, H., et al. (1999) Nature 401, 173–177. [DOI] [PubMed] [Google Scholar]
- 31.Kroslak, T., Koch, T., Kahl, E. & Hollt, V. (2001) J. Biol. Chem. 276, 39772–39778. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz, K., Lohse, M. J. & Quiterrer, U. (2003) Nature 426, 574–579. [DOI] [PubMed] [Google Scholar]
- 33.Yeung, K. C., Seitz, T., Li, S., Janosch, P., McFerran, B., Kaiser, C., Fee, F., Katsanakis, K. D., Rose, D. W., Mischak, H., et al. (2001) Mol. Cell Biol. 21, 7207–7217.11585904 [Google Scholar]
- 34.Rosso, M. G., Li, Y., Strizhov, N., Reiss, B., Dekker, K. & Weisshaar, B. (2003) Plant Mol. Biol. 53, 247–259. [DOI] [PubMed] [Google Scholar]
- 35.Horton, R. M., Ho, S. N., Pullen, J. K., Hunt, H. D., Cai, Z. & Pease, L. R. (1993) Methods Enzymol. 217, 270–279. [DOI] [PubMed] [Google Scholar]
- 36.Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S. & Mullineaux, P. M. (2000) Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- 37.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 38.Banfield, M. J., Barker, J. J., Perry, A. C. & Brady, R. L. (1998) Structure 6, 1245–1254. [DOI] [PubMed] [Google Scholar]
- 39.Serre, L., Vallee, B., Bureaud, N., Schoentgen, F. & Zelwer, C. (1998) Structure 6, 1255–1265. [DOI] [PubMed] [Google Scholar]
- 40.Banfield, M. J. & Brady, R. L. (2000) J. Mol. Biol. 297, 1159–1170. [DOI] [PubMed] [Google Scholar]
- 41.Chen, J., Anderson, J. B., DeWeese-Scott, C., Fedorova, N. D., Gerr, L.Y., He, S., Hurwitz, D. I., Jackson, J., Lanczycki, C. J., Liebert, C. A., et al. (2003) Nucleic Acids Res. 31, 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehming, N., Thanos, D., Brickman, J. M., Ma, J., Maniatis, T. & Ptashne, M. (1994) Nature 371, 175–179. [DOI] [PubMed] [Google Scholar]
- 43.Swantek, D. & Gergen, J. P. (2004) Development (Cambridge, U.K.) 131, 2281–2290. [DOI] [PubMed] [Google Scholar]
- 44.Marengere, L. E., Songyang, Z., Gish, G. D., Schaller, M. D., Parsons, J. T., Stern, M. J., Cantley, L. C. & Pawson, T. (1994) Nature 369, 502–505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.