Abstract
We have previously demonstrated that hepatitis C virus (HCV) NS5A protein promotes cell growth and transcriptionally regulates the p21/waf1 promoter, a downstream effector gene of p53. In this study, we investigated the molecular mechanism of NS5A-mediated transcriptional repression of p21/waf1. We observed that transcriptional repression of the p21/waf1 gene by NS5A is p53 dependent by using p53 wild-type (+/+) and null (−/−) cells. Interestingly, p53-mediated transcriptional activation from a synthetic promoter containing multiple p53 binding sites (PG13-LUC) was abrogated following expression of HCV NS5A. Additional studies using pull-down experiments, in vivo coimmunoprecipitation, and mammalian two-hybrid assays demonstrated that NS5A physically associates with p53. Confocal microscopy revealed sequestration of p53 in the perinuclear membrane and colocalization with NS5A in transfected HepG2 and Saos-2 cells. Together these results suggest that an association of NS5A and p53 allows transcriptional modulation of the p21/waf1 gene and may contribute to HCV-mediated pathogenesis.
Hepatitis C virus (HCV) is a major causative agent of acute and chronic hepatitis, which may lead to liver cirrhosis and hepatocellular carcinoma (2, 4, 30). The molecular mechanism of HCV persistence and pathogenesis is not well understood; however, these processes would likely require interaction of viral proteins with a cellular factor(s). HCV contains a single-stranded positive-sense RNA genome which encodes a precursor polypeptide of approximately 3,000 amino acids. This precursor polypeptide is cleaved by both host and viral proteases to at least 10 individual proteins: C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (3). The nonstructural protein 5A (NS5A) is generated as a mature product by the action of NS3 protease in conjunction with NS4A.
NS5A exists as two forms of polypeptides p56 and p58 (16) which are phosphorylated at serine residues, and phosphorylation occurs after the mature NS5A protein is released from the polyprotein (29). NS5A protein localizes in the nuclear periplasmic membrane (37). Apart from the probable role of NS5A in the virus replication cycle, it may play a critical role in determining the susceptibility of the virus to treatment with interferon (IFN). The sensitivity to IFN correlates with mutations within the discrete region of NS5A (7) and is named the IFN sensitivity-determining region. Subsequent analysis suggested that the likely mechanism of IFN resistance occurs through a direct interaction of NS5A with the IFN-induced protein kinase, PKR (9). Since PKR is a critical factor in the response to IFN (17), its inactivation by NS5A may be a possible mechanism by which HCV evades the host immune response. However, the selective pressures exerted on HCV quasispecies during IFN therapy appear to differ among different patients (26, 32). A recent study suggests that NS5A nucleotide and amino acid phylogenies did not correlate with clinical IFN responses and that the domains involved in NS5A functions in vitro were all well conserved before and during IFN treatment (24).
NS5A protein transcriptionally down-regulates the cyclin-dependent kinase inhibitor p21/waf1 gene (11) and promotes cell growth (8, 11). Induction of p21/waf1 is a common mechanism of growth arrest in different physiological situations (6). p21/waf1 may participate in apoptosis, and increased p21/waf1 expression correlates with enhanced cell death under certain conditions (6, 34). p21/waf1 is transiently induced in the course of replicative senescence, reversible and irreversible forms of damage-induced growth arrest, and terminal differentiation of postmitotic cells. The p53 tumor suppressor gene serves as a checkpoint in maintaining genomic stability (19), and p53 function is impaired in the majority of human cancers. p53 is a nuclear protein and consists of at least three functional domains: the N-terminal transcriptional activation domain, the central sequence-specific DNA binding domain, and the C-terminal oligomerization domain (18). The induction of p21/waf1 is regulated through p53-dependent and -independent mechanisms (10). p53 acts as a transcriptional activator and upregulates p21/waf1, leading to p53-dependent G1 arrest (6). Viral gene products target residues of the N terminus of p53 that are employed to interact with the transcriptional machinery of cells (20). In this study, we investigated the molecular mechanism of NS5A functions for requirement of p53 in the p21/waf1 transcriptional regulation.
MATERIALS AND METHODS
Cell lines.
NIH Swiss mouse embryo fibroblast (NIH 3T3), human hepatoma (HepG2), and human osteosarcoma (Saos-2) cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum.
Luciferase assay.
NIH 3T3 and HepG2 cells were transfected with 4 μg of reporter plasmid WWP-luc, Δp53p21-luc, or Δ1.9p21-luc (human p21/waf1 promoter or its deletion mutants in the upstream portion of the luciferase gene) and 2 μg of CMV-NS5A using Lipofectamine (Life Technologies). Empty vector or the CMV MBP-1 gene (12) was used as the control in the luciferase assay. In a different experiment, Saos-2 cells were transfected with 5 μg of PG13-LUC (reporter construct which contains an array of 13 p53 binding sites upstream of the luciferase [LUC] gene), 0.1 μg of CMV-p53 (p53 expression plasmid under the control of the CMV promoter), and different doses of CMV-NS5A (0.1 to 2 μg) by the calcium phosphate precipitation method (Life Technologies). Luciferase activity was determined as previously described (11). Briefly, cells were lysed with reporter lysis buffer (Promega), and the luciferase activity was determined using a luminometer (Optocomp II; MGM Instruments). The activities were normalized with respect to the protein concentration of the cell lysates.
GST pull-down assay.
A glutathione S-transferase (GST)-p53 fusion protein or GST–MBP-1 (13) was expressed in bacteria and immobilized onto GST-agarose beads. The 35S-methionine-labeled full-length NS5A was generated by in vitro translation and incubated with the beads. Subsequently, the beads were washed five times with NETN buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol, 0.02% NP-40, 1 mg of bovine serum albumin/ml), and proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography as previously described (13).
His pull-out assay.
The nuclei from HepG2 cells were prepared as described earlier (41). Briefly, cells were harvested, washed with ice-cold phosphate-buffered saline, and resuspended in hypotonic lysis buffer (10 mM Tris [pH 7.9], 10 mM KCl, 1.5 mM MgCl2). Cells were homogenized, and the nuclei were isolated by centrifugation. Nuclei were resuspended in phosphate-buffered saline with 0.5% NP-40 and sonicated briefly. Extracts were clarified by centrifugation and incubated with His-NS5A or His–MBP-1 beads (14) at 4°C for 2 h. The beads were washed and resuspended in SDS sample buffer. The eluted proteins were separated by SDS–8% PAGE and transferred onto nitrocellulose. The blot was probed with a mouse monoclonal antibody against p53 conjugated to horseradish peroxidase (DO-1; Santa Cruz). Proteins were detected by enhanced chemiluminescence (Amersham).
Coimmunoprecipitation.
HepG2 cells were cotransfected with 2 μg of CMV-p53 and 2 μg of CMV-NS5A, and cell lysates were prepared after 48 h using 0.3 ml of lysis buffer (150 mM NaCl, 10 mM HEPES, pH 7.6, 0.5% NP-40, 5 mM EDTA) containing a cocktail of protease inhibitors (aprotinin, leupeptin, pepstatin, and phenylmethylsulfonyl fluoride). Cell lysates were incubated with a rabbit antiserum to NS5A or pooled normal rabbit sera as a negative control for 4 h at 4°C, followed by an overnight incubation with protein G-Sepharose beads (Pharmacia). The immunoprecipitates were separated by SDS–8% polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose membrane. Immunoblotting was performed by incubation of the membrane for 1 h with a mouse monoclonal antibody against p53 conjugated to horseradish peroxidase (DO-1; Santa Cruz). Proteins were detected by enhanced chemiluminescence (Amersham). The nitrocellulose membrane was reprobed with a monoclonal antibody to NS5A for detection of the viral protein. To examine the expression of exogenous p53, a Western blot analysis was performed using p53 or mock-transfected (control) cell lysates and a monoclonal antibody to p53, followed by detection of chemiluminescence.
Mammalian two-hybrid system.
A mammalian expression plasmid encoding the VP16 transactivation domain of herpesvirus (44) fused to full-length NS5A (VP16-5A) or its deletion mutants and a Gal4 construct of either full-length p53 or different deletion mutants of p53 were used in this study. NIH 3T3 or HepG2 cells were cotransfected with 1 μg of Gal4 responsive reporter gene (G5E1b-CAT) and 2 μg of VP16-5A and p53-Gal or its deletion mutants as effector plasmid DNAs (15), and the chloramphenicol acetyltransferase (CAT) assay was performed as previously described (14). Transfection efficiencies were normalized to an internal β-galactosidase control.
Immunofluorescence study.
HepG2 cells were transfected with 2 μg of CMV-NS5A or empty vector, and after 48 h, cells were washed and fixed with 3.7% formaldehyde followed by blocking with 3% bovine serum albumin. Cells were incubated with either anti-p53 mouse monoclonal antibody (DO-1; Santa Cruz) or a rabbit antibody to NS5A for 1 h at room temperature. Cells were washed and incubated with anti-mouse immunoglobulin (Ig) conjugated with Alexa 568 or anti-rabbit Ig conjugated with Alexa 488 (Molecular Probes) for 30 min at room temperature. Finally, cells were rinsed and mounted for confocal microscopy (Bio-Rad model 1024), and the images were superimposed digitally to allow fine comparison (14, 40). Colocalization of red and green signals in a single pixel produces a yellow color, whereas separated signals remain red or green. A control cell preparation was made using only the secondary antibody conjugates. HepG2 cells expressing NS5A were also stained with antibody to p53 alone for specific detection of endogenous protein. To determine the effect of NS5A on the localization of p53 deletion mutants, HepG2 cells were cotransfected with p53(ΔN14-18)Gal4 and CMV-NS5A plasmid DNAs. Cells were fixed with formaldehyde (3.7%) and stained with a monoclonal antibody to the Gal4 DNA binding domain (Santa Cruz) and/or a rabbit polyclonal antibody to NS5A. Cells were treated with specific secondary antibody and mounted for confocal microscopy.
RESULTS
p53-dependent effect of NS5A on the p21/waf1 promoter.
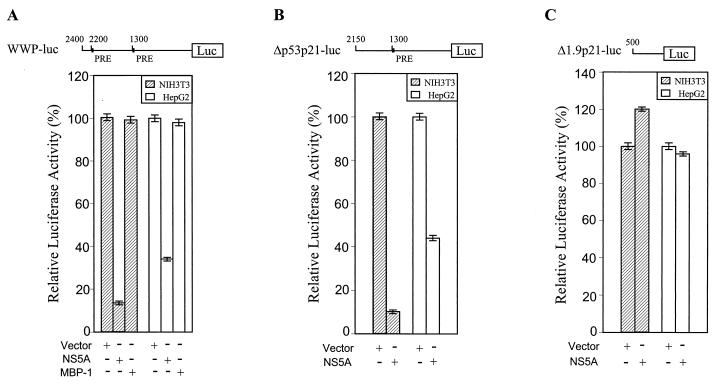
HCV NS5A protein transcriptionally down-regulates p21/waf1 promoter activity and does not bind to the promoter sequences (11). The p21/waf1 promoter is regulated by a p53-dependent or -independent mechanism (10). To determine whether the effect of NS5A on the human p21/waf1 promoter activity is dependent upon the presence of p53-responsive elements (PRE), an in vitro transient-transfection assay was performed using NIH 3T3 and HepG2 cells. Cells were transfected using Lipofectamine with a luciferase reporter construct having either full-length p21/waf1 promoter or its deletion mutant (27) and CMV-NS5A. An unrelated gene, that encoding MBP-1 (12), was used as a negative control and cotransfected with the full-length p21/waf1 promoter reporter construct. The total amount of DNA in each transfection was kept constant using empty vector. After 48 h of transfection, luciferase activity in the cell lysates was measured by a luminometer. Results suggested that NS5A represses the full-length p21/waf1 promoter activity in both cell lines (Fig. 1A). p21/waf1 promoter sequences contain two PREs (6). The extent of repression remains similar when one of the two PREs of the promoter was deleted (panel B). However, when both the responsive elements were deleted, NS5A had no significant effect on the p21/waf1 promoter (Fig. 1C). Thus, NS5A-mediated transcriptional repression of p21/waf1, at least in part, appears to be p53 dependent.
FIG. 1.
NS5A represses p21/waf1 promoters containing PREs in a reporter assay. (A) A schematic diagram of the regulatory region of a p21/waf1 promoter (WWP-luc) containing two PREs is shown at the top. p21/waf1 promoter activity is down-regulated by NS5A in NIH 3T3 and HepG2 cells. CMV MBP-1 plasmid DNA was used similarly as a negative control. (B) Repression of activity of a p21/waf1 promoter with one PRE site deleted (Δp53p21-luc) by NS5A in NIH 3T3 and HepG2 cells. (C) Absence of NS5A repressor activity in a p21/waf1 promoter lacking PRE sites (Δ1.9p21-luc). Cells were cotransfected with the indicated plasmid DNAs, and cell extracts were prepared after 48 h of transfection for determining luciferase activity. In each set of experiments, triplicate transfections were performed. The absolute values of the luciferase in vector- and unrelated-gene (MBP-1)-transfected control were ∼1.7 × 107 relative light unit in NIH 3T3 cells and ∼4 × 106 relative light unit in HepG2 cells. In all cases, the relative luciferase activity of the vector control was arbitrarily assigned a value of 100%.
NS5A represses p53-mediated gene expression.
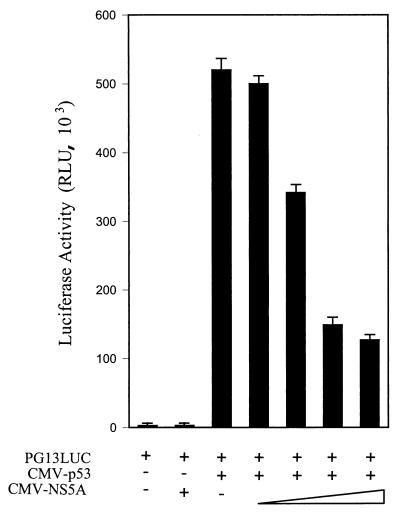
To determine whether the repressive effect of NS5A is indeed dependent on the presence of the PRE of the promoter, we have used a synthetic reporter plasmid with an array of 13 p53-binding sites (PG13-LUC). Saos-2 cells were chosen for this experiment since p53 alleles are deleted in this cell line and endogenous p53 is absent (5). Cells were transfected with the PG13-LUC reporter plasmid, CMV-p53, and various doses of CMV-NS5A. Luciferase activity was measured after 24 h of transfection. p53-dependent activation of the PG13 synthetic promoter was inhibited by NS5A in a dose-dependent manner (Fig. 2). This result suggests that NS5A interferes with p53-dependent transactivation.
FIG. 2.
NS5A modulates p53-dependent transcription. Saos-2 (p53−/−) cells were cotransfected with PG13-LUC reporter (5 μg), CMV-p53 (0.1 μg), and increasing amounts of CMV-NS5A (0.1, 0.5, 1, and 2 μg) plasmid DNAs. The total amount of plasmid DNA was kept constant by the addition of empty vector in each transfection, and luciferase activity was measured after 24 h. Triplicate transfections were performed in each set of experiments. RLU, relative light unit.
NS5A physically interacts with p53.
To examine whether NS5A physically associates with p53, an in vitro GST pull-down assay was performed. GST-p53 or GST–MBP-1 was expressed in bacteria, immobilized onto GST beads, and incubated with 35S-methionine-labeled NS5A generated by in vitro translation. Analysis of the proteins in the binding mixture, by SDS-PAGE and autoradiography, suggested retention of the NS5A polypeptide by GST-p53 beads (Fig. 3A). However, under similar experimental conditions, GST–MBP-1 did not pull down the NS5A protein. Results from this binding assay suggested an in vitro association of NS5A with p53. We also examined whether endogenous p53 protein, present in the nuclear extracts, interacts with NS5A. For this experiment, His-NS5A or His–MBP-1 protein was incubated with the nuclear extracts from HepG2 cells and His pull-out assay was performed. Results suggested that endogenous p53 protein specifically interacts with NS5A and MBP-1 did not exhibit an interaction with p53, as expected (Fig. 3B).
FIG. 3.
Physical association of NS5A with p53. (A) 35S-methionine-labeled NS5A was subjected to a pull-down analysis with GST-p53 fusion protein immobilized on agarose beads (lane 2) or GST–MBP-1 as a negative control (lane 3). Twenty percent of the in vitro-translated NS5A (lane 1) was loaded for gel electrophoresis to authenticate the position of the band derived from the experimental sample. (B) Endogenous p53 protein binds to NS5A. Nuclear extracts of HepG2 cells were incubated with His-NS5A (lane 1) and His–MBP-1 (lane 2) for binding and detection of p53 by Western blot analysis. (C) In vivo coimmunoprecipitation of NS5A with p53. HepG2 cells were cotransfected with CMV-p53 and CMV-NS5A. After 48 h of transfection, cell lysates were immunoprecipitated with a rabbit antiserum to NS5A (lane 1) or pooled normal rabbit sera (lane 2) and immunoblotted with a monoclonal antibody to p53 (DO-1). The molecular weight of the p53 protein band was ascertained from the migration of standard protein molecular weight markers (Life Technologies). The blot was reprobed with a monoclonal antibody for detection of NS5A (bottom panel). The positions of NS5A and Ig heavy chain (from experimental reagents) are shown. (D) Exogenous and endogenous expression of p53 in HepG2 cells. Cells transfected with CMV-p53 (lane 1) and vector control (lane 2) were immunoblotted with a monoclonal antibody to p53.
To further determine whether NS5A and p53 can form a complex in vivo, a coimmunoprecipitation assay was performed. HepG2 cells were transfected with CMV-p53 and CMV-NS5A. Cells were lysed after 48 h of transfection with a low-stringency lysis buffer. Cell lysates were incubated with a rabbit antiserum to NS5A or pooled normal rabbit sera as a negative control. The immunoprecipitates immobilized on protein G-Sepharose beads were separated by SDS-PAGE and blotted onto nitrocellulose membrane. The presence of p53 on nitrocellulose membrane was detected by Western blot analysis using a monoclonal antibody to p53 conjugated to horseradish peroxidase (DO-1; Santa Cruz). Coprecipitation of p53 with NS5A was evident from the specificity of the antibody and the size of the p53 protein (Fig. 3C). On the other hand, cell lysates when similarly analyzed with pooled normal rabbit sera did not exhibit precipitation of p53 protein. The blot when stripped and reprobed with a specific monoclonal antibody also detected NS5A (bottom of Fig. 3C). The levels of p53 expression from transfected and control HepG2 cells are also shown by Western blot analysis (Fig. 3D). However, in untransfected cells endogenous p53 could not be detected as coprecipitate with NS5A in several attempts, possibly for a very low level of p53 expression.
Mapping of p53 and NS5A interacting domains.
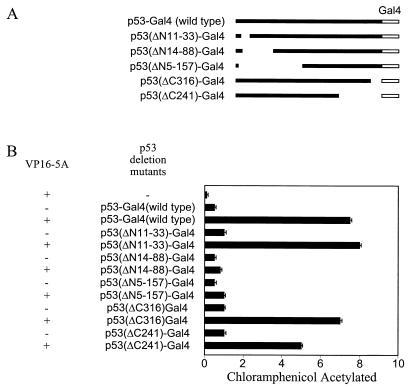
We have used Gal4-constructs of p53 deletion mutants (Fig. 4A) and VP16-5A (14) in a mammalian two-hybrid assay to initially identify the region of p53 responsible for the binding with NS5A. A significant increase in CAT activity was observed when p53-Gal4 (wild type), p53(ΔN11-33)-Gal4, p53(ΔC316)-Gal4, or p53(ΔC241)-Gal4 was cotransfected with VP16-5A. However, CAT activity was not altered following coexpression of p53 deletion mutants and VPFlag empty vector as a negative control. A significant increase in CAT activity was not observed in cell lysates from transfection with VP16-5A and p53(ΔN14-18)-Gal4 or p53(ΔN5-157)-Gal4 as compared to NS5A fusion protein and empty Gal4 chimeric vector coexpression (Fig. 4B). The results from this assay suggested that the NS5A interacting domain is localized in the N-terminal region (amino acids 33 to 88) of p53. To map the region of NS5A associating with p53, deletion mutants of the NS5A genomic region were constructed by cloning in frame downstream of the VP16 acidic transactivation domain into the vector VPFlag. These mutants were employed to identify the NS5A binding region of p53 using a mammalian two-hybrid assay. Cells were cotransfected with the deletion mutants of NS5A and the reporter gene G5E1b-CAT with or without p53-Gal4 plasmid DNA. Results suggested that the p53-interacting domain of NS5A is localized within the first 150 amino acid residues (data not shown). These findings further exhibited a specific association between p53 and NS5A through the identified regions of these two proteins. However, the precise sequences responsible for interaction between p53 and NS5A await further investigation.
FIG. 4.
Mapping of the NS5A binding domain in p53. Various amino- and carboxy-terminus deletion mutants of p53 fused with the Gal4 DNA binding domain (A) were cotransfected with VP16-5A (NS5A fused with VP16 activation domain) and reporter construct (G5E1b-CAT) for mammalian two-hybrid assay (B). The total amount of plasmid DNA in each transfection was kept constant by the addition of empty vector, and CAT activity was measured after 48 h of transfection. Results from triplicate transfections are presented.
NS5A retains p53 in the perinuclear membrane.
Since p53 is a nuclear protein (33) and NS5A localizes in the perinuclear membrane (37), the physical association of these two proteins indicated by pull-down assay, in vivo coimmunoprecipitation, and mammalian two-hybrid assay was somewhat surprising. To determine the biological significance of this interaction and to address this paradox, we investigated whether NS5A and p53 colocalize intracellularly. Initially, localization of endogenous p53 was examined by indirect immunofluorescence using empty-vector-transfected HepG2 cells and a monoclonal antibody to p53. Immunofluorescent staining of endogenous p53 exhibited a distinct nuclear localization (Fig. 5A). To compare the subcellular localization of p53 with that of NS5A, HepG2 cells were transfected with CMV-NS5A, and after 48 h, cells were stained with a mouse monoclonal antibody to p53 (Fig. 5C) and a rabbit antiserum to NS5A (Fig. 5D). Confocal microscopy suggested colocalization of the endogenous p53 with NS5A primarily in the perinuclear membrane (Fig. 5E), while control antibodies did not produce any detectable fluorescence. Specificity of p53 localization in the presence of NS5A was also examined. Sequestration of p53 protein in the perinuclear membrane of NS5A-transfected HepG2 cells was observed when cells were stained with p53 antibody alone (Fig. 5B). Similar results were obtained when Saos-2 cells were transfected with p53 alone or together with NS5A (data not shown). These results suggested that cells expressing NS5A sequester p53 on the perinuclear membrane. To further verify that perinuclear retention of p53 is indeed a result of physical interaction between p53 and NS5A, p53(ΔN14-88) construct tagged with the Gal4 DNA binding domain (for detection of mutant p53) was coexpressed with NS5A in HepG2 cells. A monoclonal antibody to the Gal4 DNA binding domain (Santa Cruz) and an antiserum to NS5A were used to stain the cells as described above. Results exhibited that these two proteins do not colocalize in the perinuclear membrane (Fig. 6B to D). Surprisingly, p53(ΔN14-18) when expressed alone localized in the perinuclear membrane (Fig. 6A) even though the nuclear localization signals of p53 reside in the carboxy terminus of the protein. A similar observation was made with the amino terminal mutants of p53 (Michael F. Clarke [University of Michigan], personal communication).
FIG. 5.
Colocalization of NS5A and endogenous p53 in HepG2 cells. Immunofluorescent staining using a monoclonal antibody to p53 exhibits nuclear localization of p53 in mock-transfected cells (A) or perinuclear localization of p53 in CMV-NS5A-transfected cells (B). Cells transfected with NS5A were stained with a monoclonal antibody to p53 (C) and a rabbit antibody to NS5A (D). Fluorescence images of panels C and D were superimposed digitally for fine comparison (E).
FIG. 6.
NS5A does not colocalize with an amino terminal deletion mutant of p53. HepG2 cells were transfected with a p53(ΔN11-88)-Gal4 construct and stained with a monoclonal antibody to the Gal4 DNA binding domain (A). Cells cotransfected with p53(ΔN11-88) and NS5A were stained together with a monoclonal antibody to Gal4 DNA binding domain (B) and a rabbit antibody to NS5A (C). Fluorescence images of panels B and C were superimposed digitally for fine comparison (D).
DISCUSSION
We have previously shown that HCV NS5A down-regulates p21/waf1 expression at the transcriptional and translational levels (11). The p21/waf1 gene has been identified as an effector of p53-mediated cell growth regulation (6). Two conserved PREs are recognized by the p21/waf1 promoter; one is located at −1.3 kb and the other is at −2.2 kb (6). In this study, we have demonstrated the requirement for a PRE in the p21/waf1 promoter for NS5A-mediated transcriptional repression. Similarly, association of NS5A and p53 reduced the transcriptional activation from a p53-dependent synthetic promoter in Saos-2 cells. NS5A physically associates with p53 both in vitro and in vivo and sequestered p53 in the perinuclear membrane. Thus, a decrease of p53 in the nucleus may in turn affect the down-regulation of the p53-mediated gene expression for normal cell growth regulation. The down-regulation of p21/waf1 promoter activity by NS5A appears to be due to an interaction between NS5A and p53, possibly blocking the access of p53 to the p21/waf1 promoter.
Sequestration of nuclear protein by viral gene products has been reported in earlier studies. Hepatitis B virus X protein interacts with and retains p53 in the cytoplasm (35) and inhibits p53-mediated transcriptional activation (42). Simian virus 40 large T antigen binds to a novel proapoptotic protein, p193, and results in cytoplasmic sequestration of both the proteins (38). Cytoplasmic sequestration of p53 has been proposed as a mechanism by which the function of this protein can be suppressed (22). Our initial study suggests that the domain of p53 that interacts with NS5A is located in the N-terminal region (amino acids 33 to 88), and the N-terminal 73 residues of p53 contain one of the strongest known activation domains (21). A number of viral and cellular proteins interact with p53 through this region and modulate its function. The adenovirus E1B 55-kDa protein, the human MDM2 protein, and hepatitis B virus X protein are known to bind to the transactivation domain of p53 for inhibition of its functional activity (19, 23, 25, 43). Recently, a novel functional region of p53 (amino acids 43 to 63) was identified as both transcriptional activation and apoptotic domains (45). The N-terminal region of p53 contains a proline-rich region (amino acids 63 to 97), and the fact that it contains five repeats of SH3-binding motifs (PXXP) makes it an interesting candidate for physical interaction with other cellular factors (21). Our initial study suggests that the N terminus of NS5A (150 amino acids) is required for physical association with p53, which also possesses proline-rich sequences. Thus, we speculate that NS5A may physically interact with p53 through proline-rich domains and modulates transcriptional regulatory functions. However, further studies are necessary to elucidate the functional association between p53 and NS5A by precise mapping of the amino acid residues.
NS5A physically interacts with Grb2 protein through its C-terminal proline-rich sequence and perturbs the mitogenic signal transduction pathway (36). NS5A also interacts with the IFN-inducible double-stranded-RNA-dependent protein kinase (PKR) and functions as a repressor of PKR (8). NS5A associates with the C terminus of hVAP-33, a SNARE-like protein, and may provide a mechanism for membrane association of the HCV RNA replication complex (40). A novel cellular transcriptional factor, SRCAP, also associates with NS5A and acts as a transcriptional corepressor (14). NS5A may have different target motifs for interaction with cellular proteins. The choice of these motifs may vary according to the transcription factors present in different cell types and/or the cellular environments. A fundamental aspect of p53 is that it has been shown to participate in cell cycle checkpoint and to be involved in apoptotic functions that regulate homoeostatic tissue renewal (21). As such, p53 appears to be the head of key cellular pathways, and the association of p53 with NS5A may disrupt the normal cell cycle. The lack of a suitable cell culture system or a small animal model for HCV infection makes it difficult to understand the virus replication and the pathogenesis of HCV-related disease. However, as other HCV proteins (core and NS3) have a role in transcriptional regulation and cell growth control (1, 28, 31, 39), the functional effect of NS5A is difficult to assess when this protein is expressed along with other HCV proteins. Furthermore, we do not know the difference between the levels of NS5A expression in transfected cells and naturally infected cells or its magnitude of incorporation during virus morphogenesis. It is possible that one or more of these viral proteins may contribute to HCV-mediated multistep disease progression. The results presented here highlight the functional role of NS5A, and its direct relationship with HCV-mediated pathogenesis remains to be elucidated.
ACKNOWLEDGMENTS
We thank M. F. Clarke for sharing results prior to publication and J. McHowat and K. Vausden for helpful discussion. We are grateful to R. Baer for providing mammalian two-hybrid system reagents, R. Brachmann for the PG13-LUC plasmid, G. Lozano for p53-GAL4 constructs, R. Padmanabhan for the CMV-NS5A construct, J. Pietenpol for GST-p53 and CMV-p53 plasmids, C. M. Rice for antiserum to NS5A, K. Shimotohno for monoclonal antibody to NS5A, B. Vogelstein for WWP-luc, and X. Wang for deletion mutants of the p21 promoter.
This research was supported by PHS grant AI45144 from the National Institutes of Health.
REFERENCES
- 1.Chang J, Yang S H, Cho Y G, Hwang S B, Hahn Y S, Sung Y C. Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J Virol. 1998;72:3060–3065. doi: 10.1128/jvi.72.4.3060-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 3.Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78:2397–2410. doi: 10.1099/0022-1317-78-10-2397. [DOI] [PubMed] [Google Scholar]
- 4.Di Bisceglie A M. Hepatitis C and hepatocellular carcinoma. Hepatology. 1997;26:34S–38S. doi: 10.1002/hep.510260706. [DOI] [PubMed] [Google Scholar]
- 5.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Moore M, Finlay C, Levine A J. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 6.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 7.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full length sequences of interferon-sensitive and resistant hepatitis C virus 1b: sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gale M, Jr, Kwieciszewski B, Dossett M, Nakao H, Katze M G. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale M J, Jr, Korth M J, Tang N M, Tan S-L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 10.Gartel A L, Tyner A L. The growth-regulatory role of p21 (WAF1/CIP1) Prog Mol Subcell Biol. 1998;20:43–71. doi: 10.1007/978-3-642-72149-6_4. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh A K, Steele R, Meyer K, Ray R, Ray R B. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol. 1999;80:1179–1183. doi: 10.1099/0022-1317-80-5-1179. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh A K, Steele R, Ray R B. Functional domains of c-myc promoter binding protein 1 involved in transcriptional repression and cell growth regulation. Mol Cell Biol. 1999;19:2880–2886. doi: 10.1128/mcb.19.4.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh A K, Steele R, Ray R B. MBP-1 physically associates with histone deacetylase for transcriptional repression. Biochem Biophys Res Commun. 1999;260:405–409. doi: 10.1006/bbrc.1999.0921. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh A K, Majumder M, Steele R, Yaciuk P, Chrivia J, Ray R, Ray R B. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor SRCAP. J Biol Chem. 2000;275:7184–7188. doi: 10.1074/jbc.275.10.7184. [DOI] [PubMed] [Google Scholar]
- 15.Hulboy D L, Lozano G. Structural and functional analysis of p53: the acidic activation domain has transforming capability. Cell Growth Differ. 1994;5:1023–1031. [PubMed] [Google Scholar]
- 16.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 17.Katze M G. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 18.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 19.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 21.May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie P P, Guichard S M, Middlemas D S, Ashmun R A, Danks M K, Harris L C. Wild-type p53 can induce p21 and apoptosis in neuroblastoma cells but the DNA damage-induced G1 checkpoint function is attenuated. Clin Cancer Res. 1999;5:4199–4207. [PubMed] [Google Scholar]
- 23.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 24.Nousbaum J-B, Polyak S J, Ray S C, Sullivan D G, Larson A M, Carithers R L, Gretch D R. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J Virol. 2000;74:9028–9038. doi: 10.1128/jvi.74.19.9028-9038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 26.Polyak S J, McArdla S, Liu S-L, Sullivan D G, Chung M, Hofgartner W T, Carithers R L, McMahon B J, Mullins J I, Corey L, Gretch D R. Evaluation of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J Virol. 1998;72:4288–4296. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray R B, Steele R, Meyer K, Ray R. Hepatitis C virus core protein represses p21WAF1/Cip1/Sid1 promoter activity. Gene. 1998;208:331–336. doi: 10.1016/s0378-1119(98)00030-4. [DOI] [PubMed] [Google Scholar]
- 28.Ray R B, Lagging L M, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarrazin C, Berg T, Lee J H, Ruster B, Kronenberger B, Roth W K, Zeuzem S. Mutations in the protein kinase-binding domain of the NS5A protein in patients infected with hepatitis C virus type 1a are associated with treatment response. J Infect Dis. 2000;181:432–441. doi: 10.1086/315263. [DOI] [PubMed] [Google Scholar]
- 33.Shaulsky G, Goldfinger N, Tosky M S, Levine A J, Rotter V. Nuclear localization is essential for the activity of p53 protein. Oncogene. 1991;6:2055–2065. [PubMed] [Google Scholar]
- 34.Sheikh S M, Rochefort H, Garcia M. Overexpression of p21WAF1/CIP1 induces growth arrest, giant cell formation and apoptosis in human breast carcinoma cell lines. Oncogene. 1995;11:1899–1905. [PubMed] [Google Scholar]
- 35.Takada S, Kaneniwa N, Tsuchida N, Koike K. Cytoplasmic retention of the p53 tumor suppressor gene product is observed in the hepatitis B virus X gene-transfected cells. Oncogene. 1997;15:1895–1901. doi: 10.1038/sj.onc.1201369. [DOI] [PubMed] [Google Scholar]
- 36.Tan S L, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai S C, Pasumarthi K B, Pajak L, Franklin M, Patton B, Wang H, Henzel W J, Stults J T, Field L J. Simian virus 40 large T antigen binds a novel Bcl-2 homology domain 3-containing proapoptosis protein in the cytoplasm. J Biol Chem. 2000;275:3239–3246. doi: 10.1074/jbc.275.5.3239. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchihara K, Hijikata M, Fukuda K, Kuroki T, Yamamoto N, Shimotohno K. Hepatitis C virus core protein regulates cell growth and signal transduction pathway transmitting growth stimuli. Virology. 1999;258:100–107. doi: 10.1006/viro.1999.9694. [DOI] [PubMed] [Google Scholar]
- 40.Tu H, Gao L, Shi S T, Taylor D R, Yang T, Mircheff A K, Wen Y, Gorbalenya A E, Hwang S B, Lai M M. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 41.Van Orden K, Giebler H A, Lemasson I, Gonzales M, Nyborg J K. Binding of p53 to the KIX domain of CREB binding protein: a potential link to human T-cell leukemia virus, type I-associated leukemogenesis. J Biol Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 42.Wang X W, Gibson M K, Vermeulen W, Yeh H, Forrester K, Sturzbecher H W, Hoeijmakers J H, Harris C C. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55:6012–6016. [PubMed] [Google Scholar]
- 43.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 44.Yu X, Wu L C, Bowcock A M, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 45.Zhu J, Zhou W, Jiang J, Chen X. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem. 1998;273:13030–13036. doi: 10.1074/jbc.273.21.13030. [DOI] [PubMed] [Google Scholar]