Abstract
A series of prion transmission experiments was performed in transgenic (Tg) mice expressing either wild-type, chimeric, or truncated prion protein (PrP) molecules. Following inoculation with Rocky Mountain Laboratory (RML) murine prions, scrapie incubation times for Tg(MoPrP)4053, Tg(MHM2)294/Prnp0/0, and Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice were ∼50, 120, and 160 days, respectively. Similar scrapie incubation times were obtained after inoculation of these lines of Tg mice with either MHM2(MHM2(RML)) or MoPrP(Δ23–88)(RML) prions, excluding the possibility that sequence-dependent transmission barriers could account for the observed differences. Tg(MHM2)294/Prnp0/0 mice displayed prolonged scrapie incubation times with four different strains of murine prions. These data provide evidence that the N terminus of MoPrP and the chimeric region of MHM2 PrP (residues 108 through 111) both influence the inherent efficiency of prion propagation.
The prion diseases are an unusual group of fatal neurodegenerative disorders which include Creutzfeldt-Jakob disease, fatal familial insomnia, and Gerstmann-Sträussler-Scheinker disease in humans as well as bovine spongiform encephalopathy in cattle and scrapie in sheep and goats. A wealth of evidence supports the hypothesis that these diseases are caused by a conformational change in the prion protein (PrP) from its normal, cellular isoform (PrPC) into a pathogenic, infectious isoform (PrPSc) (17, 18, 21).
To investigate the pathogenesis of prion diseases and to identify potential therapeutic targets, much effort is now directed toward characterizing the structural biology of PrPSc formation. Recent nuclear magnetic resonance studies of recombinant PrP molecules provide evidence for a three-α-helix bundle protein with a short β-strand region and a relatively unstructured N terminus (4, 7, 9, 23, 24). These structural data have facilitated the rational analysis of mutagenesis-specific PrP segments in order to determine their importance for prion propagation.
As part of a systematic PrP deletion mutagenesis study, we discovered that Tg(MHM2,Δ23–88)9381/Prnp0/0 mice expressing N-terminally truncated MHM2 PrP molecules were resistant to Rocky Mountain Laboratory (RML) murine prions (30). MHM2 is a chimeric construct that differs from wild-type mouse (Mo) PrP at positions 108 and 111 (28). Substitution at these positions with the homologous residues from the Syrian hamster (SHa) PrP sequence (L108M and V111M) creates an epitope for the anti-PrP 3F4 monoclonal antibody (MAb) (8). Although Tg(MHM2,Δ23–88)9381/Prnp0/0 mice failed to propagate RML prions, MHM2(Δ23–88) molecules expressed in scrapie-infected mouse neuroblastoma (ScN2a) cells successfully formed protease-resistant MHM2(Δ23–88)PrPSc (13). Furthermore, Tg(MHM2,Δ23–88)9381/Prnp+/0 mice expressing endogenous MoPrP in addition to the truncated, chimeric transgene were susceptible to RML prion infection. These heterozygote mice developed scrapie ∼260 days after inoculation with RML prions, and biochemical analysis revealed the accumulation of protease-resistant MHM2(Δ23–88)PrPSc in their brains (30). These complementary results in ScN2a cells and Prnp+/0 mice indicate that coexpression of MoPrP facilitates the conversion of MHM2(Δ23–88)PrPC to MHM2(Δ23–88)PrPSc.
In view of the foregoing results, it remained unclear why Tg(MHM2,Δ23–88)9381/Prnp0/0 mice were not susceptible to RML prions in the absence of MoPrP. Immunofluorescence studies did not reveal any significant differences in the cellular localization of truncated, chimeric, and wild-type PrP molecules in transfected mouse neuroblastoma (N2a) cells (data not shown). Possible explanations for the resistance of Tg(MHM2,Δ23–88)9381/Prnp0/0 mice to prion infection include (i) a prion transmission barrier between full-length mouse PrP and MHM2(Δ23–88), (ii) decreased prion conversion efficiency caused by removal of the N terminus, (iii) insufficient expression of the MHM2(Δ23–88) transgene, and (iv) decreased prion conversion efficiency caused by introduction of the 3F4 epitope. To distinguish between these possibilities, we conducted a series of transmission studies in transgenic mice expressing full-length and truncated PrP. These studies provide evidence that both the N terminus and the region comprising the 3F4 epitope of PrP influence scrapie incubation times.
MATERIALS AND METHODS
Construction of transgenic mice.
Tg(MoPrP)4053, Tg(MHM2)294/Prnp0/0, and Tg(MHM2,Δ23–88)9381/Prnp0/0 mice used in this study have been described previously (11, 28, 32). Tg(MoPrP,Δ23–88)Prnp0/0 mice were generated in the following manner. Plasmid pT2MoPrP (obtained from Takeshi Haga) containing the MoPrP-A open reading frame in a BglII/XhoI cassette was digested with BglII/KpnI. The vector was religated using a synthetic oligonucleotide linker (sense, GATCTCATCATGGCGAACCTTAGCTACTGGCTGCTAGCACTCTTTGTGGCTATGTGGACTGATGTTGGCCTCTGOGGCCAAGGAGGAGGTAC; antisense, CTOCTOCTTGGOOGCAGAGGOCAACATCAGTOCACATAGOCACAAAGAGTG CTAGCAGOCAGTAGCTAAGGTTOGOCATGATGA) to create pT2MoPrP(Δ23–88). The resulting plasmid was digested with BglII, and the single-strand ends were filled in using avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim). The newly created blunt ends were ligated to SalI linker (Promega), and the mixture was digested with SalI. The linearized DNA was purified by agarose electrophoresis and self-ligated to make pT2(SalI)MoPrP(Δ23–88). SalI-digested pT2(Sal I)MoPrP(Δ23–88) was inserted into the cos. SHa.Tet vector for oocyte microinjection as described previously (28). Automated screening for transgenic integration was carried out using a Beckman robotic workstation. Genomic DNA was isolated from tail tissue and screened by dot blot analysis as described previously (26), using a radioactive oligonucleotide probe that hybridizes to the 3′-untranslated region of the SHaPrP gene within the cos.SHa.Tet vector. FVB mice were obtained from Charles River Laboratories (Wilmington, Mass.). Tg(MHM2,Δ23–88)Prnp+/0 heterozygotes were generated by crossing Tg(MHM2,Δ23–88)Prnp0/0 mice with wild-type FVB mice and followed by screening the offspring for the transgene.
Determination of scrapie incubation periods.
Ten percent brain homogenates in sterile phosphate-buffered saline (PBS) without calcium or magnesium were prepared by repeated extrusion through syringe needles of successively smaller size, from 18 to 22 gauge. New, sterile, individually packaged needles, syringes, and tubes were used. All work was carried out in laminar flow hoods to avoid cross-contamination. Mice of either sex aged 7 to 10 weeks were inoculated intracerebrally with 30 μl of 1% brain homogenate in calcium- and magnesium-free PBS plus 1 mg of bovine serum albumin per ml. Inoculation was carried out with a 27-gauge disposable hypodermic needle inserted into the right parietal lobe. After inoculation, mice were examined daily for neurologic dysfunction. Standard diagnostic criteria were used to identify animals exhibiting signs of scrapie (2, 20). In each group, some animals whose deaths were imminent were sacrificed, and their brains were removed for histologic and biochemical analysis.
The full-length murine RML prion strain propagated in Swiss mice was originally provided by W. Hadlow (Rocky Mountain Laboratory, Hamilton, Mont.) and was passaged in Swiss CD-1 mice obtained from Charles River Laboratories (3). The MHM2-adapted RML prion inoculum was generated by passaging RML through Tg(MHM2)294 mice (27). Infectivity of this inoculum was verified by repassaging in Tg(MHM2)294 mice. Me7 and 139A inocula were obtained from R. Kimberlin, and the 22a inoculum was obtained from A. Dickinson. The passage histories of these strains have been reviewed (22). PrP 27–30 was prepared from the brains of CD-1 mice inoculated with RML prions, as described previously (19), utilizing both proteinase K digestion and sucrose gradient sedimentation.
Neuropathology.
Brains were removed rapidly at the time of sacrifice, immersion-fixed in 10% buffered formalin, and embedded in paraffin. Eight-micrometer-thick sections were stained with hematoxylin and eosin for evaluation of prion disease. Peroxidase immunohistochemistry with antibodies to glial fibrillary acidic protein was used to evaluate the degree of reactive astrocytic gliosis. Hydrolytic autoclaving was performed as previously described (12), using RO73 polyclonal antibody to detect PrPSc.
Protease digestion and PrP immunostaining.
Nuclei and debris were removed from brain homogenates by centrifugation at 1,000 × g for 10 min. Homogenates were adjusted to 1 mg of protein per ml in 100 mM NaCl–1 mM EDTA–2% Sarkosyl–50 mM Tris-HCl (pH 7.5). Twenty micrograms of proteinase K (Boehringer Mannheim) per ml was added to 0.5 ml of adjusted homogenate to achieve a ratio of total protein to enzyme of 50:1. After incubation at 37°C for 1 h, proteolytic digestion was terminated by the addition of 8 μl of 0.5 M phenylmethylsulfonyl fluoride in absolute ethanol. Both proteinase K-digested and undigested samples were prepared for sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis by mixing equal volumes of adjusted homogenate and 2× sample buffer.
Following electrophoresis, Western blotting was performed as previously described (26). Membranes were blocked with 5% nonfat milk protein in calcium- and magnesium-free PBS plus 0.1% Tween 20 (PBST) for 1 h at room temperature. Blocked membranes were incubated with primary 1-μg/ml D13 chimeric antibody in PBST for 1 h at 4°C. Following incubation with primary antibody, membranes were washed 3× for 10 min in PBST, incubated with horseradish peroxidase-labeled anti-human Fab secondary antibody (ICN), diluted 1:5,000 in PBST for 25 min at room temperature, and washed again 3× at 10 min in PBST. After chemiluminescent development with ECL reagent (Amersham) for 1 to 5 min, blots were sealed in plastic covers and exposed to ECL Hypermax film (Amersham). Films were processed automatically in a Konica film processor.
Human-mouse chimera (HuM) Fab preparation.
Sequence information obtained from a phage display library (34) was utilized for the D13 clone, and the variable sequences were inserted into an expression plasmid containing human immunoglobulin G (IgG) Fab framework. Escherichia coli 33B6 competent cells were transformed with plasmid containing HuM Fab sequences, and a single bacterial colony from a Luria-Bertani medium agar plate containing 100 μg of ampicillin was grown in 500 ml of Luria-Bertani media containing 100 μg of ampicillin per ml at 30°C overnight. A Biostat B controller (B. Braun, Melsungen, Germany) was used together with a 10-liter vessel for all fermentation procedures. Fermentation was carried out in medium containing (per liter) MT-8 salts (0.26 g of potassium phosphate dibasic, 0.13 g of sodium phosphate monobasic dihydrate, 0.5 g of ammonium sulfate, 0.1 g of sodium citrate dihydrate, and 0.15 g of potassium chloride per liter); 0.5 g of isoleucine, 20% NZ amines, 20% yeast extract, 1 mM magnesium sulfate, 50% glucose, trace metals, and 100 μg of ampicillin per mg and was completed in 40 to 48 h. The fermented culture was pelleted at 8,000 rpm for 1 h at 4°C in an Avanti J-20 centrifuge (Beckman), and the pellet was stored at −20°C. The frozen paste was resuspended in five volumes of 2 mM imidazole–20 mM sodium phosphate–250 mM sodium chloride (pH 7.0), homogenized in a tissue homogenizer at 9,600 rpm, and it was processed twice in a Microfluidizer M-110 EH (Microfluidics Co., Newton, Mass.). The cell paste was titrated to 0.1% polyethylenimine (PEI) (5% stock solution at pH 8.0), and it was stirred at 4°C for 30 min. The processed sample was then spun down with Sorval J-10 (Beckman) at 10,000 rpm for 30 min at 4°C; the pellet was discarded, and the supernatant was used for the purification procedure.
For purification, the sample was diluted in an equal volume of 20 mM imidazole (pH 7.0). The solution was loaded onto a Sepharose Fast Flow (Amersham Pharmacia, Uppsala, Sweden) column, and the recombinant HuM Fab was eluted with a linear gradient of five column volumes (cv) of 0 to 100% 20 mM imidazole–500 mM sodium acetate (pH 7.0). The resulting peak was directly applied onto an IMAC column, and it was eluted with 5 cv of 200 mM imidazole (pH 7.0). The peak was dialyzed with three changes of 100 volumes of 10 mM Tris-HCl (pH 7.2) at 4°C overnight. The dialyzed recombinant material was further purified through a Sepharose Fast Flow utilizing a linear gradient of 0 to 100% 10 mM Tris-HCl–500 mM sodium chloride (pH 7.2). The final purified HuM Fab peak was filtered through a 0.45-μm-pore-size sterile filter and stored at 4°C.
RESULTS
Tg(MHM2,Δ23–88)Prnp0/0 mice are resistant to prion infection.
Theoretically, an artificial, sequence-dependent prion transmission barrier could account for the resistance of Tg(MHM2,Δ23–88)9381/Prnp0/0 mice to RML mouse prions composed of full-length mouse PrPSc. We performed three experiments designed to circumvent possible transmission barriers caused by PrPC/PrPSc sequence mismatches at either the N terminus or the 3F4 epitope. First, we inoculated Tg(MHM2,Δ23–88)9381/Prnp0/0 mice with PrP 27-30 purified from the brains of CD-1 mice inoculated with RML prions. PrP 27-30 in these purified prion preparations has ∼67 amino acid residues enzymatically removed from the N terminus of PrPSc (19). Although PrP 27-30 preparations are highly infectious in CD-1 and FVB mice (1), preparations with PrPSc molecules lacking N-terminal residues did not transmit disease to Tg(MHM2,Δ23–88)9381/Prnp0/0 mice (Table 1). Second, we generated MHM2-adapted RML prions by passaging RML prions serially through Tg(MHM2)294/Prnp0/0 mice. These MHM2(MHM2(RML)) prions, composed of MHM2 PrPSc molecules, did not transmit scrapie to Tg(MHM2,Δ23–88)9381/Prnp0/0 mice (Table 1). Finally, we inoculated brain homogenates from scrapie-affected Tg(MHM2,Δ23–88)9381/Prnp+/0 mice (n = 3) into both Tg(MHM2,Δ23–88)9381/Prnp0/0 and control Tg(MoPrP)4053 mice. Although these brain homogenates contained MHM2(Δ23–88)PrPSc, they were not infectious for Tg(MHM2,Δ23–88)9381/Prnp0/0 mice (Table 1). Taken together, the results of these three experiments indicate that the inability of Tg(MHM2,Δ23–88)9381/Prnp0/0 mice to propagate prions cannot be explained by the presence of sequence-dependent transmission barriers. Therefore, we proceeded to investigate the possibility that either the N terminus or wild-type residues L108 and V111 of MoPrP might be required for efficient prion propagation.
TABLE 1.
Transmission studies in transgenic mice expressing wild-type, truncated, and chimeric PrP molecules
| PrP hosta (expression level) | Inoculum | Incubation period (days ± SEM) | n/n0b |
|---|---|---|---|
| Tg(MHM2,Δ23–88)9381 (4×) | None | >400 | 0/9 |
| RML | >500 | 0/6 | |
| MHM2(MHM2(RML)) | >400 | 0/5 | |
| (MHM2,Δ23–88)/Prnp+/0(RML) | >500 | 0/21 | |
| RML27-30 | >500 | 0/10 | |
| MoPrP(Δ23–88)(RML) | >384 | 0/9 | |
| Tg(MoPrP,Δ23–88)9949 (16×) | None | >500 | 0/12 |
| RML | 161 ± 4 | 11/11 | |
| MHM2(MHM2(RML)) | 206 ± 6 | 7/7 | |
| MoPrP(Δ23–88)(RML) | 144 ± 7 | 10/10 | |
| Tg(MHM2)294 (16×) | None | >500 | 0/9 |
| RML | 121 ± 1 | 10/10 | |
| MHM2(MHM2(RML)) | 137 ± 5 | 10/10 | |
| RML27-30 | 118 ± 8 | 8/8 | |
| Tg(MoPrP)4053 (8×) | None | >700 | 0/20 |
| RML | 50 ± 2 | 16/16 | |
| MHM2(MHM2(RML)) | 63 ± 2 | 10/10 | |
| (MHM2,Δ23–88)/Prnp+/0(RML) | 54 ± 2 | 29/29 | |
| MoPrP(Δ23–88)(RML) | 52 ± 2 | 21/21 |
All transgenes are expressed in Prnp0/0 mice. Expression levels relative to normal SHa brain were determined by immunoblots of serially diluted brain homogenates.
Number of mice developing scrapie/total number.
Tg(MoPrP,Δ23–88)Prnp0/0 mice propagate prions.
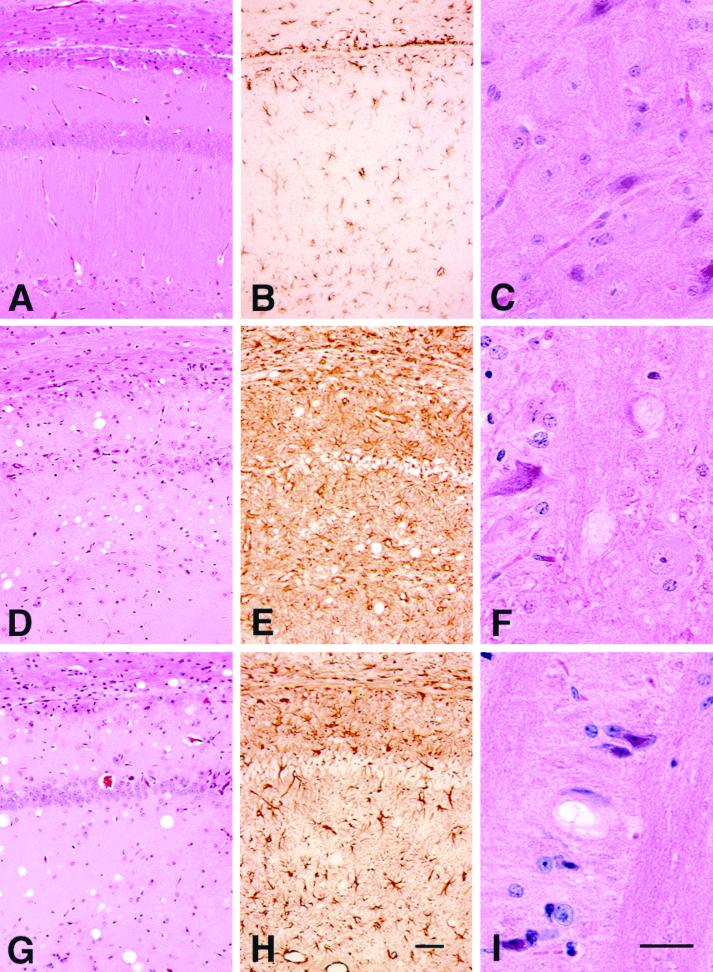
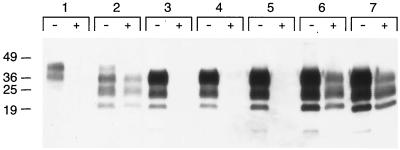
To study the effect of an isolated PrP N-terminus truncation on the efficiency of prion propagation, we generated Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice without the 3F4 epitope substitution. In contrast to Tg(MHM2,Δ23–88)9381/Prnp0/0 mice, which were resistant to prion infection, Tg(MoPrP,Δ23–88)Prnp0/0 mice propagated both RML and MHM2(MHM2(RML)) prions (Table 1). Neurological examination of Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice revealed typical signs of scrapie following inoculation with RML prions. Neuropathological studies revealed focal vacuolation with associated gliosis in the hippocampus, brain stem, and white matter (Fig. 1D through F). Intraneuronal vacuolation was seen in the brain stem (Fig. 1F). No plaques were detected with the hydrolytic autoclaving technique (data not shown). Similar findings were seen in three Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice affected with scrapie following serial transmission of MoPrP(Δ23–88)(RML) prions (Fig. 1G through I). Immunoblot analysis confirmed that brains of scrapie-affected Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice contained protease-resistant MoPrP(Δ23–88) PrPSc (Fig. 2).
FIG. 1.
Typical neuropathological features of scrapie occur in Tg(MoPrP,Δ23–88)Prnp0/0 mouse brains following inoculation with RML prions. (A, B, and C) A 300-day-old, uninoculated control Tg(MoPrP,Δ23–88)Prnp0/0 mouse shows no evidence of spongiform degeneration or reactive astrocytic gliosis. (D, E, and F) Primary inoculation of Tg(MoPrP,Δ23–88)Prnp0/0 mouse with RML prions. This brain was taken from a scrapie-affected mouse sacrificed at 130 days of age. (D) Characteristic spongiform degeneration is found in the hippocampus with loss of neurons in the CA1 region. (E) The neurodegenerative changes are associated with intense reactive astrocytic gliosis. (F) In the tegmentum of the pons, many nerve cells bodies were vacuolated. (G, H, and I) Secondary transmission of RML in Tg(MoPrP,Δ23–88)Prnp0/0 mice showed similar but less intense neuropathological changes compared to the primary passage. This brain was taken from a scrapie-affected mouse sacrificed at 130 days of age. Specimens shown in panels A, C, D, F, G, and I were stained by the hematoxylin and eosin technique. B, E, and F were immunostained for glial fibrillary acidic protein. The bar in H is 25 mm and also applies to panels A, B, D, E, and G. The bar in I is 25 μm and also applies to C and F.
FIG. 2.
Immunoblot of PrPSc in brains of Tg(MoPrP,Δ23–88)Prnp0/0 mice. Paired sample lanes are numbered as follows: (1) normal FVB mouse, (2) RML-infected FVB mouse, (3) 440-day-old, uninoculated Tg(MHM2,Δ23–88)Prnp0/0 mouse, (4) 800-day-old Tg(MHM2,Δ23–88)Prnp0/0 mouse inoculated with RML prions, (5) 300-day-old, uninoculated Tg(MoPrP,Δ23–88)Prnp0/0 mouse, (6) scrapie-affected, 130-day-old Tg(MoPrP,Δ23–88)Prnp0/0 mouse inoculated with RML prions, (7) scrapie-affected, 130-day-old Tg(MoPrP,Δ23–88)Prnp0/0 mouse inoculated with MoPrP(Δ23–88)(RML) prions. Minus (−) symbol denotes undigested control sample, and plus (+) symbol designates sample subjected to limited proteolysis. Apparent molecular sizes based on migration of protein standards are given in kilodaltons.
We compared scrapie incubation times of Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice with those of Tg(MoPrP)4053 mice expressing full-length MoPrP to assess the contribution of residues 23 through 88 towards efficient prion propagation. After inoculation with full-length RML prions, Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice developed scrapie in ∼160 days; Tg(MoPrP)4053 mice developed scrapie in only ∼50 days (Table 1). Upon serial transmission, inoculation of MoPrP(Δ23–88)(RML) prions caused scrapie with an incubation time of ∼140 days in Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice and ∼50 days in Tg(MoPrP)4053 mice (Table 1). Because Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice have longer scrapie incubation times than Tg(MoPrP)4053 mice on both primary and serial passages, the prolongation of incubation times cannot be explained by a transmission barrier. Furthermore, the prolonged scrapie incubation times obtained with Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice cannot be explained by differential transgene expression, because these mice express twice as much PrP as do Tg(MoPrP)4053 mice (16 versus 8 times the PrP level in normal SHa brain, respectively). Rather, the prolonged incubation times must result from reduced PrP conversion efficiency, caused by removal of the N terminus.
Scrapie incubation times in Tg(MHM2)Prnp0/0 mice.
To assess the influence of the 3F4 epitope on prion propagation efficiency, we compared scrapie incubation times of Tg(MHM2)294/Prnp0/0 and Tg(MoPrP)4053 mice. Following inoculation of RML prions, Tg(MHM2)294/Prnp0/0 mice developed scrapie in ∼120 days, whereas Tg(MoPrP)4053 mice developed scrapie in ∼50 days (Table 1). This difference in scrapie incubation times was not caused by a sequence-dependent prion transmission barrier since second-passage MHM2(MHM2(RML)) prions were also transmitted less efficiently in Tg(MHM2)294/Prnp0/0 mice (∼140 days) than in Tg(MoPrP)4053 mice (∼60 days) (Table 1). Furthermore, the long scrapie incubation times obtained in Tg(MHM2)294/Prnp0/0 mice cannot be attributed to low transgene expression since these mice express twice as much PrP as Tg(MoPrP)4053 mice. Therefore, we conclude that introduction of the 3F4 epitope into MoPrP, like N-terminal truncation, reduces PrP conversion efficiency.
We sought to determine whether the prolongation of scrapie incubation time caused by the 3F4 epitope might be a strain-specific phenomenon. We inoculated Tg(MHM2)294/Prnp0/0 and Tg(MoPrP)4053 mice with four different strains of murine prions. For each strain, scrapie incubation times were longer in Tg(MHM2)294/Prnp0/0 mice than in Tg(MoPrP)4053 mice (Table 2). These results indicate that the control exerted by residues L108 and V111 over scrapie incubation time is not strain specific.
TABLE 2.
Transmission of murine prion strains to transgenic mice expressing wild-type or chimeric PrP molecules
| Hosta (expression level) | Inoculum | Incubation period (days ± SEM) | n/n0 |
|---|---|---|---|
| Tg(MHM2)294 (16×) | RML | 121 ± 1 | 10/10 |
| Me7 | 157 ± 6 | 7/7 | |
| 22a | 195 ± 4 | 10/10 | |
| 301V | 204 ± 1 | 3/3 | |
| Tg(MoPrP)4053 (8×) | RML | 50 ± 2 | 16/16 |
| Me7 | 87 ± 2 | 10/10 | |
| 22a | 69 ± 4 | 5/5 | |
| 301V | 125 ± 6 | 9/9 |
All transgenes are expressed in Prnp0/0 mice. Expression levels relative to normal SHa brain were determined by immunoblots of serially diluted brain.
DISCUSSION
The N terminus of PrP is required for efficient prion propagation.
Our studies indicate that removal of the N terminus of PrP diminishes the efficiency of prion propagation. Why might N-terminally truncated PrP form PrPSc less efficiently than full-length PrP? Two independent studies suggest possible explanations. First, comparative chemical shift data from structural nuclear magnetic resonance studies of SHaPrP(29-331) and SHaPrP(90-231) indicate that residues 29 through 89 may help stabilize α-helix B (4). Second, deletion of the N terminus diminished the efficacy of MHM2(Δ23–88)Q218K as a dominant negative inhibitor of PrPSc formation in a ScN2a cells (35). These findings suggest that the N terminus of PrP may participate in intermolecular as well as intramolecular interactions necessary for PrPSc formation. Furthermore, the observation that coexpression of full-length MoPrP enables formation of MHM2(Δ23–88)PrPSc in Prnp+/0 mice and ScN2a cells indicates that these interactions can be restored in trans (30).
Others have reported that Tg(MoPrP,Δ32-80)Prnp0/0 mice propagate prions efficiently (5), whereas Tg(MoPrP,Δ32-93)Prnp0/0 mice display prolonged incubation times (6), similar to Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice. Taken together, these results argue that at least one intact octapeptide sequence is required for efficient prion propagation. It is interesting that histological features of scrapie in Tg(MoPrP,Δ32-93)Prnp0/0 mice infected with RML were limited to the spinal cord, and the brains of these animals did not contain protease-resistant PrPSc (6). In contrast, focal vacuolation and astocytic gliosis could be seen in the hippocampus, white matter, and brain stem of RML-infected Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice (Fig. 1). Furthermore, protease-resistant PrPSc was easily detected in the brains of RML-infected Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice by immunoblotting (Fig. 2). The different neuropathological and biochemical profiles obtained in RML-infected Tg(MoPrP,Δ32-93)Prnp0/0 versus Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice could be caused by (i) the absence of residues 89 through 93, (ii) the presence of residues 23 through 31, (iii) a difference between the half-genomic and cos.SHa.Tet expression vectors, or (iv) a difference in expression levels. More work will be required to distinguish among these possibilities.
Control of scrapie incubation time by the 3F4 epitope.
Our results also show that residues within the 3F4 epitope control scrapie incubation time. The combined substitutions L108M and V111M led to prolonged scrapie incubation times in Tg(MHM2)294/Prnp0/0 mice. The inability of Tg(MHM2)294/Prnp0/0 mice to propagate prions efficiently was caused neither by poor PrP expression nor by a prion transmission barrier. Furthermore, Tg(MHM2)294/Prnp0/0 mice propagated four different strains of murine prions inefficiently, indicating that the adverse effect of L108M/V111M substitution on prion propagation is not a strain-specific phenomenon. An independent finding that also implicates residue 108 in controlling scrapie incubation times is that polymorphic substitutions at residues 108 or 189 lengthen incubation times in mice (10, 33).
Residues within the 3F4 epitope might influence the efficiency of prion conversion by participating in the conformational change from PrPC to PrPSc. This concept is supported by data showing that the 3F4 epitope is accessible in PrPC molecules but hidden in PrPSc molecules (14, 25, 29).
Others have demonstrated that long-term expression of MHM2 molecules in ScN2a cells slowly reduced levels of endogenous MoPrPSc (15, 16). The authors claimed that precise interactions between homologous PrP molecules are important in PrPSc accumulation, and heterologous MHM2 molecules can block these interactions (15). We suggest a different interpretation. Our data indicate that the conversion of MHM2 PrPC molecules into MHM2 PrPSc is inherently inefficient. In chronically infected, dividing cells, slowing the rate of prion propagation would progressively dilute and eventually eliminate PrPSc. The rate at which prions are propagated must match or exceed the rate of cell division in order to maintain the levels of infection.
Finally, examination of the amino acid sequence of PrP regions 100 through 109 (KPSKPKTNMK) and 23 through 31 (KKRPKPGGW) revealed that each region contains four basic residues. It remains to be determined whether the positive charges contributed by multiple basic residues contribute to the control of scrapie incubation times exerted by these two regions. However, it is interesting that positively charged, synthetic polyamine compounds can render PrPSc molecules protease sensitive and reduce prion infectivity in ScN2a cells (31), suggesting that primary amine groups might play a role in prion propagation.
Conclusion.
We performed a series of experiments to investigate why Tg(MHM2,Δ23–88)9381/Prnp0/0 mice are resistant to prion propagation. Our results identify two regions of the PrP molecule which control scrapie incubation times. The N terminus may be necessary for the inter- as well as intramolecular interactions required for efficient prion propagation. Residues comprising the 3F4 epitope may participate in the conformational change from PrPC to PrPSc.
ACKNOWLEDGMENTS
We thank P. Tremblay, D. Groth, and C. Petromilli for helpful advice.
This work was supported by grants from the National Institutes of Health (NS14069, AG08967, AG02132, and AG10770) and a gift from the G. Harold and Leila Y. Mathers Charitable Foundation. Surachai Supattapone was supported by the Burroughs Wellcome Fund Career Development Award and an NIH Clinical Investigator Development Award (K08 NS02048-02).
REFERENCES
- 1.Bolton D C, McKinley M P, Prusiner S B. Properties and characteristics of scrapie PrP 27–30 protein. In: Prusiner S B, McKinley M P, editors. Prions—novel infectious pathogens causing scrapie and Creutzfeldt-Jakob disease. Orlando, Fla: Academic Press; 1987. pp. 173–196. [Google Scholar]
- 2.Carlson G A, Kingsbury D T, Goodman P A, Coleman S, Marshall S T, DeArmond S, Westaway D, Prusiner S B. Linkage of prion protein and scrapie incubation time genes. Cell. 1986;46:503–511. doi: 10.1016/0092-8674(86)90875-5. [DOI] [PubMed] [Google Scholar]
- 3.Chandler R L. Encephalopathy in mice produced by inoculation with scrapie brain material. Lancet. 1961;i:1378–1379. doi: 10.1016/s0140-6736(61)92008-6. [DOI] [PubMed] [Google Scholar]
- 4.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Structure of the recombinant full-length hamster prion protein PrP(29-231): the N terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 6.Flechsig E, Shmerling D, Hegyi I, Raeber A J, Fischer M, Cozzio A, von Mering C, Aguzzi A, Weissmann C. Prion protein devoid of the octapeptide repeat region restores susceptibility to scrapie in PrP knockout mice. Neuron. 2000;27:399–408. doi: 10.1016/s0896-6273(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 7.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins J. Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Farr-Jones S, Ulyanov N B, Llinas M, Marqusee S, Groth D, Cohen F E, Prusiner S B, James T L. Solution structure of Syrian hamster prion protein rPrP(90-231) Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 10.Moore R C, Hope J, McBride P A, McConnell I, Selfridge J, Melton D W, Manson J C. Mice with gene targeted prion protein alterations show that Prn-p, Sinc and Prni are congruent. Nat Genet. 1998;18:118–125. doi: 10.1038/ng0298-118. [DOI] [PubMed] [Google Scholar]
- 11.Muramoto T, DeArmond S J, Scott M, Telling G C, Cohen F E, Prusiner S B. Heritable disorder resembling neuronal storage disease in mice expressing prion protein with deletion of an α-helix. Nat Med. 1997;3:750–755. doi: 10.1038/nm0797-750. [DOI] [PubMed] [Google Scholar]
- 12.Muramoto T, Kitamoto T, Tateishi J, Goto I. The sequential development of abnormal prion protein accumulation in mice with Creutzfeldt-Jakob disease. Am J Pathol. 1992;140:1411–1420. [PMC free article] [PubMed] [Google Scholar]
- 13.Muramoto T, Scott M, Cohen F E, Prusiner S B. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc Natl Acad Sci USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peretz D, Williamson R A, Matsunaga Y, Serban H, Pinilla C, Bastidas R B, Rozenshteyn R, James T L, Houghten R A, Cohen F E, Prusiner S B, Burton D R. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 15.Priola S A, Caughey B, Race R E, Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scraple-infected murine neuroblastoma cells. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priola S A, Chesebro B. A single hamster PrP amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prusiner S B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 18.Prusiner S B. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prusiner S B, Bolton D C, Groth D F, Bowman K A, Cochran S P, McKinley M P. Further purification and characterization of scrapie prions. Biochemistry. 1982;21:6942–6950. doi: 10.1021/bi00269a050. [DOI] [PubMed] [Google Scholar]
- 20.Prusiner S B, Cochran S P, Groth D F, Downey D E, Bowman K A, Martinez H M. Measurement of the scrapie agent using an incubation time interval assay. Ann Neurol. 1982;11:353–358. doi: 10.1002/ana.410110406. [DOI] [PubMed] [Google Scholar]
- 21.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 22.Ridley R M, Baker H F. To what extent is strain variation evidence for an independent genome in the agent of the transmissible spongiform encephalopathies? Neurodegeneration. 1996;5:219–231. doi: 10.1006/neur.1996.0030. [DOI] [PubMed] [Google Scholar]
- 23.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. NMR structure of the mouse prion protein domain PrP(121–231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 24.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 25.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen F E, Prusiner S B. Eight prion strains have PrPSc molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 26.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, Westaway D, Prusiner S B. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 27.Scott M, Groth D, Foster D, Torchia M, Yang S-L, DeArmond S J, Prusiner S B. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 28.Scott M R, Köhler R, Foster D, Prusiner S B. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serban D, Taraboulos A, DeArmond S J, Prusiner S B. Rapid detection of Creutzfeldt-Jakob disease and scrapie prion proteins. Neurology. 1990;40:110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- 30.Supattapone S, Bosque P, Muramoto T, Wille H, Aagaard C, Peretz D, Nguyen H-O B, Heinrich C, Torchia M, Safar J, Cohen F E, DeArmond S J, Prusiner S B, Scott M. Prion protein of 106 residues creates an artificial transmission barrier for prion replication in transgenic mice. Cell. 1999;96:869–878. doi: 10.1016/s0092-8674(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 31.Supattapone S, Nguyen H-O B, Cohen F E, Prusiner S B, Scott M R. Elimination of prions by branched polyamines and implications for therapeutics. Proc Natl Acad Sci USA. 1999;96:14529–14534. doi: 10.1073/pnas.96.25.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telling G C, Haga T, Torchia M, Tremblay P, DeArmond S J, Prusiner S B. Interactions between wild-type and mutant prion proteins modulate neurodegeneration in transgenic mice. Genes Dev. 1996;10:1736–1750. doi: 10.1101/gad.10.14.1736. [DOI] [PubMed] [Google Scholar]
- 33.Westaway D, Goodman P A, Mirenda C A, McKinley M P, Carlson G A, Prusiner S B. Distinct prion proteins in short and long scrapie incubation period mice. Cell. 1987;51:651–662. doi: 10.1016/0092-8674(87)90134-6. [DOI] [PubMed] [Google Scholar]
- 34.Williamson R A, Peretz D, Pinilla C, Ball H, Bastidas R B, Rozenshteyn R, Houghten R A, Prusiner S B, Burton D R. Mapping the prion protein using recombinant antibodies. J Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zullanello L, Kaneko K, Scott M, Erpel S, Han D, Cohen F E, Prusiner S B. Dominant-negative inhibition of prion formation diminished by deletion mutagenesis of the prion protein. J Virol. 2000;74:4351–4360. doi: 10.1128/jvi.74.9.4351-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]