Abstract
MUC7 12-mer-l exhibits potent in vitro antifungal activity in low-ionic-strength buffers. In this study, we investigated the anticandidal activity and stability of MUC7 12-mer-l and its all-d-amino-acid isomer, along with Hsn5 12-mer (P113) and magainin-II, in human clarified and unclarified saliva in the absence or presence of protease inhibitor cocktail (PIC, which includes EDTA) or EDTA alone. In the absence of PIC or EDTA in saliva, only MUC7 peptides showed significant candidacidal activity. At a 100 μM concentration in clarified saliva and unclarified saliva, MUC7 12-mer-d demonstrated 94 versus 64% killing, respectively; MUC7 12-mer-l showed 57 versus 32% killing; Hsn5 12-mer showed 16 versus 0% killing; and magainin-II showed no killing. Addition of PIC or EDTA to either saliva caused the enhancement of antifungal activities of all peptides, although to different degrees. Taken together, the results suggest that EDTA (a metal-dependent protease inhibitor and/or divalent cation chelator) enhanced the antifungal activity of all four peptides mainly by chelation of divalent cations present in saliva (known to inhibit peptide antifungal activity), and PIC enhanced the activity of the three l peptides above that achievable by EDTA alone through inhibition of all classes of proteases. Peptide stability in saliva monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed no degradation of MUC7 12-mer-d and 23, 60, and 75% degradation of MUC7 12-mer-l, Hsn5 12-mer, and magainin-II, respectively. Cytotoxicity assays determined that, at 100 μM peptide concentrations, MUC7 12-mer-d and 12-mer-l caused 3.5 and 4.3% hemolysis in phosphate-buffered saline and no toxicity to the HOK-16B cell line (derived from normal human oral keratinocytes). In summary, MUC7 12-mer peptides appear to be excellent candidates for investigation of antifungal activity in in vivo models of oral candidiasis.
Candidiasis is the most common oral fungal infection diagnosed in humans. Due to the emergence of pathogens resistant to conventional antifungals and the toxicity of some antimycotics, intense efforts have been made to develop more effective antifungal agents for clinical use (1-3). Cationic antimicrobial peptides (CAMPs) have become attractive as novel candidates for this purpose. The native CAMPs contribute to innate host defense against a number of bacterial and fungal pathogens. They have been discovered from various natural sources, including humans, mammals, plants, insects, and bacteria (12, 13, 31). Among these, the well-characterized human salivary histatins and frog skin magainins show activity against a variety of microorganisms (18, 31, 32).
MUC7, the low-molecular-mass human salivary mucin (comprised of 357 amino acid residues), exerts antimicrobial activity by binding to and clearing microorganisms from the oral cavity (25). While the full-length MUC7 has no appreciable candidacidal activity (11), we have shown that peptides derived from its N-terminal region have significant and broad-spectrum fungicidal and bactericidal activities in vitro in low-ionic-strength buffers (6, 24, 27). MUC7 12-mer (amino acids 40 to 51 of the parent MUC7, with a net charge of +6) is a peptide of the optimal size that possesses potent activity against Candida albicans and Cryptococcus neoformans (28). MUC7 12-mer also exhibits synergistic antifungal effects in vitro with histatin 5 12-mer (Hsn5 12-mer) or miconazole (29).
It has been demonstrated in vitro that antimicrobial peptides can be degraded by proteases, resulting in a decrease or loss of their biological activity (7, 22, 26, 27). It is well known that protease inhibitors can prevent the degradation of peptides. Since proteases recognize only l amino acids, the peptide degradation can also be decreased or prevented by modifying the peptide's molecular structure by substitution of the natural l amino acids with their d forms. For example, an all-d-amino-acid magainin-II was highly resistant to proteolysis, exhibited antibacterial potency nearly identical to that of the all-l enantiomer, and showed no hemolytic activity (5). Similarly, the all-d-amino-acid isomer of Hsn5 12-mer (P113-d) was as active against C. albicans as the natural l form. In addition, P113-d but not the l peptide was active against respiratory bacteria in the presence of sputum from cystic fibrosis patients (23).
Despite the wealth of information on the antimicrobial activity of the CAMPs in vitro, in vivo studies are limited. Our own study showed that neither MUC7 16-mer nor full-length Hsn5 was effective in the treatment of a murine model of vaginal candidiasis (15). Based on the results of studies with the other d-isomer peptides, we obtained the d-amino-acid isomer of MUC7 12-mer and initiated investigation of this peptide's potential for therapeutic use against oral candidiasis. As the first step, in this study we tested the antifungal activity and stability of MUC7 12-mer-d along with that of MUC7 12-mer-l and two other well-known antimicrobial peptides, salivary Hsn5 12-mer (P113) (20) and magainin-II (32), in human saliva. Because salivary components, in particular proteases and divalent cations, may affect peptide activity, the tests were performed in the absence or presence of protease inhibitors or EDTA. Furthermore, we determined the cytotoxicity of these peptides to human erythrocytes and the cell lines of human oral epithelial origin HOK-16B (normal human oral keratinocytes) and KB (human oral epithelial carcinoma).
MATERIALS AND METHODS
Fungal strains and growth media.
C. albicans DIS was provided by M. Edgerton, University at Buffalo. C. albicans ATCC 96112 and Candida glabrata ATCC 90030 were purchased from the American Type Culture Collection. An azole-resistant clinical isolate of C. glabrata 65C was provided by John E. Bennett, National Institute of Allergy and Infectious Diseases, Bethesda, MD. A clinical isolate of Candida krusei was obtained from the Erie County Medical Center, Buffalo, NY. For each experiment, cells were cultured (from −80°C frozen stocks) on Sabouraud dextrose agar (SDA; Difco) for 24 h at 37°C. One colony was picked and resuspended in 10 mM sodium phosphate buffer (PB; pH 7.4), and the cell concentration was adjusted to 1 × 105/ml.
Peptides.
MUC7 12-mer-l (RKSYKCLHKRCR, residues 40 to 51 of the parent human salivary mucin, MUC7), MUC7 12-mer-d (with all 12 l-form amino acids replaced by the d form), Hsn5 12-mer (AKRHHGYKRKFH, also known as P113 [20], residues 4 to 15 of the human salivary histatin 5), and magainin-II (GIGKFLHSAKKFGKAFVGEIMNS, a 23-residue peptide isolated from frog skin) were custom synthesized by Bio-Synthesis (Lewisville, Texas). The company analyzed the peptide preparations by high-performance liquid chromatography and mass spectrometry. The peptide purity ranged from 70 to 100% (the “impurities” consisted of the shorter peptides). The peptides were dissolved in sterile double-distilled (dd) water at 10 mg/ml; aliquots (0.1 ml) were freeze-dried and stored at −20°C. For each experiment, the freeze-dried peptides were redissolved at 1 mg/ml in sterile dd water.
Protease inhibitors.
Protease inhibitor cocktail (PIC) P-2714 was purchased from Sigma Chemical Co. (St. Louis, MO). It was prepared as a 10× solution by dissolving the powder in 10 ml of sterile dd water and stored at −20°C [1× PIC contains 2 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, 1 mM EDTA, 130 μM bestatin, 14 μM E-64, 1 μM leupeptin, and 0.3 μM aprotinin]. EDTA was from J.T. Baker Chemical Co. (Philipsburg, NJ); a 0.2 M solution was prepared in sterile dd water.
Saliva.
The collection and use of saliva samples were approved by the Institutional Review Board of the University at Buffalo. Unclarified saliva (unstimulated whole saliva) was collected from five healthy individuals after fasting. Clarified saliva (filter sterilized) was obtained by passing unclarified saliva through the 0.2-μm-pore-size sterile syringe filter (VWR Scientific Products, West Chester, PA). The subjects were asked to salivate into a sterile 50-ml container. Immediately after collection, each specimen was divided into three equal aliquots, which were processed as follows: (i) for saliva-PIC preparation, the aliquot was mixed 10:1 with 10× PIC; (ii) for saliva-EDTA, the second aliquot was mixed with EDTA to give a final concentration of 1 mM; and (iii) for plain saliva, the third aliquot was stored at 4°C. Afterwards, all samples were placed in a −20°C freezer and stored for approximately 1 week before the analyses.
Killing assays in saliva.
Candidacidal activity assays were performed as described previously (28), except the phosphate buffer was replaced by saliva. To determine the optimal concentration of peptide for candidacidal activity in saliva, 20 μl of twofold serial dilutions of peptides in saliva (to give final peptide concentrations of 25, 50, and 100 μM), saliva containing 1× PIC, or saliva containing 1 mM EDTA was incubated in duplicate with an equal volume (20 μl) of fungal cell suspension (105 cells/ml) in PB. Saliva without the addition of any compound served as a control. After incubation at 37°C for 1.5 h, the reactions were diluted 20-fold in PB, and aliquots (50 μl, ∼120 cells) of each sample were plated on SDA. The plates were incubated at 37°C for 1 day aerobically, and the number of CFU was counted. The percentage of killing was calculated as (1 − amount of viable cells in the test group/amount of viable cells in the control group) × 100%.
Statistics.
Each value was determined from two independent experiments performed in duplicate. The Wilcoxon signed-rank test was performed by using SPSS software to compare the mean difference between PIC and non-PIC groups. The peptides are considered to have a statistically significant difference in candidacidal activity if P was <0.05.
Stability of peptides in saliva and susceptibility of peptides to trypsin.
Three micrograms (in 3 μl) of each peptide in 0.2 M Tris-HCl buffer (pH 7.5) was incubated with 11 μl of 0.2-μg/ml trypsin or saliva. The reactions, without PIC or with 1× PIC, were incubated for 0.5 h with trypsin or for 1 h with saliva at 37°C. For controls, each compound was incubated with Tris-HCl buffer in parallel. The samples were analyzed by sodium dodecyl sulfate-13% polyacrylamide gel electrophoresis (SDS-13% PAGE). Gels were stained with Coomassie brilliant blue R-250 and further analyzed for peptide degradation with a GS-700 imaging densitometer (Bio-Rad). The degradation was estimated by the following equation: [1 − (density of test band/density of control band)] × 100%.
Hemolytic assay.
Hemolysis of MUC7 12-mer-d and PIC was examined as described previously (29). Amphotericin B and MUC7 12-mer-l were used as controls.
Cell culture.
The HOK-16B cell line (originated from primary normal human keratinocytes) was kindly provided by No-Hee Park, UCLA School of Dentistry. The KB cell line was obtained from the American Type Culture Collection (CLL-17). Cells were maintained in keratinocyte growth medium (KGM; Clonetics, Cambrex, MD), which consists of keratinocyte basal medium (KBM-2) supplemented with KGM-2 SingleQuots. The cells were grown in an incubator at 37°C in an atmosphere of 5% CO2 and 95% air.
MTS cell viability assay.
The toxic effect of the compounds on the cells was determined by calorimetric assay using a CellTiter 96 AQueous One Solution cell proliferation assay (MTS) kit (Promega, WI). MTS tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] reduction is an indication of cell viability. Approximately 80% confluent cells were detached by treating with cell dissociate buffer and trypsin-EDTA solution. The cells were washed twice with trypsin neutralization solution (Clonetics) and resuspended in KGM at 2.5 × 105 cells/ml. An aliquot (50 μl) of the cell solution was incubated with an equal volume of peptides or agent (12.5 to 200 μM) in a 96-well plate at 37°C in a 5% CO2 atmosphere. After a 1.5-h exposure, 20 μl of MTS agent was added to each well, and the cells were further incubated at 37°C in a 5% CO2 atmosphere for 4 h. Absorbance at 490 nm was then measured using a microplate reader. Wells without drugs were used for cell viability, and wells without cells were used for blanking the spectrophotometer. Fifty percent inhibitory concentrations (IC50s) for each cell line were evaluated at a dose of drug causing 50% absorbance reduction in comparison to the untreated control cells.
RESULTS
Candidacidal activity of MUC7 and other peptides in saliva.
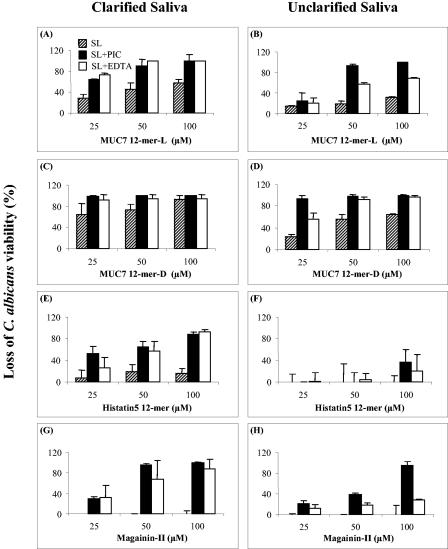
First we determined the candidacidal activities (using the C. albicans DIS isolate) of MUC7 12-mer-l and 12-mer-d and compared them to those of Hsn5 12-mer and magainin-II at three different concentrations (25, 50, and 100 μM) in clarified or unclarified whole human saliva. Saliva without peptides was used as a control. In a pilot experiment, neither Candida nor candidacidal activity was detected in saliva (data not shown). The results are shown in Fig. 1 (hatched bars in each panel). Higher peptide concentrations were required for killing activity in unclarified saliva (Fig. 1, right panels) than in clarified saliva (Fig. 1, left panels). Nevertheless, MUC7 peptides and in particular the d isomer showed significant anticandidal activity in saliva. More specifically, at a 100 μM concentration in clarified saliva, MUC7 12-mer-d exhibited 94% activity (Fig. 1C), compared to 57% exhibited by MUC7 12-mer-l (Fig. 1A), 16% by Hsn5 12-mer (Fig. 1E), and 0% by magainin-II (Fig. 1G). In unclarified saliva, MUC7 12-mer-d and -l exhibited 64 and 32% activity, respectively, while Hsn5 12-mer and magainin-II showed no activity (Fig. 1D, B, F, and H). Thus, MUC7 12-mer-d was the most active peptide in both salivas.
FIG. 1.
Candidacidal activity of MUC7 12-mer-l, MUC7 12-mer-d, Hsn5 12-mer, and magainin-II in human whole saliva and saliva supplemented with PIC or EDTA. Panels A, C, E, and G show results for clarified saliva (filter-sterilized saliva), while panels B, D, F, and H show results for unclarified saliva. Each peptide, at a final concentration of 25, 50, or 100 μM, was incubated with 20 μl of C. albicans (DIS isolate at 1 × 105 cells/ml) and 20 μl of clarified or unclarified saliva (SL; hatched bars), saliva containing PIC (SL+PIC; filled bars), or saliva containing EDTA (SL+EDTA; clear bars) at 37°C for 1.5 h. At the end of incubation, the samples were diluted 20-fold and aliquots plated on SDA. The CFU were counted after 24 h of incubation. The loss of C. albicans viability was determined by the equation (1 − CFU of the test group/CFU of the control group) × 100%. The data represent three individual trials, and in each trial the sample was duplicated; the error bars represent standard deviation.
Candidacidal activity of peptides in saliva supplemented with PIC or EDTA.
First, the amount of PIC added to saliva was optimized so that the PIC alone had very little to no effect on C. albicans cell viability (data not shown). In the presence of PIC, the activities of peptides in saliva increased at each concentration compared to those without PIC (Fig. 1, filled bars). In terms of the PIC enhancement level, magainin-II activity was most enhanced, followed by Hsn5 12-mer and MUC7 12-mer-l. Surprisingly, MUC7 12-mer-d, a peptide theoretically not susceptible to protease degradation, also showed enhanced activity (Fig. 1C and D). A closer look at the composition of PIC revealed that it contains 1 mM EDTA, a metal-dependent protease inhibitor and/or divalent cation chelator. Since saliva contains divalent cations known to inhibit peptide antifungal activity (6), we postulated that EDTA alone might play an important role in the enhancement of peptide activity in saliva by chelating divalent cations. Experimental results demonstrated that addition of 1 mM EDTA to clarified saliva achieved, in most instances, enhancement of peptide activity similar to the addition of PIC (Fig. 1A, C, E, and G). In unclarified saliva, the activity of peptides in the presence of EDTA, with the exception of MUC7 12-mer-d, was lower than in the presence of PIC (Fig. 1B, D, F, and H).
Peptide stability.
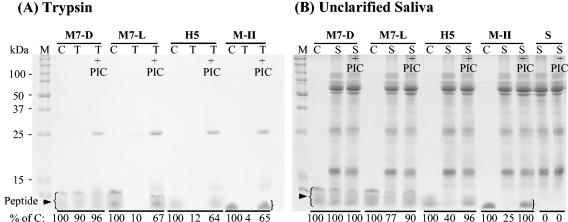
Stability was determined in trypsin and unclarified and clarified saliva, and degradation was monitored by SDS-PAGE (Fig. 2). As shown in Fig. 2A, MUC7 12-mer-d was resistant to degradation by trypsin. The other three peptides, MUC7 12-mer-l, Hsn5 12-mer, and magainin-II, were degraded by trypsin. Similarly, MUC7 12-mer-d was resistant to degradation by unclarified saliva (Fig. 2B), and the other peptides were degraded by saliva, although to different degrees. Figure 2B is a representative of several gels using whole unclarified saliva from different donors (similar patterns of peptide degradation were seen). Degradation was determined quantitatively with a GS-700 imaging densitometer. The percentage of peptide remaining was then calculated, and the values are indicated at the bottom of each lane (Fig. 2). In clarified saliva (results not shown), a much longer incubation time was needed to produce a similar but much less dramatic pattern of l-peptide degradation.
FIG. 2.
Stability of MUC7 12-mer-d (M7-D), MUC7 12-mer-l (M7-L), Hsn5 12-mer (H5), and magainin-II (M-II) in (A) trypsin and (B) unclarified saliva. Peptides were incubated with trypsin containing PIC or no PIC for 0.5 h or saliva (10 μl) for 1 h. The samples were analyzed by SDS-13% PAGE. After staining, the gel was analyzed for peptide degradation with a GS-700 imaging densitometer. The degradation was estimated by the equation [1 − (density of test band/density of control band)] × 100%. C, each peptide alone as a control; T, trypsin plus peptide; T+PIC, trypsin containing PIC plus peptide; S, saliva plus peptide; S+PIC, saliva containing PIC plus peptide. The most intense bands (the highest molecular mass) in the control (C) lanes represent full-length peptides, and the other bands (less intense) represent truncated peptides; for more detail, see the text. Densitometry data of control peptides and peptides remaining after incubation with trypsin or saliva are indicated at the bottom of each lane in each panel.
Candidacidal activity of peptides in saliva from different subjects.
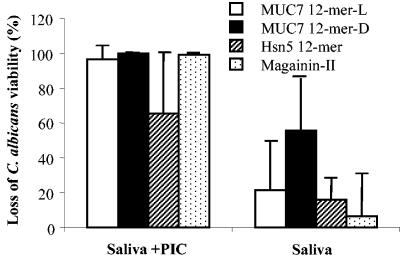
Figure 3 shows the outcome of killing assays performed in clarified whole human saliva samples from five subjects, using the peptides at a concentration of 50 μM. From 60 to 100% killing of C. albicans was observed in saliva containing PIC, while only 6 to 60% was observed in saliva without PIC. The difference is statistically significant (P < 0.05).
FIG. 3.
Candidacidal activity of peptides in saliva from different subjects and enhancement of activity by PIC. Whole human saliva from five healthy subjects was collected and clarified (filter sterilized). Peptides (at 50 μM) in 20 μl of each individual saliva or saliva containing PIC were incubated with an equal volume of C. albicans cells (105 cells/ml in 10 mM sodium phosphate buffer, pH 7.4) at 37°C for 1.5 h. At the end of the incubation, the samples were diluted 20-fold and aliquots plated on SDA. Loss of cell viability was determined as described in the legend to Fig. 1. Values represent the mean ± standard deviation (n = 5). P < 0.05 compared to those without PIC (Saliva).
Activity of peptides in saliva against different Candida strains.
Candidacidal activities of the four peptides against four Candida strains in clarified saliva are shown in Table 1. The data are presented as the percent loss of cell viability compared to a control. With the exception of MUC7 12-mer-l against both strains of C. albicans, the l peptides exhibited low activity against all strains in saliva without PIC. MUC7 12-mer-d showed high activity against all strains, even in saliva without PIC (between 47 and 100%) and was very effective against both strains of C. glabrata (∼100% killing). Hsn5 12-mer and magainin-II showed basically no activity against C. glabrata and C. krusei.
TABLE 1.
Loss of cell viability by exposure of Candida spp. to four individual peptides (50 μM) in clarified (filter-sterilized) saliva
| Strain | % Loss of cell viability
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MUC7 12-mer-d
|
MUC7 12-mer-l
|
Hsn5 12-mer
|
Magainin-II
|
|||||
| With PIC | Without PIC | With PIC | Without PIC | With PIC | Without PIC | With PIC | Without PIC | |
| C. albicans ATCC 96112 | 100.0 ± 0.0 | 74.2 ± 35.7 | 98.9 ± 1.3 | 79.9 ± 31.2 | 97.2 ± 2.3 | 19.1 ± 29.9 | 97.9 ± 0.8 | 7.2 ± 5.3 |
| C. albicans DIS | 99.9 ± 0.2 | 73.8 ± 10.5 | 91.0 ± 13.7 | 44.8 ± 13.0 | 64.6 ± 10.7 | 18.7 ± 14.0 | 94.4 ± 3.8 | 0.0 ± 0.0 |
| C. glabrata ATCC 90030 | 99.8 ± 0.3 | 100.0 ± 0.0 | 94.2 ± 0.3 | 20.6 ± 1.7 | 30.6 ± 15.6 | 1.7 ± 1.5 | 99.1 ± 0.8 | 0.0 ± 0.0 |
| C. glabrata 65C | 100.0 ± 0.0 | 99.8 ± 0.2 | 96.4 ± 1.3 | 21.6 ± 4.1 | 40.6 ± 12.4 | 0.0 ± 0.0 | 99.8 ± 0.3 | 0.0 ± 0.0 |
| C. krusei | 98.9 ± 0.0 | 47.2 ± 8.8 | 99.5 ± 0.0 | 17.6 ± 13.5 | 74.4 ± 29.5 | 0.0 ± 0.0 | 100 ± 0.0 | 0.0 ± 0.0 |
Peptide toxicity studies. (i) Hemolytic activity.
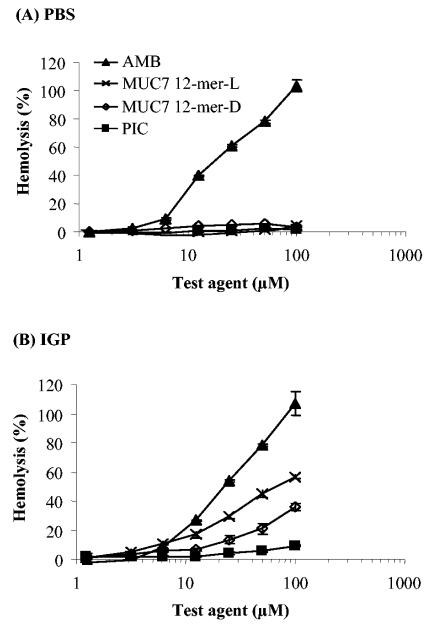
Hemolysis assays were performed in phosphate-buffered saline (PBS) (high-salt buffer, where the antifungal activity of peptides is masked by high ionic strength) and in isotonic glucose phosphate (IGP) buffer (a low-salt buffer supplemented with glucose for osmolarity, where the peptides are active). As indicated in Fig. 4, at the highest concentration tested (100 μM), MUC7 12-mer-d exhibited 3.5% hemolysis in PBS (Fig. 4A) and 36% hemolysis in IGP (Fig. 4B). These values are lower than those for MUC7 12-mer-l, which were 4.3% in PBS and 56% in IGP (29). Amphotericin B showed 100% hemolysis in both buffers.
FIG. 4.
Hemolysis caused by the antifungal agents amphotericin B (AMB, ▴), MUC7 12-mer-l (×), MUC7 12-mer-d (⋄), and PIC (▪). Human erythrocytes (final concentration, 0.5%) were incubated for 1 h at 37°C with a twofold dilution series of peptides in PBS or IGP buffer (29). Hemolysis was determined by an absorbance reading of the supernatant at 450 nm and compared to hemolysis achieved with 1% Triton X-100 (reference for 100% hemolysis). Values represent the mean of results from two independent experiments, each of which was done in duplicate.
(ii) Cytotoxic activity against oral epithelial cells.
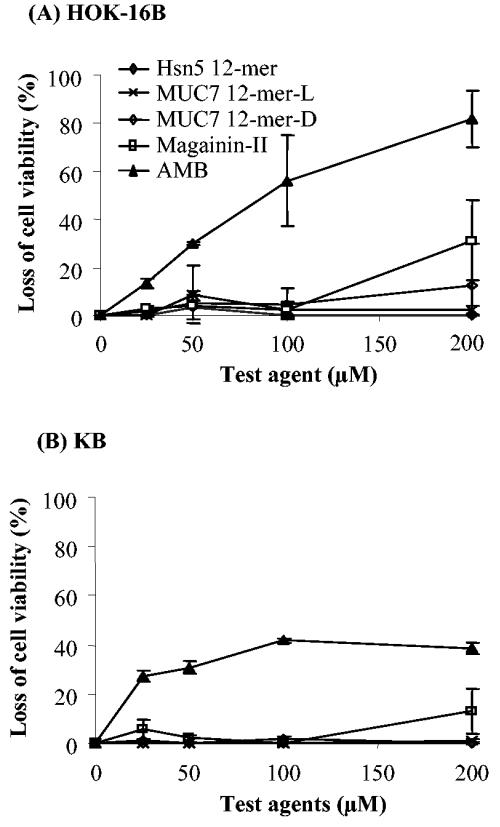
HOK-16B and KB cells were used for the toxicity studies of the four peptides. The results depicted in Fig. 5 indicate that all peptides showed very low toxicity or no toxicity to these cell lines at all concentrations tested. Specifically, at the 100 μM concentration (the highest concentration used for candidacidal killing assays), the peptides showed no toxicity to either cell line. At the highest concentration tested (200 μM), none of these peptides caused 50% cytotoxic effect (IC50) against either cell line, but magainin-II caused about 30% loss of HOK-16B cell viability and MUC7 12-mer-d caused about 10%. Amphotericin B exhibited a dose-dependent toxic activity against HOK-16B, and the IC50 was 100 μM (Fig. 5A).
FIG. 5.
Effect of peptides on viability of (A) HOK-16B cells and (B) KB cells. The toxic effect of the compounds on the cells was determined by calorimetric assay using a CellTiter 96 AQueous One Solution cell proliferation assay (MTS) kit, as described in the text. Briefly, the cells were grown to about 80% confluence, harvested, washed, resuspended in KGM, and dispensed into a 96-well microtiter plate. They were then exposed to different concentrations of peptides or amphotericin B at 37°C and 5% CO2 for 1.5 h. MTS agent was then added to each well, and the cells were further incubated for 4 h. Absorbance at 490 nm was then measured using a microplate reader. MTS reduction is an indication of cell viability. The viability is expressed as relative absorbance (percentage of no-agent control). Data represent the average and standard deviation of results from two independent experiments.
DISCUSSION
Previously, we determined the 50% effective doses (ED50s) for MUC7 12-mer-l, Hsn5 12-mer, and magainin-II in 10 mM PB to be 2.1, 3.7, and 5.7 μM, respectively (28). In this study, an ED50 of 2.6 μM was determined for MUC7 12-mer-d in PB (a value comparable to the l isomer) (results not shown). Other studies also showed that l peptides exert remarkable antifungal activity in low-ionic-strength buffers or media (4, 10, 20, 21, 27-29). However, as shown by our results, the antifungal activities of the tested peptides were substantially decreased in saliva. In unclarified saliva, 25 μM concentrations of MUC7 12-mer-l and 12-mer-d were needed to achieve mean activities of 14 and 23%, respectively, and Hsn5 12-mer and magainin-II showed no activity even at 100 μM concentrations. Salivary proteases and ionic strength are at least two factors that may account for this decrease. Human whole saliva contains a number of proteolytic enzymes, mostly derived from white blood cells and microflora but some which are produced by the salivary glands (9, 16).
PIC, which includes EDTA and has specificity for the inhibition of serine, cysteine, aspartic, and metalloproteases, enhanced antifungal activities of peptides in saliva and surprisingly also that of MUC7 12-mer-d, the peptide theoretically not susceptible to protease degradation. The use of EDTA alone facilitated determination of the roles of PIC and EDTA in this enhancement. The results suggest that in clarified saliva it is the EDTA content in the PIC that is responsible for the enhancement of MUC7 12-mer-d activity by PIC (as well as that of l peptides). EDTA serves a dual function; it not only inhibits metal-dependent proteases but also chelates divalent cations present in saliva known to inhibit peptide antifungal activity (see below). In unclarified saliva, the PIC plays a role in the l peptide antifungal activity enhancement above that achievable by EDTA alone, through inhibition of all classes of proteases.
Previous studies showed magainin-II (5) and histatins (22, 30) to be susceptible to degradation by proteolytic enzymes and histatins to be degraded by saliva; the latter is mediated mainly by trypsin-like enzyme activity. MUC7 12-mer, Hsn5 12-mer, and magainin-II contain large and almost equal numbers of lysine and arginine residues (targets of trypsin). Yet, compared to MUC7 12-mer-l, Hsn5 12-mer and magainin-II appear to be more sensitive to saliva degradation (Fig. 2), which is consistent with their lower activity in saliva (Fig. 1). These results indicate that other factors, such as other classes of proteases, may play a role in the degradation of different peptides by saliva.
Ionic strength has also been demonstrated to be one of the important factors that affect the efficiency of antimicrobial peptides in the medium (17) and in saliva (14). Both mono- and divalent ions exert inhibitory effects on some antimicrobial peptides. For example, increased amounts of Ca2+ and Mg2+ decreased the activity of MUC7 20-mer (6) and histatin 5 (8). K+, Na+, and Li+ exerted an inhibitory effect on peptide enterocin CRL35 (17). A recent study (19) determined the mean concentration of total magnesium in saliva to be 0.22 mmol/liter and that of calcium to be 1.39 mmol/liter. EDTA added to saliva was apparently able to chelate these inhibitory ions. Regarding the effect of NaCl concentration on peptide antifungal activity, we have preliminary evidence showing that MUC7 peptides are much less sensitive to NaCl than Hsn5 12-mer and magainin-II.
In conclusion, MUC7 12-mer-d exhibited activity comparable to MUC7 12-mer-l in PB, showed high candidacidal activity in clarified or unclarified saliva, and was resistant to salivary degradation. Meanwhile, the hemolysis and toxicity to oral epithelial cells caused by MUC7 12-mer-d was very low. The present study suggests that d amino acid substitution is a useful technique to improve the antifungal activity of MUC7 12-mer without increasing its toxicity. EDTA was very effective in the enhancement of antifungal activity of MUC7 12-mer-d in human saliva, suggesting that this combination may have a therapeutic potential for the treatment of oral candidal infections. For the l peptides, the combination with PIC might be more effective because PIC contains inhibitors for all classes of proteases.
Acknowledgments
This study was supported by NIH/NIDC grant DE09820.
We thank Alex Ho for his assistance with statistics, Alex Campagna and Seth Walbridge for help with killing assays, Shinshuki Onishi for help with the cell culture, and Eileen Bobek for critical reading of the manuscript.
REFERENCES
- 1.Andriole, V. T. 2000. Current and future antifungal therapy: new targets for antifungal therapy. Int. J. Antimicrob. Agents 16:317-321. [DOI] [PubMed] [Google Scholar]
- 2.Avrahami, D., and Y. Shai. 2003. Bestowing antifungal and antibacterial activities by lipophilic acid conjugation to D, L-amino acid-containing antimicrobial peptides: a plausible mode of action. Biochemistry 42:14946-14956. [DOI] [PubMed] [Google Scholar]
- 3.Avrahami, D., and Y. Shai. 2004. A new group of antifungal and antibacterial lipopeptides derived from non-membrane active peptides conjugated to palmitic acid. J. Biol. Chem. 279:12277-12285. [DOI] [PubMed] [Google Scholar]
- 4.Bessalle, R., H. Haas, A. Goria, I. Shalit, and M. Fridkin. 1992. Augmentation of the antibacterial activity of magainin by positive- charge chain extension. Antimicrob. Agents Chemother. 36:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessalle, R., A. Kapitkovsky, A. Gorea, I. Shalit, and M. Fridkin. 1990. All-D-magainin: chirality, antimicrobial activity and proteolytic resistance. FEBS Lett. 274:151-155. [DOI] [PubMed] [Google Scholar]
- 6.Bobek, L. A., and H. Situ. 2003. MUC7 20-mer: investigation of antimicrobial activity, secondary structure, and possible mechanism of antifungal action. Antimicrob. Agents Chemother. 47:643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devine, D. A., P. D. Marsh, R. S. Percival, M. Rangarajan, and M. A. Curtis. 1999. Modulation of antibacterial peptide activity by products of Porphyromonas gingivalis and Prevotella spp. Microbiology 145:965-971. [DOI] [PubMed] [Google Scholar]
- 8.Dong, J., S. Vylkova, X. S. Li, and M. Edgerton. 2003. Calcium blocks fungicidal activity of human salivary histatin 5 through disruption of binding with Candida albicans. J. Dent. Res. 82:748-752. [DOI] [PubMed] [Google Scholar]
- 9.Furuyama, M., S. Koshika, Y. Kitamura, and Y. Nakayama. 1987. Trypsin-like protease and glucose-6-phosphate dehydrogenase in the human submandibular salivary gland. Arch. Oral Biol. 32:761-762. [DOI] [PubMed] [Google Scholar]
- 10.Giacometti, A., O. Cirioni, M. S. Del Prete, F. Barchiesi, A. M. Paggi, E. Petrelli, and G. Scalise. 2000. Comparative activities of polycationic peptides and clinically used antimicrobial agents against multidrug-resistant nosocomial isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 46:807-810. [DOI] [PubMed] [Google Scholar]
- 11.Gururaja, T. L., J. H. Levine, D. T. Tran, G. A. Naganagowda, K. Ramalingam, N. Ramasubbu, and M. J. Levine. 1999. Candidacidal activity prompted by N-terminus histatin-like domain of human salivary mucin (MUC7)1. Biochim. Biophys. Acta 1431:107-119. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 14.Helmerhorst, E. J., B. Flora, R. F. Troxler, and F. G. Oppenheim. 2004. Dialysis unmasks the fungicidal properties of glandular salivary secretions. Infect. Immun. 72:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Intini, G., A. Aguirre, and L. A. Bobek. 2003. Efficacy of human salivary mucin MUC7-derived peptide and histatin 5 in a murine model of candidiasis. Int. J. Antimicrob. Agents 22:594-600. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy, S., C. Davis, W. R. Abrams, P. C. Billings, T. Nagashunmugam, H. Friedman, and D. Malamud. 1998. Submandibular salivary proteases: lack of a role in anti-HIV activity. J. Dent. Res. 77:1515-1519. [DOI] [PubMed] [Google Scholar]
- 17.Minahk, C. J., and R. D. Morero. 2003. Inhibition of enterocin CRL35 antibiotic activity by mono- and divalent ions. Lett. Appl. Microbiol. 37:374-379. [DOI] [PubMed] [Google Scholar]
- 18.Oppenheim, F. G., T. Xu, F. M. McMillian, S. M. Levitz, R. D. Diamond, G. D. Offner, and R. F. Troxler. 1988. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 263:7472-7477. [PubMed] [Google Scholar]
- 19.Rehak, N. N., S. A. Cecco, and G. Csako. 2000. Biochemical composition and electrolyte balance of “unstimulated” whole human saliva. Clin. Chem. Lab. Med. 38:335-343. [DOI] [PubMed] [Google Scholar]
- 20.Rothstein, D. M., P. Spacciapoli, L. T. Tran, T. Xu, F. D. Roberts, M. Dalla Serra, D. K. Buxton, F. G. Oppenheim, and P. Friden. 2001. Anticandida activity is retained in P-113, a 12-amino-acid fragment of histatin 5. Antimicrob. Agents Chemother. 45:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruissen, A. L., J. Groenink, E. J. Helmerhorst, E. Walgreen-Weterings, W. Van't Hof, E. C. Veerman, and A. V. Nieuw Amerongen. 2001. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 356:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruissen, A. L., J. Groenink, P. Krijtenberg, E. Walgreen-Weterings, W. van't Hof, E. C. Veerman, and A. V. Nieuw Amerongen. 2003. Internalisation and degradation of histatin 5 by Candida albicans. Biol. Chem. 384:183-190. [DOI] [PubMed] [Google Scholar]
- 23.Sajjan, U. S., L. T. Tran, N. Sole, C. Rovaldi, A. Akiyama, P. M. Friden, J. F. Forstner, and D. M. Rothstein. 2001. P-113d, an antimicrobial peptide active against Pseudomonas aeruginosa, retains activity in the presence of sputum from cystic fibrosis patients. Antimicrob. Agents Chemother. 45:3437-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satyanarayana, J., H. Situ, S. Narasimhamurthy, N. Bhayani, L. A. Bobek, and M. J. Levine. 2000. Divergent solid-phase synthesis and candidacidal activity of MUC7 D1, a 51-residue histidine-rich N-terminal domain of human salivary mucin MUC7. J. Pept. Res. 56:275-282. [DOI] [PubMed] [Google Scholar]
- 25.Schenkels, L. C. P. M., T. L. Gururaja, and M. J. Levine. 1996. Salivary mucins: their role in oral mucosal barrier function and drug delivery, p. 191-220. In M. J. Rathbone (ed.), Oral mucosal drug delivery. Marcel Dekker, New York, N.Y.
- 26.Schmidtchen, A., I. M. Frick, E. Andersson, H. Tapper, and L. Bjorck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 27.Situ, H., and L. A. Bobek. 2000. In vitro assessment of antifungal therapeutic potential of salivary histatin-5, two variants of histatin-5, and salivary mucin (MUC7) domain 1. Antimicrob. Agents Chemother. 44:1485-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Situ, H., G. Wei, C. J. Smith, S. Mashhoon, and L. A. Bobek. 2003. Human salivary MUC7 mucin peptides: effect of size, charge and cysteine residues on antifungal activity. Biochem. J. 375:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei, G. X., and L. A. Bobek. 2004. In vitro synergic antifungal effect of MUC7 12-mer with histatin-5 12-mer or miconazole. J. Antimicrob. Chemother. 53:750-758. [DOI] [PubMed] [Google Scholar]
- 30.Xu, L., K. Lal, R. P. Santarpia III, and J. J. Pollock. 1993. Salivary proteolysis of histidine-rich polypeptides and the antifungal activity of peptide degradation products. Arch. Oral Biol. 38:277-283. [DOI] [PubMed] [Google Scholar]
- 31.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 32.Zasloff, M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 84:5449-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]