Abstract
Recent studies have suggested that the herpes simplex type 1 (HSV-1) UL25 gene product, a minor capsid protein, is required for encapsidation but not cleavage of replicated viral DNA. This study set out to investigate the potential interactions of UL25 protein with other virus proteins and determine what properties it has for playing a role in DNA encapsidation. The UL25 protein is found in 42 ± 17 copies per B capsid and is present in both pentons and hexons. We introduced green fluorescent protein (GFP) as a fluorescent tag into the N terminus of UL25 protein to identify its location in HSV-1-infected cells and demonstrated the relocation of UL25 protein from the cytoplasm into the nucleus at the late stage of HSV-1 infection. To clarify the cause of this relocation, we analyzed the interactions of UL25 protein with other virus proteins. The UL25 protein associates with VP5 and VP19C of virus capsids, especially of the penton structures, and the association with VP19C causes its relocation into the nucleus. Gel mobility shift analysis shows that UL25 protein has the potential to bind DNA. Moreover, the amino-terminal one-third of the UL25 protein is particularly important in DNA binding and forms a homo-oligomer. In conclusion, the UL25 gene product forms a tight connection with the capsid being linked with VP5 and VP19C, and it may play a role in anchoring the genomic DNA.
Herpes simplex virus type 1 (HSV-1) nucleocapsids, which consist of an icosahedral capsid surrounding a viral DNA core, are assembled in the nuclei of infected cells. The icosahedral structure is composed of 162 capsomers (150 hexons and 12 pentons). The pentons have fivefold symmetry and are located at the 12 vertices, while the hexons have sixfold symmetry and occupy the edges and faces of the capsid icosahedron (10, 49). Three different capsid types, designated A (empty), B (intermediate), and C (containing DNA), have been isolated by velocity sedimentation of nuclear lysates in virus-infected cells (17, 38, 44, 56). During HSV-1 infection, the replicated concatemeric DNA is cleaved to unit-length genomes and packaged into preassembled capsids with loss of scaffolding protein. The capsid consists of the gene products of UL18 (VP23), UL19 (VP5), UL26 (VP21 and VP24), UL26.5 (VP22a), UL35 (VP26), and UL38 (VP19C) (14, 17, 23, 31, 46, 50). VP5 (major capsid protein), VP19C, VP23 (triplex proteins), and scaffolding proteins are essential for the assembly of an intact capsid (15, 16, 39). Studies on temperature-sensitive mutants have also shown that the gene products of UL6 (36, 50), UL15 (5, 40), UL17, UL25 (1, 3), UL28 (2, 13), UL32 (48), and UL33 (4) are required for the DNA cleavage and packaging process. Mutants lacking these genes have been isolated and characterized to investigate the roles of these genes in the process of DNA encapsidation (6, 21, 22, 25, 37, 47, 54). Almost all of these mutants synthesize near-wild-type levels of viral DNA but do not cleave concatemeric viral DNA into unit-length genomes and accumulate only type B capsids in infected cells. However, the UL25 null mutant is able to cleave the replicated viral DNA and produces both A and B capsids in infected cells (25). These gene products can be divided into two groups based on the DNA packaging process (57). The gene products of UL6, UL15, UL17, UL28, UL32, and UL33 play a role in DNA maturation and the packaging process, while UL25 protein may play a role in the process thereafter, which is known as the head completion process in bacteriophages.
The process of assembling HSV-1 capsids and packaging DNA is similar to that of double-stranded DNA (dsDNA) bacteriophages such as T4, P22, and lambda (9, 27, 28, 43). It is likely that the phenomenon found in UL25 null mutant-infected cells results from an abortive packaging event (25). A similar phenomenon had been observed in mutants defective in the gene products gp4, gp10, and gp26 of phage P22. In cells infected with mutants defective in these genes, the filled capsids are unstable and lose mature DNA within the cells, resulting in the accumulation of empty capsids (41, 51). A role of these gene products is thought to be “head completion”: the closing of the channel at the unique vertex and formation of a binding site for the products required for tail attachment. These products are thought to perform some valve-like function at the unique vertex for a translocation of DNA, in addition to having a DNA-stabilizing function (51). Whether UL25 protein has the valve-like function at capsid vertices is unknown; however, it may play such a role in the head completion process, as has been found with dsDNA bacteriophages. In HSV-1, if this is correct, it probably implies that UL25 protein is closely related to the packaging channel (probably penton sites) and added to the vertex after the viral DNA is packaged. However, we cannot exclude the possibility that UL25 protein is incorporated during the process of capsid assembly, because UL25 has been detected in B capsids, in which DNA is not yet packaged. To investigate the above assumption and this possibility, we attempted to determine the location of the UL25 gene product in the capsid, identify its binding patterns in the capsid, and determine when it enters the nucleus of HSV-1-infected cells.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney cells (Vero) and HeLa cells (Riken Cell Bank, Tukuba, Japan) were grown and maintained at 37°C and 5% CO2 in Eagle's minimal essential medium (EMEM) containing 10% newborn calf serum. The VR3 strain of HSV-1 was used in this study and was propagated in Vero cells in EMEM containing 2% newborn calf serum. For the growth of recombinant baculovirus, Spodoptera frugiperda (Sf9) cells were cultured in Grace's medium (Gibco/BRL) supplemented with 10% fetal bovine serum.
Plasmid construction.
The 2.3-kbp BamHI-U fragment containing the entire UL25 gene (nucleotide positions 48813 to 50552) was cloned into pBluescript II SK(−) to give pBsBam-U. The prokaryotic expression vector pETul25 encodes an amino-terminally polyhistidine (His)-tagged UL25 protein and was constructed as follows. An NdeI site (underlined) was introduced into UL25 start codon by PCR of the BamHI-U fragment. The sequence of the forward primer was 5′-CTCTCGCATATGGACCCGTACT-3′. The sequence of the reverse primer, which lies within the UL25 coding region, was 5′-CGTACACCATGTGTAGCAGAT-3′. The PCR product digested with NdeI and NotI and an NotI/BamHI fragment from pBsBam-U were ligated into the NdeI-BamHI site of the prokaryotic expression vector pET-16b to give pETul25. To construct the eukaryotic expression plasmid, a fragment containing the entire UL25 coding region was derived from pETul25 by digestion with NdeI and BsmI (position 50726). After the ends were blunted, the NdeI/BsmI fragment was ligated into the EcoRV site of the eukaryotic expression vector pcDNA3 to give pDul25. The resulting plasmid was sequenced to confirm the correct insertion of the blunt-ended fragment and to verify the region amplified by PCR. pETul6 encodes a His-tagged UL6 protein and was constructed as follows. An NdeI site (underlined) was introduced into the UL6 start codon by PCR of the genomic DNA. The sequence of the forward primer was 5′-GGTCGCGCATATGACCGCACCAC-3′. The sequence of the reverse primer, which lies within the UL6 coding region, was 5′-GCCGAGATCTCATCGTCGGCCGT-3′ (the underline indicates a BglII site). The PCR product digested with NdeI and BglII was ligated into the NdeI-BamHI site of pET-16b.
The 9.4-kb HindIII-K fragment containing the entire UL38 gene (positions 84531 to 85926) was cloned into pBluescript II SK(−), to give pBsHind-K. A fragment containing the UL38 coding region was derived from pBsHind-K by digestion with NruI and XhoI and ligated into pcDNA3 digested with EcoRV and XhoI, to give the eukaryotic expression plasmid pDul38.
The green fluorescent protein (GFP) expression vector pRSETB-GFP (S65T) was generously provided by R. Y. Tsien (San Diego, Calif.). The GFP gene was recloned into the EcoRI-BamHI site of pBluescript II SK(−). From the resulting plasmid, pBsGFP, the GFP gene was cloned into pcDNA3 using HindIII and BamHI sites (pDGFP). To delete the stop codon of the GFP gene, pBsGFP was digested with FokI (underlined), whose site is located in the stop codon (italic) of the GFP gene (5′-G GAT GAA CTA TAC AAA TAA-3′), and an FokI site was blunt ended (5′-TAC AAA TA-3′) before digestion with HindIII. The NdeI site introduced by PCR near the start codon (underlined) of pETul25 was digested with NdeI, the end was made blunt (5′-T ATG GAC CCG-3′), and then the construct was digested with BamHI. The resulting NdeI (blunt-ended)-BamHI fragment was ligated to an HindIII-FokI (blunt-ended) fragment of pBsGFP in the three-piece ligation with pcDNA3 cleaved by HindIII and BamHI. The resulting construct was designated pDGFPul25 and contains a chimeric gene encoding the GFP fused to the N terminus of UL25 with an appropriate reading frame. To construct a eukaryotic expression plasmid encoding a GFP-tagged UL38 protein, pDGFPul38 was constructed by ligating the HindIII (blunt-ended)-FokI (TAC AAA TA) fragment of pBsGFP into an HindIII (blunt-ended)-BstXI site of pDul38 in the appropriate reading frame. The BstXI site (5′-CCA GTG TGC TGG AAT TCT GCA GAT CGA TCT GGG GTC GCA ATG-3′) of pDul38 is positioned upstream of the start ATG of UL38 (double underlined). In the resulting plasmid, a sequence upstream of the UL38 start ATG was 5′-TAC AAA TAC TGG AAT TCT GCA GAT CGA TCT GGG GTC GCA ATG-3′ (the 3′ overhang blunting site of the C-terminal portion of the GFP gene is underlined).
To express the HisUL25 coding gene in baculovirus, the coding sequence was placed under the control of the polyhedron promoter as follows. The XbaI-BamHI fragment containing the HisUL25 coding region of pETul25 was subcloned into pBluescript II SK(−). The baculovirus expression plasmid pBacHisul25 was constructed by ligating the XbaI/EcoRI fragment of this subcloned plasmid into the XbaI-EcoRI site of pBacPAK8.
Antibodies.
The histidine-tagged proteins of UL25 and UL6 were expressed in Escherichia coli strain BL21(DE3) by transfection with pETul25 and pETul6. To prepare these proteins, metal chelate chromatography was performed to purify the fusion protein from the soluble form under native conditions, according to the manufacturer's instructions (nickel-nitrilotriacetic acid [Ni-NTA] resin). We prepared a mouse polyclonal antiserum to UL25 and UL6 proteins, using these preparations as immunogens.
Purification of intracellular capsids and virions.
Vero cells were infected with HSV-1 at a multiplicity of infection of 5 PFU per cell, and cells were harvested after incubation at 37°C for 12 h. Nuclear lysates were prepared by three cycles of freeze-thawing followed by sonication on ice in TNE buffer (20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, and 1 mM EDTA) containing 1% Nonidet P-40 (NP-40) and protease inhibitor cocktail (10 μM leupeptin, 10 μM pepstatin, and 0.2 mM phenylmethylsulfonyl fluoride). Three types of capsids were purified by banding on a linear 20-to-50% (wt/wt) sucrose gradient centrifugation and pelleted by centrifugation at 57,000 × g for 30 min, as described by Tatman et al. (53). Guanidine-HCl (GuHCl) and urea extraction was carried out by the procedure by Newcomb et al. (34). B capsids were treated with various concentrations of urea in Tris-borate-EDTA (TBE) buffer containing protease inhibitor cocktail for 1 h at room temperature. Extracellular virus was harvested at 18 h postinfection, purified by banding on 5-to-50% (wt/wt) sucrose gradients, and pelleted. Virions were solubilized with 1% NP-40 for 30 min on ice and centrifuged at 12,000 × g for 15 min. The resulting pellets and supernatants contain the tegument and capsid materials and the envelope materials, respectively (24).
Gel electrophoresis and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by adjusting samples to a final concentration of 1% SDS and 5% 2-mercaptoethanol in SDS sample buffer (0.32 M Tris-HCl [pH 6.8], 5% SDS, 25% 2-mercaptoethanol, and 35% glycerol) before boiling them for 3 min. Proteins were developed on 10% or 4-to-12% gradient SDS-polyacrylamide gels. All gels were stained with Coomassie brilliant blue (CBB) or silver. The B capsid extracts with urea were applied to the gels and electrophoresed under nondenaturing conditions (0.025 M Tris and 0.192 M glycine [pH 8.3]).
For Western blot analysis, the separated proteins were electrophoretically transferred to polyvinylidene difluoride (PVDF) sheets. The blotted sheets were then blocked with 5% (wt/vol) skim milk in TBS (10 mM Tris-HCl [pH 7.4] and 0.15 M NaCl) and were incubated with the primary antibody (UL25 or UL6 antiserum) diluted in 1% skim milk and TBS-T buffer (TBS buffer containing 0.1% Tween 20). After a wash in TBS-T buffer, the immunoreactive protein was detected using anti-mouse antibody conjugated to horseradish peroxidase (HRP) and a luminogenic reagent (for enhanced chemiluminescence [ECL]) prior to exposure to Kodak BioMax-MR film.
Immunogold labeling and EM.
Immunogold labeling was used to determine the localization of UL25 protein in gradient-purified B capsids. Capsids were adsorbed to carbon-coated electron microscope grids in TBS and fixed in 2% glutaraldehyde–TBS buffer for 2 min. The immunostaining of UL25 protein was carried out in a manner similar to Western blotting. Briefly, fixed capsids on the grids were incubated with UL25 antiserum diluted in TBS-T buffer, washed in TBS-T buffer, and then incubated with protein A conjugated to 10-nm-diameter gold particles. After a wash in TBS-T buffer, capsids were stained with 2% uranyl acetate and examined by electron microscopy (EM). For the negative control, the UL25 antiserum was replaced with nonimmune mouse serum.
Transfections and immunofluorescence analysis.
HeLa cells were cultured at 37°C and 5% CO2 in phenol red-free EMEM containing 10% newborn calf serum. Transient DNA transfections were performed using cationic lipid reagent, and cells were maintained in growth medium for 2 days prior to fluorescence analysis or selection with Geneticin (G418). To obtain the stable transfectant, cells transfected with pDul38 were maintained in selective medium containing 0.5 mg of G418 per ml. After 14 days, G418-resistant colonies (VP19C-expressing cells) were isolated and maintained in selective medium.
For fluorescence analysis, transfected cells were fixed in 4% paraformaldehyde–TBS buffer. After being washed in TBS, the GFP-tagged proteins on slides mounted in 50% glycerol–PBS were analyzed by a Bio-Rad MRC600 laser scanning confocal microscope. In addition to detection of GFP fluorescence, the expression of UL25 protein in cells transfected with pDul25 was visualized in a manner similar to Western blotting. Briefly, fixed cells on a chamber slide were blocked in TBS containing 5% normal goat serum and reacted with UL25 antiserum diluted in TBS-T buffer. Immunoreactive protein was detected by incubation with Texas red-labeled anti-mouse antibody and a subsequent confocal microscopic analysis.
Protein-protein interaction analysis.
Recombinant baculovirus was constructed with the BacPAK baculovirus expression system (Clontech) as recommended by the manufacturer. Sf9 cells infected with recombinant baculovirus (BacHisUL25) were prepared for purification of the native form of UL25 protein. Sf9 cells were infected with BacHisUL25 at 5 PFU per cell and harvested at 48 h postinfection. Cells were suspended in hypotonic buffer (10 mM Tris-HCl, 50 mM NaCl, and 0.1% Tween 20 [pH 8.0]) and centrifuged at 4,000 × g after undergoing lysis by three cycles of freezing and thawing. Under native conditions, His-tagged UL25 (HisUL25) protein was purified from the resulting supernatants with a Ni-NTA column. To remove a His tag, HisUL25 protein was again applied to the column after overnight incubation in TBS buffer containing 1 μg of factor Xa at 4C°. The flowthrough fraction was dialyzed in 50 mM bicarbonate buffer (pH 8.6) at 4C° before being blotted to a sheet or subjected to biotinylation. To investigate potential interactions of UL25 protein with other virus proteins, two analyses using a soluble form of UL25 protein were carried out, as described below.
(i) Identification of the virus proteins reconstituted with UL25 on the sheet.
Native UL25 protein was dot blotted to a PVDF sheet (total area, 0.12 cm2) by vacuum filtration and blocked with 1% (wt/vol) polyvinylpyrrolidone 40 in TBS. Virions were solubilized with 8.0 M urea in TNE buffer containing 1% (wt/vol) bovine serum albumin (BSA) and protease inhibitor cocktail after treatment with 0.5% NP-40 on ice for 30 min. Following an overnight incubation at room temperature, the remaining insoluble proteins were removed by centrifugation prior to reaction with the UL25-blotted sheet. A mixture of solubilized virions and the blotted sheet was renatured by slow stepwise dialysis with a decreasing urea concentration and finally dialyzed with TBS-T buffer. After 3 days of dialysis, the blotted sheet was harvested and washed five times with TBS-T buffer. The bound proteins were eluted with SDS sample buffer from the blotted sheet, developed by SDS-PAGE, and transferred to a nitrocellulose sheet. The stained protein band with 0.1% Ponceau S was excised and digested with TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin. Following fractionation by reverse-phase high-pressure liquid chromatography (5C18-AR-300 column), the resulting peptides were subjected to amino-terminal sequencing.
(ii) Far Western blotting.
Purified UL25 protein was biotinylated, using an N-hydroxy-succinimide-biotin ester, with an ECL biotinylation module (Amersham). Capsid preparations developed on SDS–10% polyacrylamide gels were electrophoretically transferred to PVDF sheets. The proteins were renatured by washing three times with TBS-T buffer (containing 5 mM 2-mercaptoethanol and 10% glycerol), followed by an overnight incubation at 4°C. The sheet was then blocked with 5% skim milk in TBS buffer, and reacted overnight with biotin-labeled UL25 in TBS-T buffer containing 5% BSA at 4°C. Excess probe was removed by five washes in excess TBS-T buffer. Detection was by incubation with HRP-probed streptavidin and ECL.
Gel mobility shift analysis.
To investigate whether UL25 protein has the inherent capacity to bind DNA, we constructed a two-dimensional gel electrophoresis system. The soluble form of UL25 protein was prepared from Sf9 cells infected with BacHisUL25. Plasmid pQul32(c191) is a pQE30-derived UL32 expression plasmid in which the BamHI/SalI fragment of UL32 is inserted into BamHI/SalI sites. The UL32(c191) protein, which expressed in BL21(DE3) cells, was made up of the C-terminal 191 amino acids (residues 274 to 464) of UL32 protein with a His tag. These two proteins were purified with a Ni-NTA column and analyzed by SDS-PAGE (12% polyacrylamide gel). To prepare genomic DNA of HSV-1 or baculovirus (BacPAK6), the extracellular virions purified by the sucrose gradients were incubated for 12 h at 50°C in 0.1 M Tris-HCl (pH 8.0) containing 1% SDS, 5 mM EDTA, and 0.1 mg of proteinase K/ml after treatment with RNase (final concentration of 10 μg/ml), and organic extraction and ethanol precipitation were subsequently carried out. Baculovirus DNA was digested with Bsu36I.
The genomic DNAs were resuspended in binding buffer (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 2 mg of BSA/ml, 0.1 mg of factor Xa/ml, and 5 μg of sonication-degenerated salmon DNA/μl) and then incubated overnight with UL25 or UL32(c191) protein at 4°C. These reaction mixtures (20 μl) were applied to a 1.5% agarose gel in 0.5× TBE buffer (45 mM Tris [pH 8.0], 45 mM boric acid, and 2 mM EDTA). After gel electrophoresis, gels were stained with ethidium bromide and photographed. The genomic DNA-containing bands and bands corresponding to those positions were excised from each lane. Proteins bound to DNA were electroeluted from these gels and incubated in DNase I (1 U/50 μl) solution (10 mM Tris, 10 mM MgCl2, and 10 mM CaCl2 [pH 7.5]). The resulting extracts were analyzed by dot immunoblotting using Ni-NTA HRP conjugate. Bands excised from lanes to which reaction mixtures of UL25 with or without HSV genome had been applied were loaded onto 12% SDS-polyacrylamide gels, sealed with agarose in SDS sample buffer, and electrophoresed in the second dimension. The separated proteins were blotted onto PVDF sheets and subjected to Western blot analysis using UL25 antiserum.
RESULTS
UL25 protein is part of the penton and the hexon structures.
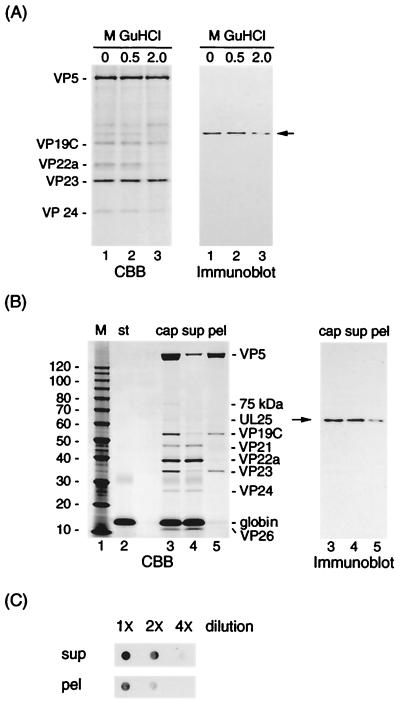
If the HSV-1 capsid vertex (penton) serves as the site for DNA entry, one of the DNA-packaging proteins, UL25, is probably associated with the penton. Figure 1 shows SDS-PAGE (CBB stain) and Western blot (immunoblot) analyses of B capsid extracts with GuHCl and urea. To determine the strength of the association between UL25 protein and HSV-1 capsid, B capsids were treated with 0, 0.5, and 2.0 M GuHCl. As shown in Fig. 1A, the amounts of stained VP5 bands among B capsid extracts did not differ much, whereas UL25 protein with significant loss of signal intensity was detected at 2.0 M. The UL25 protein may be part of the penton, on the basis of the previous reports (30, 34) that 2.0 M GuHCl extraction results in the removal of the penton of capsids. However, if UL25 is bound loosely to regions other than the pentons, some UL25 proteins may be stripped off by this extraction. The result of separation of B capsid extracts (Fig. 1B) shows that the 6.0 M urea extraction resulted in the complete removal of the scaffold proteins (VP22a) (Fig. 1B, lane 5 in the CBB-stained gel) and in some extraction of VP5, VP19C, and VP23 to the supernatants (lane 4). Both extracts contained UL25 protein, as shown in the immunoblot. For an accurate comparison of UL25 protein between supernatants and pellets, the globin proteins (Fig. 1B, lane 2), as internal standards, were added to B capsid samples (lane 3), and extraction with 6.0 M urea was carried out. The globin was almost undetectable in the pellets (lane 5). The amount of samples applied to a PVDF sheet was corrected by the amount of VP5 between these two extracts, and dot immunoblotting was carried out after serial dilution of these extracts. This dot immunoblot analysis (Fig. 1C) shows that the supernatants contain 1.5 to 2 times more UL25 protein than the pellets.
FIG. 1.
Detection of UL25 protein in B capsids extract with GuHCl. (A) B capsids were extracted with various concentrations of GuHCl. The preparations were subjected to SDS-PAGE, stained with CBB, and analyzed by Western blotting using UL25 antiserum (immunoblot). (B) The 18-kDa globin, as an internal standard (st), was added to B capsids (lane 3; cap) prior to the 6.0 M urea extraction. The B capsid samples with urea were centrifuged through a 20% sucrose cushion to give supernatants (lane 4; sup) and pellets (lane 5; pel). The resulting samples were subjected to SDS-PAGE and analyzed by Western blotting using UL25 antiserum. (C) These two samples were solubilized with urea at a final concentration of 8.0 M and serially diluted in TBS containing 8.0 M urea, prior to dot blotting on PVDF sheet. The UL25 on the sheet was detected by Western blotting after a washing in TBS-T buffer. The positions of molecular mass markers (M; in kilodaltons), capsid proteins, and of UL25 (arrows) are indicated.
The proteins stained with CBB in lane 3 of Fig. 1B were scanned in a densitometer, and a quantitative determination was made with National Institutes of Health Image software. Table 1 shows the amounts and the copy numbers calculated for the 75-kDa protein, UL25 protein, and capsid proteins. It was calculated that there were approximately 42 ± 17 copies of UL25 and 44 ± 13 copies of the 75-kDa protein per B capsid. The 75-kDa band is probably UL6 protein (see the immunoblot in Fig. 6A). These results suggest that UL25 protein is a minor capsid protein and may be part of the penton in addition to the hexon structure.
TABLE 1.
Protein composition of HSV-1 B capsida
| Measurement | VP5 | 75-kDa protein | UL25 | VP19C | VP21 | VP22a | VP23 | VP24 | VP26 |
|---|---|---|---|---|---|---|---|---|---|
| Mol wt | 149,075 | 75,000 | 62,666 | 50,260 | 45,000 | 33,765 | 34,268 | 26,618 | 12,095 |
| Fraction (%, wt/wt)b | 52.6 ± 3.4 | 1.5 ± 0.3 | 1.2 ± 0.4 | 8.2 ± 0.9 | 2.4 ± 0.9 | 13.9 ± 1.1 | 10.8 ± 3.5 | 2.2 ± 0.9 | 5.7 ± 1.6 |
| Corr.fc | 58.4 | 1.3 ± 0.3 | 1.1 ± 0.4 | 7.2 ± 0.8 | 2.1 ± 0.8 | 12.2 ± 1.0 | 9.5 ± 3.1 | 1.9 ± 0.8 | 5.0 ± 1.4 |
| Copy no.d | 960 | 44 ± 13 | 42 ± 17 | 351 ± 38 | 114 ± 44 | 885 ± 70 | 679 ± 223 | 175 ± 76 | 1,013 ± 282 |
Proteins stained by Coomassie brilliant blue in lane 3 of Fig. 1B were scanned in a densitometer, and a quantitative determination was made using National Institutes of Health Image software. Values are the means ± standard deviations for nine independent measurements in B capsids.
Fraction of total protein.
Corr.f, corrected percentages calculated assuming that B capsid contains 960 copies of VP5.
a Copy numbers (N) were calculated from the capsid masses (245 MDa [37]), the corrected percentages (Coff.f), and molecular masses (m) of each capsid, as follows: N = (245 Corr.f/m) × 106.
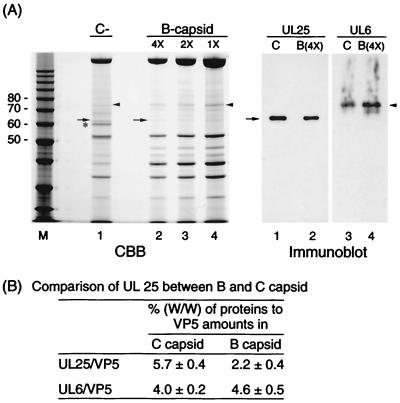
FIG. 6.
Comparison of the amount of UL25 in B and C capsids. The two types of capsid were prepared by two cycles of a linear sucrose gradient. (A) B and C capsids were subjected to SDS-PAGE, stained with CBB, and analyzed by Western blotting using UL25 or UL6 antiserum (immunoblot). The B capsid samples were serially diluted in TBS buffer (4× to 1×) prior to SDS-PAGE. Lanes 1 and 2 and lanes 3 and 4 of the immunoblot correspond to lanes 1 and 2 of the CBB-stained gel. Arrowheads and arrows indicate the position of 75-kDa (UL6) and UL25 proteins, respectively. M, molecular mass markers (masses, in kilodaltons, are on the left). (B) The amounts of proteins (VP5, UL6, and UL25) were measured by densitometric scanning of each lane in the CBB-stained gel. Values are the means ± standard deviations for three independent measurements in B and C capsids.
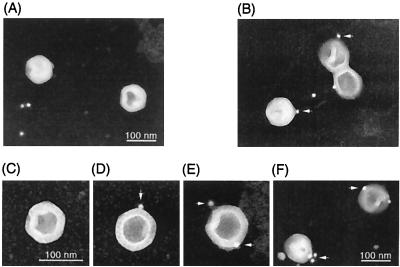
To clarify the possibility that UL25 protein is associated with pentons, we examined immunolocalization of this protein by EM (Fig. 2). When purified B capsids were incubated with UL25 antiserum, binding of gold particles (conjugated with protein A) to capsid vertices was observed (Fig. 2B). Counting of gold particles associated with capsids yielded one to four particles per capsid. In many cases, the distribution of immunogold particles among 12 vertices was one or two total (Fig. 2D and E) and was symmetric (Fig. 2E). As shown in Fig. 2F, the cluster of particles on the vertex was observed in some cases. When B capsids were incubated with normal mouse serum, binding of particles to capsid vertices was not observed (Fig. 2A and C). Thus, it is likely that UL25 protein is associated with pentons.
FIG. 2.
Electron microscopic immunostaining. B capsids were purified by gradient ultracentrifugation. B capsids were adsorbed to carbon grids and fixed in 2% glutaraldehyde. The grids were incubated with UL25 antiserum (B, D, E, and F) or normal mouse serum (A and C) and washed. Following incubation with 10-nm-gold-conjugated protein A, capsids were stained with uranyl acetate and examined by transmission electron microscopy. Arrows indicate gold particles on capsid vertices. Images of negative film are shown.
Interaction of UL25 protein with other virus proteins.
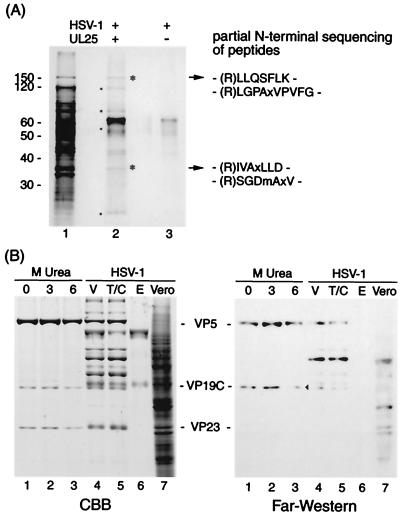
The association of UL25 protein with capsids indicates that UL25 has to interact with other proteins of virions (probably capsid proteins). To clarify if this is the case, we analyzed potential interactions of UL25 protein with virion proteins, by reconstitution of virions solubilized with 8.0 M urea on the UL25 protein immobilized on a PVDF sheet. SDS-PAGE analysis of the proteins bound on the UL25 sheet reveals six proteins of 150, 120, 80, 52, 34, and <30 kDa (asterisks and dots) as shown in Fig. 3A. The 150- and 34-kDa proteins were subjected to amino-terminal sequencing after trypsin digestion. The amino-terminal sequences of these peptides correspond to the predicted fragments (residues 288 to 294 and residues 1157 to 1167) of VP5 and the predicted fragments (residues 83 to 89 and residues 308 to 314) of VP23. The 52-kDa protein is probably VP19C, as determined by far-Western analysis (Fig. 3B). The 120- and 80-kDa bands were phosphorylated proteins of the serine or tyrosine type (data not shown). It is likely that these two are tegument proteins. The bands of about 45 kDa and less than 30 kDa could not be characterized. It is unknown whether renatured capsid proteins form capsid-like complexes. This result, however, suggests that the reconstitution of some capsid substructures on the UL25 sheet may have occurred by renaturation from these solubilized proteins (VP5, VP19C, VP23, and others) in 8.0 M urea.
FIG. 3.
Protein-protein interaction of UL25 with virus proteins. (A) Identification of virus proteins reconstituted with UL25 on sheets; (B) far-Western analysis of UL25 binding to virus proteins. (A) A mixture of virus proteins solubilized with 8.0 M urea and PVDF sheets blotted with UL25 (lane 2) or not (lane 3) were renatured by stepwise dialysis. This dialysis allows virus proteins to bind UL25 protein on PVDF by reconstitution of interactive proteins. Virion proteins (lane 1) and proteins reconstituted on UL25 sheets (lane 2) were analyzed by SDS-PAGE. Asterisks and dots indicate the proteins reconstituted with UL25. Following trypsin digestion of bound proteins (asterisks in lane 2), the result obtained by amino-terminal sequencing of the digested peptides is shown on the right. The 65-kDa band appears to be more intense in lane 2 than in lane 3, since bands of UL25 and BSA are overlapping. The migration of molecular mass markers is shown on the left. (B) HSV-1 B capsids were extracted with 0 (lane 1), 3.0 (lane 2), and 6.0 (lane 3) M urea. These extracts (lane 1 to 3), Vero cells (lane 7), purified virions (V, lane 4), and their tegument-capsid (T/C, lane 5) and envelope (E, lane 6) components were subjected to SDS-PAGE (CBB stain) and far-Western analysis using biotin-labeled UL25. An arrowhead indicates the VP19C band.
To identify the proteins associated with UL25, we examined the direct interaction between UL25 protein and virus proteins by far-Western analysis using biotin-labeled UL25. Gradient-purified capsids and virions developed on CBB-stained SDS gels (Fig. 3B) were transferred to a PVDF sheet. The blotted sheet was reacted with biotin-labeled UL25 after renaturation by washing the sheets with TBS-T buffer. The far-Western analysis in Fig. 3B shows that this UL25 protein reacted with the 150-kDa protein (VP5) and the 52-kDa protein (VP19C) of B capsids without urea extraction (lane 1). However, a significant decrease in binding activity was observed upon treatment with 6.0 M urea (Fig. 3B, lane 3), compared with the level observed upon 3.0 M urea treatment (lane 2). Treatment of capsids with 6.0 M urea (34) results in the removal of the penton and releases more VP19C (16.5%) than VP5 (6.1%). The loss of binding to these proteins, especially VP19C (Fig. 3B, lane 3), therefore, reflects the reduced amount of VP19C present in penton and peripentonal triplexes. To examine the interaction of UL25 with envelope and tegument components, virions treated with NP-40 were divided into envelope and tegument-capsid fractions by centrifugation. In addition to VP5 and VP19C, UL25 protein interacts with an 80-kDa protein in the tegument and capsid (Fig. 3B, lane 5) but not with proteins in the envelope (lane 6). The UL25 protein may not specifically bind to this protein, since there is the nonspecific binding of the 80-kDa protein in Vero cells (lane 7). Taken together, these observations indicate that UL25 protein is associated with VP19C in addition to VP5 and an 80-kDa tegument protein.
Relocation of UL25 protein into the nucleus.
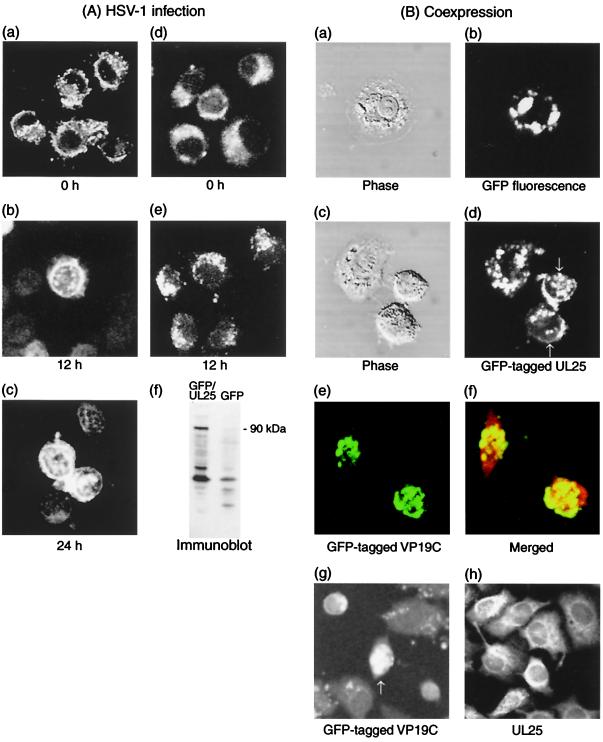
Since capsid assembly takes place in the nucleus, UL25 protein must be localized in the nuclei of HSV-1-infected cells. In a number of heterogenous expression systems, the use of GFP as a tag appears to not interfere with properties of proteins tagged with GFP. To obtain a UL25 chimeric protein possessing a fluorescent tag, we fused GFP to the N terminus of UL25. The deduced molecular masses of UL25 and GFP are 62.5 kDa (580 amino acids) and 27 kDa (238 amino acids), respectively. Fusion of the two sequences gave a chimera of about 89.5 kDa (819 amino acids). As shown in Fig. 4A, panel f, GFP-tagged UL25 protein (GFP-UL25) with a molecular mass of about 90 kDa was detected in pDGFPul25-transfected cells but not in pDGFP-transfected cells. Cells transfected with pDGFPul25 or pDGFP were analyzed with a confocal microscope. When native GFP is expressed, fluorescent GFP can be visualized as a diffuse distribution in the cytoplasm (Fig. 4A, panel d). Similarly, when GFP-UL25 is expressed, fluorescence exhibits a cytoplasmic pattern (Fig. 4A, panel a). To investigate the influence of HSV-1 infection on the distribution of GFP-UL25, transfected cells were infected and fixed at various times after infection (Fig. 4A). The expression of GFP alone exhibits a diffuse or punctate distribution throughout the cytoplasm at 12 h postinfection (Fig. 4A, panel e). The GFP also showed no changes in its distribution at 24 h after infection (data not shown). However, HSV-1 infection had the effect of relocating GFP-UL25 into the nucleus long after infection (Fig. 4A, panels b and c). These results suggested that UL25 protein does not have a nuclear localization signal but may be transported into the nucleus by an association with other virus proteins in the late period of HSV-1 infection.
FIG. 4.
Intracellular localization of UL25. (A) Nuclear relocation of UL25 by HSV-1 infection. Cells were transfected with pDGFPul25 (a, b, and c) or pDGFP (d and e). After 60 h of transfection, cells were infected with HSV-1 at 10 PFU of virus per cell and then fixed at 0 h (a and d), 12 h (b and e), and 24 h (c) after infection. GFP fluorescence of these fixed cells was analyzed by a confocal microscope. HeLa cells (60-mm culture dishes) transfected with pDGFPul25 or pDGFP were cultured for 3 days and harvested. Cells were sonicated in TNE buffer on ice and centrifuged at 3,500 × g for 10 min. The resulting supernatants of cells expressing GFP-tagged UL25 (GFP-UL25) and GFP were analyzed by Western blotting using UL25 antiserum (f). The position of a chimeric protein (90 kDa) is indicated. (B) Coexpression of UL25 with VP19C. VP19C-expressing cells were further transfected with pDGFP (a and b) or pDGFPul25 (c and d). Phase, phase-contrast microscopy of tagged protein. Cells were cotransfected with pDGFPul38 and pDul25. Merged, merged image with UL25 immunostaining using a Texas red probe. (g and h) Expression of GFP-tagged VP19C alone (pDGFPul38) and unmodified UL25 protein (pDul25). Arrows (d and g) indicate the fluorescence in the nucleus.
As demonstrated by far-Western analysis (Fig. 3B), UL 25 protein is directly associated with VP5 and VP19C. The interaction with these proteins may cause the relocation of UL25 protein into the nucleus after HSV-1 infection. pDGFPul38 and pDGFPul25 encode GFP-tagged versions of VP19C and UL25 protein, respectively. VP19C-expressing cells were transfected with pDGFP (Fig. 4B, panels a and b) or pDGFPul25 (4B, panels c and d). Coexpression of VP19C and GFP (4B, panel b) exhibits a cytoplasmic pattern of GFP distribution. However, coexpression with VP19C causes GFP-UL25 to relocate into the nucleus (4B, panel d), resulting in a punctate pattern in the nucleus (arrows). Coexpression of GFP-VP19C and unmodified UL25 shows the colocalization of VP19C and UL25 protein in the nucleus (Fig. 4B, panels e and f). The expression of these each proteins alone exhibits a diffuse distribution throughout the nucleus (VP19C) (Fig. 4B, panel g) or the cytoplasm (UL25) (panel h). These results indicate that UL25 protein is associated with VP19C in the cytoplasm and then moves to the nucleus and forms a complex with VP19C. In Fig. 4, the remaining or punctate pattern in cytoplasm is likely to be due to overexpression of GFP-UL25.
Potential capacity of UL25 protein to bind DNA.
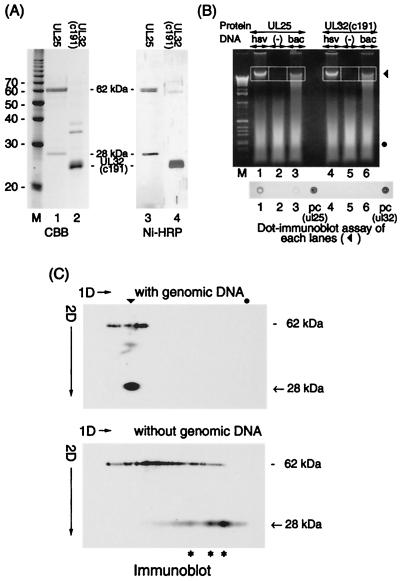
The functional role of UL25 protein is to retain DNA in the capsid (25). The UL25 protein may achieve this through an inherent capacity to bind DNA. We investigated the interaction between UL25 protein and genomic DNA prepared from virions. The UL25 protein was purified from BacHisUL25-infected Sf9 cells with a Ni-NTA column under native conditions. The proteins with molecular masses of 62 kDa (full-length UL25 protein with an N-terminal histidine tag) and 28 kDa were purified with the Ni-NTA column (Fig. 5A). This 28-kDa protein probably corresponds to a proteolytic fragment of the N-terminal portion of UL25 protein, since this protein has a His tag as shown in Fig. 5A, lane 3 (Ni-HRP). The mixtures of UL25 proteins with and without genome DNA were subjected to agarose gel electrophoresis (Fig. 5B, lanes 1 to 3), and the proteins bound to DNA in gels (indicated by frames) were analyzed by dot immunoblotting. The UL25 protein was detected in the gel extract of HSV-1 DNA (Fig. 5B, dot 1) but not in the extract of baculovirus DNA (dot 3) or the extract with no DNA (dot 2). Moreover, when UL32(c191) with a His tag (made up of the C-terminal 191 amino acid residues of UL32) was used in place of UL25 protein as a control (Fig. 5B, lanes 4 to 6), UL32(c191) was not detected in gel extracts of either viral DNA (dots 4 to 6). This suggests that the interaction of UL25 protein with HSV-1 DNA may be specific.
FIG. 5.
Interaction between UL25 and genomic DNA. (A) SDS-PAGE analysis of UL25 and UL32(c191) proteins. These His-tagged proteins were prepared from HisUL25-expressing Sf9 cells and UL32(c191)-expressing BL21(DE3) cells and purified with a Ni-NTA column. The molecular masses (in kilodaltons) are on the left. The two proteins were analyzed by Western blotting using a Ni-NTA HRP conjugate (Ni-HRP). (B) Mixtures of UL25 with HSV-1 DNA (lane 1; hsv) and with baculovirus DNA digested by Bsu36 I (lane 3; bac) and protein without genomic DNA (lane 2) in binding buffer were applied to a 1.5% agarose gel in the first dimension. Then UL32(c191) was used in place of UL25 and applied to a gel (lanes 4 to 6). Lane M, 1-kb DNA ladder marker. Genomic DNA (▾) and the degenerated salmon DNA (●) are indicated. (Bottom) Dot immunoblot assay of the extracts obtained from each lane (outlined). The His-tagged proteins included in these extracts were detected with the Ni-NTA HRP conjugate. The dot number corresponds to the lane number in the ethidium bromide-stained gel. In the dots labeled pc (ul25) and pc (ul32), UL25 and UL32(c191) were directly dot blotted to the sheet as positive controls. (C) Gel mobility shift analysis of UL25 by two-dimensional (2D) gel electrophoresis as described in Materials and Methods. The bands from lanes 1 and 2 in panel B were subjected to SDS-PAGE in the second dimension and analyzed by Western blotting using UL25 antiserum. The arrowhead and dot indicate the positions of genomic DNA and degenerated salmon DNA, respectively. Asterisks indicate oligomers of the 28-kDa protein. The 28-kDa band marked with an arrow likely represents a proteolytic fragment of UL25 (see the text).
We further investigated this interaction by the modified gel mobility shift analysis using a two-dimensional gel electrophoresis system. Bands excised from lanes 1 and 2 in Fig. 5B were loaded onto SDS gels and analyzed by Western blotting using UL25 antiserum (Fig. 5C). In the absence of genomic DNA, the 62-kDa protein is found in smeared bands extending across the gel (Fig. 5C, bottom panel). The 28-kDa protein, however, appears as a ladder spot extending across the gel. We interpret the ladder spot as an oligomer of the 28-kDa protein. The mobility shift of UL25 proteins (62 and 28 kDa) to a position corresponding to the DNA band was observed in the mixture with genomic DNA (Fig. 5C, top panel) but not without DNA (Fig. 5C, bottom panel), indicating the formation of UL25-DNA complexes. The UL25 protein does not bind a degenerated salmon DNA (a dot). This indicates that UL25 protein has an inherent capacity to bind dsDNA, and the 28-kDa region in the N-terminal half of the protein likely forms a homo-oligomer. As shown in Fig. 5C, the mobility shift and the migration pattern of the 62-kDa protein are not clear compared to those of the 28-kDa protein. This difference in migration of both proteins may result from a difference in hydrophobicity of these proteins, since the migration in electrophoresis under nondenaturing conditions is affected by the hydrophobicity of the protein.
Comparison of UL25 protein between B and C capsids.
If UL25 protein plays a role in head completion similar to that found with dsDNA bacteriophage, C capsids may contain more UL25 protein than B capsids. Therefore, we compared the amount of UL25 or the 75-kDa protein (as a control protein) in B and C capsids. This 75-kDa band was UL6 protein, and its amount may differ somewhat between the two types of capsid. The immunoblot in Fig. 6A shows that C capsids contain somewhat more UL25 protein than B capsids. Quantitative analyses of the stained gels (Fig. 6A) indicate that the percentages of UL25 relative to VP5 in C capsids (5.7%) were two to three times larger than those in B capsids (2.2%), as shown in Fig. 6B. However, there was not much difference in the amount of 75-kDa protein (UL6) between the two types of capsid (4.6% in B and 4.0% in C capsids). There may be much more UL25 protein in C than in B capsids. As shown in Fig. 6A, C capsid fractions contained some B capsids (less than 15%), since the band of about 40 kDa (VP22a) is observed in lane 1. However, since the difference in protein composition between B and C capsids is observed in lane 1 of Fig. 6A (60-kDa band), further investigation of this difference will be necessary.
DISCUSSION
The DNA packaging machinery of HSV-1 is likely to be analogous to that of dsDNA bacteriophages (30, 42). The bacteriophage prohead also includes a portal vertex that serves as the site for DNA entry (7) as well as for DNA injection into the host cell (20). Such a unique vertex has not been defined in herpesviruses; however, the pentons may serve as the site for a translocation of DNA. In HSV-1, the penton channels (with a minimum diameter of ∼40 Å) may be better suited to the transport of genomic DNA than the hexon channels (with a diameter of ∼20 Å), since dsDNA has a diameter of approximately 22 Å (12, 59). For this reason, capsid proteins required for the translocation of DNA have to exist at penton sites or sites near the penton channel.
EM analysis has shown that 6.0 M urea extraction of B capsids results in the removal of the pentons (30, 34). On the basis of this finding, the results of separation of B capsid extracts (Fig. 1) indicate that UL25 may also exist at the penton sites. This study was begun to clarify the presence of the UL25 in pentons. For this aim, we examined immunolocalization of this protein by EM. As shown in Fig. 2, it is probable that UL25 is located in capsid vertices and is associated with pentons. Although there are approximately 42 copies of UL25 per B capsid, the amount of immunogold associated with capsid was small (one to four particles per 12 vertices). Approximately half the UL25 should be associated with capsid substructures beside pentons (Fig. 1C). However, the binding of gold particles to these substructures (probably hexons) was not observed in many cases. These findings indicate that the UL25, a minor capsid protein, is associated with both pentons and hexons, but many UL25 molecules may not be exposed to the outer surface of the capsid shell.
The assembly of HSV-1 capsids is known to take place in the nuclei of HSV-1-infected cells. However, the UL25 itself does not move into the nucleus until the late stage of HSV-1 infection (Fig. 4A). Moreover, both UL6 and UL25 are incorporated into capsids assembled in the infected cells along with recombinant baculoviruses that express a series of capsid proteins (25). These results suggest an association of UL25 with other capsid proteins expressed at later stages of infection. In capsid assembly, capsid proteins move into the nucleus through interactions with VP19C (which forms heterotrimeric triplexes) and with the scaffolding protein (which forms pre-VP22a-VP5 complexes) (32, 35, 45, 55). Figure 3B shows the direct interactions of UL25 with VP5 and VP19C. This indicates the possibility that UL25 is transported to the nucleus and arrives at the procapsid through these interactions. However, UL25 could be bound indirectly to VP5, VP19C, and VP23, as shown in Fig. 3A. Coexpression with VP19C relocates UL25 into the nucleus in transfected cells (Fig. 4B). The fact that VP19C is a late or true late (γ2) HSV-1 protein and has the capacity for nuclear localization (45) makes it suited to the relocation of UL25 at the late stage of infection. Thus, the association with VP19C is likely to be involved at least in the early incorporation of UL25 into the capsid. The UL25 must be incorporated into the capsid (and pentons) through at least two routes, because there was a difference in amount between B and C capsids (Fig. 6B). One route is through the assembly process of the capsid as described above; another is through the DNA-packaging process after the capsid is complete.
How does UL25 function to maintain the genomic DNA within the capsid? The abortive packaging event in UL25 null mutant-infected cells (25) had also been shown in the cases of mutants defective in gp4, gp16, and gp26 of P22 phages (41, 42). These head completion proteins of P22 phages are added at a unique vertex composed of the portal protein for plugging the channel after DNA packaging. If UL25 plays a role analogous to the role of these proteins, it may be incorporated into pentons when DNA is packaged. The larger amount of UL25 in C than in B capsids (Fig. 6) suggests that this is possible and that it may function to plug the packaging channel. A unique vertex and a plugging device have not been defined for HSV-1 (29, 33, 58). The addition of UL25 after DNA packaging, however, may be specific to pentons (especially the packaging channel). The portal proteins of dsDNA bacteriophages are required for initiation of procapsid assembly and are assembled into a unique vertex (9, 43). However, some of those (for example, gp1 of phage P22) are not required for this initiation and are incorporated into the procapsid by interacting either with coat proteins or with the growing procapsid shell (7, 8). There is little sequence homology of the portal proteins among dsDNA bacteriophages. However, the portal proteins of some phages appear to contain DNA binding motifs such as a helix-turn-helix sequence (19). One study reported that the portal protein (p10) of phage φ29 has non-sequence-specific DNA binding capacity and binds preferentially to the ends of DNA in the presence of the terminal protein p3 (18). Moreover, p10 is involved in the closure of packaging channels after DNA translocation (26, 52). Thus, p10 plays a role in docking the terminase-DNA complex to a portal vertex and in some head completion processes. Both UL6 and UL25 may play a role analogous to that of p10. Since UL6 appears not to be added after DNA is packaged (Fig. 6), it is likely that UL6 and UL25 principally function in the docking and in the completion event, respectively. These proteins may function independently or in concert in the encapsidation process. All capsid vertices (12 pentons) may have the potential to serve as the translocation site of viral DNA in HSV-1 (29, 58). However, the DNA has to enter the capsid through only one vertex. According to gel mobility shift analysis, the UL25 protein (especially its amino-terminal residue) appears to have the potential to bind DNA. Moreover, UL25 is associated with the capsid vertices, as shown in Fig. 2.
An assumption of the potential roles of these proteins can be made on the basis of these findings. When one of the 12 vertices encounters the terminase-DNA complex, UL6 works to dock this complex at the vertex. Once initiation has occurred, this vertex functions as the packaging channel and packaging is polarized. However, it is possible that 1 or 2 of the 12 vertices function as packaging vertices, since there may be differences in the amount of UL25 among 12 vertices (Fig. 2D to F). A model for the potential roles of the packaging proteins proposed by Yu and Weller suggests that UL25 also enhances the turnover ratio of the terminase (UL15) by dissociating it from the capsids, after viral DNA is packaged (57). This potential interaction between UL25 and UL15 is thought to be analogous to that between p10 (portal protein) and p3 (terminal protein) in phage φ29. Taken together, these results indicate that when the DNA fills the capsid and is cut in relation to a sequence-specific mechanism (“a” sequences), UL25 may bind the terminase-DNA complex, disengage the DNA from this complex, and trap the DNA at this vertex. Thus, UL25 may play a role in the termination of packaging by trapping the DNA and then closing the packaging channel by a new addition of this protein (as found in C capsids). It cannot be concluded from the present results alone whether UL25 binds to DNA in a sequence-specific fashion. Much of the UL25 in B capsids is not likely to be exposed to the outer shell surface (Fig. 2), and some of the proteins may be exposed to the capsid floor. Cryomicroscopic analysis of C capsids suggests that the outermost layer of the packaged DNA has been in contact with the floor of pentons (58). This contact may be due to the triplexes forming the capsid floor (11) or the UL25 exposed to the floor. Moreover, DNA fibers that can be traced to the capsid of origin are rarely observed in B capsid fractions by the immunogold-EM analysis. In this case, UL25 has bound to some of three portions, the origin (at the vertex), the end, and the bending site of the DNA fiber (data not shown). Whether this binding takes place through the terminase is unknown; however, this binding is likely to be sequence specific. These results suggest that UL25 may nonspecifically bind the DNA condensed within the capsid, while it may recognize the DNA end by replacing the terminase of the DNA complex at the packaging vertex.
UL25 is incorporated at an early stage of capsid assembly and then waits for viral DNA to be packaged. To complete the packaging process, the UL25 may anchor the DNA to the vertex that initiated DNA packaging. Thus, the UL25 gene product may be a DNA-anchoring protein.
REFERENCES
- 1.Addison C, Rixon F J, Palfreyman J W, O'Hara M, Preston V G. Characterization of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 3.Ali A M, Forghani B, Cantin E M. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology. 1996;216:278–283. doi: 10.1006/viro.1996.0061. [DOI] [PubMed] [Google Scholar]
- 4.Al-Kobaisi F M, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 5.Baines J D, Poon A P, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines J D, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazinet C, King J. Initiation of P22 procapsid assembly in vivo. J Mol Biol. 1988;202:77–86. doi: 10.1016/0022-2836(88)90520-7. [DOI] [PubMed] [Google Scholar]
- 8.Bazinet C, Benbasat J, King J, Carazo J M, Carrascosa J L. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27:1849–1856. doi: 10.1021/bi00406a009. [DOI] [PubMed] [Google Scholar]
- 9.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 10.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun D K, Batterson W, Roizman B. Identification and genetic mapping of a herpes simplex virus capsid protein that binds DNA. J Virol. 1984;50:645–648. doi: 10.1128/jvi.50.2.645-648.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantor C R, Schimmel P R. Biophysical chemistry. W. H. San Francisco, Calif: Freeman and Company; 1980. [Google Scholar]
- 13.Cavalcoli J D, Baghian A, Homa F L, Kousoulas K G. Resolution of genotypic and phenotypic properties of herpes simplex virus type 1 temperature-sensitive mutant (KOS) tsZ47: evidence for allelic complementation in the UL28 gene. Virology. 1993;197:23–34. doi: 10.1006/viro.1993.1563. [DOI] [PubMed] [Google Scholar]
- 14.Cohen G H, Ponce de Leon M, Diggelmann H, Lawrence W C, Vernon S K, Eisenberg R J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai P, Watkins S C, Person S. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J Virol. 1994;68:5365–5374. doi: 10.1128/jvi.68.9.5365-5374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson W, Roizman B. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herranz L, Salas M, Carrascosa J L. Interaction of the bacteriophage phi 29 connector protein with the viral DNA. Virology. 1986;155:289–292. doi: 10.1016/0042-6822(86)90191-1. [DOI] [PubMed] [Google Scholar]
- 19.Herranz L, Bordas J, Towns-Andrews E, Mendez E, Usobiaga P, Carrascosa J L. Conformational changes in bacteriophage φ 29 connector prevents DNA-binding activity. J Mol Biol. 1990;213:263–273. doi: 10.1016/s0022-2836(05)80189-5. [DOI] [PubMed] [Google Scholar]
- 20.King J, Lenk E V, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- 21.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 23.Liu F Y, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLauchlan J, Rixon F J. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J Gen Virol. 1992;73:269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- 25.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller D J, Engel A, Carrascosa J L, Velez M. The bacteriophage φ29 head-tail connector imaged at high resolution with the atomic force microscope in buffer solution. EMBO J. 1997;16:2547–2553. doi: 10.1093/emboj/16.10.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murialdo H. Bacteriophage lambda DNA maturation and packaging. Annu Rev Biochem. 1991;60:125–153. doi: 10.1146/annurev.bi.60.070191.001013. [DOI] [PubMed] [Google Scholar]
- 28.Murialdo H, Becker A. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol Rev. 1978;42:529–576. doi: 10.1128/mr.42.3.529-576.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb W W, Brown J C. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J Virol. 1994;68:433–440. doi: 10.1128/jvi.68.1.433-440.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newcomb W W, Brown J C. Use of Ar+ plasma etching to localize structural proteins in the capsid of herpes simplex virus type 1. J Virol. 1989;63:4697–4702. doi: 10.1128/jvi.63.11.4697-4702.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newcomb W W, Homa F L, Thomsen D R, Trus B L, Cheng N, Steven A, Booy F, Brown J C. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 35.Nicholson P, Addison C, Cross A M, Kennard J, Preston V G, Rixon F J. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J Gen Virol. 1994;75:1091–1099. doi: 10.1099/0022-1317-75-5-1091. [DOI] [PubMed] [Google Scholar]
- 36.Patel A H, MacLean J B. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology. 1995;206:465–478. doi: 10.1016/s0042-6822(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 37.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 38.Perdue L M, Cohen J C, Randall C C, O'Callaghan D J. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology. 1976;74:2169–2179. [PubMed] [Google Scholar]
- 39.Pertuiset B, Boccara M, Cebrian J, Berthelot N, Chousterman S, Puvion-Dutilleul F, Sisman J, Sheldrick P. Physical mapping and nucleotide sequence of a herpes simplex virus type 1 gene required for capsid assembly. J Virol. 1989;63:2169–2179. doi: 10.1128/jvi.63.5.2169-2179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poon A P, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poteete A R, King J. Functions of two new genes in Salmonella phage P22 assembly. Virology. 1977;76:725–739. doi: 10.1016/0042-6822(77)90254-9. [DOI] [PubMed] [Google Scholar]
- 42.Preston V G, Coates J A, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prevelige P E, Jr, King J. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog Med Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- 44.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 45.Rixon F J, Addison C, McGregor A, Macnab S J, Nicholson P, Preston V G, Tatman J D. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 46.Rixon F J, Cross A M, Addison C, Preston V G. The products of herpes simplex virus type 1 gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J Gen Virol. 1988;69:2879–2891. doi: 10.1099/0022-1317-69-11-2879. [DOI] [PubMed] [Google Scholar]
- 47.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaffer P A, Aron G M, Biswal N, Benyesh-Melnick M. Temperature-sensitive mutants of herpes simplex virus type 1: isolation, complementation and partial characterization. Virology. 1973;52:57–71. doi: 10.1016/0042-6822(73)90398-x. [DOI] [PubMed] [Google Scholar]
- 49.Schrag J D, Prasad B V, Rixon F J, Chiu W. Three-dimensional structure of the HSV-1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 50.Sherman G, Bachenheimer S L. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 51.Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]
- 52.Tao Y, Olson N H, Xu W, Anderson D L, Rossmann M G, Baker T S. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 54.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 56.Weller S K. Herpes simplex virus DNA replication and genome maturation. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus: Howard Temin's scientific legacy. Washington, D.C.: ASM Press; 1995. pp. 189–213. [Google Scholar]
- 57.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Z H, Chen D H, Jakana J, Rixon F J, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Z H, Prasad B V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400-kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]