Abstract
Oxidative stress caused by chronic lung inflammation in patients with cystic fibrosis (CF) and chronic lung infection with Pseudomonas aeruginosa is characterized by the reactive oxygen species (ROS) liberated by polymorphonuclear leukocytes (PMNs). We formulated the hypothesis that oxidation of the bacterial DNA by ROS presents an increased risk for the occurrence of hypermutable P. aeruginosa. The occurrence of hypermutable P. aeruginosa isolates was investigated directly in the sputum of 79 CF patients and among 141 isolates collected from 11 CF patients (10 to 15 isolates/patient) collected from the 1st and up to the 25th year of their chronic lung infection. The level of oxidized guanine moiety 8-oxo-2′-deoxyguanosine (8-oxodG), which is a frequently investigated DNA oxidative lesion, was measured. Hypermutable P. aeruginosa isolates were found in the sputum bacterial population of 54.4% of the CF patients. The earliest mutator P. aeruginosa isolates were found after 5 years from the onset of the chronic lung infection, and once they were present in the CF lung, the prevalence increased with time. The hypermutable isolates were significantly more resistant to antipseudomonal antibiotics than nonhypermutable isolates (P ≤ 0.001). The level of 8-oxodG/106 deoxyguanosine (dG) was significantly higher in hypermutable P. aeruginosa isolates (87 ± 38) than in nonhypermutable P. aeruginosa isolates (59.4 ± 17) (P = 0.02), and an increase to 86.84 from 21.65 8-oxodG/106 dG was found after exposure of the reference strain PAO1 to activated PMNs. Our results suggest that the chronic PMN inflammation in the CF lung promotes oxidative stress and is associated with the occurrence of hypermutable bacteria in the lung. The hypermutable phenotype can associate with mutations that confer adaptation of the bacteria in the lung and persistence of the infection.
The reduced volume of the epithelial lining fluid and viscous mucus in the lungs of patients with cystic fibrosis (CF) leads to a dysfunction of the mucociliary clearance, and as a consequence, the patients suffer from recurrent and chronic respiratory tract infections caused mainly by bacteria such as Staphylococcus aureus, Haemophilus influenzae, Burkholderia cepacia, and especially Pseudomonas aeruginosa.
A high percentage of H. influenzae, S. aureus, and P. aeruginosa strains isolated from patients with CF and chronic lung infection show higher frequencies of mutations (“hypermutable strains”) than bacterial strains isolated from non-CF patients (26, 28a, 29a). Acquisition of hypermutability confers an advantage to the bacteria in stressful and fluctuating environments, as high mutation rates and recombination allow faster adaptation (17). The CF lung is a highly stressful and fluctuating environment, as, in addition to the challenge imposed by the host defenses, P. aeruginosa has to cope with the high doses of different antibiotics administered for prolonged periods of time (25). The frequency of resistant strains was found to be roughly double in hypermutable strains than in nonhypermutable bacteria (26).
The chronic P. aeruginosa lung infection causes a chronic inflammatory response in the lung, dominated by polymorphonuclear leukocytes (PMNs). Analysis of bronchoalveolar lavage fluid has shown that the number of PMNs recovered from the lungs of patients with CF is 1,000 times higher than that recovered from the lungs of controls (2).
PMNs release leukocyte proteases, myeloperoxidase, and reactive oxygen species (ROS), which are the main mechanisms of lung tissue damage in CF (6, 13). The ROS liberated by leukocytes, together with the endogenous oxygen species that occur during replication, may increase the oxidative stress of the bacterial DNA. Oxidation of the guanine 8-oxo-2′-deoxyguanosine (8-oxodG) is a frequently encountered lesion. 8-oxodG promotes the misincorporation of adenine in the replication round, producing G:C to T:A transversions, thus making unrepaired 8-oxodG a highly mutagenic lesion.
Several mechanisms are involved in the repair of this mutagenic lesion. The main pathway for the removal of 8-oxodG in DNA involves the formamidopyrimidine DNA glycosylase (Fpg or MutM), encoded by the mutM gene (36). If the 8-oxodG is not removed from the DNA by Fpg and mispairs with an adenine, a mismatch-correction glycosylase, encoded by mutY, removes the mispaired adenine from the DNA (1). Besides the oxidation of DNA, other bacterial components can be oxidized, like the nucleotide pool. The oxidation product of GTP, 8-oxodGTP, is removed by hydrolysis from the nucleotide pool by a protein encoded by mutT (18).
Mutations in genes involved in the repair of the DNA oxidative damage (GO system) increase the number of 8-oxodG molecules and the mutation frequencies of the isolates (20, 21, 35).
Besides the repair of the oxidative damage, bacteria posses a major mismatch correction pathway, a methyl-directed pathway specified by proteins encoded by mutS, mutL, and uvrD (the MMR system). Inactivation of the MMR and/or the GO system can give rise to the mutator phenotype (22).
Our study tries to address the role of chronic inflammation in the occurrence of the mutagenic 8-oxodG lesion in P. aeruginosa isolates from CF patients and the implication for the occurrence of hypermutable isolates. To determine if there is any association between the chronic inflammation and the occurrence of hypermutable isolates, we analyzed in a longitudinal study the occurrence and the prevalence of hypermutable isolates during chronic lung infection with P. aeruginosa, and we determined in a cross-sectional study the frequency of hypermutable P. aeruginosa strains directly in the sputum of CF patients.
As a marker of the degree of oxidative damage of the DNA, we measured the concentration of 8-oxodG in hypermutable and nonhypermutable clinical isolates of P. aeruginosa and in the reference P. aeruginosa strain PAO1 exposed in vitro to activated PMNs.
MATERIALS AND METHODS
CF patients and P. aeruginosa isolates.
Three hundred CF patients, both children and adults, are monitored at the CF Center of Copenhagen University Hospital. All CF patients have been seen in the outpatient clinic on a monthly basis since 1968 (7).
Since 1976 all CF patients chronically infected with P. aeruginosa have been admitted every third month to the center for a 2-week course of intravenous antipseudomonal antibiotics.
P. aeruginosa isolates have been collected from the sputum of CF patients since 1973 and stored in nutrient broth containing 5% glycerol at −80°C. The selection criteria were phenotypic (mucoid and nonmucoid) and different patterns of resistance to antibiotics, as determined on the basis of direct sensitivity testing of isolates from plated sputum. Chronic P. aeruginosa infection is defined as the continuous growth of P. aeruginosa for ≥6 months or the presence of two or more precipitating antibodies against P. aeruginosa (7).
(i) Cross-sectional study.
To study the occurrence of hypermutable P. aeruginosa directly in the sputum bacterial population, sputum samples were collected from 79 consecutive CF patients with chronic lung infection during the period from October 2002 to March 2003 and were investigated further, as described below. The mean age of the patients was 31.5 ± 7.3 years, and the mean duration of the chronic lung infection was 21 ± 6.8 years.
(ii) Longitudinal study.
To study the occurrence and the dynamics of the hypermutable P. aeruginosa isolates during the course of the chronic lung infection, 141 P. aeruginosa isolates collected from 1973 to 1999 from 11 CF patients were investigated.
For practical purposes the duration of the chronic lung infection was expressed in five 5-year periods, from 0 to 5, 5 to 10, 10 to 15, 15 to 20, and 20 to 25 years from the onset of the chronic lung infection.
Determination of antibiotic resistance.
The in vitro activities of piperacillin-tazobactam, ceftazidime, aztreonam, meropenem, ciprofloxacin, tobramycin, and colistin (Rosco, Copenhagen, Denmark) against all P. aeruginosa isolates from the cross-sectional and the longitudinal studies were tested by the diffusion method.
The MICs of piperacillin-tazobactam, ceftazidime, aztreonam, meropenem, ciprofloxacin, tobramycin, and colistin were determined by the agar plate dilution method, as described previously (5).
MIC determinations were done on all 141 P. aeruginosa isolates collected from 1973 to 1999 from 11 CF patients included in the longitudinal study, 100 P. aeruginosa isolates collected from the 43 CF patients with hypermutable isolates, and 100 isolates collected from the 36 CF patients without hypermutable isolates included in the cross-sectional study.
Precipitating antibodies against P. aeruginosa in CF sera.
P. aeruginosa precipitins were determined by crossed immunoelectrophoresis, as described previously (11), with the normal range being zero to one precipitin.
Measurement of mutation frequencies of P. aeruginosa isolates.
To determine the mutation frequencies after exposure to rifampin and streptomycin, the bacterial isolates were grown overnight in 20 ml Luria-Bertani (LB) medium, centrifuged at 3,000 rpm for 10 min, and resuspended in 1 ml LB medium. A 100-μl volume of 0, 10−1, and 10−2 dilutions were plated on LB plates containing 300 μg/ml rifampin and on LB plates containing 500 μg/ml streptomycin. A 100-μl volume of 10−7 to 10−10 dilutions were plated on LB plates, and the numbers of CFU were counted after incubation at 37°C for 48 h. An isolate was considered hypermutable if the mutation frequency after exposure to rifampin and streptomycin was 20 times higher than the mutation frequency of the reference strain PAO1, as published previously (26).
Direct bacteriological examination of sputum.
Quantification of the P. aeruginosa isolates in the sputum samples was performed by a previously described population method (8). This technique allowed us to investigate the sputum samples directly without subculture and detect the presence of rifampin- and streptomycin-resistant isolates in the bacterial population from the sputum. In short, sputum samples collected from CF patients were stored at 4°C. All sputum samples were Gram stained and examined by microscopy in order to ensure that the samples were of sputum rather than saliva. Homogenization with Sputulycin (dithiothreitrol; Sigma, St. Louis, Mo.) at a final concentration of 50 mg/liter was performed by a previously described method (10).
Sputum was diluted 10−1, 10−2, 10−3, 10−4, and 10−5 in saline. An aliquot of 0.1 ml of the undiluted sputum and of the first two dilutions was inoculated on modified Conradi-Drigalski medium (“blue plates”; State Serum Institute, Copenhagen, Denmark) containing 300 μg/ml rifampin and on plates containing 500 μg/ml streptomycin, while 0.1 ml of the last three higher dilutions were inoculated on “blue plates” without antibiotics. After 48 h of incubation at 35°C, the P. aeruginosa colonies on the various plates were counted. For this study only CF sputum samples with ≥104 CFU/ml were considered. The frequency of rifampin- or streptomycin-resistant bacteria was calculated by dividing the number of colonies grown on plates containing antibiotics by the number of colonies grown on plates without antibiotics. Only plates containing ≥20 colonies/plate were considered.
The colonial morphotypes and the antibiotic resistance patterns were used to discriminate between several isolates. For each sputum sample at least two and up to six different isolates from plates with and without antibiotics were stored in nutrient broth containing 5% glycerol at −80°C. For each patient, at least two isolates from plates with and without antibiotics, respectively, were tested for their mutation frequencies after exposure to rifampin and streptomycin, as described above.
Typing of the isolates by PFGE.
The P. aeruginosa isolates included in the longitudinal study and the P. aeruginosa isolates collected from the cross-sectional study were typed by pulsed-field gel electrophoresis (PFGE), as described previously (24, 30), by using the Spe enzyme. The patterns were analyzed visually.
Measurement of 8-oxodG in P. aeruginosa DNA. (i) DNA purification.
Bacterial DNA was purified with a PUREGENE DNA purification system (Gentra Systems, Minneapolis, Minn.), according to the instructions of the manufacturer, with some modifications.
Briefly, the bacterial lysate from an overnight culture (500 μl) was treated with RNase A solution (4 mg/ml) for 60 min at 52°C. After protein precipitation, the DNA was precipitated with 100% isopropanol and treated for a second time with RNase (10 mg/ml) for 60 min at 52°C. Sodium acetate (3 M; 10 μl; pH 5.2) was added, and the DNA was precipitated with 2 volumes absolute ethanol and washed with 70°C ethanol. The purified DNA was resuspended in 10 mM Tris-0.1 mM desferrioxamine buffer, pH 7.
(ii) DNA hydrolysis.
The purified DNA was hydrolyzed by nuclease P1 (1 U/μl in 30 mM sodium acetate, 1 mM ZnCl2, pH 5.3; Z-0152; Sigma) for 120 min at 37°C and then incubated with alkaline phosphatase from Escherichia coli (1 U/μl in purified water; P-5931; Sigma) for 60 min at 37°C. The protein was extracted with 50 μl chloroform, and the digested DNA was transferred to the analysis vials.
(iii) Measurement of 8-oxodG.
Quantification of 8-oxodG was done by high-performance liquid chromatography with electrochemical and UV detection by use of a Prodigy 5 μm octyldecyl silane column (Phenomenex, Torrance, Calif.) and 3% acetonitrile in phosphate buffer (pH 6) as the mobile phase. The samples were extracted in duplicate, and each was injected twice. The 8-oxodG was quantified in an electrochemical detector (Coulochem II and model 5011 Analytical Cell; both from ESA, Inc., Chelmsford, Mass.), while dG was quantified by determination of the UV absorbance (LaChrom UV detector L-7400; Merck Hitachi). Peak areas were used for the calculations. Separate calibration curves for 8-oxodG and dG were run together with each batch of samples.
The reproducibility of the assay for the measurement 8-oxodG was assessed by measuring in duplicate 10 different DNA purifications of reference strain PAO1. The variation coefficient was determined by using the formula standard deviation (SD) = 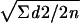 (where Σd2 is the sum of squared differences of double determinations of the same sample and n is the number of observations) and was found to be 2.1%.
(where Σd2 is the sum of squared differences of double determinations of the same sample and n is the number of observations) and was found to be 2.1%.
PMN bactericidal assay.
A bactericidal assay was performed by a previously described method (9). Human PMNs and the reference strain PAO1 at a ratio of 1:20 were incubated in the presence of 10% serum for 2 h at 37°C. The bacteria recovered from this experiment represented bacteria that survived phagocytosis (i.e., bacteria that were present both inside and outside the PMNs). In order to obtain only bacteria that survived inside the PMNs, the extracellular bacteria were killed by the addition of ceftazidime and tobramycin (200 μg/ml) to another mixture of PMNs and bacteria after 15 min, and the tubes were further incubated for 2 h 45 min. The bacteria that survived the phagocytosis by PMNs were cultured overnight, and the DNA was purified as described above for the 8-oxodG measurements.
Statistical analysis.
The description and analysis of the data were carried out by using StatView 5 software for personal computers. The data are given as means ± SDs or geometric means. The unpaired t test was used for comparison of the data for the hypermutable isolates and the nonhypermutable isolates.
RESULTS
Hypermutable P. aeruginosa in the bacterial population from CF sputum samples (cross-sectional study).
The number of P. aeruginosa CFU in the 79 sputum samples was (1.4 ± 5.6) × 108 CFU/ml sputum (mean value ± SD).
The frequency of rifampin-resistant isolates in the sputum samples was 1.8 × 10−5, and that of streptomycin-resistant isolates was 2 × 10−5 (geometric mean).
The hypermutable phenotype of colonies isolated from the rifampin-containing plates was confirmed by testing the mutation frequencies after exposure to streptomycin.
Hypermutable P. aeruginosa isolates were found in the sputum of 43 of the 79 CF patients (54.4%). Genotyping showed that the rifampin- and streptomycin-resistant isolates and the sensitive subpopulation present in the sputum samples had similar PFGE patterns.
At the time of bacterial isolation from sputum, the 43 CF patients with hypermutable P. aeruginosa isolates were significantly older (mean age, 33.7 ± 6.5 years), had significantly longer durations of chronic lung infection (mean duration, 23.5 ± 4.8 years), and higher number of precipitins in serum (mean number, 30 ± 9) compared to the 36 CF patients from whom hypermutable P. aeruginosa could not be isolated (mean age, 29.3 ± 7.5 years; duration of chronic lung infection, 18.4 ± 7.7 years; mean number of serum precipitins, 26 ± 11) (P = 0.007, P = 0.0007, and P = 0.036, respectively).
The MICs of P. aeruginosa isolates from the sputum of the 36 CF patients without hypermutable isolates to colistin (MIC50 = 3.1 μg/ml), piperacillin-tazobactam (MIC50 = 25 μg/ml), aztreonam (MIC50 = 25 μg/ml), and ceftazidime (MIC50 = 12.5 μg/ml) were significantly lower than the MICs of isolates from the sputum of the 43 CF patients with hypermutable isolates (MIC50 of colistin = 6.2 μg/ml, MIC50 of piperacillin-tazobactam = 100 μg/ml, MIC50 of aztreonam = 100 μg/ml, MIC50 of ceftazidime = 200 μg/ml).
Hypermutable P. aeruginosa isolates during chronic lung infection (longitudinal study).
Hypermutable P. aeruginosa isolates were found in 8 of the 11 CF patients. Forty-three mutators were identified among the 141 P. aeruginosa isolates. The mutation frequency after exposure to rifampin was 1.5 × 10−6 (geometric mean), and that after exposure to streptomycin was 1.4 × 10−6 (geometric mean); these frequencies are similar to a previously published frequency of 3.6 × 10−6 (26).
In comparison, the mutation frequencies of the nonhypermutable isolates after exposure to rifampin and streptomycin were 8.7 × 10−9 and 3.2 × 10−8, respectively.
The distribution of the hypermutable isolates in the five time periods of the chronic lung infection were 0%, 7%, 29%, 32%, and 65% (Fig. 1). The mean numbers of precipitating antibodies to P. aeruginosa in the patient's sera during the five time periods were 15, 28, 28, 33, and 38, respectively.
FIG. 1.
Mutation frequency after exposure to rifampin of 141 P. aeruginosa isolates collected in five periods of chronic lung infection (0 to 5, 5 to 10, 10 to 15, 15 to 20, and 20 to 25 years) from 11 patients with CF. The results for hypermutable (hp; ▴) and nonhypermutable (nhp; ○) P. aeruginosa isolates are presented. The mutation frequency after exposure to rifampin of reference strain PAO1 is shown.
The cumulative numbers of 2-week antibiotic courses/patient received by the 11 CF patients during the treatment of the chronic lung infection in the five time periods were 17.5, 35.5, 58, 82, and 97, respectively.
Hypermutable isolates had significantly higher MICs than the nonhypermutable isolates against all the antibiotics tested except colistin (P ≤ 0.001) (Table 1).
TABLE 1.
MIC50s, MIC90s, and MIC ranges for various antimicrobial agents of P. aeruginosa isolates with hypermutable and nonhypermutable phenotypesa
| Phenotype | MIC (μg/ml)
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ceftazidimeb
|
Piperacillin-tazobactamb
|
Aztreonamb
|
Meropenemb
|
Tobramycinb
|
Colistin
|
Ciprofloxacinb
|
|||||||||||||||
| 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | |
| Hypermutable | 200 | 400 | 1.6-400 | 400 | 400 | 12.5-400 | 400 | 400 | 0.8-400 | 50 | 200 | 0.4-400 | 12.5 | 25 | 1.6-50 | 0.8 | 5 | 0.4-400 | 12.5 | 25 | 0.4-50 |
| Nonhypermutable | 6.2 | 200 | 0.8-400 | 100 | 400 | 0.8-400 | 12.5 | 200 | 0.4-400 | 3.1 | 50 | 0.1-200 | 3.1 | 12.5 | 0.4-25 | 1.6 | 6.2 | 0.2-100 | 1.6 | 6.2 | 0.05-12.5 |
Hypermutable isolates had frequencies of mutations after exposure to rifampin and streptomycin of 1.6 × 10−6 and 1.4 × 10−6, respectively. For the nonhypermutable isolates, the frequencies were 9 × 10−9 and 3.3 × 10−8, respectively.
Statistically significant difference (P < 0.001) between the MICs for the hypermutable and nonhypermutable isolates.
Genotyping of the consecutive P. aeruginosa isolates showed that the CF patients maintained the same strain in their lungs during the chronic lung infection (data not shown).
Bacterial DNA oxidation (measurement of 8-oxodG) in clinical P. aeruginosa isolates.
The levels of 8-oxodG/106 dG were measured in P. aeruginosa isolates with the hypermutable and the nonhypermutable phenotypes but with identical genotypes. The measurements were done with isolates from the sputum samples of seven CF patients (from rifampin- and streptomycin-containing plates and from control plates) and 15 isolates collected during the chronic lung infection from two CF patients (Table 2).
TABLE 2.
Levels of 8-oxodG/106 dG in hypermutable and nonhypermutable P. aeruginosa isolates from CF patientsa
| CF patient no. | 8-oxodG/106 dG
|
|
|---|---|---|
| Hypermutable isolates | Nonhypermutable isolates | |
| CF 1 | 52 | 39.6 |
| CF 2 | 62.3 | 60.1 |
| CF 3 | 71.9 | 57.8 |
| CF 4 | 70.2 | 40.8 |
| CF 5 | 105.4 | 37.7 |
| CF 5 | 49 | |
| CF 6 | 51.4 | NDb |
| CF 6 | 70.6 | |
| CF 6 | 90.2 | |
| CF 7 | 50.3 | ND |
| CF 7 | 56 | |
| CF 8 | 117.9c | 75d |
| CF 9 | 67.4d | 58.8e |
For patients CF 1 to CF 7 the 8-oxodG levels were measured by the use of single isolates. These isolates were collected from the population analysis of the sputum samples (cross-sectional study). For patients CF 8 and CF 9 the levels are means of the levels for several isolates from the longitudinal study.
ND, not determined.
Mean for five isolates.
Mean for four isolates.
Mean for two isolates.
Large variations in the levels of 8-oxodG were found among different P. aeruginosa isolates, but the hypermutable isolates showed significantly higher (P = 0.02) levels of 8-oxodG/106 dG (87 ± 38) than the nonhypermutable isolates (59.4 ± 17), which was sign of the oxidative damage of their DNA.
Mutation frequency of P. aeruginosa reference strain PAO1 exposed to activated PMNs.
Exposure of PAO1 to activated PMNs led to a slight increase in the mutation frequency after exposure to rifampin, from 3 × 10−9 for the controls to 7.4 × 10−9 for the bacteria that survived intra- and extracellularly and to 1.2 × 10−8 for the bacteria that survived intracellularly.
Bacterial DNA oxidation (measurement of 8-oxodG) in P. aeruginosa reference strain PAO1 exposed to activated PMNs.
Exposure of reference strain PAO1 to PMNs in a bactericidal assay led to a 90% reduction in the bacterial count. Higher levels of 8-oxodG/106 dG were measured for the DNA of PAO1 isolates that survived the phagocytosis outside and inside the PMNs (37.78) and for the DNA of PAO1 isolates that survived in the PMNs (40.39) than for the DNA of the control (21.65).
Reexposure to PMNs of the PAO1 isolates that have survived inside the PMNs increased the levels of 8-oxodG/106 dG in the bacteria present outside and inside the PMNs to 61.95 and in bacteria recovered from the PMNs to 86.84, which shows the increased oxidative damage to the bacterial DNA.
DISCUSSION
We demonstrated an association between oxidative DNA modifications in P. aeruginosa and the development of hypermutable bacteria resistant to antibiotics. This confirms that the chronic lung infection in CF patients is a state of chronic oxidative stress (16, 37) that promotes DNA changes in the P. aeruginosa genome, which is linked to an increased possibility of adaptation in the CF lung, for example, the development of resistance to antibiotics, and, in addition, to growth in biofilms and to the production of alginate (19).
Investigation of the occurrence of mutators directly in the P. aeruginosa population present in the CF sputum showed that more than half of the 79 Danish CF patients included in the study had hypermutable isolates in their lungs. This proportion is higher than the proportion of 36% of 30 CF patients described by Oliver et al. (26). This difference can be explained by the different methods for the detection of hypermutable isolates used, namely, population analysis in our study and the detection of mutation frequencies among the isolated colonies in the study of Oliver et al. (26). The long duration of the chronic lung infection in the CF population included in our study might also explain the higher proportion of patients with hypermutable isolates compared to the proportion published previously (26).
The earliest hypermutable P. aeruginosa isolates occurred after a relatively long period of time from the onset of the chronic lung infection (5 years). It has been shown that the rate of PMN oxidative burst in response to a biofilm is reduced to about 30 to 80% of the response to planktonic bacteria (14). Moreover, it has also been shown that H2O2 cannot penetrate to the bottom of a P. aeruginosa biofilm (34). Thus, the biofilm-growing bacteria in the CF lung might need to be exposed to oxygen radicals from the PMNs for a relatively long period of time before an effect on hypermutability can be measured. The detection of hypermutable isolates after 5 years of chronic lung infection might be also a consequence of the criteria used for the selection of the isolates included in the longitudinal study, namely, the mucoid or nonmucoid phenotype and their resistance to antibiotics. In the beginning of the chronic phase of infection there were no or few resistant strains, but due to the development of antibiotic resistance, the number of resistant strains increased with time (4). However, once they were present in the CF lung, the prevalence of hypermutable P. aeruginosa isolates increased during the chronic lung infection. Our results are in agreement with the anticipated accumulation of mutants that occur in the course of chronic infection due to bacterial replication (27).
It is important that stable hypermutability does not confer an advantage by itself, due to the high number of deleterious mutations, but the association (hitchhiking) with rare favorable mutations confers a survival advantage (29).
Both in the cross-sectional and in the longitudinal studies, hypermutable isolates had significantly higher MICs of antipseudomonal antibiotics compared to those of nonhypermutable isolates. These data support previously published results reporting that resistance to antibiotics was roughly twofold greater in frequency in hypermutable strains than in nonhypermutable strains (26).
The number of serum precipitating antibodies to P. aeruginosa, which correlates to inflammatory parameters (12), also increased during the chronic infection, which suggests that the development of hypermutable isolates is associated with the degree of inflammation in the CF lung. This is also supported by the results of the cross-sectional study, in which we found that CF patients with P. aeruginosa hypermutable isolates in their sputum were significantly older, had a longer duration of chronic lung infection, and a higher number of precipitating antibodies to P. aeruginosa compared to the age, duration of infection, and number of precipitating antibodies in the group of patients from whom no hypermutable isolates were found.
The level of oxidative damage of the bacterial DNA was increased in the hypermutable P. aeruginosa isolates compared to that in the nonhypermutable P. aeruginosa isolates, as shown by measurement of the levels of 8-oxodG/106 dG. In addition, in vitro exposure of reference strain PAO1 to activated PMNs led to increased levels of DNA oxidation after exposure and reexposure to PMNs compared to those for the controls.
In PAO1 the repair mechanisms of the DNA oxidative lesions are functional, and the high levels of 8-oxodG measured are probably due to the overburden imposed by the ROS liberated by PMNs to the repair system. After a single exposure of 2.5 h to PMNs, a slight increase in the mutation frequencies of strain PAO1 was observed, probably due to unrepaired 8-oxodG lesions.
Measurements of DNA oxidation in the hypermutable and nonhypermutable isolates showed no linear correlation between the levels of 8-oxodG and the mutation frequency. Hypermutability requires the occurrence of mutations in the genes that have been shown to be responsible for DNA repair, the GO system (mutT, mutY, and mutM) and/or the MMR system (mutS, mutL, and uvrD) (22). It has been shown in E. coli that inactivation of mutM, mutY, and mutT gives rise to weak, moderate, and strong mutator phenotypes, respectively, while inactivation of the MMR system genes gives rise to the strong mutator phenotype.
Oliver et al. (25) showed that alterations in the genes of the MMR system were responsible for the mutator phenotype in 7 of 11 mutator P. aeruginosa strains from CF patients. Complementation of the same 11 mutator P. aeruginosa strains with cloned mutT and mutM, not surprisingly, failed to decrease the mutation frequencies (28). However, mutator P. aeruginosa CF isolates in which mutY genes failed to be amplified by PCR were also reported by the same group (26). The coexistence of alterations in both MMR and GO systems in strong mutators cannot be excluded. A mutator cascade, in which one type of mutator (mutT) generates a second mutator (mutHLS), which then allows stepwise frameshift mutations, has been described in E. coli (23).
We hypothesize that oxidation of DNA by activated PMNs during chronic inflammation in the CF lung is an initial event in the development of a hypermutable strain.
Strains with high levels of 8-oxodG will have an increased risk of occurrence of mutations in genes involved in DNA repair and, subsequently, the emergence of hypermutable isolates.
Different hypermutable isolates from the same patient presented various levels of 8-oxodG. This probably reflects the heterogeneity of the P. aeruginosa strains in the CF lung, in which bacteria adapted to the different macrocompartments in the lung and the microcosmos of the biofilm are present.
The association of hypermutability with mutations in genes that improve the adaptation of the bacteria and the presence of selective pressure are necessary for the survival of these strains.
These data suggest that the chronic PMN inflammation in the lungs of CF patients may contribute to the occurrence of hypermutable isolates in this group of patients.
Our hypothesis may have implications for other conditions characterized by chronic lung inflammation, such as chronic obstructive pulmonary disease (COPD). In COPD the ROS from inflammatory cells as well as from the inhaled cigarette smoke play a major role in the pathogenesis and progression of this disease (32). Patients with COPD have chronic lung colonization with H. influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae; and other pathogens, like S. aureus, P. aeruginosa, and members of the family Enterobacteriacae, can also be isolated.
Chronic colonization of the lower respiratory tract by bacterial pathogens amplifies the chronic inflammatory response present in COPD and leads to progressive airway obstruction (32). The persistence of the same bacterial strains in the lungs of patients with COPD is a consequence of the good adaptation of the bacteria in the lungs, and we suggest that this might be due to the occurrence of hypermutable strains under the oxidative stress present in the lungs of patients with COPD.
An obvious strategy for prevention of the occurrence of hypermutable isolates due to the oxidative damage is the use of antioxidants. Chopra et al. (3) showed that addition of antioxidants to cultures of hypermutable E. coli reduced the mutation frequency.
The oxidative stress in the CF lung is determined by the large number of activated PMNs in the respiratory airways and also by the deficiency in the antioxidant systems, like reduced amounts of glutathione. It has been shown that the administration of glutathione aerosols to patients with CF suppresses the oxidative burden on the surface of lung epithelial cells (31). We have previously shown that N-acetylcysteine can decrease the oxidative burst of PMNs and monocytes and that this drug has a positive influence on the clinical condition of CF patients with chronic P. aeruginosa infection (15, 33)
Treatment with antioxidants would probably also decrease the oxidative damage to the bacterial DNA and prevent the occurrence of hypermutable isolates.
Our results support the hypothesis that the chronic PMN inflammation in the lungs of patients with CF may contribute to the occurrence of P. aeruginosa hypermutable isolates that play an important role in the development of resistance to antibiotics under the selection pressure of the heavy use of antibiotics in these patients. The use of early and aggressive antibiotic treatment for prevention of the chronic infection with P. aeruginosa and therapeutic approaches with antioxidants such as N-acetylcysteine to prevent the development of hypermutable bacteria during the chronic lung infection should be considered in the treatment of patients with CF.
Acknowledgments
The excellent technical assistance of Tina Wassermann, Lis Kjær Hansen, and Ulla Johansen is highly appreciated.
This study was supported by the Danish Medical Research Council (to N.H. and H.E.P.).
REFERENCES
- 1.Au, K. G., S. Clark, J. H. Miller, and P. Modrich. 1989. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc. Natl. Acad. Sci. USA 86:8877-8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, R. K., and F. J. Kelly. 1994. Evidence for increased oxidative damage in patients with cystic fibrosis. Pediatr. Res. 36:487-493. [DOI] [PubMed] [Google Scholar]
- 3.Chopra, I., A. J. O'Neill, and K. Miller. 2003. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist. Update 6:137-145. [DOI] [PubMed] [Google Scholar]
- 4.Ciofu, O., B. Giwercman, S. S. Pedersen, and N. Høiby. 1994. Development of antibiotic resistance in Pseudomonas aeruginosa during two decades of antipseudomonal treatment at the Danish CF Center. APMIS 102:674-680. [PubMed] [Google Scholar]
- 5.Ciofu, O., T. Jensen, T. Pressler, H. K. Johansen, C. Koch, and N. Høiby. 1996. Meropenem in cystic fibrosis patients infected with resistant Pseudomonas aeruginosa or Burkholderia cepacia and with hypersensitivity to beta-lactam antibiotics. Clin. Microbiol. Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 6.Döring, G., W. Goldstein, K. Botzenhart, A. Kharazmi, P. O. Schiøtz, N. Høiby, and M. Dasgupta. 1986. Elastase from polymorphonuclear leucocytes: a regulatory enzyme in immune complex disease. Clin. Exp. Immunol. 64:597-605. [PMC free article] [PubMed] [Google Scholar]
- 7.Frederiksen, B., C. Koch, and N. Høiby. 1999. Changing epidemiology of Pseudomonas aeruginosa infection in Danish cystic fibrosis patients (1974-1995). Pediatr. Pulmonol. 28:159-166. [DOI] [PubMed] [Google Scholar]
- 8.Giwercman, B., P. A. Lambert, V. T. Rosdahl, G. H. Shand, and N. Høiby. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in-vivo selection of stable partially derepressed beta-lactamase producing strains. J. Antimicrob. Chemother. 26:247-259. [DOI] [PubMed] [Google Scholar]
- 9.Hammer, M. C., A. L. Baltch, N. T. Sutphen, R. P. Smith, and J. V. Conroy. 1981. Pseudomonas aeruginosa: quantitation of maximum phagocytic and bactericidal capabilities of normal human granulocytes. J. Lab. Clin. Med. 98:938-948. [PubMed] [Google Scholar]
- 10.Hammerschlag, M. R., L. Harding, A. Macone, A. L. Smith, and D. A. Goldmann. 1980. Bacteriology of sputum in cystic fibrosis: evaluation of dithiothreitol as a mucolytic agent. J. Clin. Microbiol. 11:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Høiby, N. 1976. The serology of Pseudomonas aeruginosa analysed by means of quantitative immunoelectrophoretic methods. Acta Pathol. Microbiol. Scand. Sect. C 84C:372-382. [PubMed] [Google Scholar]
- 12.Høiby, N., L. Jacobsen, B. A. Jorgensen, E. Lykkegaard, and B. Weeke. 1974. Pseudomonas aeruginosa infection in cystic fibrosis. Occurrence of precipitating antibodies against Pseudomonas aeruginosa in relation to the concentration of sixteen serum proteins and the clinical and radiographical status of the lungs. Acta Paediatr. Scand. 63:843-848. [DOI] [PubMed] [Google Scholar]
- 13.Hull, J., P. Vervaart, K. Grimwood, and P. Phelan. 1997. Pulmonary oxidative stress response in young children with cystic fibrosis. Thorax 52:557-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, E. T., A. Kharazmi, K. Lam, J. W. Costerton, and N. Høiby. 1990. Human polymorphonuclear leukocyte response to Pseudomonas aeruginosa grown in biofilms. Infect. Immun. 58:2383-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, T., A. Kharazmi, P. O. Schiøtz, H. Nielsen, S. Stenvang Pedersen, G. Stafanger, C. Koch, and N. Høiby. 1988. Effect of oral N-acetylcysteine administration on human blood neutrophil and monocyte function. APMIS 96:62-67. [DOI] [PubMed] [Google Scholar]
- 16.Lagrange-Puget, M., I. Durieu, R. Ecochard, F. Abbas-Chorfa, J. Drai, J. P. Steghens, Y. Pacheco, D. Vital-Durand, and G. Bellon. 2004. Longitudinal study of oxidative status in 312 cystic fibrosis patients in stable state and during bronchial exacerbation. Pediatr. Pulmonol. 38:43-49. [DOI] [PubMed] [Google Scholar]
- 17.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 18.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355:273-275. [DOI] [PubMed] [Google Scholar]
- 19.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Høiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 20.Michaels, M. L., C. Cruz, A. P. Grollman, and J. H. Miller. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89:7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1996. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 50:625-643. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H., A. Suthar, J. Tai, A. Yeung, C. Truong, and J. L. Stewart. 1999. Direct selection for mutators in Escherichia coli. J. Bacteriol. 181:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ojeniyi, B., N. Høiby, and V. T. Rosdahl. 1991. Genome fingerprinting as a typing method used on polyagglutinable Pseudomonas aeruginosa isolates from cystic fibrosis patients. APMIS 99:492-498. [PubMed] [Google Scholar]
- 25.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 26.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 27.Oliver, A., B. R. Levin, C. Juan, F. Baquero, and J. Blazquez. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 48:4226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver, A., J. M. Sanchez, and J. Blazquez. 2002. Characterization of the GO system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 217:31-35. [DOI] [PubMed] [Google Scholar]
- 28a.Prunier, A. L., B. Malbruny, M. Laurans, J. Brouard, J. F. Duhamel, and R. Leclercq. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187:1709-1716. [DOI] [PubMed] [Google Scholar]
- 29.Radman, M., F. Taddei, and I. Matic. 2000. Evolution-driving genes. Res. Microbiol. 151:91-95. [DOI] [PubMed] [Google Scholar]
- 29a.Roman, F., R. Canton, M. Perez-Vazquez, F. Baquero, and J. Campos. 2004. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 42:1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romling, U., and B. Tummler. 2000. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:464-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roum, J. H., Z. Borok, N. G. McElvaney, G. J. Grimes, A. D. Bokser, R. Buhl, and R. G. Crystal. 1999. Glutathione aerosol suppresses lung epithelial surface inflammatory cell-derived oxidants in cystic fibrosis. J. Appl. Physiol. 87:438-443. [DOI] [PubMed] [Google Scholar]
- 32.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stafanger, G., and C. Koch. 1989. N-Acetylcysteine in cystic fibrosis and Pseudomonas aeruginosa infection: clinical score, spirometry and ciliary motility. Eur. Respir. J. 2:234-237. [PubMed] [Google Scholar]
- 34.Stewart, P. S., F. Roe, J. Rayner, J. G. Elkins, Z. Lewandowski, U. A. Ochsner, and D. J. Hassett. 2000. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 66:836-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tajiri, T., H. Maki, and M. Sekiguchi. 1995. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 336:257-267. [DOI] [PubMed] [Google Scholar]
- 36.Tchou, J., H. Kasai, S. Shibutani, M. H. Chung, J. Laval, A. P. Grollman, and S. Nishimura. 1991. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. USA 88:4690-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood, L. G., D. A. Fitzgerald, P. G. Gibson, D. M. Cooper, C. E. Collins, and M. L. Garg. 2001. Oxidative stress in cystic fibrosis: dietary and metabolic factors. J. Am Coll. Nutr. 20:157-165. [DOI] [PubMed] [Google Scholar]



