Abstract
We analyzed the resistance to cefotaxime of a Salmonella enterica serovar Enteritidis isolate from a stool culture of a 4-year-old boy. It produced a β-lactamase CTX-M-14, encoded by two related R plasmids. The region surrounding the blaCTX-M-14 gene had an original mosaic structure containing insertion sequences (IS26 and IS903D).
Nontyphoidal salmonellae are important causative agents of food-borne diseases in Japan as well as other developed countries. Infections with nontyphoidal salmonellae are generally self-limiting, and antimicrobial therapy is not needed. However, they sometimes cause systemic infections which need treatment with antimicrobials. Extended-spectrum cephalosporins (ESCs) are commonly used to treat children with invasive salmonellosis.
There have been no reports of salmonellae resistant to ESCs in Japan so far. The frequency of nontyphoidal salmonellae resistant to ESCs seems to be low in other countries as well, though reports on such organisms are increasing (2, 4, 7).
In July 2003, Salmonella enterica serovar Enteritidis (isolate Hd63) was isolated from a stool culture of a 4-year-old outpatient with diarrhea and fever. Neither he nor members of his family reported traveling abroad prior to the infection. Hd63 was found to be resistant to cefotaxime and nalidixic acid by the disk diffusion method (9). In this work we investigated this isolate for antimicrobial resistance determinants.
Antimicrobial susceptibility of Hd63 was determined with an E test (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations. Of the β-lactams tested, Hd63 was resistant to ampicillin, cephalothin, cefotaxime, cefuroxime, ceftriaxone, and cefoperazone and showed decreased susceptibility to aztreonam and ceftazidime but was susceptible to cefoxitin and cefotetan (Table 1). The resistance to cefotaxime was significantly reduced by clavulanic acid (Table 1), suggesting production of an extended-spectrum β-lactamase. The MIC testing also confirmed resistance to nalidixic acid and showed decreased susceptibility to fluoroquinolones (Table 1).
TABLE 1.
Strains used in this study and their relevant phenotypes
| Strain | Species | Source or derivation | Phage type | MIC (μg/ml)a
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CEF | CXM | CTX | CRO | CFP | ATM | CAZ | FOX | CTT | ESBL test
|
NAL | CIP | SPX | NOR | OFX | |||||
| CTX | CTL | |||||||||||||||||||
| Hd63 | S. Enteritidis | Clinical isolate | 6a | >256 | >256 | >256 | 256 | >256 | >256 | 2 | 1.5 | 3 | 0.094 | >16 | 0.125 | >256 | 0.19 | 0.25 | 1 | 1 |
| Hd92 | S. Enteritidis | Hd63 derivative lacking pS12 and pS12-1 | 1 | 1 | 2 | 2 | 0.047 | 0.047 | 0.5 | 0.047 | 0.19 | 2 | 0.094 | NTb | NT | >256 | 0.19 | 0.19 | 0.5 | 1 |
| DH10B | E. coli | Purchased from Invitrogen | 2 | 8 | 2 | 0.032 | 0.023 | 0.047 | 0.032 | 0.19 | 4 | 0.19 | NT | NT | 1.5 | 0.002 | <0.002 | 0.016 | 0.008 | |
| Hd90 | E. coli | DH10B transformant of pS12 | >256 | >256 | >256 | 256 | 256 | 256 | 4 | 1.0 | 3 | 0.19 | >16 | 0.064 | 1.5 | 0.002 | <0.002 | 0.016 | 0.008 | |
| Hd91 | E. coli | DH10B transformant of pS12-1 | >256 | >256 | >256 | 256 | 256 | 256 | 3 | 0.75 | 3 | 0.19 | >16 | 0.064 | 0.75 | 0.002 | <0.002 | 0.016 | 0.008 | |
| Hd47 | S. Enteritidis | Clinical isolate | 1 | 1.5 | 3 | 3 | 0.094 | 0.094 | 0.75 | 0.032 | 0.25 | 2 | 0.094 | NT | NT | >256 | 0.25 | 0.25 | 0.75 | 0.75 |
| Hd93 | S. Enteritidis | Transconjugant of Hd47 with Hd90 | 6a | >256 | >256 | >256 | 256 | >256 | >256 | 3 | 2 | 2 | 0.094 | >16 | 0.094 | >256 | 0.19 | 0.19 | 0.5 | 0.75 |
Determined by E test. AMP, ampicillin; CEF, cephalothin; CXM, cefuroxime; CTX, cefotaxime; CRO, ceftriaxone; CFP, cefoperazone; ATM, aztreonam; CAZ, ceftazidime; FOX, cefoxitin; CTT, cefotetan; CTL, cefotaxime plus clavulanic acid; NAL, nalidixic acid; CIP, ciprofloxacin; SPX, sparfloxacin; NOR, norfloxacin; OFX, ofloxacin.
NT, not tested.
DNA sequences of the quinolone resistance-determination region of the gyrA, gyrB, parC, and parE genes determined as described previously (5, 8) revealed that only GyrA had a point mutation at amino acid 87 from Asp to Asn (GAC to AAC). This mutation is often reported in Salmonella strains with resistance to nalidixic acid (3), including those from Japan (6).
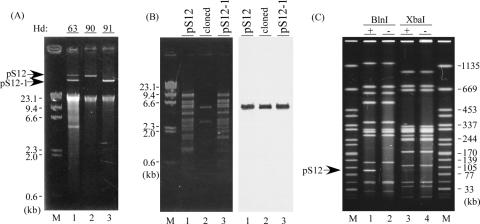
A plasmid profile analysis of Hd63, using Wizard plus SV Minipreps DNA purification systems (Promega Corporation, Madison, WI), revealed two plasmid bands (Fig. 1A, lane 1). Both plasmids (named pS12 and pS12-1) were recovered after the transformation of Escherichia coli DH10B (Invitrogen Corporation, Carlsbad, CA) with DNA prepared from Hd63 (Fig. 1A). The resulting transformants, named Hd90 and Hd91, showed patterns of β-lactam resistance similar to the original strain Hd63 (Table 1), suggesting that most probably the same resistance genes were carried by the two plasmids. The β-lactamase genes present in pS12 and pS12-1 were screened by PCR specific for genes coding for SHV, TEM, CTX-M, CTX-M-2, PER-1, and AmpC-type enzymes by using primers described in reference 10. Only a single primer pair for blaCTX-M, designed for detection of blaCTX-M-1-, blaCTX-M-2-, and blaCTX-M-9-like genes, could amplify a DNA fragment of the expected size from each plasmid. Plasmid pS12-1 shared similar profiles of restriction fragment length polymorphisms with pS12, and indeed, it contained the same blaCTX-M gene, as determined by Southern hybridization and PCR (Fig. 1B and data not shown).
FIG. 1.
Plasmid and PFGE profiles of Hd63 and its derivatives. (A) Plasmid profiles of Hd63 (lane 1) and its transformants, Hd90 and 91 (lanes 2 and 3, respectively). Lane M, lambda DNA digested with HindIII. Numbers at the side indicate the sizes of the bands in lanes M. (B) Southern blot analysis of pS12 (lane 1) and pS12-1 (lane 3). Purified pS12, pS12-1, and pSTV28 plasmids containing the 5.5-kb EcoRI-SphI fragment (“cloned,” lane 2) were digested with EcoRI and SphI. DNAs were separated in 1% agarose gel (left panel) and probed with a DNA fragment amplified by the pair of primers for blaCTX-M (right panel). Lane M, lambda DNA digested with HindIII. Numbers at the side indicate the sizes of the bands in lanes M. (C) PFGE profiles of Hd63 (lanes 1 and 3) and its plasmid-cured derivative, Hd92 (lanes 2 and 4). Endonucleases used were BlnI (lanes 1 and 2) and XbaI (lanes 3 and 4). The arrow on the left indicates the band of the BlnI-digested profile of Hd63 putatively corresponding to that of pS12. Lanes M, Salmonella enterica serovar Braenderup H9812 digested with XbaI.
While pS12 was transferred from Hd90 to another serovar Enteritidis strain, Hd47, pS12-1 was not. The resulting transconjugant, named Hd93, was identified by bacteriophage typing as type PT6a (14), whereas the parental Hd47 strain was PT1. Hd63 was also identified as PT6a, and its plasmid-cured derivative Hd92, generated by negative resistance screening, was identified as PT1 (Table 1). This indicates the ability of pS12 to convert phage types. A comparison of pulsed-field gel electrophoresis (PFGE) profiles between Hd63 and Hd92, done as described in reference 12, identified pS12 as the most likely DNA band migrating between the linear DNA fragments of 77 and 105 kb (Fig. 1C). These results suggest that Hd63 might have emerged from a certain PT1 strain by acquisition of pS12 and that pS12-1 is a recombined version of pS12 missing certain genes, including those responsible for plasmid transfer.
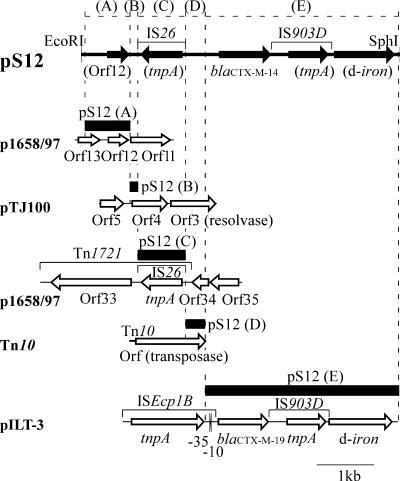
The 5.5-kb EcoRI-SphI DNA fragment of pS12, containing the CTX-M-type β-lactamase gene (Fig. 1B), was cloned in the plasmid vector pSTV28 (Takara Bio Inc., Shiga, Japan) and sequenced. Sequence analysis revealed the presence of a blaCTX-M gene identical to blaCTX-M-14 (GenBank accession no. AF252622), belonging to the blaCTX-M-9 group (1). An approximately 3.4-kb region encompassing the blaCTX-M-14 gene (Fig. 2, segment E) was 99% identical to a region of the Klebsiella pneumoniae plasmid pILT-3, containing ISEcp1B, blaCTX-M-19, IS903D, and a truncated gene for a putative iron transporter (11). However, in pS12, an IS26 was found upstream of the blaCTX-M gene, instead of ISEcp1B (although a putative promoter was still present upstream of blaCTX-M-14) (Fig. 2). The upstream region of segment E consists of four discrete segments (A to D); segments A and C (0.8 kb and 0.9 kb, respectively) are 84% and 98% identical to different parts of the E. coli plasmid p1658/97 (GenBank accession no. AF550679). Segment B (0.2 kb) is 99% identical to a part of the E. coli plasmid pTJ100 (GenBank accession no. AY214164). Segment D (0.3 kb) is 100% identical to a part of Tn10 (Fig. 2). These results imply a surprisingly high level of mosaicism in the pS12 region flanking the blaCTX-M gene.
FIG. 2.
A schematic map of a 5.5-kb EcoRI-SphI fragment of pS12 surrounding blaCTX-M-14. An arrow indicates the position and orientation of each open reading frame (Orf) or putative gene presumed by its homology. The line bar in the upper part indicates the location of each insertion sequence. Shown in the lower part are schematic maps containing regions of the plasmids and transposon homologous to the segments (A to E) of the 5.5-kb fragment of pS12. Names are indicated on the left. Putative products of Orfs are indicated in parentheses. Black bars indicate the locations of sequences homologous to those of the corresponding regions of pS12. tnpA, sequence encoding the transposase of each insertion sequence. -35 and -10 represent -35 and -10 sequences of the ISEcp1B-mediated promoter. d-iron, truncated gene encoding a putative outer membrane protein for iron transport.
In conclusion, we isolated and characterized a serovar Enteritidis isolate, resistant to ESCs, which carried blaCTX-M-14. A serovar Enteritidis strain producing CTX-M-14 was recently reported in Spain (13), but in that case the bla gene had apparently been acquired in the hospital setting, while the isolate analyzed in this study was from an outpatient. This report raises concern about the emergence of blaCTX-M-14 in serovar Enteritidis outside hospitals.
Nucleotide sequence accession number.
The nucleotide sequence data for the 5.5-kb EcoRI-SphI fragment have been submitted and registered in DDBJ/GenBank/EMBL with accession no. AB180674.
Acknowledgments
We thank Linda R. Ward, HPA, United Kingdom, for kindly providing the typing phages and the scheme. We thank Bala Swaminathan, PulseNet, Centers for Disease Control and Prevention, for kindly providing Salmonella enterica serovar Braenderup H9812.
REFERENCES
- 1.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eaves, D. J., L. Randall, D. T. Gray, A. Buckley, M. J. Woodward, A. P. White, and L. J. Piddock. 2004. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 48:4012-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 5.Giraud, E., A. Brisabois, J. L. Martel, and E. Chaslus-Dancla. 1999. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 43:2131-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izumiya, H., J. Terajima, S. Matsushita, K. Tamura, and H. Watanabe. 2001. Characterization of multidrug-resistant Salmonella enterica serovar Typhimurium isolated in Japan. J. Clin. Microbiol. 39:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulvey, M. R., G. Soule, D. Boyd, W. Demczuk, and R. Ahmed. 2003. Characterization of the first extended-spectrum β-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 41:460-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakaya, H., A. Yasuhara, K. Yoshimura, Y. Oshihoi, H. Izumiya, and H. Watanabe. 2003. Life-threatening infantile diarrhea from fluoroquinolone-resistant Salmonella enterica Typhimurium with mutations in both gyrA and parC. Emerg. Infect. Dis. 9:255-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests; approved standard—7th ed. NCCLS document M2-A7. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 10.Paterson, D. L., K. M. Hujer, A. M. Hujer, B. Yeiser, M. D. Bonomo, L. B. Rice, and R. A. Bonomo. 2003. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob. Agents Chemother. 47:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero, L., L. Lopez, L. Martinez-Martinez, B. Guerra, J. R. Hernandez, and A. Pascual. 2004. Characterization of the first CTX-M-14-producing Salmonella enterica serotype Enteritidis isolate. J. Antimicrob. Chemother. 53:1113-1114. [DOI] [PubMed] [Google Scholar]
- 14.Ward, L. R., J. D. de Sa, and B. Rowe. 1987. A phage-typing scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]