Abstract
Resistance of Streptococcus pneumoniae to fluoroquinolones is caused predominantly by amino acid substitutions at positions Ser79 of ParC and Ser81 of GyrA to either Phe or Tyr encoded in the quinolone resistance-determining regions of the parC topoisomerase IV and gyrA DNA gyrase genes. Analysis of highly resistant clinical isolates identified novel second-step substitutions, Ser79Leu (ParC) and Ser81Ile (GyrA). To determine contributions of these new mutations to fluoroquinolone resistance either alone or in combination with other Ser79/81 alleles, the substitutions Ser79Leu/Phe/Tyr in ParC and Ser81Ile/Phe/Tyr in GyrA were introduced into the R6 background, resulting in 15 isogenic strains. Their level of fluoroquinolone resistance was determined by susceptibility testing for ciprofloxacin, levofloxacin, moxifloxacin, gatifloxacin, gemifloxacin, garenoxacin, and norfloxacin. Leu79 and Ile81 alone as well as 79/81Phe/Tyr substitutions did not contribute significantly to resistance, with fluoroquinolone MICs increasing two- to fourfold compared to wild type for all agents tested. Fluoroquinolone MICs for double transformants ParC Ser79Phe/Tyr/Leu-GyrA Ser81Phe/Tyr were uniformly increased by 8- to 64-fold regardless of pairs of amino acid substitutions. However, combinations including Ile81 conferred two- to fourfold-higher levels of resistance than did combinations including any other Ser81 GyrA substitution, thus demonstrating the differential effects of diverse amino acid substitutions at particular hotspots on fluoroquinolone MICs.
Streptococcus pneumoniae is an important human pathogen that causes an array of diseases including otitis media, meningitis, and bacteremia and is the leading causative agent for community-acquired pneumonia. Fluoroquinolones with antipneumococcal activity, such as levofloxacin, moxifloxacin, gatifloxacin, and gemifloxacin, play an important role in the management of pneumococcal disease (34, 37).
Two mechanisms that decrease susceptibilities to fluoroquinolones have been identified so far among clinical isolates: target alteration and reduced drug accumulation due to efflux. The main cellular targets for fluoroquinolones are two closely related type II topoisomerases: DNA gyrase and DNA topoisomerase IV (10, 19). Topoisomerases are involved in the regulation of chromosome supercoiling and decatenation (11). Gyrase, composed of the GyrA and GyrB subunits, is the only topoisomerase known to catalyze negative supercoiling of DNA (11, 32). A major function of DNA gyrase is to relieve the torsional stress that accumulates ahead of the transcription and replication complexes. Topoisomerase IV, composed of the ParC and ParE subunits, is the principal chromosomal decatenase that acts during replication (32). DNA gyrase and topoisomerase IV share extensive amino acid sequence homology, including highly conserved regions in all subunits (28).
DNA gyrase has been determined to be the primary target of fluoroquinolones in gram-negative bacteria (3, 11) and the only target in mycobacteria (1, 46). On the other hand, DNA topoisomerase IV appears to be the primary target of fluoroquinolones in most gram-positive bacteria, including such important clinical pathogens as Staphylococcus aureus (14) and enterococci (26). In S. pneumoniae some fluoroquinolones appear to target preferentially DNA topoisomerase IV (ciprofloxacin, levofloxacin, and norfloxacin), others target DNA gyrase (gatifloxacin and garenoxacin), and a third group has been assumed to target both (gemifloxacin and moxifloxacin) (6, 7, 43, 48).
In S. pneumoniae development of resistance to the respiratory fluoroquinolones requires a combination of two or more mutations in the quinolone resistance-determining region (QRDR) of the genes encoding the DNA gyrase and DNA topoisomerase IV subunits (6, 7). A single mutation in the type II topoisomerase genes' QRDRs results in reduced fluoroquinolone susceptibility, but the affected strain is still considered to be clinically susceptible (8, 33, 51). Secondary substitutions affecting the paralog topoisomerase further reduce susceptibility, often in excess of synergy, and can result in overt fluoroquinolone resistance.
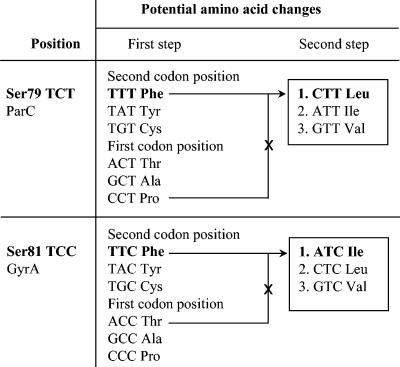
Currently, resistance to fluoroquinolones among pneumococci remains rare (7, 27). For example, TRUST 6 and 7 (2001 to 2003) surveillance data showed that only 0.9% of clinical isolates of S. pneumoniae in the United States were resistant to levofloxacin (27, 42). Predominantly, the fluoroquinolone resistance of S. pneumoniae clinical isolates is attributed to amino acid substitutions at positions Ser79 of ParC and Ser81 of GyrA to either Phe or Tyr (8, 33). Recent analysis of highly resistant isolates revealed the novel substitutions of Ser79 to Leu in ParC and Ser81 to Ile in GyrA (T. A. Davies, S. Pfleger, R. Goldschmidt, K. Bush, and A. Evangelista, Abstr. 40th Annu. Meet. Infect. Dis. Soc. Am., abstr. 78, 2002). These amino acid substitutions cannot be explained by single-base modification of the appropriate codons. Instead, they represent second-step substitutions that result from 2-bp modifications of the codons for the serine at ParC (TCT to CTT) or GyrA (TCC to ATC), as shown in Fig. 1.
FIG. 1.
Amino acid substitutions resulting from first-step mutations within the codons for Ser79 in parC and Ser81 gyrA and second-step mutations leading to the Ser79Leu ParC and Ser81Ile GyrA substitutions identified among clinical isolates of S. pneumoniae. Amino acids in the hypothesized pathways to the identified substitutions are indicated in bold. Crossed-out pathways are deemed unlikely since they involve intermediate substitutions that have never been reported among pneumococci.
To evaluate the contributions of these newly found mutations to fluoroquinolone resistance either alone or in combination with other Ser79 (ParC) or Ser81 (GyrA) substitutions in the paralog genes, we introduced the respective mutated alleles into the susceptible pneumococcal strain R6. Testing the MICs of a panel of fluoroquinolones against these isogenic strains allowed us to evaluate the impact of these new mutations on fluoroquinolone resistance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A set of clinical isolates collected during the TRUST 4 to 6 studies (2000 to 2002) were characterized and used as the source for the different QRDR alleles encoding the amino acid substitutions used in this work (see Table 2). The susceptible laboratory strain R6 (American Type Culture Collection, Manassas, VA; no. BAA-255) was used as a recipient (see Table 3). Strains were grown in Todd-Hewitt broth (Difco, Becton Dickinson and Company, Sparks, MD) supplemented with 0.5% yeast extract (Difco) or on Trypticase soy agar (BBL, Becton Dickinson and Company) supplemented with 5% sheep blood (Rockland, Gilbertsville, PA) at 35°C in 5% CO2.
TABLE 2.
QRDR mutations and MICs for selected clinical isolates collected during TRUST 4 to 6 surveillance studies (2000 to 2002), containing Ser79 ParC and Ser81 GyrA substitutions
| Isolate | Amino acid substitution in protein
|
MICa (μg/ml)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ParC
|
GyrA
|
ParE
|
CIP | LVX | GAT | MXF | NOR | GEM | GAR | ||||
| Ser79 | Lys137 | Ser81 | Glu85 | Asp435 | Ile460 | ||||||||
| OC 6621 | Leu | WTb | WT | Lys | WT | WT | 64 | 32 | 16 | 8 | 128 | 2 | 2 |
| OC 6610 | Phe | WT | Ile | WT | Asn | WT | 128 | 128 | 32 | 16 | 128 | 2 | 2 |
| OC 6582 | Phe | Asn | Phe | WT | WT | Val | 32 | 8 | 4 | 2 | 64 | 0.5 | 0.5 |
| OC 5477 | Tyr | Asn | Tyr | WT | WT | Val | 16 | 16 | 4 | 2 | 64 | 0.25 | 0.25 |
| OC 6755 | Tyr | WT | Phe | WT | WT | WT | 16 | 16 | 4 | 4 | 4 | 0.25 | 0.5 |
| OC 6806 | Phe | WT | Tyr | WT | WT | WT | 32 | 16 | 4 | 2 | 64 | 0.25 | 0.5 |
MICs determined in the presence and in the absence of reserpine (10 μg/ml) were the same. CIP, ciprofloxacin; LVX, levofloxacin; GAT, gatifloxacin; MXF, moxifloxacin; NOR, norfloxacin; GEM, gemifloxacin; GAR, garenoxacin.
WT, wild type.
TABLE 3.
Fluoroquinolone susceptibilities of R6 derivatives with single Ser79 ParC and Ser81 GyrA substitutions
| Strain | Amino acid substitutiona
|
MICb (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ParC79 | GyrA81 | CIP | LVX | GAT | MXF | NOR | GEM | GAR | |
| R6WTc | Ser | Ser | 0.5 | 0.5 | 0.12 | 0.06 | 2 | 0.03 | 0.03 |
| OC 7451 | Phe | Ser | 2 | 1 | 0.25 | 0.12 | 16 | 0.06 | 0.06 |
| OC 7452 | Tyr | Ser | 2 | 1 | 0.25 | 0.12 | 16 | 0.06 | 0.06 |
| OC 7453 | Ser | Phe | 1 | 1 | 0.25 | 0.25 | 2 | 0.06 | 0.125 |
| OC 7454 | Ser | Tyr | 1 | 1 | 0.5 | 0.25 | 2 | 0.06 | 0.125 |
| OC 7455 | Leu | Ser | 2 | 1 | 0.25 | 0.12 | 16 | 0.06 | 0.06 |
| OC 7456 | Ser | Ile | 0.5 | 0.5 | 0.25 | 0.25 | 2 | 0.06 | 0.125 |
No mutations detected in QRDRs of parE and gyrB genes.
MICs determined in the presence and in the absence of reserpine (10 μg/ml) were the same. See Table 2 for definitions of drug abbreviations.
R6, susceptible pneumococcal ATCC strain (BAA-255), used as a recipient in transformation experiments.
MIC determinations.
Fluoroquinolones tested were ciprofloxacin, levofloxacin, moxifloxacin, and gatifloxacin, using panels manufactured by Trek Diagnostics (Westlake, OH); gemifloxacin and garenoxacin (Johnson & Johnson Pharmaceutical Research & Development); and norfloxacin (Sigma, St. Louis, MO). MICs were determined in triplicate by broth microdilution according to National Committee for Clinical Standards recommendations (35). Quality control strain S. pneumoniae ATCC 49619 was assayed in each MIC determination. MICs were also determined in the presence and absence of 10 μg/ml of reserpine (Sigma) to evaluate the presence of an efflux mechanism.
DNA techniques.
PCR amplicons of QRDRs containing the appropriate mutations from clinical isolates were used as donor DNA and were introduced into the isogenic R6 background. DNA templates for PCR were obtained from supernatants of crude extracts of harvested cells that had been prewashed and boiled for 5 min. Fragments encompassing the QRDRs of parC (367 bp) and gyrA (502 bp) were amplified with PfuUltra Hotstart DNA polymerase (Stratagene, La Jolla, CA). The pairs of forward/reverse primers (IDT Inc, Coralville, IA) for the parC QRDR and the gyrA QRDR are shown in Table 1.
TABLE 1.
Oligonucleotides employed to amplify or sequence the QRDRs of S. pneumoniae DNA gyrase and topoisomerase IV genes
| Oligonucleotide | Primer application | Sequence (5′ to 3′) | Codons amplified or sequenceda | Reference |
|---|---|---|---|---|
| parCQRDR(M0363) | Forward PCR | TGGGTTGAAGCCGGTTCA | 35-157 | 38 |
| parCQRDR(M4271) | Reverse PCR | TGCTGGCAAGACCGTTGG | 38 | |
| parC S | Sequence | TGGGTTGAAGCCGGTTCA | 54-143 | 38 |
| gyrA QRDR1 | Forward PCR | GATGAAGGCAAGTTTTATCG | 14-182 | This study |
| gyrA QRDR2 | Reverse PCR | GTGGAATATTGGTTGCCATC | This study | |
| gyrA S | Sequence | TGGCTTAAAACCTGTTCACC | 55-167 | This study |
| parE QRDR1 | Forward PCR | CGGCCGTTCTTTCTATCTTAGTTC | 325-542 | 47 |
| parE QRDR2 | Reverse PCR | AGAGGTGGGAGGGCAATATAGAC | 47 | |
| parE S | Sequence | TGGGGAATTAGCTTCTAACCTCATC | 386-527 | 47 |
| gyrB QRDR1 | Forward PCR | AGATGTTCGCGAAGGATTAAC | 322-637 | 47 |
| gyrB QRDR2 | Reverse PCR | GATAAACTCACGACGAGGCT | 47 | |
| gyrB S | Sequence | CAGATTGCCAAACGTATCGTAG | 394-621 | This study |
Sequencing codons are given for reliable sequence data starting from ∼30 nucleotides downstream of the sequencing primer and ∼20 nucleotides upstream of the 5′ end of the fragment.
After gel purification (QIAGEN, Inc., Valencia, CA), the PCR-generated fragments were transformed at approximately 10 μg/ml into pneumococci (R6 strain) (52) that had been exposed to competence factor CSP-1 (16). The reaction mixtures were incubated first for 20 min at 30°C and then for 1 h at 37°C to allow the expression of the resistance determinants before plating on selective medium. With these small PCR-generated amplicons as donor DNA, transformants were generally obtained at a frequency of 10−4 at the optimal concentration for the selective agents.
Selection for transformants with the appropriate parC and gyrA mutations was done on media containing ciprofloxacin and gemifloxacin, respectively. For selection of first-step transformants the ranges of concentrations for ciprofloxacin and gemifloxacin in twofold dilutions were 0.5 to 4 μg/ml and 0.016 to 0.064 μg/ml, respectively; for second-step transformants the respective ranges were 1 to 16 μg/ml and 0.032 to 0.256 μg/ml. Colonies were picked after 18 to 24 h of incubation at 35°C. In order to confirm the presence of the expected mutations, PCR amplicons encompassing the QRDRs of parC and/or gyrA were generated from all transformants studied. Purified PCR fragments were sequenced bidirectionally using the primers in Table 1 (ACGT Inc., Wheeling, IL).
To exclude the potential introduction of mutations into parE and gyrB genes during the two rounds of selection, transformants chosen for further characterization had their parE and gyrB QRDRs amplified and sequenced using the primers described in Table 1.
RESULTS
Fluoroquinolone-resistant S. pneumoniae clinical isolates.
Analysis of levofloxacin-resistant S. pneumoniae clinical isolates collected during the TRUST 5 surveillance study (2000 to 2001) (Davies et al., 40th IDSA) identified two strains with amino acid substitutions at Ser79 of ParC and Ser81 of GyrA that had not been described previously in S. pneumoniae. In one of these isolates (OC 6621) the TCT codon for Ser79 in parC was replaced by CTT for Leu; in the other (OC 6610) the TCC codon for Ser81 in gyrA was replaced by ATC for Ile. In each case substitutions of two nucleotides were required for the observed amino acid change. Thus, the nature of the nucleotide changes suggested that those mutations did not originate directly from the respective wild-type codons but instead arose in both cases from already existing Ser-to-Phe first-step substitutions: TCT (Ser) to TTT (Phe) to CTT (Leu) for parC and TCC (Ser) to TTC (Phe) to ATC (Ile) for gyrA, as explained in Fig. 1.
Clinical isolates containing the ParC Ser79Leu and the GyrA Ser81Ile substitutions demonstrated high fluoroquinolone resistance against a broad range of fluoroquinolones (Table 2). Indeed, the susceptibilities of these isolates to fluoroquinolones were among the lowest displayed by all levofloxacin-resistant S. pneumoniae isolates thus far characterized in the TRUST studies (9) (Davies et al., 40th IDSA; T. A. Davies, unpublished data). Both OC 6621 and OC 6610 contained additional known QRDR amino acid substitutions: Glu85Lys in GyrA, for the former, and Asp435Asn in ParE along with Ser79Phe in ParC, for the latter. However, those additional substitutions are not usually associated with decreases in fluoroquinolone susceptibility as high as those shown by isolates OC 6621 and OC 6610. Thus, it appeared that the substitutions Leu79 in ParC and Ile81 in GyrA might be contributing to the high level of fluoroquinolone resistance of these two strains.
The poorly defined genetic background of clinical isolates usually does not allow one to discern the contributions to resistance of individual mutations. Therefore, the Leu79 ParC and Ile81 GyrA substitutions were introduced into susceptible pneumococcal strain R6 for the purpose of evaluating their contributions to fluoroquinolone resistance in an isogenic background and comparing them to Tyr and Phe substitutions at the same sites.
Strains with single mutations in parC or gyrA.
Transformation of R6 with PCR amplicons of QRDRs from parC and gyrA genes of clinical isolates containing mutations at Ser79/Ser81 hotspots (Table 2) yielded six independent sets of transformants with low-level fluoroquinolone resistance. Transformants containing the ParC Ser79 substitutions were obtained at 4 × 10−4 to 6 × 10−4 with 1-μg/ml ciprofloxacin selection. Similarly, transformants containing the GyrA Ser81 substitutions were obtained at 1 × 10−4 to 2 × 10−4 with 0.032-μg/ml gemifloxacin selection. In contrast, mutation frequencies obtained in control experiments for parC and gyrA upon ciprofloxacin and gemifloxacin selection, respectively, were in the range of 10−8 to 10−9 and were significantly lower than transformation frequencies.
Representative transformants OC 7451, OC 7452, and OC 7455 (Table 3), containing the Ser79 ParC Phe, Tyr, and Leu substitutions, and OC 7453, OC 7454, and OC 7456, containing the Ser81 GyrA Phe, Leu, and Ile substitutions were selected for further analysis.
The results in Table 3 indicated that the MICs of the various tested fluoroquinolones against the single-substitution transformants were mostly allele independent. Thus, neither the Ser79Leu ParC (OC 7455) nor the Ser81Ile GyrA substitution (OC 7456) conferred lower fluoroquinolone susceptibilities than did the alternative Phe or Tyr substitutions. Indeed, OC 7456 had wild-type-like susceptibilities to both ciprofloxacin and levofloxacin.
Generally, all Ser79 ParC and Ser81 GyrA substitutions decreased by twofold the susceptibilities of the transformants to levofloxacin, gemifloxacin, and gatifloxacin. However, for gatifloxacin the Ser81Tyr GyrA substitution produced a fourfold susceptibility decrease. For ciprofloxacin the ParC and GyrA substitutions produced fourfold and twofold decreases, respectively, whereas for both moxifloxacin and garenoxacin the decreases were twofold and fourfold and for norfloxacin were eightfold and zero, respectively.
In addition to the Leu79 or Ile81 substitution, clinical isolates OC 6621 and OC 6610 contained other mutations: Ser79Phe in ParC, Glu85Lys in GyrA, and Asp435Asn in ParE (Table 2). Thus, though Leu79 and Ile81 were not associated with higher decreases in fluoroquinolone susceptibilities when present as single QRDR substitutions, it was possible that their activities were dependent on the presence of other amino acid substitutions at the paralog proteins. To simplify the analysis, we limited ourselves to double mutations and among them to those most frequently found among clinical isolates, namely, combinations associated with substitutions at Ser79 of ParC and Ser81 of GyrA. Therefore, we examined next the effect of the presence of a combination, Leu79 in ParC or Ile81 in GyrA together with Tyr or Phe substitutions at Ser81 or Ser79 of the respective paralog type II topoisomerase position, on possible differential reductions in fluoroquinolone susceptibilities.
Strains with mutations in both parC and gyrA.
Each of the six isogenic transformants was used in turn to construct nine double mutants where the Ser of the paralog protein was replaced by one of the three alleles at frequencies in the range 1 × 10−4 to 8 × 10−4. Representative double transformants (OC 7457-OC 7465) were picked for further analysis (Table 4).
TABLE 4.
Fluoroquinolone susceptibilities of R6 derivatives with double combinations of Ser79 ParC and Ser81 GyrA substitutions
| Strain | Amino acid substitutiona:
|
MICb (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ParC79 | GyrA81 | CIP | LVX | GAT | MXF | NOR | GEM | GAR | |
| R6WT | Ser | Ser | 0.5 | 0.5 | 0.12 | 0.06 | 2 | 0.03 | 0.03 |
| OC 7457 | Phe | Phe | 32 | 8 | 4 | 2 | 64 | 0.5 | 0.5 |
| OC 7458 | Tyr | Tyr | 32 | 16 | 4 | 2 | 64 | 0.25 | 0.5 |
| OC 7459 | Phe | Tyr | 32 | 16 | 4 | 2 | 64 | 0.25 | 0.5 |
| OC 7460 | Tyr | Phe | 32 | 16 | 4 | 2 | 64 | 0.25 | 0.5 |
| OC 7461 | Phe | Ile | 32 | 32 | 8 | 4 | 64 | 1 | 2 |
| OC 7462 | Leu | Tyr | 32 | 16 | 4 | 2 | 64 | 0.25 | 0.5 |
| OC 7463 | Leu | Phe | 32 | 16 | 4 | 2 | 64 | 0.25 | 0.5 |
| OC 7464 | Tyr | Ile | 32 | 64 | 16 | 8 | 64 | 1 | 2 |
| OC 7465 | Leu | Ile | 32 | 64 | 16 | 8 | 128 | 1 | 2 |
No mutations detected in QRDRs of parE and gyrB genes.
MICs determined in the presence and in the absence of reserpine (10 μg/ml) were the same. See Table 2 for definitions of drug abbreviations.
To rule out the accidental introduction of active efflux effects into the mutant strains as a consequence of the selection exerted during strain construction, the fluoroquinolone MICs were also determined in the presence of reserpine; no effects of reserpine on MICs were observed for any of the tested drugs (Tables 3 and 4).
To exclude the effect on susceptibilities of spontaneous parE and gyrB mutations that might have been acquired during single- and double-step selections on fluoroquinolone-containing media, the appropriate QRDRs were sequenced for selected transformants. No additional mutations were detected. In general, fluoroquinolone susceptibilities in these double-mutant isogenic strains were decreased 8- to 64-fold compared to wild type (Table 4). Susceptibilities to levofloxacin, gatifloxacin, moxifloxacin, and garenoxacin were reduced 32- to 128-fold and to gemifloxacin were reduced 8- to 32-fold. For ciprofloxacin all double mutants showed the same 64-fold allele-independent increase in MICs. For norfloxacin all but one of the allele combinations led to a 32-fold reduction in susceptibility relative to wild type.
The presence of Leu79 ParC in double mutants (OC 7462 and OC 7463) did not confer any higher fluoroquinolone resistance compared to double mutants (OC 7457-OC 5760) with Phe or Tyr at the same position. At the same time, double mutants containing Ile81 GyrA in combination with any of the Ser79 ParC substitutions (OC 7461, OC 7464, and OC 7465) generally yielded decreases in fluoroquinolone susceptibilities two- to fourfold higher than those of the other double mutants. Thus, double mutants containing Ile81 GyrA displayed fourfold-lower susceptibilities to gemifloxacin and garenoxacin and two- to fourfold-lower susceptibilities to levofloxacin, gatifloxacin, and moxifloxacin than double mutants with either Phe81 or Tyr81 at GyrA (Table 4). The particular combination of Leu79 ParC/Ile81 GyrA conferred twofold-higher resistance to norfloxacin than did any other combination of substitutions.
DISCUSSION
Development of a higher level of fluoroquinolone resistance by means of secondary intracodon mutations within the QRDR.
The novel mutations identified in clinical isolates of S. pneumoniae at the Ser79 codon of parC and the Ser81 codon of gyrA appear to have originated in two steps from the respective wild-type codons. Most probably in both instances the first-step mutation involved transitions that replaced the original codons of Ser79 of parC (TCT) and Ser81 of gyrA (TCC) with codons specifying Phe (TTT and TTC, respectively; Fig. 1). It follows from Fig. 1 that alternative first-step mutations at these sites are the TCT-to-CCT parC transition and TCC-to-ACC gyrA transversion and would lead to Pro and Thr substitutions, respectively. However, no Pro substitutions have ever been reported among resistant laboratory mutants or clinical isolates at Ser79 of ParC of S. pneumoniae or at equivalent positions in other bacteria. Because a Pro substitution at that site may cause a major disruption of protein conformation, such a change might be lethal. Thr, on the other hand, is closely related to Ser, with the extension of a methyl group. To our knowledge Thr substitutions at the Ser81 GyrA in S. pneumoniae or at homologous residues in other gram-positive bacteria have never been reported in resistant clinical isolates. However, Thr does occur as the 83rd amino acid residue in the wild-type GyrA subunit of Pseudomonas aeruginosa (18), a site homologous to Ser81 in S. pneumoniae. Thus, a Thr substitution at Ser79 ParC or Ser81 GyrA may confer essentially a wild-type phenotype.
Figure 1 predicts that, in addition to Thr and Pro, Ala and Cys may also occur as potential amino acid substitutions resulting from first-step mutations at the Ser79 parC and Ser81 gyrA codons. Though rare, both of these substitutions have been observed among levofloxacin-resistant clinical isolates of S. pneumoniae (Davies, unpublished data). According to Fig. 1, the possible second-step mutations at the Ser79 (parC) and Ser81 (gyrA) codons derived from the first-step Phe substitutions should lead to Leu, Ile, and Val substitutions in both ParC and GyrA. Of these, here we report the identification of Leu in ParC and Ile in GyrA. The other substitutions remain to be detected in clinical isolates.
Alone, Leu79 (ParC) and Ile81 (GyrA) encoding mutations introduced by transformation into the susceptible R6 strain conferred essentially similar levels of resistance to fluoroquinolones as did Phe and Tyr substitutions. However, when present together with other Ser79 or Ser81 alleles, the Ile81 allele conferred higher levels of resistance than that observed for any Phe and Tyr combinations. At the same time, the Leu79 allele behaved indistinguishably from Phe and Tyr alleles.
The fluoroquinolone MICs for clinical isolates OC 6621 and OC 6610 were higher than those for all relevant double transformants examined. This may be at least partly explained by the additional presence of Glu85Lys (GyrA) and Asp435Asn (ParE) substitutions. Glu85Lys GyrA is known to contribute to fluoroquinolone resistance based on comparisons between isogenic laboratory strains (48). Asp435Asn is assumed to contribute to fluoroquinolone resistance based on comparisons of susceptibilities between clinical isolates (15, 51). Our results suggest that neither the Ile460Val (ParE) nor the Lys137Asn (ParC) substitutions found in other clinical isolates, OC 6582 and OC 5477 (Table 2), contributed to resistance. Indeed, their fluoroquinolone MIC profiles are similar to those of transformants OC 7457 and OC 7458 (Table 4), which do have the same Ser79 (ParC) and Ser81 (GyrA) substitutions but lack the Ile460Val (ParE) and Lys137Asn (ParC) substitutions.
Though rare in S. pneumoniae, Leu and Ile replacements of Ser79 ParC and Ser81 GyrA are frequently found at equivalent positions of other fluoroquinolone-resistant gram-positive (41, 50) and gram-negative (3, 5, 31, 40) pathogens. In these bacteria Leu and Ile result from single-step mutations, and along with other mutations in the paralog genes, both usually confer high levels of resistance, though in S. pneumoniae only the effect of Ile was observed.
Thus, in S. aureus the Ser84Leu substitution results from a TCA-to-TCT transition. Ser84Leu is the most prevalent GyrA substitution found among fluoroquinolone-resistant S. aureus laboratory mutants and clinical isolates. Similar to S. pneumoniae, the single Ser84Leu encoding gyrA mutation in S. aureus is responsible for only twofold increases in MICs for ciprofloxacin, moxifloxacin, gemifloxacin, and garenoxacin (23-25). Compared to wild type the Ser84Leu substitution in GyrA, along with other mutations in GrlA, contributed to a 64- to 128-fold increase in fluoroquinolone MICs for gemifloxacin, moxifloxacin, garenoxacin, and gatifloxacin and a 128- to 256-fold increase for ciprofloxacin (23, 50). Furthermore, in another example, the highest levofloxacin MICs (8 to 32 μg/ml, representing 64- to 256-fold decreases in susceptibility compared to a reference wild type) among a group of fluoroquinolone-resistant and methicillin-resistant clinical isolates of S. aureus were predominantly associated with Ser84Leu substitutions in GyrA in combination with Ser80Phe/Tyr in GrlA and a third mutation in GrlA/B (20).
In laboratory double and triple mutants of Bacillus anthracis (41) high-level resistance to ciprofloxacin was associated with a TCA-to-TCT transition leading to a Ser85Leu substitution in GyrA, at a site homologous to Ser81 of S. pneumoniae.
In Escherichia coli the homologous sites for Ser79 ParC and Ser81 GyrA occur at Ser80 and Ser83, respectively. Ser83Leu GyrA and Ser80Ile ParC substitutions have been associated with the highest level of fluoroquinolone resistance in either isogenic laboratory mutants (4) or clinical isolates (3, 31). A detailed analysis of QRDR mutations associated with norfloxacin (MICs of ≥16 μg/ml) and ciprofloxacin (MICs of ≥4 μg/ml) resistance in clinical isolates demonstrated the presence of Ser83Leu and Ser80Ile substitutions in 100% and 74% of cases, respectively, though additional mutations in both genes were also detected (31).
In Haemophilus influenzae, the Ser84Leu QRDR ParC substitution, which is homologous to the S. pneumoniae Ser79 ParC site, is the predominant substitution reported among fluoroquinolone-resistant clinical isolates (5, 40).
The previous examples point to the strong association between Leu and Ile substitutions at the Ser79 ParC and Ser81 GyrA equivalent positions and high levels of fluoroquinolone resistance. In all these instances, a nucleophilic amino acid (Ser) was replaced by a hydrophobic one (Leu or Ile) by a single-step mutation. In contrast, the same substitutions require two steps in S. pneumoniae via an intermediate aromatic amino acid substitution (Phe). The appearance of Leu and Ile substitutions at the hotspots Ser79 in ParC and Ser81 in GyrA and homologous positions in other bacteria, resulting in the subsequent rise of fluoroquinolone resistance, is favored, most likely because these polypeptide positions directly interact with the fluoroquinolone molecule in the ternary complex of enzyme, DNA, and the drug.
Correlations with primary targets.
The effects on fluoroquinolone susceptibility of single substitutions at Ser79 ParC or Ser81 GyrA were agent dependent. From Table 3 it follows that susceptibility to norfloxacin was affected only by ParC substitutions. In contrast, for all other examined fluoroquinolones, susceptibilities decreased with both ParC and GyrA substitutions, though to a different extent. Thus, fourfold-higher reductions in susceptibility were observed with ParC substitutions for ciprofloxacin or with GyrA substitutions for moxifloxacin and garenoxacin. For gemifloxacin, both ParC and GyrA substitutions resulted in a twofold susceptibility reduction. Gatifloxacin and levofloxacin behaved similarly to gemifloxacin, with the exception of a fourfold decrease in susceptibility associated with the Ser81Tyr substitution for the former and no effect of the Ser81Ile GyrA substitution for the latter. Our results agree qualitatively but not quantitatively with those described for ciprofloxacin and moxifloxacin by Varon et al. (48). Thus, for these authors Tyr and Phe substitutions at both Ser79 ParC and Ser81 GyrA resulted in uniform fourfold decreases in susceptibilities to moxifloxacin. The observed discrepancies are possibly due to the different methods used for MIC determinations: whereas Varon et al. used an agar dilution method (48), the results obtained in this work were obtained through broth microdilution. The enhanced decrease in susceptibilities associated with ParC over GyrA substitutions observed for ciprofloxacin and with GyrA over ParC for moxifloxacin and garenoxacin are in agreement with the specificity displayed by these fluoroquinolones in selecting spontaneous drug-resistant mutants in S. pneumoniae. The almost homogeneous decrease in susceptibilities to levofloxacin and gatifloxacin upon either ParC or GyrA substitution was surprising in view of the target specificity that these fluoroquinolones display in selecting for drug-resistant mutants. Thus, it appears that small differences in susceptibility may result in selective differences that are amplified when mutants are selected on solid media.
Mechanistic considerations.
Ciprofloxacin and all other fluoroquinolones developed thereafter target both DNA gyrase and DNA topoisomerase IV and are assumed to form topoisomerase-drug-DNA ternary complexes that eventually lead to lethal lesions (32). The relative affinities of fluoroquinolones for those two enzymes vary among bacterial species. Mutations that reduce susceptibility to fluoroquinolones commonly map to the QRDR of the particular subunits of those enzymes. Whereas in S. pneumoniae single-step mutations in the QRDR reduce susceptibilities by two- to fourfold relative to wild type, secondary mutations in the QRDR of the paralog topoisomerase genes generally result in reductions in excess of 8- to 16-fold (48). The detailed cause or causes for this hypersynergistic response are still only partially understood. The two DNA topoisomerases have similar biochemical activities (13). However, inside the cell DNA gyrase and DNA topoisomerase IV are both essential enzymes that have distinct functions in dealing with the various topological challenges imposed by DNA replication and chromosomal partition (29, 53), even though some complementary activities have been reported (30).
Experiments with isogenic constructs of S. aureus have led to an interpretative hypothesis of the patterns of susceptibility to fluoroquinolones of mutants with single-step or secondary QRDR mutations. Such a hypothesis states that interactions of fluoroquinolones with either DNA gyrase or DNA topoisomerase IV may result in cell death, with cell susceptibilities being determined directly by the most sensitive enzyme (36). This hypothesis accounts well for the patterns of susceptibility of S. aureus to the DNA topoisomerase IV targeting ciprofloxacin. Unlike what occurs upon introduction of a single grlA mutation, introduction of a gyrA mutation into wild-type S. aureus does not change the susceptibility of the strain to ciprofloxacin. However, gyrA mutations produce an incremental decrease in susceptibility when introduced into a strain with a grlA mutation (36). Thus, this hypothesis accounts for the facts and is both simple and biochemically sensible.
The results in Table 4 can be explained in general terms by the above hypothesis, assuming that among the substitutions of Ser81 of GyrA the one containing Ile has the lowest affinity for the various fluoroquinolones. However, this hypothesis, based only on the affinities of the various fluoroquinolones for ternary complexes with DNA, does not strictly predict the susceptibilities of isogenic strains of S. pneumoniae into which have been singly transformed diverse parC or gyrA alleles. Thus, in the case of Table 3, the hypothesis would predict that, as seen for S. aureus, GyrA Ser81 substitutions should confer wild-type susceptibilities to fluoroquinolones that target DNA topoisomerase IV (ciprofloxacin, levofloxacin, and norfloxacin), whereas ParC Ser79 substitutions should confer wild-type susceptibilities to fluoroquinolones that target DNA gyrase (gatifloxacin, moxifloxacin, gemifloxacin, and garenoxacin). With one major and two minor exceptions, the results of Table 3 do not conform to these predictions. Only norfloxacin, which in S. pneumoniae targets exclusively DNA topoisomerase IV, conforms to the hypothesis. The two minor exceptions are the wild-type susceptibilities to ciprofloxacin and levofloxacin for the strains harboring the Ile substitution of Ser81 GyrA. Furthermore, the results in Table 3 cannot be explained by assuming that but for norfloxacin all of the other fluoroquinolones had approximately similar affinities for both DNA topoisomerases, for in that instance wild-type susceptibilities would have been expected for all isogenic constructs (54). Our results are consistent with those obtained previously by others using a similar system (48). Because both the previous study (48) and the present one used isolates from the genetically well characterized strain R6 (2, 21) as isogenic background, strain effects cannot be excluded. The results in Table 3 suggest that in S. pneumoniae (at least in strain R6) the susceptibilities to fluoroquinolones are not entirely determined by the affinity of the most sensitive target. The less sensitive target also appears to exert some influence. Thus, the susceptibility determined by the most sensitive wild-type target is higher in the presence of the wild-type version of the less sensitive paralog than in the presence of a mutated allele (Table 3).
Explanations for how the less sensitive target could influence the susceptibility of S. pneumoniae R6 to fluoroquinolones are largely speculative and fall into the regulatory, functional, or structural categories. For example, in E. coli DNA supercoiling is under tight homeostatic control involving alterations of expression of topoisomerase genes (44), and mutations in topoisomerase genes can in turn affect the abundance levels of many bacterial proteins (45). It has been shown recently that decreased expression of a topoisomerase gene may reduce susceptibilities to fluoroquinolones, presumably by diminishing formation of topoisomerase-drug-DNA ternary complexes leading to lethal events (22). It is unclear whether such a scenario could explain the influence of the less sensitive target on reduced susceptibility of S. pneumoniae to fluoroquinolones, given that in this organism Ser81Phe and Ser81Tyr substitutions in GyrA do not appear to affect topoisomerase expression (39). Alternatively, functional complementation such as that observed between topoisomerases in E. coli (17, 30, 32) might contribute to the effect of the less susceptible target on fluoroquinolone susceptibility. Finally, one intriguing possibility is a protein-protein interaction between DNA gyrase and DNA topoisomerase IV with the possible participation of additional proteins. It is still unclear whether DNA topoisomerase IV can act during fork progression or whether its activity is circumscribed to the end of the DNA replication cycle, during segregation and partition of the daughter chromosomes (13). Nonetheless, it has been recently shown in both E. coli (12) and Caulobacter crescentus (49) that ParC is closely associated with the replisome, and hence there might be opportunity for subunits of DNA gyrase and DNA topoisomerase to interact physically. In such a model the less sensitive target might be physically associated with the target of higher affinity and thereby affect fluoroquinolone susceptibility. We are presently using both genetic and structural approaches to determine if such putative protein-protein interactions between DNA gyrase and DNA topoisomerase IV do exist.
In summary, we have examined the contribution to fluoroquinolone resistance of two novel S. pneumoniae QRDR mutations (coding for Ser79-to-Leu ParC and Ser81-to-Ile GyrA amino acid substitutions) when placed either alone or with other common QRDR mutations into the R6 genetic background. When present alone, all alleles had a similar effect on fluoroquinolone susceptibilities. However, when present as combinations, allele-dependent effects became manifest, with GyrA 81Ile yielding the highest fluoroquinolone MICs.
Acknowledgments
We thank Karen Bush for valuable suggestions and critical comments on the manuscript.
This work was supported by Johnson & Johnson Pharmaceutical Research & Development, L.L.C., Raritan, NJ.
REFERENCES
- 1.Aubry, A., X. S. Pan, L. M. Fisher, V. Jarlier, and E. Cambau. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 48:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachoual, R., J. Tankovic, and C. J. Soussy. 1998. Analysis of the mutations involved in fluoroquinolone resistance of in vivo and in vitro mutants of Escherichia coli. Microb. Drug Resist. 4:271-276. [DOI] [PubMed] [Google Scholar]
- 4.Bagel, S., V. Hullen, B. Wiedemann, and P. Heisig. 1999. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biedenbach, D. J., and R. N. Jones. 2003. Five-year analysis of Haemophilus influenzae isolates with reduced susceptibility to fluoroquinolones: prevalence results from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 46:55-61. [DOI] [PubMed] [Google Scholar]
- 6.Browne, F. A., C. Clark, B. Bozdogan, B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2002. Single and multi-step resistance selection study in Streptococcus pneumoniae comparing ceftriaxone with levofloxacin, gatifloxacin and moxifloxacin. Int. J. Antimicrob. Agents 20:93-99. [DOI] [PubMed] [Google Scholar]
- 7.Critchley, I. A., R. S. Blosser-Middleton, M. E. Jones, J. A. Karlowsky, E. A. Karginova, C. Thornsberry, and D. F. Sahm. 2002. Phenotypic and genotypic analysis of levofloxacin-resistant clinical isolates of Streptococcus pneumoniae collected in 13 countries during 1999-2000. Int. J. Antimicrob. Agents 20:100-107. [DOI] [PubMed] [Google Scholar]
- 8.Davies, T. A., A. Evangelista, S. Pfleger, K. Bush, D. F. Sahm, and R. Goldschmidt. 2002. Prevalence of single mutations in topoisomerase type II genes among levofloxacin-susceptible clinical strains of Streptococcus pneumoniae isolated in the United States in 1992 to 1996 and 1999 to 2000. Antimicrob. Agents Chemother. 46:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, T. A., R. Goldschmidt, S. Pfleger, M. Loeloff, K. Bush, D. F. Sahm, and A. T. Evangelista. 2003. Cross-resistance, relatedness and allele analysis of fluoroquinolone-resistant US clinical isolates of Streptococcus pneumoniae (1998-2000). J. Antimicrob. Chemother. 52:168-175. [DOI] [PubMed] [Google Scholar]
- 10.Drlica, K. 1999. Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 2:504-508. [DOI] [PubMed] [Google Scholar]
- 11.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espeli, O., C. Levine, H. Hassing, and K. J. Marians. 2003. Temporal regulation of topoisomerase IV activity in E. coli. Mol. Cell 11:189-201. [DOI] [PubMed] [Google Scholar]
- 13.Espeli, O., and K. J. Marians. 2004. Untangling intracellular DNA topology. Mol. Microbiol. 52:925-931. [DOI] [PubMed] [Google Scholar]
- 14.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 15.Grohs, P., S. Houssaye, A. Aubert, L. Gutmann, and E. Varon. 2003. In vitro activities of garenoxacin (BMS-284756) against Streptococcus pneumoniae, viridans group streptococci, and Enterococcus faecalis compared to those of six other quinolones. Antimicrob. Agents Chemother. 47:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heddle, J. G., T. Lu, X. Zhao, K. Drlica, and A. Maxwell. 2001. gyrB-225, a mutation of DNA gyrase that compensates for topoisomerase I deficiency: investigation of its low activity and quinolone hypersensitivity. J. Mol. Biol. 309:1219-1231. [DOI] [PubMed] [Google Scholar]
- 18.Higgins, P. G., A. C. Fluit, D. Milatovic, J. Verhoef, and F. J. Schmitz. 2003. Mutations in GyrA, ParC, MexR and NfxB in clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 21:409-413. [DOI] [PubMed] [Google Scholar]
- 19.Hooper, D. C. 1998. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Biochim. Biophys. Acta 1400:45-61. [DOI] [PubMed] [Google Scholar]
- 20.Horii, T., Y. Suzuki, A. Monji, M. Morita, H. Muramatsu, Y. Kondo, M. Doi, A. Takeshita, T. Kanno, and M. Maekawa. 2003. Detection of mutations in quinolone resistance-determining regions in levofloxacin- and methicillin-resistant Staphylococcus aureus: effects of the mutations on fluoroquinolone MICs. Diagn. Microbiol. Infect. Dis. 46:139-145. [DOI] [PubMed] [Google Scholar]
- 21.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ince, D., and D. C. Hooper. 2003. Quinolone resistance due to reduced target enzyme expression. J. Bacteriol. 185:6883-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ince, D., X. Zhang, and D. C. Hooper. 2003. Activity of and resistance to moxifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1410-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2002. Dual targeting of DNA gyrase and topoisomerase IV: target interactions of garenoxacin (BMS-284756, T-3811ME), a new desfluoroquinolone. Antimicrob. Agents Chemother. 46:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ince, D., X. Zhang, L. C. Silver, and D. C. Hooper. 2003. Topoisomerase targeting with and resistance to gemifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:274-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanematsu, E., T. Deguchi, M. Yasuda, T. Kawamura, Y. Nishino, and Y. Kawada. 1998. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 42:433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlowsky, J. A., C. Thornsberry, M. E. Jones, A. T. Evangelista, I. A. Critchley, D. F. Sahm, and T. S. Program. 2003. Factors associated with relative rates of antimicrobial resistance among Streptococcus pneumoniae in the United States: results from the TRUST Surveillance Program (1998-2002). Clin. Infect. Dis. 36:963-970. [DOI] [PubMed] [Google Scholar]
- 28.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Hiraga, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 29.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:27668-27677. [DOI] [PubMed] [Google Scholar]
- 30.Khodursky, A. B., B. J. Peter, M. B. Schmid, J. DeRisi, D. Botstein, P. O. Brown, and N. R. Cozzarelli. 2000. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci. USA 97:9419-9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komp Lindgren, P., A. Karlsson, and D. Hughes. 2003. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob. Agents Chemother. 47:3222-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine, C., H. Hiasa, and K. J. Marians. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400:29-43. [DOI] [PubMed] [Google Scholar]
- 33.Lim, S., D. Bast, A. McGeer, J. de Azavedo, and D. E. Low. 2003. Antimicrobial susceptibility breakpoints and first-step parC mutations in Streptococcus pneumoniae: redefining fluoroquinolone resistance. Emerg. Infect. Dis. 9:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandell, L. A., J. G. Bartlett, S. F. Dowell, T. M. File, Jr., D. M. Musher, and C. Whitney. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin. Infect. Dis. 37:1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 36.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niederman, M. S., L. A. Mandell, A. Anzueto, J. B. Bass, W. A. Broughton, G. D. Campbell, N. Dean, T. File, M. J. Fine, P. A. Gross, F. Martinez, T. J. Marrie, J. F. Plouffe, J. Ramirez, G. A. Sarosi, A. Torres, R. Wilson, and V. L. Yu. 2001. Guidelines for the management of adults with community-acquired pneumonia. Diagnosis, assessment of severity, antimicrobial therapy, and prevention. Am. J. Respir. Crit. Care Med. 163:1730-1754. [DOI] [PubMed] [Google Scholar]
- 38.Pan, X., and L. Fisher. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 178:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan, X. S., G. Yague, and L. M. Fisher. 2001. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Vazquez, M., F. Roman, B. Aracil, R. Canton, and J. Campos. 2004. Laboratory detection of Haemophilus influenzae with decreased susceptibility to nalidixic acid, ciprofloxacin, levofloxacin, and moxifloxacin due to GyrA and ParC mutations. J. Clin. Microbiol. 42:1185-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price, L. B., A. Vogler, T. Pearson, J. D. Busch, J. M. Schupp, and P. Keim. 2003. In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob. Agents Chemother. 47:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahm, D. F. 2003. Resistance issues and community-acquired respiratory infections. Clin. Cornerstone Suppl. 3:S4-S11. [DOI] [PubMed] [Google Scholar]
- 43.Smith, H. J., K. A. Nichol, D. J. Hoban, and G. G. Zhanel. 2002. Dual activity of fluoroquinolones against Streptococcus pneumoniae: the facts behind the claims. J. Antimicrob. Chemother. 49:893-895. [DOI] [PubMed] [Google Scholar]
- 44.Snoep, J. L., C. C. Van der Weijden, H. W. Andersen, H. V. Westerhoff, and P. R. Jensen. 2002. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur. J. Biochem. 269:1662-1669. [DOI] [PubMed] [Google Scholar]
- 45.Steck, T. R., R. J. Franco, J. Y. Wang, and K. Drlica. 1993. Topoisomerase mutations affect the relative abundance of many Escherichia coli proteins. Mol. Microbiol. 10:473-481. [DOI] [PubMed] [Google Scholar]
- 46.Takiff, H. E., L. Salazar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs, and A. Telenti. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urban, C., N. Rahman, X. Zhao, N. Mariano, S. Segal-Maurer, K. Drlica, and J. J. Rahal. 2001. Fluoroquinolone-resistant Streptococcus pneumoniae associated with levofloxacin therapy. J. Infect. Dis. 184:794-798. [DOI] [PubMed] [Google Scholar]
- 48.Varon, E., C. Janoir, M. D. Kitzis, and L. Gutmann. 1999. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, S. C., and L. Shapiro. 2004. The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc. Natl. Acad. Sci. USA 101:9251-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, T., Y. Onodera, Y. Uchida, and K. Sato. 2000. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. J. Infect. Chemother. 6:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weigel, L. M., G. J. Anderson, R. R. Facklam, and F. C. Tenover. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:3517-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yother, J., L. S. McDaniel, and D. E. Briles. 1986. Transformation of encapsulated Streptococcus pneumoniae. J. Bacteriol. 168:1463-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zechiedrich, E. L., and N. R. Cozzarelli. 1995. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 9:2859-2869. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, X., C. Xu, J. Domagala, and K. Drlica. 1997. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc. Natl. Acad. Sci. USA 94:13991-13996. [DOI] [PMC free article] [PubMed] [Google Scholar]