Abstract
Many steps in the replication cycle of cytomegalovirus (CMV), like cell entry, capsid assembly, and egress of newly synthesized virions, have not been completely analyzed yet. In order to facilitate these studies, we decided to construct a recombinant CMV that incorporates the green fluorescent protein (GFP) into the nucleocapsid. A comparable herpes simplex virus type 1 (HSV-1) mutant has recently been generated by fusion of the GFP open reading frame (ORF) with the HSV-1 ORF encoding small capsid protein (SCP) VP26 (P. Desai and S. Person, J. Virol. 72:7563–7568, 1998). Recombinant CMV genomes expressing a fusion protein consisting of GFP and the SCP were constructed by the recently established bacterial artificial chromosome mutagenesis procedure. In transfected cells, the SCP-GFP fusion protein localized to distinct foci in the nucleus that may represent sites for capsid assembly (assemblons). However, no viable progeny was reconstituted from these mutant CMV genomes. CMV genomes with deletion of the SCP ORF also did not give rise to infectious virus. Rescue of the mutation by insertion of the SCP gene at an ectopic position in an SCP knockout genome indicates that, in contrast to the HSV-1 SCP, the CMV SCP is essential for viral growth. Expression of the SCP-GFP fusion protein together with the authentic SCP blocked the CMV infection cycle, suggesting that the SCP-GFP fusion protein exerts a dominant-negative effect on the assembly of new virions. The results of this study are discussed with regard to recently published data about the structure of the CMV virion and its differences from the HSV-1 virion.
Human cytomegalovirus (HCMV) is a pathogen of worldwide importance that is the leading infectious cause of birth abnormalities and a major risk factor for immunocompromised patients, e.g., transplant patients (6). Although the DNA sequence of the HCMV genome was determined more than 10 years ago (10) and many of the structural proteins have been identified and characterized in the meantime (for reviews, see references 17, 21, and 28), many questions remain about the replication cycle of HCMV in tissue culture and all the more in vivo. In particular, the early events of the replication cycle, like binding of the virion to the not yet identified receptor, uptake into the cell, uncoating, transport of the capsid to the nuclear pores, and release of the viral genome into the nucleus, have been only poorly examined. Similarly, egress of nucleocapsids from the nucleus and especially the site of envelopment of the tegumented nucleocapsids have been matters for debate although there is growing evidence that the HCMV nucleocapsids acquire their final envelope in a cytoplasmic compartment that may be connected to the trans-Golgi network (26, 27). We reasoned that a fluorescence-labeled HCMV virion will greatly facilitate studies on the viral infection cycle in living cells. Such a tool can also help in the re-evaluation of the tropism of CMV for various cell types.
The HCMV virion has a complex architecture that is common to all herpesviruses (25). The large, double-stranded DNA genome (230 kbp) is packed within the capsid, which has icosahedral symmetry. The nucleocapsid is enclosed by a proteinaceous layer, which is referred to as the tegument, surrounded by a lipid-rich envelope containing various viral glycoproteins. Since the envelope is lost upon entry into the cell and the tegument proteins may be rapidly distributed in the cytoplasm, it was desirable to label the capsid, although there might be structural constraints concerning the possibility of modifying capsid proteins. The HCMV capsid is made up of at least four proteins (17): the major capsid protein (UL86, 154 kDa), the minor capsid protein (UL85, 35 kDa), the minor capsid protein-binding protein (UL46, 33 kDa), and the small capsid protein (SCP, UL48/49), which is only about 8.5 kDa in size (2, 18). The HCMV SCP is the homologue of the herpes simplex virus type 1 (HSV-1) VP26 protein (18). As with HSV-1 and other human herpesviruses (HHVs), like HHV-6 and HHV-7, the open reading frame (ORF) encoding the SCP (UL48/49) is located directly adjacent but in complementary orientation to the largest ORF of the viral genome (UL48) and gives rise to a small protein with a basic pI value (18). Ultrastructural analyses of CMV capsids (8) and virions (11, 29) showed that the HCMV SCP is located at the tips of the hexameric capsomers, like its counterpart VP26 on the HSV-1 capsid (3, 34). VP26 is nonessential for growth of HSV-1 in cell culture (14), and the ORF for the green fluorescent protein (GFP) was successfully fused in frame to the HSV-1 ORF encoding VP26 (UL35), resulting in a recombinant HSV-1 that expresses a VP26-GFP fusion protein (15). This fusion protein was incorporated into the HSV-1 capsid and into infectious virions. Using the recently established bacterial artificial chromosome (BAC) mutagenesis technique (1, 4, 7, 19, 30), we constructed HCMV and mouse CMV (MCMV) genomes that encode a fusion protein consisting of SCP and GFP. Surprisingly, these CMV genomes did not give rise to infectious virus upon transfection into permissive cells. A more thorough genetic analysis indicated that, in contrast to VP26 of HSV-1, the CMV SCP is essential for viral growth.
MATERIALS AND METHODS
Virus and cells.
The HCMV and MCMV strains used in this study were BAC-derived recombinant viruses RV-HB5 (4) and MW97.01 (30), respectively. Human foreskin fibroblasts (HFF) were prepared from surgical material and cultured as previously described (4). The telomerase-immortalized human retina pigment epithelial (RPE) cell line (Clontech, Palo Alto, Calif.) was cultured in Dulbecco's modified Eagle medium/Nut-Mix F12 with 15 mM HEPES (GIBCO-BRL) supplemented with 5% fetal calf serum (GIBCO-BRL), 2 mM glutamine, 0.348% sodium bicarbonate, 100 U of penicillin per ml, and 100 μg of streptomycin sulfate per ml. Mouse NIH 3T3 cells (ATCC CRL1658) were grown in Dulbecco's modified Eagle medium supplemented with 10% newborn calf serum (GIBCO-BRL), 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin sulfate per ml.
Plasmids.
For construction of plasmid pUC-CKgfp, a 1,688-bp KpnI fragment (nucleotides [nt] 69635 to 71323 of the published HCMV sequence [10]) was excised from cosmid pCM1049 (16) and cloned into the KpnI site of pUC19, resulting in plasmid pUC1049. The kanamycin resistance marker from plasmid pCP15 (12) was then inserted into a StyI site (equivalent to nt position 70170 of the HCMV genome [10]) of pUC1049, yielding plasmid pUC-CK. The GFP ORF without the ATG start codon was amplified from plasmid pEGFP-C1 (Clontech) using primers capsidgfp.for (5′-CCG TGG GTC CCC CGG ACT TGT ACA GCT CGT CCA T-3′) and capsidgfp.rev (5′-CGC CGG GAC CCG TGA GCA AGG GCG AGG AGC TGT T-3′), which contain PpuMI restriction enzyme sites (underlined). The PCR fragment was cloned into the PpuMI restriction site of pUC-CK (equivalent to HCMV nt position 70385 [10]), resulting in pUC-CKgfp.
In order to insert the gene for SCP at an ectopic position in the HCMV genome, plasmid pCapek was constructed. An 8.7-kbp HindIII fragment (nt 17243 to 25921 of the HCMV genome [10]) containing UL13 was isolated from cosmid pCM1050 (16) and cloned into the HindIII restriction site of pUC19, yielding pUC1050. A PCR fragment containing the gene for SCP and the kanamycin resistance marker was amplified from pUC-CK using primers capek.for (5′-GGG CTC GAG TTA CAA AAC AAC GTA TCA CTT TCA CGG TGA-3′) and capek.rev (5′-GCG CTC GAG CAA AAC TTT CCG CTC AAC TCG ATG TTC TA-3′), which provide XhoI restriction enzyme sites (underlined), and cloned into the XhoI site (equivalent to nt position 20705 of the HCMV genome [10]) of pUC1050.
Plasmid pCapgfpek was constructed for ectopic expression of the SCP-GFP ORF. Primers capek.for and capek.rev (see above) were used to amplify a DNA fragment that contains the GFP-SCP ORF and the kanamycin resistance marker from pUC-CKgfp. The resulting PCR product was cloned in the XhoI restriction site of pUC1050.
For construction of an MCMV genome encoding an SCP-GFP fusion protein, two PCR fragments (nt 73051 to 73560 and nt 73561 to 74530 of the MCMV genome [23]) were amplified from MCMV BAC plasmid pSM3fr (30) using primer pairs 73051.for-73560.rev (5′-CCG GAA TTC ACG ATG CGG ATG ATC CCC CAC-3′ and 5′-GCG GGA TCC GAT ATC GAC ACA CAT ACA GAA AAA TAA AAC-3′) and 73561.for-74530.rev (5′-CGC GGA TCC TTA TTG TAT GAC GGT GGC TTT TTT AG-3′ and 5′-GGG AAG CTT GTG CTC GAG GCC ATC CTC TGT GAC-3′) and cloned between the EcoRI and HindIII sites of pUC19. The GFP ORF without the start codon was amplified by PCR from pEGFP-C1 (Clontech) using primers EGFPSalI.for (5′-ACG CGT CGA CCA ACG TGA GCA AGG GCG AGG AGC TG-3′) and EGFPSalI.rev (5′-GCG TGT CGA CTT GTA CAG CTC GTC CAT GCC GAG-3′), both of which contain SalI sites (underlined), and cloned into the SalI site at the beginning of ORF m48.2 (nt position 73860 of the MCMV genome [23]). The kanamycin resistance marker from plasmid pCP15 was inserted into an EcoRV site a few nucleotides downstream of the stop codon of the SCP-GFP ORF, resulting in plasmid pSCP-GFP-Kan.
Mutagenesis.
CMV BAC plasmids were mutated by homologous recombination in Escherichia coli using a recently described mutagenesis procedure (1, 22, 32). Briefly, linear recombination fragments were isolated from plasmids pUC-CKgfp, pCapek, and pCapgfpek after digestion with KpnI and PshAI, respectively, and electroporated into E. coli JC8679 that contained the parental HCMV BAC plasmid. E. coli clones were selected on agar plates containing chloramphenicol (20 μg/ml) and kanamycin (25 μg/ml). Mutated CMV BAC plasmids were analyzed by digestion with at least two different restriction enzymes, followed by separation of the DNA fragments on a 0.7% agarose gel and staining with ethidium bromide. Successfully mutated BAC plasmids were transformed into recA-negative E. coli strain DH10B for stable maintenance. For deletion of the UL48/49 ORF from HCMV BAC plasmids pHB5 (4) and pHB5-GFP (5), a PCR fragment with a kanamycin resistance marker from pACYC177 (9) was amplified using primers capko.for (5′-CAA AAC AAC GTA TCA CTT TCA CGG TGA TTT ATT CTT GCT ATT CCT TTT CCC CTT GGG CTG CCA CGT CGT GGA ATG CCT TCG AAT T-3′) and capko.rev (5′-GCA GCG GCT TCC TCT TCG TCC TCC CCC CAC GGC CTG CCC CAT GTC TAA CAC CGC GCC GGG A CT ACA AGG ACG ACG ACG ACA AGT AA-3′). The primers provided 60 nt of viral sequences at their 5′ ends that were required for homologous recombination between the PCR fragment and the HCMV BAC plasmid. Mutagenesis of MCMV BAC plasmid pSM3fr (30) was performed in an analogous way using an EcoRI/XhoI fragment from plasmid pSCP-GFP-Kan that contained the SCP-GFP ORF and the kanamycin resistance marker flanked by viral sequences required for recombination. The Flp recognition target site (FRT)-flanked kanamycin resistance marker was excised from the CMV BAC plasmids by site-specific recombination in E. coli DH10B using Flp recombinase essentially as previously described (1, 12).
Viral nucleic acid isolation and cell transfection.
Large quantities of BAC plasmid DNA were obtained from 500-ml E. coli overnight cultures using Nucleobond PC 500 columns (Macherey-Nagel, Düren, Germany) and following the manufacturer's instructions. Viral DNA was isolated from HCMV virions as follows. The supernatant of infected HFF cells was harvested when the cultures reached a 90% cytopathic effect (CPE), and cellular debris was separated by centrifugation in a Heraeus centrifuge (10 min, 2,000 rpm). The supernatant was filtered through 0.45-μm-pore-size filters, and virions were pelleted by centrifugation for 2 h at 25,000 rpm in an SW28 rotor. The pellet was resuspended in 50 mM Tris-HCl (pH 8.0)–1 mM MgCl2–100 μg of bovine serum albumin per ml, and 100 U of Benzonase (Merck, Darmstadt, Germany) per ml was added. After 1 h of incubation at room temperature, EDTA was added to a final concentration of 20 mM and virions were disrupted by addition of sodium dodecyl sulfate (final concentration, 0.5%), followed by proteinase K digestion (500 μg/ml) for 3 h at 56°C. DNA was extracted with phenol-chloroform and precipitated with isopropanol.
Two micrograms of HCMV BAC plasmid DNA was transfected into RPE cells that had been seeded into six-well dishes 1 day before transfection (105 cells per well) using the Superfect transfection reagent (Qiagen, Hilden, Germany) in accordance with the instructions of the manufacturer. At 7 days after transfection, the RPE cells were split into 10-cm-diameter dishes and cultured until the cells became confluent. The supernatant of the transfected cells was then transferred to HFF cells, and the cells were cultivated until a complete CPE was observed in the samples that received supernatant from the control transfections. MCMV BAC plasmids were transfected into NIH 3T3 fibroblasts using the Superfect transfection reagent. All transfection experiments were done in triplicate.
RESULTS
Construction of an HCMV genome encoding an SCP-GFP fusion protein.
For HSV-1, it has been shown that the GFP ORF can be fused in frame with the HSV-1 UL35 ORF (15). The resulting fusion of GFP and the smallest capsid protein (VP26) of HSV-1 was incorporated into the HSV-1 capsid and into infectious virions. We wanted to generate a similar HCMV mutant because a GFP-labeled CMV capsid would greatly facilitate the monitoring of various steps in the replication cycle of HCMV in living cells.
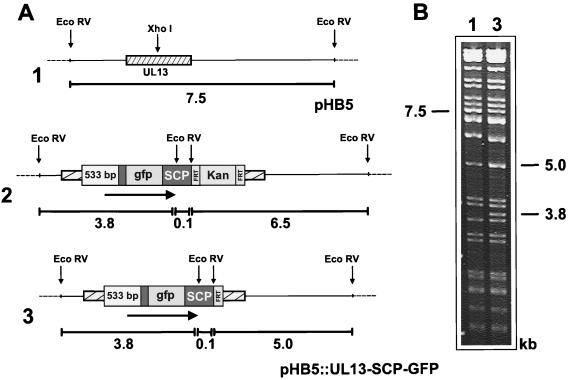
Assuming that the SCP of HCMV has a structure similar to that of the HSV-1 VP26 protein, we decided to insert the GFP at the amino terminus of the HCMV SCP (Fig. 1) because GFP was successfully inserted at this position of HSV-1 VP26 (15). Construction of an HCMV genome that encodes an SCP-GFP fusion protein was performed in E. coli using the BAC mutagenesis procedure (4, 19) with the recently described modifications (1, 22, 32). To this end, we constructed plasmid pUC-CKgfp, which contains the HCMV UL48/49 ORF and flanking homologies required for recombination into the HCMV genome. The GFP ORF without the ATG start codon was inserted in frame at a PpuMI restriction enzyme site after codon 8 of the UL48/49 ORF (Fig. 1). Thus, the encoded SCP-GFP fusion protein contains 8 amino acids (aa) of the SCP at the N terminus, followed by 240 aa of GFP and 69 aa of the SCP (Fig. 1). A kanamycin resistance marker flanked by FRT sites was inserted a few nucleotides downstream of the stop codon of the ORF in order to provide a selection principle for integration of the SCP-GFP ORF into the cloned HCMV genome. The SCP-GFP-kanamycin resistance fragment was transferred to BAC plasmid pHB5 (Fig. 2A, line 1) (4) by homologous recombination in E. coli as described in Materials and Methods, resulting in plasmid pHB5::SCP-GFP-Kan (Fig. 2A, line 2). The mutant HCMV BAC plasmid was analyzed by restriction enzyme digestion with EcoRV (Fig. 2B). After insertion of the SCP-GFP ORF and the kanamycin resistance cassette, two EcoRV fragments of 4.2 and 9.4 kbp of parental BAC plasmid pHB5 were replaced with 5.5- and 10.2-kbp fragments (Fig. 2B, compare lanes 1 and 2). The new 10.2-kbp fragment migrates together with another EcoRV fragment of 10.1 kbp, leading to a double band in Fig. 2B, lane 2. Since insertion of the kanamycin resistance marker may result in destabilization of viral transcripts or interfere with correct termination, we wanted to keep the modification of the mutant HCMV genome as small as possible. Therefore, the kanamycin resistance marker was excised by site-specific recombination using Flp recombinase (1, 12). Flp recombinase-mediated excision in E. coli utilizing the FRT sites that flank the kanamycin resistance marker (12) led to BAC plasmid pHB5::SCP-GFP (Fig. 2A, line 3). Accordingly, the 5.5-kbp EcoRV fragment of BAC plasmid pHB5::SCP-GFP-Kan was replaced with a 4.1-kbp fragment in BAC plasmid pHB5::SCP-GFP (Fig. 2B, compare lanes 2 and 3). Sequencing of pHB5::SCP-GFP verified that no additional mutations had occurred in the SCP ORF and the flanking regions provided for homologous recombination during the cloning-and-mutagenesis procedure. Taken together, these data confirm the successful generation of an HCMV BAC plasmid encoding an SCP-GFP fusion protein.
FIG. 1.
Alignment of the amino acid sequences of the HCMV (upper line) and MCMV (lower line) SCPs. Identical (vertical lines) and conserved (colons) residues and the sites of GFP insertion into the SCPs are indicated. Amino acid positions are on the right. Dots indicate gaps that were introduced into the MCMV SCP sequences to achieve optimal alignment.
FIG. 2.
Construction of HCMV BAC plasmids encoding an SCP-GFP fusion protein (A) and structural analysis of the mutated BAC plasmids (B). (A) Structure of the genomic region of the HCMV BAC plasmids containing the SCP ORF (diagram 1) or the SCP-GFP ORF (diagrams 2 and 3). The arrow indicates the orientation of the SCP ORF. The kanamycin resistance marker (Kan) was excised from BAC plasmid pHB5::SCP-GFP-Kan (diagram 2) by Flp recombinase-mediated site-specific recombination, resulting in BAC plasmid pHB5::SCP-GFP (diagram 3). Fragment sizes are indicated in kilobase pairs below each diagram. (B) DNA of the HCMV BAC plasmids was digested with EcoRV, separated on a 0.7% agarose gel, and stained with ethidium bromide. The sizes of the EcoRV fragments characteristic of parental BAC plasmid pHB5 (lane 1) and mutant BAC plasmids pHB5::SCP-GFP-Kan (lane 2) and pHB5::SCP-GFP (lane 3) are indicated.
The HCMV BAC plasmid expressing an SCP-GFP fusion protein is not infectious.
For reconstitution of mutant HCMV, recombinant BAC plasmid pHB5::SCP-GFP encoding the SCP-GFP fusion protein was transfected into an RPE cell line. We used RPE cells because we found that they can be transfected much more efficiently with HCMV BAC plasmids than can the primary cells, like HFF, that are commonly used for propagation of HCMV. In addition, we observed that RPE cells can be productively infected with HCMV and that infectious virus is released into the supernatant (data not shown). Since plaque formation is sometimes difficult to discover on RPE cells, we usually transfer the supernatant of the transfected RPE cells to HFF cells in order to detect any infectious virus and to grow the virus reconstituted from the BAC plasmids to high titers.
Two days after transfection, RPE cells that took up the BAC plasmid displayed a punctate green fluorescence that was located exclusively in the nucleus (Fig. 3). The fluorescence was stable for up to 2 weeks but neither changed to form a homogeneous pattern throughout the cells nor spread to adjacent cells. When plasmid pUC-CKgfp containing the gene for the SCP-GFP fusion protein was transfected, a faint green fluorescence, distributed throughout the cell, was seen (data not shown). We concluded from this observation that in the absence of other viral proteins, the SCP-GFP fusion protein is not retained in the nucleus. The punctate green fluorescence seen after transfection of BAC plasmid pHB5-SCP-GFP suggested that the SCP-GFP fusion protein is still able to interact with other viral proteins. The labeled structures may represent sites where proteins involved in capsid assembly aggregate and assemble to form new viral capsids (assemblons [31]). However, assembly or egress of nucleocapsids may be blocked since no fluorescence was observed in the cytoplasm of transfected cells.
FIG. 3.
Subnuclear localization of the SCP-GFP fusion protein. HCMV BAC plasmid pHB5::SCP-GFP was transfected into RPE cells, and expression of the SCP-GFP protein was analyzed 2 days later by confocal laser scanning microscopy.
In order to examine if infectious virus was released from the transfected cells, the supernatant of the RPE cells was transferred to HFF. By microscopical inspection, we could not detect any green fluorescence in HFF cells and plaque formation was not observed on the fibroblasts. In contrast, when the supernatant of control transfections with parental BAC plasmid pHB5 was transferred to HFF cells, viral plaques appeared and the infection readily spread throughout the cultures. These findings suggested that BAC plasmid pHB5::SCP-GFP does not give rise to a viable HCMV mutant.
An MCMV genome expressing an SCP-GFP fusion protein is not infectious.
The MCMV SCP and the HCMV SCP show amino acid homology, especially at their carboxy termini (Fig. 1). The amino terminus of the MCMV SCP is less well conserved and contains a cluster of glycine and serine residues predicting some flexibility in this part of the protein. We reasoned, therefore, that there might be a better chance for incorporation of an SCP-GFP fusion protein into the MCMV capsid if the GFP were inserted in front of this putative flexible region of the SCP. For this purpose, a plasmid was constructed containing the MCMV SCP gene (ORF m48.2 [23]) flanked by homologous MCMV DNA sequences necessary for recombination into the MCMV genome. The GFP ORF was inserted in frame into a SalI restriction site at the beginning of the SCP ORF, resulting in expression of a fusion protein containing four N-terminal amino acids of the SCP followed by the GFP and ending with the C terminus of the SCP (Fig. 1). The mutation was introduced into MCMV BAC genome pSM3fr (30) as described in Materials and Methods, resulting in MCMV BAC plasmid pSM3fr::SCP-GFP (Table 1). Correct mutagenesis of the BAC plasmid was verified by restriction enzyme digestion (data not shown). NIH 3T3 fibroblasts were transfected with mutant MCMV BAC plasmid pSM3fr:SCP-GFP in order to reconstitute the mutant virus. However, no fluorescent plaques indicative of a recombinant virus could be detected at any time and only a punctate green fluorescence located in the nucleus was seen in single cells. In conclusion, we showed that recombinant MCMV and HCMV expressing the GFP fused to the amino terminus of the SCP are not viable.
TABLE 1.
CMV BAC plasmids used in this study
| BAC plasmid | Genetic modification | Reference | Virus production upon transfection into permissive cells |
|---|---|---|---|
| HCMV | |||
| pHB5 | Parental BAC-cloned HCMV AD169 genome | 4 | Yes |
| pHB5::SCP-GFP | GFP ORF fused to UL48/49 | This study | No |
| pHB5-ΔSCP | Deletion of UL48/49 | This study | No |
| pHB5::GFP | AD169 BAC plasmid with insertion of GFP gene in US region | 5 | Yes |
| pHB5::GFP-ΔSCP | BAC plasmid pHB5::GFP with deletion of UL48/49 | This study | No |
| pHB5::UL13-SCP | Disruption of UL13 ORF by insertion of SCP gene; authentic UL48/49 not modified | This study | Yes |
| pHB5-ΔSCP::UL13-SCP | Deletion of UL48/49; SCP gene inserted within UL13 ORF | This study | No |
| pHB5::UL13-SCP-GFP | Gene encoding SCP-GFP fusion protein inserted within UL13; UL48/49 not modified | This study | No |
| MCMV | |||
| pSM3fr | Parental BAC-cloned MCMV genome | 30 | Yes |
| pSM3fr::SCP-GFP | GFP ORF fused to m48.2 (23) | This study | No |
The SCP is essential for infectivity of HCMV DNA.
Our data are in contrast to results obtained with HSV-1, where expression of a VP26-GFP fusion protein did not interfere with viral growth (15). One could think of several reasons why an SCP-GFP CMV mutant exhibits a growth defect. (i) The SCP may be essential for CMV. If this were true, the SCP might be difficult to manipulate because small modifications could already abrogate its function. (ii) Insertion of the GFP, which is relatively large (27 kDa) compared with the SCP (8.5 kDa) (2, 18), may change the structure of the SCP in such a way that interaction with other capsid and tegument proteins is hindered, thereby blocking the assembly of viral particles. (iii) The defect may not be due to alteration of the SCP itself, but neighboring genes could be affected. Since many of the CMV ORFs are in close proximity to each other or even overlap (10, 23), one must consider the possibility that manipulation of one viral gene can also influence the expression of adjacent ORFs. Analysis of the DNA sequence suggests that UL48/49 probably shares its polyadenylation signal with UL49 (10, 18). Thus, insertion of the GFP ORF into UL48/49 could possibly alter the expression of UL49 ORF. In order to differentiate between these possibilities, we constructed and tested several additional mutant HCMV BAC plasmids (Table 1).
First, we examined whether the SCP is essential for HCMV. To this end, the UL48/49 ORF was deleted from the HCMV BAC plasmid and replaced with a kanamycin resistance marker (Fig. 4A). Except for the first seven codons, the complete ORF encoding the SCP (UL48/49) was deleted in the resulting mutant HCMV BAC plasmid, pHB5-ΔSCP-Kan. Since the 5′ end of the UL48/49 ORF overlaps the 3′ end of the UL49 ORF (10, 18), these codons were not deleted in order to preserve the integrity of the UL49 ORF. The mutant BAC plasmids were again characterized by EcoRV digestion. Two EcoRV bands of 4.2 and 9.4 kbp characteristic of parental BAC plasmid pHB5 (Fig. 4A, line 1, and B, lane 1) were replaced with a 14.3-kbp fragment in the BAC plasmid with UL48/49 deleted (Fig. 4A, line 2, and data not shown). The kanamycin resistance marker was then excised from the mutant BAC plasmid with Flp recombinase in order to minimize any potential impact on the neighboring ORFs. The resulting BAC plasmid, pHB5-ΔSCP, contains a 13.3-kbp EcoRV fragment instead of the 4.2- and 9.4-kbp fragments in the parental BAC plasmid (Fig. 4B, compare lanes 1 and 3). Only one FRT site remained in the 13.3-kbp fragment next to the deletion of the UL48/49 ORF (Fig. 4A, line 3). The DNA profile obtained indicated that the intended deletion was introduced into BAC plasmid pHB5-ΔSCP.
FIG. 4.
Deletion of the ORF encoding the SCP. (A) Structures of the HCMV BAC plasmids with the EcoRV fragments containing the SCP ORF (diagram 1), after replacement of the SCP ORF with a kanamycin resistance marker (Kan) (diagram 2), and after excision of the kanamycin resistance marker (diagram 3). Fragment sizes in kilobase pairs are shown below the diagrams. (B) Structural analysis of HCMV BAC plasmids pHB5 and pHB5-ΔSCP. DNA of the BAC plasmids was subjected to EcoRV digestion, followed by gel electrophoresis. Relevant DNA bands are indicated.
Mutant BAC plasmid pHB5-ΔSCP was then transfected into permissive RPE cells. After transfer of the supernatant to HFF cells, plaque formation was not observed on the fibroblasts. When parental BAC plasmid pHB5 was transfected as a positive control, infectious virus could be recovered. This result suggested that an HCMV genome with a deletion of the ORF encoding the SCP is unable to generate infectious viral progeny. Still, we had to demonstrate that the failure to generate infectious virus was due to the deletion of the SCP ORF and did not result from poor transfection efficiency. To this end, the UL48/49 ORF was deleted in an HCMV BAC plasmid that contains the GFP gene under the control of the major immediate-early promoter inserted in the unique short (US) region of the viral genome (5). Deletion of the SCP ORF in this HCMV GFP BAC plasmid was performed as described for BAC plasmid pHB5 and was confirmed by restriction enzyme digestion (data not shown). Successful transfection of the parental HCMV GFP BAC plasmid into either RPE or human fibroblast cells resulted in a homogeneous bright green fluorescence easily detected by fluorescence microscopy. Transfer of the supernatant of transfected RPE cells led to green plaques on HFF cells and to a complete CPE in about 1 week. Following transfection of the UL48/49 knockout BAC plasmid into RPE cells, expression of the GFP marker could be observed in single cells only. However, in contrast to the results obtained with the parental HCMV GFP BAC plasmid, the fluorescence did not spread to neighboring cells and did not result in green plaques. When the supernatant of the transfected cells was transferred to HFF, neither green cells nor viral plaques were generated on the HFF (data not shown). The result of this experiment indicated that the BAC plasmid with UL48/49 deleted was taken up by the cells, resulting in GFP expression, and that the block in the replication cycle occurs at a later stage.
As a second control, we tested whether we can rescue the SCP knockout mutation in BAC plasmid pHB5-ΔSCP by cotransfection with plasmid pUC1049, which contains an insert which spans the deletion, thus allowing reinsertion of the UL48/49 ORF into the HCMV genome at its original location by homologous recombination in permissive eukaryotic cells. When the supernatant of RPE cells cotransfected with pHB5-ΔSCP and pUC1049 was transferred to HFF, viral plaques appeared on the HFF, resulting in a complete CPE after about 1 week. The overlapping fragment was probably reinserted into the SCP knockout genome, thereby restoring the SCP ORF and the replication competence of the resulting genome. Altogether, the results of these experiments showed that the failure to reconstitute infectious virus from BAC plasmid pHB5-ΔSCP is a consequence of deletion of the UL48/49 ORF and is not due to technical failure of the transfection procedure.
Rescue of the SCP knockout by ectopic insertion of the SCP ORF.
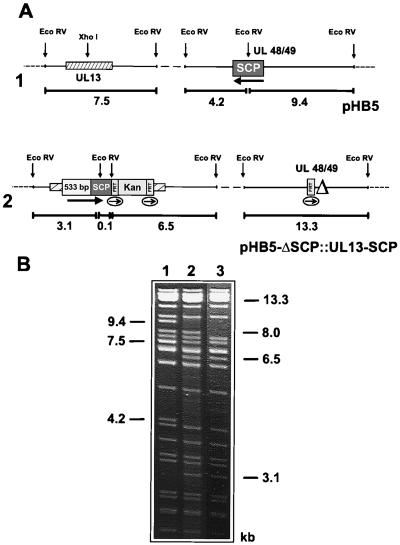
Although the results obtained so far suggested that the SCP is essential for HCMV, we could not formally exclude the possibility that deletion of UL48/49 influenced the expression of the neighboring ORFs in a way that is incompatible with generation of infectious virus. To demonstrate that the ORF encoding the SCP is essential, we decided to reinsert the SCP gene at an ectopic position in the HCMV genome. A series of ORFs located at the termini of the unique long and unique short regions have been shown to be nonessential for HCMV growth in cell culture (reviewed in reference 20). We chose the UL13 ORF for ectopic insertion of the SCP gene. Since the function of the UL13 gene product has not been elucidated, we first tested whether the UL13 ORF can be disrupted without interfering with the viability of the virus. To this end, the gene for SCP was inserted into the UL13 ORF on the backbone of HCMV BAC plasmid pHB5 (data not shown). Following transfection into permissive cells, BAC plasmid pHB5::UL13-SCP gave rise to infectious virus, indicating that the UL13 ORF is not essential for replication of HCMV in cell culture and that foreign sequences can be inserted at this locus.
Next, we examined whether expression of the SCP ORF from an ectopic position in the genome will rescue the growth phenotype of the SCP knockout mutant. To this end, the SCP ORF plus 533 bp of upstream sequences containing the putative promoter of the gene for SCP were inserted into the UL13 gene of SCP knockout BAC plasmid pHB5-ΔSCP (Fig. 5A). Mutant BAC plasmid pHB5-ΔSCP::UL13-SCP was characterized by digestion with restriction enzyme EcoRV (Fig. 5B). The site containing the deletion of the authentic SCP ORF is characterized by a 13.3-kbp fragment (Fig. 5A, line 2, and B, lane 2), as described above for SCP knockout genome pHB5-ΔSCP (Fig. 4A, line 3) instead of the two fragments of 4.2 and 9.4 kbp in parental BAC plasmid pHB5 (Fig. 5A, line 1, and B, lane 1). Insertion of the gene for SCP and the kanamycin resistance marker into UL13 generated three new fragments of 0.1, 3.1, and 6.5 kbp instead of the 7.5-kbp fragment present in the parental BAC plasmid (Fig. 5A, lines 1 and 2, and B, lanes 1 and 2). In this mutant BAC plasmid, it was not possible to excise the kanamycin resistance marker because expression of Flp recombinase led to recombination among all three FRT sites (Fig. 5A, line 2) in the BAC plasmid and resulted in loss of the kanamycin resistance gene, as well as the UL14 to UL48 ORFs (data not shown). Therefore, mutant BAC plasmid pHB5-ΔSCP::UL13-SCP still harboring the kanamycin resistance marker was used for further experiments.
FIG. 5.
Rescue of the SCP knockout mutation by ectopic insertion of the gene for SCP. (A) Structures of the genomic regions containing the UL13 and UL48/49 ORFs of BAC plasmid pHB5 (diagram 1) and of BAC plasmid pHB5-ΔSCP::UL13-SCP after deletion of the UL48/49 ORF and ectopic insertion of the gene for SCP (diagram 2). The black arrows indicate the orientation of the SCP ORF at its original (diagram 1) and ectopic (diagram 2) positions, and the thin arrows display the orientation of the FRT sites. Fragment sizes are indicated below the diagrams in kilobase pairs. (B) DNA of HCMV BAC plasmids pHB5 (lane 1) and pHB5-ΔSCP::UL13-SCP (lane 2) and of the corresponding virus reconstituted from BAC plasmid pHB5-ΔSCP::UL13-SCP (lane 3) was digested with EcoRV and analyzed by gel electrophoresis. The sizes of the DNA bands characteristic of pHB5 (lane 1), pHB5-ΔSCP::UL13-SCP (lane 2), and the viral DNA (lane 3) are indicated.
BAC plasmid pHB5-ΔSCP::UL13-SCP was tested for the potential to give rise to infectious virus after transfection into permissive cells. Indeed, viral plaques were generated on HFF cells, finally resulting in a complete CPE. Viral DNA was isolated from purified virions and analyzed by EcoRV digestion (Fig. 5B, lane 3) to test whether the virus obtained had the expected genome structure. Except for an 8.0-kbp fragment, all of the bands that were seen in the DNA pattern of BAC plasmid pHB5-ΔSCP::UL13-SCP were also present in the DNA of the mutant virus (Fig. 5B, compare lanes 2 and 3). The 8.0-kbp fragment of BAC plasmid pHB5-ΔSCP::UL13-SCP spans the termini of the HCMV genome and indicates the circular nature of the BAC plasmid. The abundance of this band is diminished in the viral DNA due to generation of the different isomeric forms of the HCMV genome (4) and because the viral DNA in the virions is present in a linear form. The DNA profile of the mutant virus confirmed that the gene for SCP was inserted within the UL13 ORF and that the SCP ORF at the original position was deleted. Rescue of the SCP knockout mutation by ectopic insertion and expression of the SCP ORF clearly demonstrates that the failure of BAC plasmid pHB5-ΔSCP to generate infectious progeny is due to deletion of the ORF encoding the SCP and not to any potential impact of the deletion on the expression of adjacent ORFs. Thus, the SCP ORF is essential for HCMV growth.
The SCP-GFP fusion protein exerts a dominant-negative effect on the formation of infectious virus.
Since the SCP is required for the life cycle of HCMV, it is probably quite difficult to modify the protein without affecting its essential function. Our failure to generate a mutant expressing an SCP-GFP fusion protein suggested that integration of the GFP changes the structure of the SCP in a way that sterically hinders the interaction of the SCP with either other viral capsid proteins or tegument proteins. The formation of assembly sites that are labeled by the SCP-GFP fusion protein (Fig. 3) and the absence of any spread of the fluorescence to the cytoplasm point to the possibility that assembly and egress of infectious viral particles are inhibited. In order to test the hypothesis that the SCP-GFP fusion protein interferes with capsid assembly, we finally generated an HCMV genome that encodes the SCP-GFP fusion protein in addition to the authentic SCP.
A DNA fragment comprising the SCP-GFP ORF plus the upstream regulatory sequences of the gene for SCP was inserted into the UL13 ORF of HCMV BAC plasmid pHB5 (Fig. 6A, lines 2 and 3). New EcoRV fragments of 0.1, 3.8, and 6.5 kbp and 0.1, 3.8, and 5.0 kbp arose in mutant BAC plasmids pHB5::UL13-SCP-GFP-Kan and pHB5::UL13-SCP-GFP, respectively (Fig. 6A, lines 2 and 3, and B, lane 3). Mutant BAC plasmid pHB5::UL13-SCP-GFP was then analyzed for the ability to generate infectious virus after transfection into permissive cells. The typical green fluorescent dots were observed in the nuclei of transfected RPE cells. However, as was found before for the other mutant BAC plasmids showing this fluorescence pattern, the fluorescence did not spread to the cytoplasm or to neighboring cells. Also, after transfer of the supernatant to HFF cells, no green fluorescence was detectable and no plaques appeared on these cells. These data suggest that the SCP-GFP fusion protein is expressed and traffics to the site of capsid maturation but then provokes a stop in virus assembly. Although the authentic SCP is also expressed by this mutant BAC plasmid, it obviously could not overcome the effects caused by the SCP-GFP fusion protein. Hence, expression of the SCP-GFP fusion protein exerts a dominant-negative effect on the formation of infectious HCMV particles.
FIG. 6.
Insertion of the SCP-GFP gene within the UL13 ORF. (A) Structures of the region of the HCMV BAC plasmids containing the UL13 ORF prior to (diagram 1) and after (diagram 2) insertion of the SCP-GFP gene. The kanamycin resistance marker was excised by Flp recombinase, resulting in BAC plasmid pHB5::UL13-SCP-GFP (diagram 3). The arrows indicate the orientation of the ectopic SCP-GFP gene. Fragment sizes in kilobase pairs are indicated below the diagrams. (B) Ethidium bromide-stained agarose gel showing the EcoRV-digested DNA of BAC plasmids pHB5 (lane 1) and pHB5::UL13-SCP-GFP (lane 3). Relevant DNA fragments are marked. Note that the 7.5-kb band of pHB5 and the 5.0-kb band of pHB5::UL13-SCP-GFP are double bands.
DISCUSSION
In this communication, we report on our attempts to generate CMV mutants that express an SCP-GFP fusion protein. Infectious virus could not be reconstituted from CMV BAC plasmids encoding the fusion protein upon transfection into permissive cells. Deletion of the SCP ORF from the HCMV BAC plasmid revealed that the SCP is essential for HCMV growth. The growth defect of the SCP knockout mutant could be rescued by ectopic expression of the SCP ORF, resulting in production of infectious virus. Interestingly, expression of the SCP-GFP ORF in addition to the authentic SCP blocked the generation of a viable HCMV mutant, implying a dominant-negative effect of the SCP-GFP fusion protein on the formation of infectious particles.
The mutant CMV genomes were constructed by the BAC mutagenesis procedure that we have recently established for the MCMV and HCMV genomes (4, 7, 19, 30). This technique allows us to insert, in a first step, any kind of mutation into the cloned CMV genomes by homologous recombination in E. coli and to examine, in a separate second step, the phenotypic consequences of the mutation after transfection of the mutated CMV genomes into permissive cells. Thus, construction of the mutant genome is completely independent of the viability or growth properties of the corresponding mutant virus. In addition, we showed that consecutive rounds of mutagenesis can be performed on the cloned HCMV genome without the need to reconstitute viral intermediates. Intermediate steps can be used to construct growth-deficient genomes, as demonstrated for the HCMV BAC plasmids with deletion of the SCP ORF. In a second mutagenesis step, the gene for SCP was reinserted at an ectopic position, leading to rescue of the growth defect of the SCP knockout mutant. This experiment showed unequivocally that the inability of the SCP knockout genome to generate infectious virus was due to deletion of the UL48/49 ORF and not to any influence of the deletion on the expression of neighboring genes or to any adventitious mutation that might have been introduced accidentally during mutagenesis of the cloned CMV genome. The result of this experiment clearly indicates that the SCP gene of CMV is essential for viral growth.
The data obtained with our mutant CMV genomes suggest differences in the architecture of the CMV virion in comparison to the HSV-1 virion. The smallest capsid protein of HSV-1, VP26, turned out to be nonessential for viral replication in cell culture, and an HSV-1 mutant with a null mutation of the UL35 ORF showed a twofold decrease in virus yields from infected cells only (14). It has also been possible to generate an HSV-1 mutant expressing a VP26-GFP fusion protein that grows almost as well as the wild-type virus in cell culture and that incorporates the fusion protein into nucleocapsids and infectious virions (15). Electron cryomicroscopy and computer reconstruction of HSV-1 wild-type capsids and capsids which lack VP26 demonstrated that VP26 molecules form a hexameric ring structure which is attached to all of the hexons on the HSV-1 wild-type capsid (3, 34). Although the ring structure is probably disrupted after insertion of GFP at the amino terminus of VP26, the fusion protein can probably still interact through one of its two domains with an epitope on the hexon (15, 34). The subcellular localization of the CMV SCP-GFP fusion proteins may give some hints to why the formation of infectious CMV particles containing the SCP-GFP fusion protein is not possible. After transfection of the HCMV and MCMV BAC plasmids into permissive cells, a protein was expressed that displayed green fluorescence and localized to distinct foci in the cell nucleus. When the fusion protein was expressed from a plasmid in the absence of other viral proteins, a faint green fluorescence was observed in the cytoplasm and nuclei of transfected cells. We conclude from these observations that the SCP-GFP fusion protein expressed from the CMV BAC plasmids is still able to interact with other viral proteins. This interaction is responsible for transport of the protein into the nucleus and/or its retention in this discrete nuclear compartment. Comparable interactions between the HSV-1 VP26 or the VP26-GFP fusion protein and other HSV-1 capsid proteins have been described (13, 15, 24). The structures labeled by the CMV SCP-GFP fusion proteins may represent sites for assembly of new capsids. For HSV-1, these structures have been named assemblons (31), and localization of HSV-1 VP26 to such structures has recently been demonstrated (13). In order to learn at which step the formation of infectious CMV particles is blocked by the SCP-GFP fusion protein or by the absence of the SCP, we have to generate complementing cell systems that provide these proteins in trans.
The absence of any spread of the green fluorescence to the cytoplasm let us speculate that the block in formation of infectious particles may be in a step past the assembly of capsids, namely, during attachment of the tegument. Recent ultrastructural analyses using electron cryomicroscopy of HCMV and simian CMV virions (11, 29) suggest that attachment of the tegument to the capsid differs fundamentally between HSV-1 and CMV. In HSV-1, attachment of filamentous tegument structures seems to be exclusively localized to the region around the pentons (11, 33). In HCMV and simian CMV, an icosahedrally ordered tegument layer was identified that interacts not only with pentons but also with hexons and triplexes (11, 29). Since the HSV-1 VP26 protein is located exclusively on the tips of the hexons and there seems to be no ordered interaction of HSV-1 tegument proteins with the hexons, this might explain why in HSV-1 the probably rather bulky VP26-GFP protein can be incorporated into the HSV-1 virion. In the HCMV particle, there is probably much less space for incorporation of an SCP-GFP protein because tegument proteins bind to every capsomer. The triplex-binding tegument proteins probably also contact the tips of the hexons (11), where the SCP has been located (8, 11, 29). Likewise, the capsomer-capping tegument protein that has been identified for the simian CMV (29) binds to the top of all of the capsomers. The failure to generate infectious virus from the SCP knockout genomes can thus be easily explained if there is a requirement for interaction of these tegument proteins with the SCP. The results obtained with the HCMV BAC plasmid that expresses both the SCP-GFP fusion protein and the authentic SCP suggest that the assembly of infectious CMV particles is efficiently blocked by incorporation of the SCP-GFP fusion protein, perhaps by disrupting the interaction between capsid and tegument proteins. Thus, this step of the virion assembly process may represent a target for antiviral drugs.
In summary, we provide genetic evidence that the SCP of HCMV is essential for viral growth. Further studies are required to understand the interaction of CMV capsid and tegument proteins at the molecular level and to learn at which step the assembly process is blocked in the absence of the SCP or by expression of the SCP-GFP fusion protein. Our data indicate that there are differences between the assembly processes of HCMV and HSV virions.
ACKNOWLEDGMENTS
E.-M.B. and M.M. designed the experiments and wrote the manuscript. E.-M.B. and S.M. constructed the recombinant HCMV and MCMV BAC plasmids and analyzed their properties, respectively. W.M. contributed the fluorescence experiment, and M.W. established the ET mutagenesis procedure in our department.
This study was supported by grants from the Bundesministerium für Bildung und Forschung (project 01GE9918) and the Deutsche Forschungsgemeinschaft (project A2 of Sonderforschungsbereich 455) to M.M.
REFERENCES
- 1.Adler H, Messerle M, Wagner M, Koszinowski U H. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol. 2000;74:6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick C J, Jr, Shenk T. Proteins associated with purified human cytomegalovirus particles. J Virol. 1996;70:6097–6105. doi: 10.1128/jvi.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst E M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borst E M, Messerle M. Development of a cytomegalovirus vector for somatic gene therapy. Bone Marrow Transplant. 2000;25(Suppl. 2):S80–S82. doi: 10.1038/sj.bmt.1702361. [DOI] [PubMed] [Google Scholar]
- 6.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2524. [Google Scholar]
- 7.Brune W, Messerle M, Koszinowski U H. Forward with BACs: new tools for herpesvirus genomics. Trends Genet. 2000;16:254–259. doi: 10.1016/s0168-9525(00)02015-1. [DOI] [PubMed] [Google Scholar]
- 8.Butcher S J, Aitken J, Mitchell J, Gowen B, Dargan D J. Structure of the human cytomegalovirus B capsid by electron cryomicroscopy and image reconstruction. J Struct Biol. 1998;124:70–76. doi: 10.1006/jsbi.1998.4055. [DOI] [PubMed] [Google Scholar]
- 9.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Chen D H, Jiang H, Lee M, Liu F, Zhou Z H. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- 12.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 13.Chi J H, Wilson D W. ATP-dependent localization of the herpes simplex virus capsid protein VP26 to sites of procapsid maturation. J Virol. 2000;74:1468–1476. doi: 10.1128/jvi.74.3.1468-1476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai P, DeLuca N A, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 15.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleckenstein B, Muller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 17.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 18.Gibson W, Clopper K S, Britt W J, Baxter M K. Human cytomegalovirus (HCMV) smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J Virol. 1996;70:5680–5683. doi: 10.1128/jvi.70.8.5680-5683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mocarski E S, Kemble G W. Recombinant cytomegaloviruses for study of replication and pathogenesis. Intervirology. 1996;39:320–330. doi: 10.1159/000150503. [DOI] [PubMed] [Google Scholar]
- 21.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publisher; 1996. pp. 2447–2492. [Google Scholar]
- 22.Muyrers J P, Zhang Y, Testa G, Stewart A F. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rixon F J, Addison C, McGregor A, Macnab S J, Nicholson P, Preston V G, Tatman J D. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 25.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Lippincott-Raven Publisher. 1996. pp. 2221–2230. Philadelphia, Pa. [Google Scholar]
- 26.Sanchez V, Greis K D, Sztul E, Britt W J. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez V, Sztul E, Britt W J. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J Virol. 2000;74:3842–3851. doi: 10.1128/jvi.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spaete R R, Gehrz R C, Landini M P. Human cytomegalovirus structural proteins. J Gen Virol. 1994;75:3287–3308. doi: 10.1099/0022-1317-75-12-3287. [DOI] [PubMed] [Google Scholar]
- 29.Trus B L, Gibson W, Cheng N, Steven A C. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner M, Jonjic S, Koszinowski U H, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Buchholz F, Muyrers J P, Stewart A F. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z H, Chen D H, Jakana J, Rixon F J, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]