Abstract
Community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) infection occurs commonly in children. Clindamycin resistance may be inducible or constitutive, and the rates of inducible resistance in CA-MRSA that could produce clindamycin treatment failures vary worldwide. The double-disk test was performed in 197 erythromycin-resistant and clindamycin-susceptible CA-MRSA strains from children in Dallas, Texas, from 1999 to 2002 to determine inducible clindamycin resistance. Resistance mechanisms were studied by PCR; epidemiologic trends were studied by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). Inducible resistance was demonstrated in 28 (93% ±6%) of 30 tested isolates in 1999, 21 (64%, ±11%) of 33 in 2000, 12 (23% ±7%) of 52 in 2001, and 6 (7% ±3%) of 82 in 2002. All noninducible strains had the msr(A) gene. Among inducible resistant strains, 31 had erm(B), 24 had erm(C), and 12 had erm(A) genes. Two distinct pulsed types were the most prevalent; one of them was the most common pulsed type in 1999, whereas in 2002 a different pulsed type was prevalent. MLST analyses determined that ST-8 was the most common type, with 76% ±5% found in 2002. All but one of these clindamycin-susceptible, erythromycin-resistant ST-8 strains showed no induction of clindamycin resistance. We conclude that, among erythromycin-resistant, clindamycin-susceptible CA-MRSA strains isolated from children in Dallas, inducible methylase resistance became less common from 1999 to 2002 (P < 0.001). The phenotype of strains was associated with their sequence type. Our results demonstrate a clonal shift in CA-MRSA in Dallas children from 1999 to 2002.
The rates of methicillin-resistant Staphylococcus aureus (MRSA) infection have dramatically increased in recent years in children and adult populations (12). In some areas, methicillin resistance constitutes 40% or more of S. aureus isolates from patients (13, 25) and many are now recognized as community-acquired MRSA (CA-MRSA), based on a lack of known risk factors for acquisition of this organism (5, 7, 22; J. D. Siegel, N. B. Cushion, L. C. Roy, and K. K. Krisher, Abstr. 37th IDSA Meet., abstr. 597, 1999). Invasive infections with CA-MRSA can occur with significant morbidity and mortality in children (6), but the majority are associated with mild to moderate skin and soft tissue infections (15).
Clindamycin, a lincosamide antibiotic, is among the limited choices of antimicrobials effective against MRSA. There is concern about use of this antibiotic in the presence of erythromycin resistance because of the possibility of induction of cross-resistance among members of the macrolide, lincosamide, streptogramin B (MLSB) group (14). Demonstration of the inducible MLSB phenotype in isolates that are susceptible to clindamycin and resistant to erythromycin is possible by using the double-disk diffusion agar inhibitory assay, or D-test (32).
Because the rates of inducible clindamycin resistance (MLS B phenotype) have ranged from 8% to 95% in different areas and at different times in the United States, we hypothesized that there had been a shift in the phenotypic mechanism of resistance in the past 4 to 5 years. The objectives of our study were (i) to determine the rate of CA-MRSA isolates that had inducible resistance to clindamycin at Children's Medical Center of Dallas from 1 January 1999 to 31 December 2002; (ii) to compare the phenotypic mechanisms of resistance evidenced by the D-test with the presence of resistance genes determined by PCR; and (iii) to ascertain whether the rate of inducible clindamycin resistance changed over time and, if so, whether there was an expansion of a specific clone during that period.
MATERIALS AND METHODS
Laboratory specimens.
The clinical microbiology laboratory at Children's Medical Center of Dallas routinely saves all MRSA isolates recovered from pediatric patients seeking outpatient and inpatient medical attention. Staphylococcus aureus isolates were initially identified by standard laboratory methods followed by screening for oxacillin resistance using agar plates containing 6 μg/ml of oxacillin (16). After October 2002, MRSA isolates were identified by the penicillin binding protein latex agglutination test (Oxoid Inc., Ogdensburg, NY). Further susceptibility tests of MRSA isolates were performed using the MicroScan dried gram-positive panels with the Micro Scan Walk Away 96 system (Dade MicroScan Inc., West Sacramento, CA). Isolates collected from 1 January 1999 to 31 December 2002 were retrospectively identified, and the medical records of these patients were reviewed by a single investigator (N.C.) to collect demographic and clinical data. Isolates were classified as CA-MRSA if they were obtained from patients without a history of MRSA infection at our institution and included isolates from outpatients and inpatients within the first 72 h of hospitalization. Only the initial isolate from each patient in the study period was used in our studies. Isolates from patients with a known diagnosis of cystic fibrosis and those isolates obtained for infection control purposes were excluded. Microbiology records were reviewed to determine the reported antimicrobial susceptibilities of these CA-MRSA strains to clindamycin and erythromycin. CA-MRSA isolates that were both susceptible to clindamycin (Clis) and resistant to erythromycin (Eryr) were identified, and out of a total of 468 isolates, a randomly selected sample of 197 isolates was used to determine the presence of inducible resistance to clindamycin (see Table 1). The random selection of samples was based on the number of available isolates each year and the desirable precision for the expected proportions.
TABLE 1.
MRSA isolates, Children's Medical Center of Dallas, 1999 to 2002
| Isolate type | No. (%) of MRSA isolates in study yr:
|
|||
|---|---|---|---|---|
| 1999 | 2000 | 2001 | 2002 | |
| Total MRSA | 85 | 89 | 177 | 374 |
| CA-MRSA | 79 (93) | 79 (89) | 167 (94) | 358 (96) |
| Available with susceptibility | 76 | 78 | 163 | 330 |
| Eryr | 56 (74) | 62 (79) | 122 (75) | 305 (92) |
| Clir Eryr | 16 (21) | 17 (22) | 18 (11) | 26 (8)a |
| Clis | 60 (78) | 61 (78) | 145 (89) | 303 (90) |
| Clis Eryr | 40 | 45 | 104 | 279 |
| Tested by D-test | 30 | 33 | 52 | 82 |
One isolate in 2002 was resistant to clindamycin and suceptible to erythromycin.
The study was approved by the Institutional Review Board of the University of Texas Southwestern Medical Center of Dallas. Informed consent was not required.
Phenotypic inducible resistance to clindamycin by D-test.
The presence of inducible clindamycin resistance was sought in a sample of pediatric CA-MRSA isolates that were Clis Eryr, using the D-test as previously described (32). Briefly, isolates were plated on a Mueller-Hinton agar plate at a MacFarland concentration of 0.5 to evenly cover the agar surface. Clindamycin and erythromycin disks, containing 2 μg and 15 μg of each antibiotic, respectively, were placed in the center of the plate separated by a distance of 1.5 cm between the edges (23). Plates were incubated at 37°C for 24 h. Inducible resistance to clindamycin was defined as blunting of the clear circular area of no growth around the clindamycin disk on the side adjacent to the erythromycin disk and was designated D-test positive. Absence of a blunted zone of inhibition was designated D-test negative, which shows that the strain is truly susceptible to clindamycin.
PFGE.
Isolates studied by D-test in 1999 (n = 30) and 2002 (n = 82) were also tested by pulsed-field gel electrophoresis (PFGE) to assess their genetic relatedness. PFGE was also performed in a randomly selected sample of isolates from 2000 (21 of 33) and 2001 (19 of 52). PFGE was performed using digestion of the genomic DNA by SmaI as described elsewhere (29). After staining with ethidium bromide, DNA fragments were photographed and analyzed. The guidelines of Tenover et al. were used to interpret the PFGE patterns. Isolates were considered indistinguishable if they had the same electrophoretic pattern and clonally related if they showed differences of ≤3 bands (31).
Mechanisms of resistance by PCR.
The macrolide resistance mechanisms of the 197 Clis Eryr isolates included in this study were tested at the Hershey Medical Center by PCR using specific primers to detect the presence of erm(A), erm(B), erm(C), and msr(A) genes as described by Sutcliffe et al. (30).
MLST.
Multilocus sequence typing (MLST) of representative clones was done as described at www.mlst.net. Template DNA for PCR was prepared using InstaGen Matrix, as recommended by the manufacturer (Bio-Rad Laboratories, Hercules, CA). After amplification, PCR products were purified from excess primers and nucleotides using a QIAquick PCR purification kit (QIAGEN, Valencia, CA) and sequenced directly using the CEQ8000 genetic analysis system (Beckman Coulter, Fullerton, CA).
Statistical analysis.
To compare differences between groups, data were analyzed using the chi-square and Fisher exact tests with the statistical software package Sigma Stat version 3.0. In addition, Mantel-Haenszel extension of the chi-square test for trend analysis was performed to test the significant linear changes in proportions over time. All proportions are reported along with their standard errors (SE).
RESULTS
Changes in macrolide resistance over the years.
The erythromycin resistance rates were 74 to 80% from 1999 to 2001. In 2002, the resistance rate climbed to 92%. Among erythromycin-resistant strains, clindamycin resistance rates dropped from 22% to 10% over 4 years. Before being studied for the possibility of inducible clindamycin resistance, 90% of the strains appeared susceptible to clindamycin among CA-MRSA strains in 2002 (Table 1).
Inducible resistance to clindamycin.
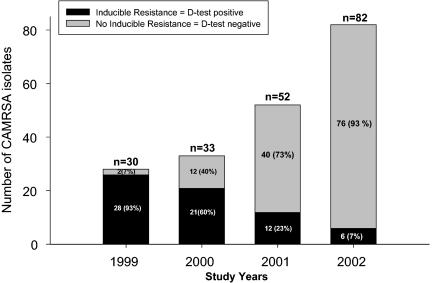
To determine the change in frequency of inducible clindamycin resistance in CA-MRSA strains over time, phenotypic evaluation of inducible resistance to clindamycin was performed by D-test in a total of 197 Clis Eryr CA-MRSA pediatric isolates obtained from 1999 to 2002. As shown in Fig. 1, inducible clindamycin resistance decreased from 93% (SE, 6%) (n = 28/30) in 1999 to 7% (SE, 3%) (n = 6/82) in 2002, representing an about 92% decrease over the 4-year study period (P < 0.001; chi-square test with Yates' correction). The proportion of D-test-positive isolates also showed a significant linear downward trend for each consecutive year from 1999 to 2002 (P < 0.001; chi-square test for trend analysis).
FIG. 1.
Inducible resistance to clindamycin in pediatric CA-MRSA isolates. The proportion of D-test-positive isolates showed a significant linear downward trend for each consecutive year from 1999 to 2002 (P < 0.001; chi-square test for trend analysis).
CA-MRSA genetic relatedness over time by PFGE.
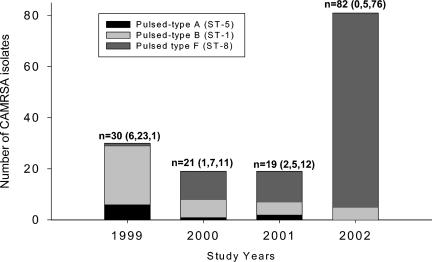
To evaluate the possibility of a clonal shift during the 4-year study, PFGE was performed for all of the strains included in our study in 1999 and 2002 (n = 30 and 82, respectively). A subset of samples from the years 2000 and 2001 (n = 12 and 19, respectively) were also included. Among the 152 strains tested, PFGE analyses determined six pulsed types, designated PT-A through -F. The most common pulsed types were PT-B (26%; n = 40, including 3 isolates distributed in two subtypes) and PT-F (66%; n = 100, including 17 isolates distributed in five subtypes). The frequency of these common pulsed types significantly changed with time; of the 30 isolates tested in 1999, 23 (77%; SE, 10%) were classified as PT-B, whereas in 2002, 76 (93%; SE, 3%) of the 82 isolates tested were PT-F (P < 0.001; chi-square test with Yates' correction). A single isolate in 1999 demonstrated PT-F, whereas five strains in 2002 belonged to PT-B.
Resistance gene analysis by PCR.
The macrolide mechanism of resistance of all the isolates studied by D-test was determined by PCR. All 130 noninducible strains carried the msr(A) gene. Among the 67 inducible resistant strains, 31 had the erm(B) gene, 24 had erm(C), and 12 had erm(A). Twelve strains had both erm and msr(A) genes; the combination of both erm(B) and msr(A) genes was the most frequent, with 7 strains uniformly distributed over the 4-year study period. The three strains carrying both the erm(C) and msr(A) genes were found only in 2002. None of the 2002 tested strains had the erm(A) gene.
MLST.
To better define the epidemiology of the most common CA-MRSA strains in our population, representative strains for the most common PFGE-defined clones were studied at the molecular level by MLST. These clones included PT-A (n = 9), PT-B (n = 40), and PT-F (n = 100) (Fig. 2). PT-A strains belonged to ST-5 and PT-B strains to ST-1, and PT-F isolates corresponded to ST-8. Figure 3 depicts the distribution of the most common pulsed types and the corresponding sequence types during the 4-year study period. All 40 ST-1 strains and 8 of 9 ST-5 strains had inducible clindamycin resistance. In 99 of the 100 ST-8 strains, clindamycin resistance was not inducible. The one ST-8 strain that showed inducible resistance to clindamycin was obtained in 2002 and had a PFGE pattern identical to the dominant PT-F strains. PCR demonstrated the presence of both erm(B) and msr(A) genes in this isolate.
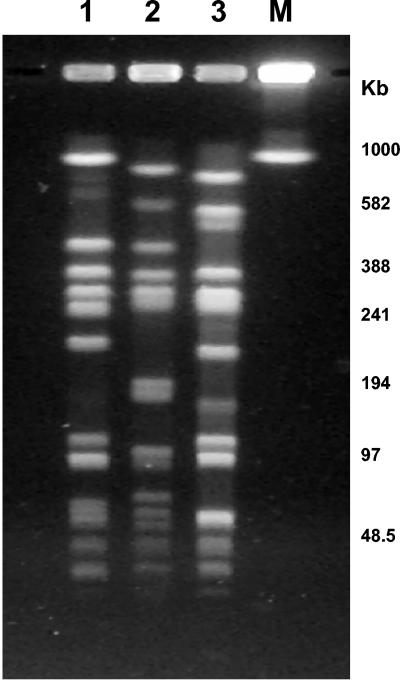
FIG. 2.
PFGE patterns of common pediatric CA-MRSA clones. Lane 1, PT-F; lane 2, PT-B; lane 3, PT-A.
FIG. 3.
PFGE-defined common clones an d their MLST sequence types in pediatric CA-MRSA isolates, Children's Medical Center of Dallas, 1999 to 2002. One hundred fifty-two isolates were studied by PFGE, and the most representative clones underwent MLST analysis (see text). Numbers in parentheses correspond to PT-A, -B, and -F, respectively. Not included are one PT-C isolate and one PT-D isolate found in 2000 and one PT-E isolate from 2002.
DISCUSSION
The increased incidence of disease caused by CA-MRSA in Dallas children as well as in many other cities has created a dilemma regarding appropriate initial empirical antibiotic therapy. Because greater than 50% of S. aureus isolates at Children's Medical Center of Dallas are CA-MRSA, the usual antistaphylococcal penicillins are ineffective. Clindamycin is an appealing option because of its proven efficacy and safety and convenience of parenteral and oral administration in pediatric patients, but the possibility of inducible resistance is a concern.
Inducible clindamycin resistance in our pediatric CA-MRSA strains, evidenced by a positive D-test and confirmed by the presence of erm genes by PCR, markedly decreased from 1999 to 2002. The resistance pattern change we observed also was associated with the predominance over time of a clonally related strain that we classified as pulsed type F by PFGE and further identified by MLST as ST-8. ST-8 strains have shown at least two different PFGE patterns, classified as USA500 and USA300 in previous reports; the latter has been associated with community-acquired infections in Mississippi, Texas, Tennessee, and Georgia, whereas the epidemiology of USA500 has shown these strains to be health care associated. Therefore, our predominant strain is likely to belong to the USA300 group, by this classification (19). The susceptibility rates of CA-MRSA to clindamycin have been studied by other authors. Hussein et al. showed that 23% of all S. aureus isolates from hospitalized pediatric patients in Chicago from 1998 to 1999 were MRSA, and of these, CA-MRSA comprised 43% (n = 10) (13). Seven of their 10 isolates were clindamycin susceptible, but no D-test was performed. PFGE analyses showed diverse clonality in this small isolate sample.
Frank et al. in Chicago also reported an increase of CA-MRSA isolates from the total MRSA strains in their pediatric population (9). They studied 91 available MRSA isolates from 1987 to 1997 and found that 59 were CA-MRSA. CA-MRSA acquisition strongly correlated with clindamycin susceptibility, although inducible resistance was not investigated (8). PFGE analyses of some of these isolates distinguished two predominant PFGE groups concordant with the presence or absence of clindamycin susceptibility. The PFGE group with the greater number of clindamycin-susceptible isolates was the one with the greater proportion of CA-MRSA isolates and was the prevalent group in the latter part of their study period (1). In a later report that included samples up to and including the year 2000, these researchers focused on clindamycin-susceptible MRSA isolates and found that, contrary to our data, the majority of strains were also erythromycin susceptible (10). Among the 33 isolates that were Clis Eryr, 31 (94%) were D-test positive.
In Corpus Christi, Texas, CA-MRSA in children increased from 12% of total MRSA isolates in 1990 to 80% in 2000; 92% of these CA-MRSA isolates were susceptible to clindamycin, as determined by routine susceptibility testing, but the D-test was not performed (7).
In Houston, Sattler et al. prospectively studied pediatric patients with community-acquired S. aureus infection from February to November 2000 and found that the percentage of MRSA from all S. aureus infections increased from 35% to 51%. Of 61 CA-MRSA isolates with available antimicrobial susceptibilities, 52 were Clis Eryr and only 4 (8%) showed inducible resistance to clindamycin (25). From the same center in Houston, Martinez-Aguilar et al. studied 59 children with community-acquired S. aureus musculoskeletal infections from February 2000 to December 2002. MRSA was found in 31. Thirty of these isolates were clindamycin susceptible, 25 were erythromycin resistant, and only 1 had inducible clindamycin resistance by D-test (18).
The clonality of different CA-MRSA isolates has been studied in different regions. In Chicago, the majority of their pediatric Clis CA-MRSA from 1987 to 2000 belonged to a single pulsed type, and the majority were also Erys. These authors noted the appearance of three new pulsed subtypes among the Clis Eryr groups that were not observed before 1998 (10). The predominant clonal group in Chicago was also the most common among 174 CA-MRSA strains from a study in Minnesota from 1996 to 1998, which included 105 isolates from patients <16 years old (21). Clonally related pediatric CA-MRSA strains have been studied in a different hospital in Chicago, where two strains with identical PFGE patterns were assigned to MLST type 1 (20). PFGE patterns of these strains were identical to the fully sequenced strain MW2 (4), which caused fatal MRSA disease in a child in North Dakota (6). The spread of the MW2 strain in postpartum women has also been documented in New York (24). In a study of 581 MRSA strains in Wisconsin from 1989 to 1999, researchers found that none of the major clonal groups belonged to ST-8 and one of the predominant clones was identical to strain MW2, and therefore, MLST type 1. These authors however, did not have clinical data to define CA- versus hospital-acquired MRSA (28).
The striking differences in inducible clindamycin resistance between the northwestern and southwestern areas of the United States could likely be explained by the prevalence of different CA-MRSA clones in different areas of the country. Alternatively, these contrasting results might also be a result of differences in study periods, since our study included strains recovered up to 2002 and the inducible resistance reports from northwestern areas discussed above do not address data regarding inducible clindamycin rates from recent years. Recent data from Texas Children's Hospital in Houston (3) indicate that the predominant clone reported in our study was also the most prevalent among their CA-MRSA isolates evaluated until July 2003, perhaps suggesting that once these clones become prevalent they are able to spread rapidly and predominate in large geographic areas.
The reasons for the predominance of certain MRSA clones over others are unclear, and selective antibiotic pressure is one possibility. The genetic determinants of macrolide resistance vary in different regions of the world. In a European study from 24 countries, 93% of macrolide-resistant MRSA strains were constitutive and 7% were inducible, but none carried the msr(A) gene (26). In another study in the United States, none of 465 MRSA isolates from adult outpatients had the msr(A) gene (2). Taken together, these studies indicate that the prevalence of clindamycin-susceptible, erythromycin-resistant CA-MRSA isolates among U.S. children might be different from the phenotype observed in adult patients in this and other countries. Additionally, in these countries, erm-type resistance in Streptococcus pneumoniae is predominant, in contrast to the United States, where efflux-mediated resistance rates are high (27). Increased macrolide use among children may be the reason for the increased macrolide resistance of the CA-MRSA strains isolated from pediatric patients in the United States (11).
There is ongoing research directed to defining the most appropriate antibiotic therapy for CA-MRSA (17). When clindamycin treatment is considered, laboratories should perform a D-test to try to avoid treatment failures in the presence of inducible resistance. The data discussed above would support the use of clindamycin for empirical therapy for CA-MRSA infections in areas where low rates of both constitutive and inducible clindamycin resistance are prevalent. However, the finding in the latter part of our study of a strain belonging to a predominant clone with the presence of an erm gene is worrisome because these strains could possibly acquire these genetic determinants of resistance, which are known to be carried in mobile genetic elements, such as plasmids and transposons.
Acknowledgments
We wish to thank Jane Siegel for allowing us to study the MRSA strains used in this report.
REFERENCES
- 1.Abi-Hanna, P., A. L. Frank, J. P. Quinn, S. Kelkar, P. C. Schreckenberger, M. K. Hayden, and J. F. Marcinak. 2000. Clonal features of community-acquired methicillin-resistant Staphylococcus aureus in children. Clin. Infect. Dis. 30:630-631. [DOI] [PubMed] [Google Scholar]
- 2.Almer, L. S., V. D. Shortridge, A. M. Nilius, J. M. Beyer, N. B. Soni, M. H. Bui, G. G. Stone, and R. K. Flamm. 2002. Antimicrobial susceptibility and molecular characterization of community-acquired methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 43:225-232. [DOI] [PubMed] [Google Scholar]
- 3.Avalos Mishaan, A. M., E. O. Mason, G. Martinez-Aguilar, W. Hammerman, J. J. Propst, J. R. Lupski, P. Stankiewicz, S. L. Kaplan, and K. Hulten. 2005. Emergence of a predominant clone of community-acquired Staphylococcus aureus among children in Houston, Texas. Pediatr. Infect. Dis. J. 24:201-206. [DOI] [PubMed]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Buckingham, S. C., L. K. McDougal, L. D. Cathey, K. Comeaux, A. S. Craig, S. K. Fridkin, and F. C. Tenover. 2004. Emergence of community-associated methicillin-resistant Staphylococcus aureus at a Memphis, Tennessee Children's Hospital. Pediatr. Infect. Dis. J. 23:619-624. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 20. August 1999, posting date. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus: Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. [Online.] http://www.cdc.gov. [PubMed] [Google Scholar]
- 7.Fergie, J. E., and K. Purcell. 2001. Community-acquired methicillin-resistant Staphylococcus aureus infections in south Texas children. Pediatr. Infect. Dis. J. 20:860-863. [DOI] [PubMed] [Google Scholar]
- 8.Frank, A. L., J. F. Marcinak, P. D. Mangat, and P. C. Schreckenberger. 1999. Community-acquired and clindamycin-susceptible methicillin-resistant Staphylococcus aureus in children. Pediatr. Infect. Dis. J. 18:993-1000. [DOI] [PubMed] [Google Scholar]
- 9.Frank, A. L., J. F. Marcinak, P. D. Mangat, and P. C. Schreckenberger. 1999. Increase in community-acquired methicillin-resistant Staphylococcus aureus in children. Clin. Infect. Dis. 29:935-936. [DOI] [PubMed] [Google Scholar]
- 10.Frank, A. L., J. F. Marcinak, P. D. Mangat, J. T. Tjhio, S. Kelkar, P. C. Schreckenberger, and J. P. Quinn. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21:530-534. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Rey, C., L. Aguilar, F. Baquero, J. Casal, and J. E. Martin. 2002. Pharmacoepidemiological analysis of provincial differences between consumption of macrolides and rates of erythromycin resistance among Streptococcus pyogenes isolates in Spain. J. Clin. Microbiol. 40:2959-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 13.Hussain, F. M., S. Boyle-Vavra, C. D. Bethel, and R. S. Daum. 2000. Current trends in community-acquired methicillin-resistant Staphylococcus aureus at a tertiary care pediatric facility. Pediatr. Infect. Dis. J. 19:1163-1166. [DOI] [PubMed] [Google Scholar]
- 14.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 15.Lee, M. C., A. M. Rios, M. F. Aten, A. Mejias, D. Cavuoti, G. H. McCracken, Jr., and R. D. Hardy. 2004. Management and outcome of children with skin and soft tissue abscesses caused by community-acquired methicillin-resistant Staphylococcus aureus. Pediatr. Infect. Dis. J. 23:123-127. [DOI] [PubMed] [Google Scholar]
- 16.Lennette, E. H., A. Balows, W. J. Hansler, and H. J. Shadomy (ed.). 1995. Manual of clinical microbiology, 4th ed. American Society for Microbiology, Washington, D.C.
- 17.Marcinak, J. F., and A. L. Frank. 2003. Treatment of community-acquired methicillin-resistant Staphylococcus aureus in children. Curr. Opin. Infect. Dis. 16:265-269. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Aguilar, G., A. Avalos-Mishaan, K. Hulten, W. Hammerman, E. O. Mason, Jr., and S. L. Kaplan. 2004. Community-acquired, methicillin-resistant and methicillin-susceptible Staphylococcus aureus musculoskeletal infections in children. Pediatr. Infect. Dis. J. 23:701-706. [DOI] [PubMed] [Google Scholar]
- 19.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mongkolrattanothai, K., S. Boyle, M. D. Kahana, and R. S. Daum. 2003. Severe Staphylococcus aureus infections caused by clonally related community-acquired methicillin-susceptible and methicillin-resistant isolates. Clin. Infect. Dis. 37:1050-1058. [DOI] [PubMed] [Google Scholar]
- 21.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 22.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 23.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing: 14th informational supplement. NCCLS document M100-S14. NCCLS, Wayne, Pa.
- 24.Saiman, L., M. O'Keefe, P. L. Graham III, F. Wu, B. Said-Salim, B. Kreiswirth, A. LaSala, P. M. Schlievert, and P. Della-Latta. 2003. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin. Infect. Dis. 37:1313-1319. [DOI] [PubMed] [Google Scholar]
- 25.Sattler, C. A., E. O. Mason, Jr., and S. L. Kaplan. 2002. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr. Infect. Dis. J. 21:910-917. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz, F. J., R. Sadurski, A. Kray, M. Boos, R. Geisel, K. Kohrer, J. Verhoef, and A. C. Fluit. 2000. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45:891-894. [DOI] [PubMed] [Google Scholar]
- 27.Shortridge, V. D., G. V. Doern, A. B. Brueggemann, J. M. Beyer, and R. K. Flamm. 1999. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the United States in 1994-1995. Clin. Infect. Dis. 29:1186-1188. [DOI] [PubMed] [Google Scholar]
- 28.Shukla, S. K., M. E. Stemper, S. V. Ramaswamy, J. M. Conradt, R. Reich, E. A. Graviss, and K. D. Reed. 2004. Molecular characteristics of nosocomial and Native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J. Clin. Microbiol. 42:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strandén, A., R. Frei, and A. F. Widmer. 2003. Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J. Clin. Microbiol. 41:3181-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisblum, B., and V. Demohn. 1969. Erythromycin-inducible resistance in Staphylococcus aureus: survey of antibiotic classes involved. J. Bacteriol. 98:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]