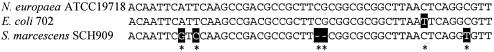Abstract
An Escherichia coli strain, isolated from wild reindeer in a remote mountain area, contained a class 1 integron with two unusual features: a group II intron and a cassette with homology to a superintegron cassette. Alignments indicate that attC sites of gene cassettes may be insertion sites for introns.
Integrons play an important role in the development of antibiotic resistance in gram-negative pathogens. Class 1 and class 2 integrons, also referred to as multiresistance integrons (MRIs), have a worldwide distribution and are described from bacteria colonizing humans, animals, and farmed fish (7, 14, 15, 18). The backbone structure of an integron contains a conserved region encoding an integrase (intI) and a variable region with integrated gene cassettes (16). A gene cassette usually contains a single open reading frame and a recombination site, the attC site (59-base element). The attC sites consist of an inverse core site and a core site separated by an intervening palindrome of variable length. The inverse core site is defined as RYYYAAC and the core site as GTTRRRY (R = A or G, Y = C or T) (4, 17).
The ancestors of multiresistance integrons (MRIs) and their resistance genes are presumed to be the superintegrons (SIs) (11, 12). A multitude of gene cassettes is inserted within each SI. The attC sites of cassettes within a specific SI show extended homology, whereas attC sites of cassettes located within a MRI share little sequence homology.
Gene cassettes are probably ancient structures (10), but their origins and evolution are not clear. It has been proposed that the coding DNA and the attC site originally had separate origins and that these elements have been joined through a specific assembly process (10, 13). Recently it has been discussed whether bacterial group II introns might have played a role in this assembly process (1). Group II introns are considered novel genetic elements, first discovered from organelles of plants, fungi, and other lower eucaryotes. Almost all identified bacterial group II introns encode reverse transcriptase open reading frames and are mobile genetic elements able to translocate via RNA intermediates (retro-elements) (2, 3).
The Escherichia coli strain investigated, strain 2003-10-702 (hereafter termed E. coli 702), was recovered from a fecal sample of a free-ranging reindeer (Rangifer tarandus tarandus) from a remote mountain area in mid-Norway (Forelhogna; 4,370 feet above sea level). The reindeers in this area are completely wild living and have no contact with humans. They are never provided feed or medical treatment such as antimicrobial therapy. Fecal samples were collected, as part of the Health Surveillance Program for Cervids in Norway, for microbiological investigation (9). E. coli strains were tested for susceptibility to oxytetracycline, chloramphenicol, florfenicol, ampicillin, amoxicillin-clavulanate, ceftiofur, trimethoprim, sulfamethoxazole, streptomycin, gentamicin, neomycin, enrofloxacin, and nalidixic acid (9). Of 42 strains, all from different animals, 10 were resistant to one or more of the tested antimicrobials (9). E. coli 702 was resistant to sulfonamides (MIC, >2,048 μg/ml) and streptomycin (MIC, 32 μg/ml). The sul1 and intI genes of class 1 integrons were detected using PCR with primers and conditions previously reported (6). Conjugation experiments and plasmid analysis showed that the integron resided on an approximately 80-kb conjugative plasmid. The variable region of the integron was amplified using a method described previously (7). The primers (5′-TGATGTTATGGAGCAGCAACGATG-3′ and 5′-CGCACAACCTCGTCGATATCACC-3′) hybridized to the conserved segments of the integron (illustrated in Fig. 1). The sequence of the variable region was subsequently determined. Sequencing was performed on a model 3100-Avant genetic analyzer (Applied Biosystems). Sequences were analyzed using the BioEdit program, the NCBI GeneBlast2 program, and the ClustalW program via the Internet.
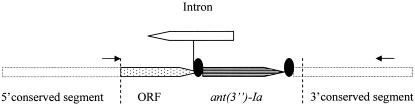
FIG. 1.
Organization of the variable region of the class 1 integron reported. The arrows indicate the direction of transcription. The primers used for amplification of the variable region are indicated by small arrows. The two broken vertical lines indicate the region sequenced (shown under accession number AY785243).
The variable region of the integron contained two unusual features: a group II intron and a novel gene cassette (Fig. 1). The ant(3”)-Ia cassette (5) was also detected, which is one of the most frequently found cassettes in MRIs. The novel gene cassette contained a 402-bp open reading frame of unknown function. The sequence showed 31% identity at protein level to a hypothetical protein encoded by a cassette found in the superintegron of Xanthomonas sp. strain CIP 102397 (11) (accession number AF324484).
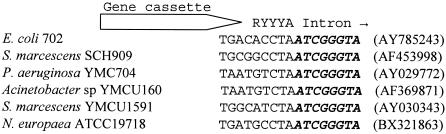
A 1,970-kb-sized group II intron, encoding a reverse transcriptase-like open reading frame on the complementary strand, was inserted downstream of the reading frame. The intron was equal to an intron found within a class 1 integron in a multiresistant clinical isolate of Serratia marcescens (SCH88050909) isolated in 1988 in Greece (1). Alignments showed four nucleotide differences, one of them producing an amino acid alteration (G187S). The intron was identically inserted in E. coli 702 and in S. marcescens SCH909, between the RYYYA and the AC of an attC site, as illustrated in Fig. 2. Another group II intron, distinct from the one reported, has been found within class 1 integrons in a Pseudomonas aeruginosa strain (8), in a S. marcescens strain (19), and in an Acinetobacter sp. strain (20). The group II intron was identically inserted in all three strains, in the same way as reported here (illustrated in Fig. 2). The presence of group II introns, inserted identically at the 5′ end of attC sites of gene cassettes, might indicate that this site is a preferred insertion site for such genetic elements.
FIG. 2.
Alignment of intron insertion points within gene cassettes in class 1 integrons. All introns are inserted to the 5′ end of an attC site. The same intron was found in the two upper strains. The same intron, but different from the first, is found in the three next strains. A third intron is present in the Nitrosomonas europaea ATCC19718 strain; noncoding DNA is present at the left side of the attC site in this strain.
A gene cassette from the SI of a Vibrio fischeri strain contains a 56-codon segment with homology to intron maturase (13) (accession number AY177199, cassette c667-2). This indicates that introns can be involved with cassettes in SIs as well.
It is suggested that some environmental organisms, like Nitrosomonas europaea, can serve as reservoirs of integron components since these species contain integrase-like genes and attC sites not associated with a gene cassette (1). An attC site located adjacent to a group II intron in an N. europaea strain (Acc no BX321863) differs by only one nucleotide when aligned with the attC of E. coli 702 (Fig. 3). This may be a further indication of a connection between environmental bacteria, introns and integron components.
FIG. 3.
Alignment of attC sites associated with introns in E. coli 702, S. marcescens SCH909, and N. europaea ATCC19718. The same intron was inserted adjacent to the attC sites in E. coli and S. marcescens, whereas a different intron was found in the N. europaea strain.
The intron of E. coli 702 was still present at the attC site in both the isolated strain and its transconjugant, following 3 months of continuous culture in nutrient broth at room temperature (recultured every fortnight). Further investigations will soon be carried out in our laboratory to investigate possible intron mobility to attC sites of gene cassettes.
Most MRIs so far characterized originate from bacteria isolated from environments where antibiotics are heavily used. This study shows that class 1 integrons can be found in bacteria occurring in “antibiotic-free” environments, located far away from human activities. This may indicate that class 1 integrons are more prevalent in nature than originally thought and that they have existed since before the antibiotic era.
During the last 15 years, since the discovery of the integron structure, hundreds of integrons have been investigated. Integron-borne group II introns have been detected only five times. Most integrons characterized to date originate from clinical isolates subjected to antibiotic selection force. The search for integrons in environments with no history of antibiotic exposure could perhaps lead to further findings of group II introns, and hopefully their role could be fully determined.
Nucleotide sequence accession number.
The sequence of the variable region of the integron was submitted to GenBank under accession number AY785243.
Acknowledgments
Atle Lillehaug (National Veterinary Institute), Henning Sørum, and Jostein Bjorland (The Norwegian School of Veterinary Science) are acknowledged for collaboration and constructive comments on this paper.
This work was supported by a grant (153080/120) from the Norwegian Research Council.
REFERENCES
- 1.Centrón, D., and P. H. Roy. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cousineau, B., S. Lawrence, D. Smith, and M. Belfort. 2000. Retrotransposition of a bacterial group II intron. Nature 404:1018-1021. [DOI] [PubMed] [Google Scholar]
- 3.Dai, L., and S. Zimmerly. 2002. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 30:1091-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 5.Hollingshead, S., and D. Vapnek. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenyltransferase. Plasmid 13:17-30. [DOI] [PubMed] [Google Scholar]
- 6.Kerrn, M. B., T. Klemmensen, N. Frimodt-Møller, and F. Espersen. 2002. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 50:513-516. [DOI] [PubMed] [Google Scholar]
- 7.L'Abèe-Lund, T. M., and H. Sørum. 2001. Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microb. Drug Resist. 7:253-262. [DOI] [PubMed] [Google Scholar]
- 8.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lillehaug, A., B. Bergsjø, J. Schau, T. Bruheim, T. Vikøren, and K. Handeland. 2005. Campylobacter spp., Salmonella spp., verocytotoxic Escherichia coli, and antibiotic resistance in indicator organisms in wild cervids. Acta Vet. Scand. 46:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recchia, G. D., and R. M. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5:389-394. [DOI] [PubMed] [Google Scholar]
- 11.Rowe-Magnus, D. A., A.-M. Guerout, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multi-resistant integrons. Proc. Natl. Acad. Sci. USA 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowe-Magnus, D. A., A.-M. Guerout, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43:1657-1669. [DOI] [PubMed] [Google Scholar]
- 13.Rowe-Magnus, D. A., A.-M. Guerout, L. Biskri, P. Bouige, and D. Mazel. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 13:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sallen, B., A. Rajoharison, S. Desvarenne, and C. Mabilat. 1995. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb. Drug Resist. 1:195-202. [DOI] [PubMed] [Google Scholar]
- 15.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 16.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 17.Stokes, H. W., D. B. O' Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 18.Sunde, M., and H. Sørum. 1999. Characterization of integrons in Escherichia coli of the normal intestinal flora of swine. Microb. Drug Resist. 5:279-287. [DOI] [PubMed] [Google Scholar]
- 19.Yum, J. H., D. Yong, K. Lee, H. S. Kim, and Y. Chong. 2002. A new integron carrying VIM-2 metallo-β-lactamase gene cassette in a Serratia marcescens isolate. Diagn. Microbiol. Infect. Dis. 42:217-219. [DOI] [PubMed] [Google Scholar]
- 20.Yum, J. H., K. Yi, H. Lee, D. Yong, K. Lee, J. M. Kim, G. M. Rossolini, and Y. Chong. 2002. Molecular characterization of metallo-β-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the blaVIM-2 gene cassettes. J. Antimicrob. Chemother. 49:837-840. [DOI] [PubMed] [Google Scholar]