Abstract
Resistance to fluoroquinolones in urinary tract infection (UTIs) caused by Escherichia coli is associated with multiple mutations, typically those that alter DNA gyrase and DNA topoisomerase IV and those that regulate AcrAB-TolC-mediated efflux. We asked whether a fitness cost is associated with the accumulation of these multiple mutations. Mutants of the susceptible E. coli UTI isolate Nu14 were selected through three to five successive steps with norfloxacin. Each selection was performed with the MIC of the selected strain. After each selection the MIC was measured; and the regions of gyrA, gyrB, parC, and parE, previously associated with resistance mutations, and all of marOR and acrR were sequenced. The first selection step yielded mutations in gyrA, gyrB, and marOR. Subsequent selection steps yielded mutations in gyrA, parE, and marOR but not in gyrB, parC, or acrR. Resistance-associated mutations were identified in almost all isolates after selection steps 1 and 2 but in less than 50% of isolates after subsequent selection steps. Selected strains were competed in vitro, in urine, and in a mouse UTI infection model against the starting strain, Nu14. First-step mutations were not associated with significant fitness costs. However, the accumulation of three or more resistance-associated mutations was usually associated with a large reduction in biological fitness, both in vitro and in vivo. Interestingly, in some lineages a partial restoration of fitness was associated with the accumulation of additional mutations in late selection steps. We suggest that the relative biological costs of multiple mutations may influence the evolution of E. coli strains that develop resistance to fluoroquinolones.
Treatment of urinary tract infections (UTIs) is a major community indication for antibiotic usage (10, 14, 26) and affects 40 to 50% of women at some point during their lifetimes. Escherichia coli is the cause of more than 80% of community-acquired cases of uncomplicated UTIs (20). Fluoroquinolones are potent broad-spectrum antibacterial agents that are frequently used for the treatment of UTIs (14, 26, 27). Fluoroquinolones bind to DNA gyrase (topoisomerase II) and/or DNA topoisomerase IV and inhibit their activities in the control of DNA unwinding and supercoiling (25).
The management of UTIs is complicated by increasing rates of antimicrobial resistance. In the case of fluoroquinolone resistance among E. coli isolates, important mechanisms of resistance include mutations that reduce the affinity for the antibiotic targets, DNA gyrase and/or DNA topoisomerase IV (12, 13, 23, 31, 33, 34), and mutations that reduce fluoroquinolone accumulation in the cell, either by decreased uptake through outer membrane porins or by increased expression of drug efflux systems (1, 2, 29, 30, 32, 36). Target alterations due to mutations in the gyrA, gyrB, parC, and parE genes have been described, most of which are located in a small region of either gyrA or parC, known as the quinolone resistance-determining region.
Individual resistance mutations isolated experimentally confer only low-level resistance to fluoroquinolones. Experimental evidence from in vitro selection studies shows that higher-level fluoroquinolone resistance can be built up successively through the accumulation of several independently acquired mutations, including both target and efflux mutations. Similarly, evidence from studies with clinical isolates supports the conclusion that higher-level resistance is associated with multiple resistance mutations (23). This raises the question of whether the accumulation of multiple mutations is associated with a fitness cost for the resistant bacteria. For several other antibiotic resistance phenotypes, it has been shown that single mutations are frequently associated with fitness costs both in vitro and in vivo (5, 6, 28). Accordingly, we selected several lineages of E. coli for reduced susceptibility to successively higher levels of the fluoroquinolone norfloxacin and measured the associated changes in fitness. The exposure of bacteria to low levels of antibiotic has been postulated to be a significant factor in the development of clinical resistance (3, 4, 41). To mimic this situation we made each selection at the MIC of the selected strain.
MATERIALS AND METHODS
Bacterial strains.
E. coli cystitis isolate Nu14 (15, 23, 24) was used for the in vitro selection of norfloxacin-resistant mutants. Nu14 (15, 24) is susceptible to norfloxacin (Etest MIC = 0.094 μg/ml) and to other antibiotics, including nalidixic acid, ciprofloxacin, tetracycline, chloramphenicol, amdinocillin, and trimethoprim (Table 1). E. coli ATCC 25922, Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains in antibiotic resistance testing.
TABLE 1.
Genetic identification, MIC, and in vitro fitness of first-step mutants
| Strain | Mutationa | MIC (μg/ml)b
|
Fitnessc | |||||
|---|---|---|---|---|---|---|---|---|
| NOR | CIP | NAL | TET | CHL | MEC | |||
| Nu14 | Wild type | 0.094 | 0.016 | 1.5 | 0.75 | 3 | 0.064 | 1.000 |
| Nu98 | gyrA D82G (1)d | 0.38 | 0.047 | 16 | 0.75 | 3 | 0.064 | ND |
| Nu118 | gyrA S83A (1) | 0.25 | 0.094 | 16 | 0.5 | 2 | 0.064 | 1.001 |
| Nu41 | gyrA D87Y (1) | 0.5 | 0.19 | 96 | 0.75 | 4 | 0.047 | 0.995 |
| Nu79 | gyrA D87G (2) | 1 | 0.125 | 96 | 0.75 | 3 | 0.064 | 0.999 |
| Nu91 | gyrA A119E (1) | 0.5 | 0.125 | 64 | 0.75 | 3 | 0.047 | ND |
| Nu87 | gyrB R389S (1) | 0.25 | 0.032 | 2 | 0.75 | 6 | 0.094 | ND |
| Nu71 | gyrB S464F (1) | 0.50 | 0.094 | 6 | 1.5 | 3 | 0.064 | 1.023 |
| Nu141 | gyrB S464Y (1) | 0.75 | 0.38 | 8 | 1.5 | 6 | 0.064 | ND |
| Nu17 | gyrB Δ9 nte (2) | 0.25 | 0.064 | 3 | 0.75 | 4 | 0.064 | 1.009 |
| Nu96 | marR + 1 ntf (1) | 0.25 | 0.032 | 3 | 1.5 | 12 | 0.094 | ND |
| Nu93 | marR R77L (1) | 0.5 | 0.064 | 3 | 2 | 12 | 0.125 | 0.987 |
| Nu18 | marR R94H (2) | 0.38 | 0.064 | 3 | 2 | 24 | 0.125 | 0.997 |
| Nu135 | marOR Δ13 ntg (1) | 0.19 | 0.047 | 3 | 1.5 | 16 | 0.094 | 1.003 |
Six genes or regions (gyrA, gyrB, parC, parE, marOR, and acrR) were sequenced as described in Materials and Methods, resulting in each case in the identification of the one mutation listed. Amino acid substitution mutations, e.g., D82G, are written in the form original amino acid (D), amino acid position (82), mutant amino acid (G).
The MIC is the modal value for at least five measurements made by Etest. MICs for trimethoprim were also measured but were not altered in any mutants (Nu14 trimethoprim MIC, 0.5). NOR, norfloxacin; CIP, ciprofloxacin; NAL, nalidixic acid; TET, tetracycline; CHL, chloramphenicol; MEC, amdinocillin.
Fitness is expressed as 1 + S, the selection coefficient per generation determined in growth competition assays in urine (see Materials and Methods). ND, not determined. The mean standard error of the fitness values is 0.014. Values can be read as proportional to the growth rate of Nu14.
Values in parentheses are the number of independent isolates (from different cultures) with each mutation.
The mutation is a deletion of 9 nucleotides (nt; nucleotides 1427 to 1435) within the gyrB-coding sequence, where nucleotide 1 is the first nucleotide of the initiation codon. The deletion removes the underlined nucleotides from the sequence TGT GGT ATC GGT; and as a result, three amino acids, G477, I478, and G479, are predicted to be lost from the GyrB protein.
Insertion of G into a run of 3 G residues at nucleotides 203 to 205, which results in a predicted reading frame shift and an altered protein sequence from amino acid 70 (A70R) in MarR.
The mutation is a deletion of 13 nucleotides from position −3 to +10, where position 1 is the first nucleotide (underlined) of the marR-coding sequence (GTG). The deleted nucleotides are numbered 1617140 to 1617152 in the E. coli K-12 MG1655 genome sequence (note that the initiation codon of marR [GTG] is incorrectly identified in the genome annotation of MG1655 [GenBank accession number NC_000913] as ATG [codon 21]).
Media and growth conditions.
The liquid medium used for bacterial growth was Luria broth (LB) or sterile filtered urine. Midstream urine, collected from several healthy individuals at different time points during a day, was pooled, centrifuged at 4,000 × g, filtered through a 0.2-μm-pore-size filter (Sarstedt, Nümbrecht, Germany), stored at 4°C, and used within 2 weeks. Only urine with a pH of >5 was used in these experiments. The solid medium used was either Luria broth supplemented with 1.5% agar (LA; Oxoid Ltd., Basingstoke, England) or Mueller-Hinton medium (Sigma Aldrich, Stockholm, Sweden). Strains were grown at 37°C, and liquid cultures were incubated without shaking.
Determination of MICs.
The MICs of norfloxacin, ciprofloxacin, nalidixic acid, chloramphenicol, tetracycline, amdinocillin, and trimethoprim were determined for each mutant by Etest, according to the instructions of the manufacturer (AB BIODISK, Solna, Sweden). Etests were performed on Mueller-Hinton agar plates, and the plates were incubated for 16 to 18 h at 37°C. In order to check the performance of the Etest with respect to the correct handling of the system, the recommended National Committee for Clinical Laboratory Standards (NCCLS) quality control reference strains for tests with the antibiotics concerned were tested on all occasions (see bacterial strains above). The variations in the Etest results for the American Type Culture Collection (ATCC) strains were measured on at least 10 independent occasions for the antibiotics tested and were always within ±1 concentration step.
Stepwise selection of norfloxacin-resistant mutants.
Several independent cultures of the strain to be selected (initially Nu14) were grown overnight in LB. From each culture, approximately 108 CFU was spread on LA plates containing, for the first-step selection, norfloxacin at 0.1 μg/ml (the approximate MIC of Nu14). In this initial selection, colonies appeared at a frequency of 5 × 10−8 cells. All subsequent selections were made on norfloxacin at the approximate MIC of the strain being selected. The plates were incubated overnight at 37°C. Colonies were counted, picked, and streaked for the selection of single colonies on the selective medium. After each selection step, mutants were collected (10 from each independent culture) and frozen at −80°C in LB containing 7% dimethyl sulfoxide (Sigma Aldrich). The resistance patterns among the mutants were initially screened by streaking them on LB plates containing different levels of norfloxacin, nalidixic acid, and chloramphenicol. On the basis of the differences in antibiotic susceptibilities and their independence in origin from one another, a few mutants were chosen after each selection step for further analysis and future selections. Successive stepwise selections were continued for three to five steps.
PCR amplification and DNA sequencing.
The oligonucleotide primers used for PCR amplification and DNA sequencing, together with details of the specific regions sequenced in each gene, have been described previously (23). The PCR primers were designed to amplify the quinolone resistance-determining regions (7, 11, 12, 21, 35, 38-40) of gyrA, gyrB, parC, and parE and the whole of the marR and acrR genes. Conditions for PCR amplification and DNA sequencing were as described previously (23).
OST.
Tolerance to the organic solvents hexane and cyclohexane is indicative of above-normal efflux by the AcrAB-TolC efflux pump (37). Organic solvent tolerance (OST) was measured in the selected mutants, as described previously (23).
Measurement of fitness in vitro.
To determine the biological cost associated with different resistance mutations in vitro, each norfloxacin-resistant mutant was competed against Nu14 in urine. The experiment is a growth competition at 37°C in sterile urine, in which the outcome is evaluated as the change in the ratios of the competing strains as a function of the number of generations. Each competition was performed in triplicate by using independent starting cultures of each of the competing strains. In cases in which one of the outcomes differed significantly from the others, the experiment was performed again. The results are thus an average of at least three independent experiments in each case. Each competition was initiated from overnight cultures grown in urine, diluted in 0.9% NaCl, and mixed at a cell density of ∼5,000 CFU/ml in 2 ml urine. The tubes were incubated without shaking overnight (24 h) to complete one cycle of competition (approximately 18 cell doublings). After each competition cycle the mixed culture was serially diluted and ∼300 CFU was plated onto LB and LB plus 3 μg/ml nalidixic acid. This level of nalidixic acid was determined empirically to inhibit the growth of Nu14 but to permit the growth of each of the mutants with reduced norfloxacin susceptibility. After overnight incubation at 37°C, the ratio of the number of sensitive Nu14 cells to the number of cells of the resistant mutant was calculated. Successive cycles of growth competition were made by transferring on each day ∼5,000 CFU from a competition tube to 2 ml urine and performing another overnight incubation. Up to six successive competition cycles were made in each experiment. The in vitro selection coefficient (S) per generation of each mutant was calculated by linear regression as the slope [(number of mutant cells/number of wild-type cells)/(number of generations/number of cycles)] × ln 2. Relative fitness (with respect to that of the starting strain Nu14) was defined as 1 + S.
Measurement of fitness in a mouse UTI model.
All strains were checked for type 1 fimbriae by a mannose-sensitive hemagglutination assay (9) prior to in vivo testing. Single-strain infections showed that the individual strains could cause infections. The relative fitness of the wild-type and selected strains was measured in an ascending UTI model in mice by a published protocol (16, 22). Briefly, wild-type Nu14 and selected norfloxacin-resistant bacteria were cultured overnight on 5% horse blood agar plates (no. 677; SSI Diagnostica, Hillerød, Denmark). One colony from each culture was suspended in 0.5 M phosphate-buffered saline (pH 7.4; no. 3892; SSI Diagnostica) to a cell density of 109 to 1010 CFU/ml determined with a colorimeter (Sherwood colorimeter 254). Nu14 was mixed with each of the norfloxacin-resistant bacteria in a 1:1 ratio. Infections were made in outbred female albino Ssc-CF1 mice (weight, 30 ± 2 g; Hvidesten, Allerød, Denmark). Anesthetized mice (five mice per competition) were inoculated transurethrally with 0.05 ml of the bacterial suspension by the use of plastic catheters, with 3 × 107 to 5 × 108 CFU/mouse placed into the bladder. Mice were killed by cervical dislocation 1, 3, or 7 days after infection. The bladder was removed and homogenized in 0.5 ml sterile 0.5 M phosphate-buffered saline. The bladder suspension was diluted in 0.9% saline (0.9% NaCl; no. 8901; Diagnostica) and plated on LA and on LA plus 3 μg/ml nalidixic acid to distinguish Nu14 from the norfloxacin-resistant mutant. A competition index was calculated as the ratio of the number of mutant cells/number of wild-type cells on day 3. Animal experiments were carried out at Statens Serum Institut, Copenhagen, Denmark, in compliance with institutional and national regulations.
RESULTS
Selection and characterization of first-step mutants.
Norfloxacin-resistant colonies were selected from six independent cultures of Nu14. A total of 102 colonies were purified on the selection media and tested for norfloxacin MICs by Etest. On the basis of the increased MICs, 33 mutants were chosen for DNA sequencing of the genes and regions associated with mutations to fluoroquinolone resistance. A single mutation was identified in each of these 33 mutants (Table 1). In all, 13 different mutations were identified: 5 in gyrA, 4 in gyrB, and 4 in marOR. None were identified in parC, parE, or acrR. Each mutant type was subjected to tests for the MICs of several different antibiotics (Table 1). The norfloxacin MICs of the mutants were increased 2- to 10-fold (mean, 4-fold) over that of Nu14, and all mutants showed similar relative increases in the MIC for ciprofloxacin. The MICs for nalidixic acid were also increased, in particular, for gyrA mutants. A correlation between single mutations in gyrA and high-level resistance to nalidixic acid has been noted previously for clinical UTI isolates (23). The MIC for chloramphenicol was increased in each of the four marOR mutants, as expected for efflux mutants (Table 1). OST was also increased in the four marOR mutants but not in any of the gyrA or gyrB mutants, supporting the suggestion that the four marOR mutations increase multidrug efflux. The MICs for amdinocillin, trimethoprim, and tetracycline were unaltered in the mutants. The relative fitness of eight of the mutants was measured in vitro (in urine) in growth competition assays against Nu14. None of the mutants showed any significant deviation in fitness from Nu14 (Table 1). Six mutants, two gyrA, two gyrB, and two marR mutants, were chosen as starting points for subsequent selections and are referred to as lineages 1 to 6, respectively.
MICs, mutations, and fitness in lineages 1 to 6.
The genealogies of lineages 1 to 6, together with the corresponding MICs for norfloxacin and other antibiotics, are shown in Table 2. Each of the selected strains listed in Table 2 was subject to DNA sequence analysis of genes and regions associated with reduced susceptibility to fluoroquinolones and to competition experiments against the standard wild-type, Nu14 (Tables 3 to 5). Tables 3 to 5 include details for each of the mutations identified. All strains in which a mar mutation was identified had an increased MIC for chloramphenicol and were also positive for OST, supporting the suggestion that they have increased drug efflux. There are several interesting correlations between decreased susceptibility to norfloxacin and changes in susceptibilities to unrelated antibiotics (see Discussion). Fitness in vitro is presented as the growth rate per generation relative to that of Nu14. Each of the six first-step mutants had fitness values very close to the value for parental strain Nu14 (Table 1). Among the nine second-step mutants, six were fit within ±3% of the value for Nu14, close to the standard error of these measurements, while three were less fit, with generation times 13 to 16% slower than those of Nu14. Of 18 third-step mutants, only 3 retained fitness close to that of Nu14 (±2 to 3%), while the remaining 15 mutants had severe or very severe fitness losses, with generation times increased by between 6% and 55% (mean increase in generation time, 31%). The six fourth-step mutants tested all had reduced fitness, with generation times increased by between 4% and 44%. Two fifth-step mutants tested had generation times that increased by 23% and 43%, respectively, relative to that of Nu14.
TABLE 2.
MICs of strains selected stepwise for reduced susceptibility to norfloxacin
| Lineage and strain (step no.)b | MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|
| NOR | CIP | NAL | TET | CHL | MEC | |
| Parent (Nu14) | 0.064 | 0.016 | 1.5 | 0.75 | 3 | 0.064 |
| Lineage 1 | ||||||
| Nu118 (1) | 0.25 | 0.094 | 16 | 0.5 | 2 | 0.064 |
| PL668 (2) | 1 | 0.25 | 16 | 0.75 | 8 | 0.094 |
| PL3637 (3) | 2 | 0.38 | 24 | 0.25 | 1.5 | 0.032 |
| PL3622 (3) | 3 | 0.5 | 64 | 0.75 | 4 | 0.125 |
| PL2090 (3) | 1.5 | 0.38 | 256 | 1 | 6 | 0.25 |
| PL2055 (3) | 1.5 | 0.38 | 256 | 1 | 6 | 0.25 |
| PL787 (2) | 0.5 | 0.125 | 32 | 1 | 8 | 0.064 |
| PL3665 (3) | 1 | 0.25 | 32 | 1.5 | 8 | 0.094 |
| PL3687 (3) | 1 | 0.19 | 32 | 1.5 | 8 | 0.094 |
| Lineage 2 | ||||||
| Nu41 (1) | 0.5 | 0.19 | 96 | 0.75 | 4 | 0.047 |
| PL213 (2) | 1.5 | 0.5 | >256 | 0.5 | 4 | 0.064 |
| PL1660 (3) | 6 | 2 | >256 | 0.75 | 8 | 0.125 |
| PL1965 (4) | 16 | 3 | >256 | 1 | 24 | 0.19 |
| PL3511 (5) | 64 | 8 | >256 | 0.75 | 48 | 0.38 |
| PL3515 (5) | 32 | 3 | >256 | 0.75 | 16 | 0.125 |
| PL1710 (3) | 8 | 4 | >256 | 0.75 | 3 | 0.064 |
| Lineage 3 | ||||||
| Nu71 (1) | 0.5 | 0.094 | 6 | 0.75 | 4 | 0.047 |
| PL136 (2) | 1.5 | 0.38 | 8 | 1 | 12 | 0.094 |
| PL3543 (3) | 2 | 0.38 | 12 | 1 | 12 | 0.5 |
| PL3547 (3) | 3 | 0.5 | 8 | 1 | 12 | 0.094 |
| Lineage 4 | ||||||
| Nu17 (1) | 0.25 | 0.064 | 3 | 0.75 | 4 | 0.064 |
| PL490 (2) | 0.75 | 0.094 | 3 | 0.38 | 2 | 0.094 |
| PL1270 (3) | 1 | 0.25 | 64 | 0.38 | 3 | 0.064 |
| PL3563 (4) | 4 | 0.75 | >256 | 1 | 24 | 0.125 |
| Lineage 5 | ||||||
| Nu18 (1) | 0.38 | 0.064 | 3 | 2 | 24 | 0.125 |
| PL845 (2) | 2 | 0.38 | >256 | 3 | 24 | 0.125 |
| PL1773 (3) | 3 | 0.75 | >256 | 1.5 | 12 | 0.094 |
| PL1801 (3) | 4 | 1 | >256 | 2 | 64 | 4 |
| PL1900 (3) | 3 | 0.5 | >256 | 2 | 24 | 0.094 |
| PL3425 (4) | 4 | 0.75 | >256 | 2 | 32 | 0.094 |
| PL3460 (4) | 16 | 1.5 | >256 | 2 | 16 | 0.125 |
| PL858 (2) | 0.75 | 0.125 | 6 | 2 | 32 | 0.5 |
| PL1385 (3) | 4 | 1.5 | >256 | 2 | 24 | 0.5 |
| PL2113 (4) | 12 | 3 | >256 | 2 | 16 | 0.38 |
| PL2127 (4) | 16 | 4 | >256 | 1.5 | 12 | 0.38 |
| Lineage 6 | ||||||
| Nu135 (1) | 0.19 | 0.047 | 3 | 1.5 | 8 | 0.094 |
| PL923 (2) | 1 | 0.25 | >256 | 1 | 8 | 0.094 |
| PL1570 (3) | 1.5 | 0.38 | >256 | 1 | 6 | 0.064 |
| PL1645 (3) | 3 | 0.5 | >256 | 0.75 | 1.5 | 1 |
The MIC for trimethoprim was also measured for all selected strains and showed no significant variation as a function of selection from the value in the starting strain Nu14 (trimethoprim MIC, 0.5). See footnote b of Table 1 for definitions of the abbreviations.
The selection step at which a particular mutant was isolated is provided in parentheses.
TABLE 3.
Mutations selected and associated fitness in gyrA lineages
| Lineage and strain | Mutation at the following selection step:
|
Fitnessa | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| Lineage 1 | ||||||
| Nu118 | gyrA S83A | 1.001 | ||||
| PL668 | marR + 14 ntb | 0.996 | ||||
| PL3637 | nim-1c | 0.518 | ||||
| PL3622 | nim-2 | 0.512 | ||||
| PL2090 | nim-3 | 0.823 | ||||
| PL2055 | nim-4 | 0.845 | ||||
| PL787 | marR + 1 ntd | 1.000 | ||||
| PL3665 | nim-5 | 1.017 | ||||
| PL3687 | nim-6 | 1.022 | ||||
| Lineage 2 | ||||||
| Nu41 | gyrA D87Y | 0.995 | ||||
| PL213 | parE S458P | 0.834 | ||||
| PL1660 | marR + 1 nte | 0.688 | ||||
| PL1965 | gyrA D72G | 0.563 | ||||
| PL3511 | nim-7 | 0.770 | ||||
| PL3515 | nim-8 | 0.568 | ||||
| PL1710 | gyrA S83L | 0.940 | ||||
Fitness is expressed as 1 + S, the selection coefficient per generation determined in growth competition assays in urine (see Materials and Methods). The mean standard error of the fitness values is 0.014. Values can be read as proportional to the growth rate of Nu14.
A 14-nucleotide (nt) duplication (nucleotides 228 to 241) in the marR-coding sequence predicted to cause a reading frame shift.
nim, nonidentified mutation.
An additional A residue in a run of five A residues (nucleotides [nt] 181 to 185) in the marR-coding region predicted to cause a reading frame shift.
An additional G residue in a run of five G residues (nucleotides [nt] 204 to 208) in the marR-coding region predicted to cause a reading frame shift from codon 70 onwards.
TABLE 5.
Mutations selected and associated fitness in marR lineages
| Lineage and strain | Mutation at the following selection step:
|
Fitnessa | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Lineage 5 | |||||
| Nu18 | marR R94H | 0.997 | |||
| PL845 | gyrA D87G | 0.981 | |||
| PL1900 | marR + 1 ntb | 0.503 | |||
| PL3425 | nim-12 | 0.954 | |||
| PL3460 | nim-13 | 0.965 | |||
| PL858 | nim-14 | 0.869 | |||
| PL1385 | gyrA S83L | 0.848 | |||
| PL2113 | nim-15 | 0.767 | |||
| PL2127 | nim-16 | 0.835 | |||
| PL905 | gyrA D87G | 0.964 | |||
| PL1773 | nim-17d | 0.584 | |||
| PL1801 | nim-18 | 0.649 | |||
| PL1860 | nim-19 | 0.869 | |||
| Lineage 6 | |||||
| Nu135 | marR Δ13 ntc | 1.003 | |||
| PL923 | gyrA D87Y | 0.994 | |||
| PL1570 | nim-21 | 0.971 | |||
| PL1645 | nim-22 | 0.449 | |||
Fitness is expressed as 1 + S, the selection coefficient per generation determined in growth competition assays in urine (see Materials and Methods). The mean standard error of the fitness values is 0.014. Values can be read as growth rate proportional to the growth rate of Nu14.
An additional G in a run of five G residues (nucleotides [nt] 204 to 208) within the marR-coding sequence predicted to cause a reading frame shift.
Deletion of 13 nucleotides (nt; nucleotides −3 to +10 inclusive) spanning the start of the marR coding sequence.
nim, nonidentified mutation.
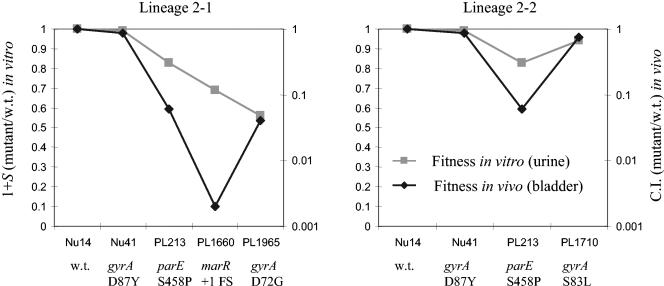
Relative fitness of lineage 2 strains in vitro and in vivo.
In lineage 2 we identified specific resistance-associated mutations for each of selection steps 1 to 4 and therefore chose lineage 2 for the measurement of relative fitness both in vitro and in vivo. In lineage 2-1 (Table 6; Fig. 1), an increasing loss of fitness in vivo was associated with the first three selection steps, but this was partially reversed by the fourth selection step. In contrast, in vitro the same strains showed a progressive decline in fitness with each of the four selection steps. In lineage 2-2, the pattern both in vivo and in vitro was a small loss of fitness associated with the first selection step, a further loss with the second step, and finally, a partial restoration of fitness associated with the third selection step (Table 6; Fig. 1).
TABLE 6.
Relative fitness in vitro and in vivo for lineage 2
| Lineage and strain | CIa in the bladder (in vivo) (SEb) | Fitnessc in urine (in vitro) |
|---|---|---|
| Lineage 2-1 | ||
| Nu41 | 0.86 (0.22) | 0.995 |
| PL213 | 0.06 (0.02) | 0.834 |
| PL1660 | 0.002 (0.002) | 0.688 |
| PL1965 | 0.04 (0.02) | 0.563 |
| Lineage 2-2 | ||
| Nu41 | 0.86 (0.22) | 0.995 |
| PL213 | 0.06 (0.02) | 0.834 |
| PL1710 | 0.74 (0.2) | 0.940 |
CI, competition index, calculated as the ratio of the number of mutant cells/number of wild-type Nu14 cells in mouse bladders 3 days after infection with equal numbers of each competing strain. Competition indices are the mean of the ratios for several bladders (Nu41, n = 12; PL213, n = 6; PL1660, n = 4; PL1965, n = 7; PL1710 n = 5).
SE, standard error of the mean for the competitions in mice.
Fitness in vitro is 1 + S (selection coefficient per generation) calculated from competitions between Nu14 and the mutant strain in urine, as described in Materials and Methods. The mean standard error of the fitness values is 0.014.
FIG. 1.
Fitness costs (and compensation), in vitro (squares) and in vivo (diamonds), associated with the accumulation of mutations causing a loss of susceptibility to norfloxacin. Each successive point on the x axis represents a successive selection step, with Nu14 being the parental strain starting point. The new mutation associated with each successive selection step is given on the x axis below the strain name. C.I., competition index; w.t., wild type.
DISCUSSION
Increases in norfloxacin MICs and breakpoints.
After three successive selection steps no strain (0 of 16 strains) had crossed the NCCLS-defined breakpoint for resistance to norfloxacin (≥16 μg/ml). Resistant strains were selected only after a fourth selection step (three of six strains) and a fifth selection step (two of two strains). This suggests that there is a reservoir of potential genetic alterations capable of causing small reductions in susceptibility to norfloxacin before the current breakpoint is reached.
Cross-resistance to unrelated antibiotics.
We found evidence for cross-resistance to other antibiotic classes among the norfloxacin-selected mutants. In lineage 1 third-step mutants PL2090 and PL2055, in which the norfloxacin MIC increased slightly from 1 to 1.5 μg/ml, have an increase in the amdinocillin MIC from 0.094 to 0.25 μg/ml (Table 2). The mutations responsible were not identified. In lineage 2, the second-, third-, fourth-, and fifth-step mutants (mutants PL213, PL1660, PL1965, and PL3511, respectively) showed parallel increases in the MICs for norfloxacin (1.5, 6, 16, and 64 μg/ml, respectively), chloramphenicol (4, 8, 24, and 48 μg/ml, respectively), and amdinocillin (0.064, 0.125, 0.19, and 0.38 μg/ml, respectively) (Table 2). The mutations associated with these selection steps are in parE, marR, gyrA, and nim-7, respectively (Table 3). Correlations between decreased susceptibility to norfloxacin and amdinocillin were also found among mutants (mutants PL845 → PL1801, Nu18 → PL858, and PL923 → PL1645) in lineages 5 and 6 (Table 2). The mutations causing these parallel changes in MICs were not identified. These correlations show that selection for decreased susceptibility to a fluoroquinolone may coselect for decreased susceptibility to an unrelated class of antibiotic. One possible explanation is that mutations that affect cell wall biosynthesis and that decrease susceptibility to amdinocillin may reduce the transport of quinolones into the cell.
Genetic locations of resistance mutations.
Among 16 independent first-step mutants, we identified one mutation in gyrA (n = 6), gyrB (n = 5), and marOR (n = 5) in each strain (Table 1). Among second-step mutants we identified new mutations in gyrA (n = 3), marR (n = 3), and parE (n = 1) in seven of nine strains. Among third-step mutants we identified mutations in gyrA (n = 3) and marR (n = 2) in only 5 of 18 strains. Mutations were also identified in two of six fourth-step mutants, one each in gyrA and marOR. No mutation was identified in either of the two fifth-step mutants tested. In these last two mutants the entire coding sequences of gyrA, gyrB, parC, and parE were sequenced, ruling out the possibility that a novel mutation within these coding sequences was responsible for their high norfloxacin MICs. Thus, mutations in genes and regions known to be associated with fluoroquinolone resistance were identified in virtually all (23 of 25) first- and second-step mutants but in less than one-third (7 of 26) of third-, fourth-, and fifth-step mutants. The paucity of mutations identified in the later selection steps suggests that the reservoir of potential mutations capable of reducing susceptibility to fluoroquinolones may be significantly larger than is currently appreciated. Nu14 is refractive to transduction with bacteriophage P1, and so we have not attempted to genetically map these unknown mutations.
Genetic spectrum of norfloxacin resistance mutations.
Approximately one-third of the first-step mutations arose in gyrB, a class which almost never appears in fluoroquinolone-resistant clinical isolates. A possible explanation for their absence in clinical isolates is that they have a high fitness cost, although this was not apparent, at least in vitro, among the mutants selected here (Table 1). A second possibility is that gyrB mutations are selected only at very low antibiotic concentrations, like those used here, and that in the clinical situation, selection at a high antibiotic concentration is more usual, precluding their emergence. In support of the latter possibility, no gyrB mutations were selected after the first selection step. A second feature of the mutation spectrum is the absence of mutations in parC. Among the fluoroquinolone-resistant E. coli UTI isolates, parC mutations were identified in 25 of 30 resistant isolates (23). One possibility is that the frequency of parC mutations may increase when selection is for higher-level resistance and that the selections in this study do not meet this criterion. As defined by the NCCLS breakpoint, only five of the mutants selected here would be defined as resistant to norfloxacin. An alternative possibility is that parC mutations arise more frequently in strains that already carry a double mutation in gyrA. Thus, among 25 fluoroquinolone-resistant UTI isolates with parC mutations, all but 2 also carried double mutations at positions 83 and 87 in gyrA (23). Only one of the strains isolated in this study had acquired double mutations in gyrA at positions 83 and 87 (PL1710, lineage 2-2; Table 3). A third feature of the mutation spectrum is that mutations in marR arose frequently (in all six lineages), whereas no mutations arose in acrR, although such mutations are associated with fluoroquinolone resistance in clinical isolates (23, 35, 36). Finally, most mutations selected in the third and subsequent steps were not identified, suggesting the existence of other common classes of mutations that reduce E. coli susceptibility to norfloxacin.
Fitness of mutant strains.
Our starting strain for lineage selection, Nu14, belongs to the B2 phylogenetic group of E. coli, a group associated with a high degree of virulence (8). Previous studies of clinical E. coli isolates have shown a negative correlation between fluoroquinolone resistance and bacterial virulence (17-19). Analysis of those data suggests that the negative correlation may be due to the preferential emergence of resistance in phylogenetically distinct strains with an intrinsically low level of virulence, and there is no evidence for any loss of virulence associated with the acquisition of resistance (17, 18). Furthermore, it should be noted that virulence and fitness are not directly comparable parameters.
To determine whether the accumulation of resistance-associated mutations is associated with a loss of fitness, each of the mutants was tested in a growth competition assay in urine against the parental Nu14 strain. In addition, strains from lineage 2 were also tested against Nu14 in a mouse UTI model. In general, both in vitro and in vivo, there was little or no difference in fitness between Nu14 and any of the first-step mutants (Tables 1 and 6). In contrast, 3 of 9 second-step mutants, 15 of 18 third-step mutants, and 6 of 8 fourth- and fifth-step mutants suffered large decreases in growth rates relative to that of Nu14 (Tables 3 to 5). In many cases the loss of fitness was extreme, with the generation times being doubled. One can therefore conclude that the accumulation of mutations that decrease susceptibility to norfloxacin, in particular, three or more mutations, is strongly associated with a severe loss of bacterial fitness, defined as a decrease in the growth rate. However, a closer examination of the data reveals a more interesting and complicated picture (Fig. 2). Thus, in lineages 2-1 and 2-2, the loss of fitness, both in vitro and in vivo (Fig. 1), associated with the initial accumulation of mutations is partially reversed by later mutations (Tables 3 and 6). A similar pattern of a loss of fitness followed by a partial restoration of fitness was also seen in lineage 5 (Fig. 2). This suggests the possibility that particular combinations of resistance-associated mutations can effect a partial compensation for the fitness losses associated with individual mutations.
FIG. 2.
Graphic representation of the relative fitness in vitro of each of the mutants selected and tested as a function of the number of successive selection steps. Different symbols (squares, diamonds, triangles, etc.) represent different sublineages (Tables 3, 4, and 5).
The reversal of fitness costs associated with the accumulation of mutations is interesting in terms of the clinical development of fluoroquinolone resistance. Thus, if the multiple mutations required for resistance impose a severe fitness cost, resistant bacteria will be under selection pressure to avoid, minimize, or compensate for these costs. A naïve expectation is that as resistance-associated mutations accumulate, there could be a parallel accumulation of fitness-compensating mutations either within or outside of the genes associated with resistance. In the natural setting, these fitness-compensatory mutations might be acquired in the time intervals between antibiotic selections, as has been shown to occur in selections made in the laboratory in the absence of antibiotic selection (5, 28). A general conclusion from these published experiments is that fitness compensation is not necessarily associated with any loss of antibiotic resistance. Our experiments differed from those described above, in that we made each successive selection in the presence of the antibiotic. Thus, we may have selected for a subset of the potential fitness-compensatory mutations, i.e., those that are also associated with increased antibiotic resistance. Whether the apparent coselection of fitness-compensating and resistance mutations observed in this study is relevant to the clinical evolution of fluoroquinolone resistance will require further genetic analysis.
TABLE 4.
Mutations selected and associated fitness in gyrB lineages
| Lineage and strain | Mutation at the following selection step:
|
Fitnessa | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Lineage 3 | |||||
| Nu71 | gyrB S464F | 1.023 | |||
| PL136 | marR Δ1 ntb | 1.030 | |||
| PL3543 | nim-9 | 0.488 | |||
| PL3547 | nim-10 | 0.832 | |||
| Lineage 4 | |||||
| Nu17 | gyrB Δ9 nt | 1.009 | |||
| PL490 | nim-11c | 0.868 | |||
| PL1270 | gyrA D87G | 0.846 | |||
| PL3563 | marR Δ113 ntd | 0.841 | |||
Fitness is expressed as 1 + S, the selection coefficient per generation determined in growth competition assays in urine (see Materials and Methods). The mean standard error of the fitness values is 0.014. Values can be read as proportional to the growth rate of Nu14.
Deletion of an A residue in a run of two A residues (codons 133 and 134) in marR predicted to cause a reading frame shift. nt, nucleotide.
PL490 has an OST phenotype, suggesting that the nim-11 mutation causes increased drug efflux, although its chloramphenicol MIC was not increased. nim, nonidentified mutation.
Deletion of 113 nucleotides (nt; nucleotides −34 to +79 inclusive) spanning the start of marR.
Acknowledgments
This work was supported by grants from Vetenskapsrådet (The Swedish Research Council) to D.H. and from the European Union 5th Framework (DEAR project) to D.H. and N.F.-M. D.S. is on a postdoctoral stipend supported by the European Union.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baquero, F. 2001. Low-level antibiotic resistance, p. 117-136. In D. Hughes and D. I. Andersson (ed.), Antibiotic development and resistance. Taylor and Francis, London, United Kingdom.
- 4.Baquero, F., and M. C. Negri. 1997. Strategies to minimize the development of antibiotic resistance. J. Chemother. 9(Suppl. 3):29-37. [PubMed] [Google Scholar]
- 5.Bjorkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 7.Breines, D. M., S. Ouabdesselam, E. Y. Ng, J. Tankovic, S. Shah, C. J. Soussy, and D. C. Hooper. 1997. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob. Agents Chemother. 41:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eden, C. S., L. Hagberg, L. A. Hanson, T. Korhonen, H. Leffler, and S. Olling. 1981. Adhesion of Escherichia coli in urinary tract infection. Ciba Found. Symp. 80:161-187. [DOI] [PubMed] [Google Scholar]
- 10.Fihn, S. D. 2003. Clinical practice. Acute uncomplicated urinary tract infection in women. N. Engl. J. Med. 349:259-266. [DOI] [PubMed] [Google Scholar]
- 11.Friedman, S. M., T. Lu, and K. Drlica. 2001. Mutation in the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob. Agents Chemother. 45:2378-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heisig, P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40:879-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heisig, P., and R. Tschorny. 1994. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob. Agents Chemother. 38:1284-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooton, T. M. 2003. The current management strategies for community-acquired urinary tract infection. Infect. Dis. Clin. N. Am. 17:303-332. [DOI] [PubMed] [Google Scholar]
- 15.Hultgren, S. J., W. R. Schwan, A. J. Schaeffer, and J. L. Duncan. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 54:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hvidberg, H., C. Struve, K. A. Krogfelt, N. Christensen, S. N. Rasmussen, and N. Frimodt-Moller. 2000. Development of a long-term ascending urinary tract infection mouse model for antibiotic treatment studies. Antimicrob. Agents Chemother. 44:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J. R., M. A. Kuskowski, T. O'Bryan, R. Colodner, and R. Raz. 2005. Virulence genotype and phylogenetic origin in relation to antibiotic resistance profile among Escherichia coli urine sample isolates from Israeli women with acute uncomplicated cystitis. Antimicrob. Agents Chemother. 49:26-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. R., M. A. Kuskowski, K. Owens, A. Gajewski, and P. L. Winokur. 2003. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J. Infect. Dis. 188:759-768. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. R., C. van der Schee, M. A. Kuskowski, W. Goessens, and A. van Belkum. 2002. Phylogenetic background and virulence profiles of fluoroquinolone-resistant clinical Escherichia coli isolates from The Netherlands. J. Infect. Dis. 186:1852-1856. [DOI] [PubMed] [Google Scholar]
- 20.Kahlmeter, G. 2003. An international survey of the antimicrobial susceptibility of pathogens from uncomplicated urinary tract infections: the ECO.SENS Project. J. Antimicrob. Chemother. 51:69-76. [DOI] [PubMed] [Google Scholar]
- 21.Kern, W. V., M. Oethinger, A. S. Jellen-Ritter, and S. B. Levy. 2000. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 44:814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerrn, M. B., N. Frimodt-Moller, and F. Espersen. 2003. Effects of sulfamethoxazole and amdinocillin against Escherichia coli strains (with various susceptibilities) in an ascending urinary tract infection mouse model. Antimicrob. Agents Chemother. 47:1002-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komp Lindgren, P., A. Karlsson, and D. Hughes. 2003. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob. Agents Chemother. 47:3222-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langermann, S., R. Mollby, J. E. Burlein, S. R. Palaszynski, C. G. Auguste, A. DeFusco, R. Strouse, M. A. Schenerman, S. J. Hultgren, J. S. Pinkner, J. Winberg, L. Guldevall, M. Soderhall, K. Ishikawa, S. Normark, and S. Koenig. 2000. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 181:774-778. [DOI] [PubMed] [Google Scholar]
- 25.Marians, K. J., and H. Hiasa. 1997. Mechanism of quinolone action. A drug-induced structural perturbation of the DNA precedes strand cleavage by topoisomerase IV. J. Biol. Chem. 272:9401-9409. [DOI] [PubMed] [Google Scholar]
- 26.Naber, K. G. 2000. Survey on antibiotic usage in the treatment of urinary tract infections. J. Antimicrob. Chemother. 46(Suppl. 1):49-52. [PubMed] [Google Scholar]
- 27.Naber, K. G. 2000. Treatment options for acute uncomplicated cystitis in adults. J. Antimicrob. Chemother. 46(Suppl. 1):23-27. [PubMed] [Google Scholar]
- 28.Nagaev, I., J. Bjorkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 29.Oethinger, M., I. Podglajen, W. V. Kern, and S. B. Levy. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan, X. S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, Y. H., J. H. Yoo, D. H. Huh, Y. K. Cho, J. H. Choi, and W. S. Shin. 1998. Molecular analysis of fluoroquinolone-resistance in Escherichia coli on the aspect of gyrase and multiple antibiotic resistance (mar) genes. Yonsei Med. J. 39:534-540. [DOI] [PubMed] [Google Scholar]
- 33.Perichon, B., J. Tankovic, and P. Courvalin. 1997. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1166-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitz, F. J., M. E. Jones, B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, A. Fluit, J. Verhoef, U. Hadding, H. P. Heinz, and K. Kohrer. 1998. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob. Agents Chemother. 42:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webber, M. A., and L. J. Piddock. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob. Agents Chemother. 45:1550-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamagishi, J., H. Yoshida, M. Yamayoshi, and S. Nakamura. 1986. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol. Gen. Genet. 204:367-373. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, X., and K. Drlica. 2002. Restricting the selection of antibiotic-resistant mutant bacteria: measurement and potential use of the mutant selection window. J. Infect. Dis. 185:561-565. [DOI] [PubMed] [Google Scholar]