Abstract
Quinolone-resistant Streptococcus agalactiae bacteria were recovered from single-patient isolates and found to contain mutations in the gyrase and topoisomerase IV genes. Pulsed-field gel electrophoresis demonstrated that four isolates from the same long-term care facility were closely related; in seven cases, quinolone-resistant Haemophilus influenzae and S. agalactiae bacteria were isolated from the same patient.
Although disease caused by Streptococcus agalactiae (group B streptococcus [GBS]) in infants and during pregnancy is well documented, the epidemiology of patients infected with this bacterium is changing to include nonpregnant and elderly adults (1-3, 8, 14, 16). These patients often have significant underlying illness and reside in long-term care facilities (LTCFs) (5). While GBS remains fully susceptible to penicillin, antibiotic susceptibility surveys demonstrate increasing prevalences of resistance to erythromycin and clindamycin (4, 8, 17). Recently, quinolone-resistant GBS was detected (6). Since LTCFs can also have problems with quinolone-resistant Haemophilus influenzae (7, 9), we have looked for and found coisolation of quinolone-resistant H. influenzae with GBS from the same patients.
Antimicrobial susceptibility and clinical sites of all GBS isolates processed by our clinical microbiology laboratory from 1999 to 2002 were recorded. This laboratory receives clinical specimens from patients in our hospital and from those in several affiliated LTCFs. Levofloxacin resistance in GBS was initially identified by disk diffusion methodology. The isolates were further characterized by the infectious disease research laboratory using Etest methodology (AB Biodisk, Piscataway, N.J.) and broth microdilution (STP1 and GPN Sensititre plates; Trek Diagnostic Systems, Inc., Westlake, OH) for susceptibility to erythromycin, clindamycin, ampicillin, penicillin, levofloxacin, moxifloxacin, ciprofloxacin, gatifloxacin, gemifloxacin, and garenoxacin according to manufacturers' recommendations. NCCLS susceptibility breakpoints were used for all antibiotics (10). Since only urinary isolates were routinely tested for levofloxacin susceptibility by our clinical microbiology laboratory, we retrospectively measured levofloxacin susceptibility on all blood isolates collected from 1999 to 2002. Levofloxacin susceptibility was determined with all GBS isolates coisolated with levofloxacin-resistant H. influenzae isolates. Patient charts corresponding to levofloxacin-resistant GBS (LR-GBS) isolates were reviewed for demographics, comorbid conditions, antibiotic use, invasive device use, antibiotic susceptibility of organisms isolated, and determination of colonization or infection.
Pulsed-field gel electrophoresis (PFGE) analysis was performed on 23 LR-GBS isolates following Sma1 digestion, program 15 for 19.7 h, with analysis using Genepath (Bio-Rad Laboratories, Hercules, CA) and pulsotypes as described previously (15). Nucleotide sequence determination was as described in reference 7 except that cell lysates were incubated at 4°C overnight prior to boiling.
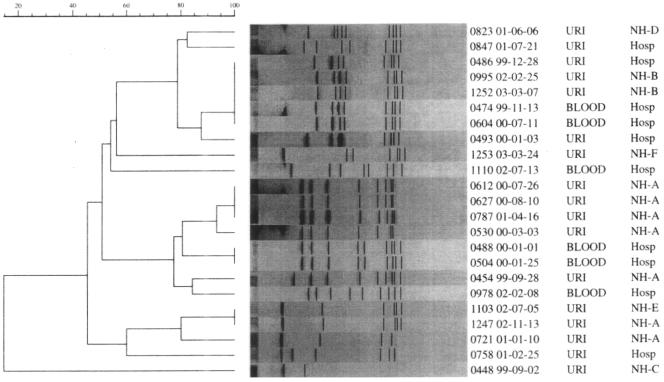
The total of 914 GBS isolates obtained during the survey period consisted of 462 urine isolates, 46 blood isolates, 172 respiratory isolates, 204 skin and soft tissue isolates, and 30 isolates from other body fluids. Resistances to clindamycin and erythromycin, determined by disk diffusion methodology, were 34.6% and 16%, respectively (Table 1). Of 508 urine and blood isolates, 5% (23 isolates; 17/462 from urine and 6/46 from blood) were LR-GBS isolates. Thirteen of the 23 LR-GBS isolates were from patients residing in LTCFs; 7 of those were from the same LTCF, 4 of which had highly related PFGE profiles (Fig. 1). Analysis of the 23 LR-GBS isolates revealed two major clusters representing 9/23 isolates examined, which suggested nosocomial spread.
TABLE 1.
Clindamycin and erythromycin resistance of 914 group B streptococci by disk diffusiona
| Source or type of isolate | No. of isolates with indicated resistance/ total no. of isolates (%)
|
|
|---|---|---|
| Clindamycin | Erythromycin | |
| Urine | 65/462 (14) | 167/462 (36) |
| Blood | 7/46 (15) | 9/46 (19.5) |
| Respiratory | 40/172 (23) | 77/172 (44.7) |
| Skin/soft tissue | 29/204 (14) | 56/204 (27) |
| Other | 6/30 (20) | 8/30 (26.6) |
| Total | 147/914 (16) | 317/914 (34.6) |
Resistance was defined by the presence of a zone diameter of ≤15 mm for erythromycin and clindamycin (10).
FIG. 1.
Dendrograms of 23 LR-GBS isolates. The Molecular Analyst Fingerprinting and Fingerprinting Plus software programs (Bio-Rad) were used to compare PFGE patterns by utilizing unweighted-pair group arithmetic averages clustering techniques and Dice correlation with 1.9% band tolerance to generate dendrograms. Abbreviations: URI, urine; Hosp, New York Hospital Queens; NH, long-term care facility.
Female gender (19/23; median age, 80) was the only demographic parameter associated with LR-GBS. All had multiple comorbid conditions, including dementia, stroke, carcinoma, and diabetes mellitus. Information obtained from available charts revealed that 12/18 patients received prior quinolone therapy. Seven charts obtained from the same LTCF also revealed the coisolation of LR-GBS and quinolone-resistant H. influenzae organisms, as well as other quinolone-resistant bacteria (Table 2). All 23 LR-GBS isolates were resistant to levofloxacin, trovafloxacin, and gatifloxacin but susceptible to penicillin and ampicillin. MICs for quinolones lacking NCCLS guidelines with six selected LR-GBS strains are listed in Table 3. Clindamycin and erythromycin MICs ranged from ≤0.25 to >2 and ≤0.12 to >4, respectively.
TABLE 2.
Coisolation of levofloxacin-resistant H. influenzae, group B streptococci, and other gram-negative bacteria from sputum
| Patient (age [yr]/sex)a | Date (mo/day/yr) of isolation; quinolone-resistant species | Date(s) (mo/day/yr) of antibiotic administration (antibiotic[s] administered prior to isolation) |
|---|---|---|
| 1 (81/F) | 5/03/04; GBS, H. influenzae, Serratia marcescens, and Morganella morganii | 4/02/04, (500 mg levofloxacin); 2/21/04 (500 mg levofloxacin) |
| 2 (77/F) | 6/23/04; GBS, H. influenzae, Providencia stuartii, and Pseudomonas aeruginosa | 5/09/03 (500 mg levofloxacin) |
| 3 (53/F) | 4/16/01; GBS, H. influenzae, P. aeruginosa, and P. stuartii | 8/18/00 (500 mg ciprofloxacin); 8/30/00 (500 mg levofloxacin); 12/19/00 (500 mg levofloxacin); 4/13/01 (500 mg ciprofloxacin) |
| 4 (59/F) | 6/14/02; GBS, H. influenzae, and P. aeruginosa | 5/28/02 (250 mg ciprofloxacin); 6/04/02 (500 mg levofloxacin) |
| 5 (80/F) | 6/5/04; GBS, H. influenzae, Acinetobacter baumannii, and P. stuartii | 5/24/04 (500 mg levofloxacin) |
| 6 (46/F) | 9/16/03; GBS, H. influenzae, P. aeruginosa, and M. morganii | 3/17/03 (500 mg levofloxacin); 4/10/03 (500 mg levofloxacin) |
| 7 (81/M) | 8/17/03; GBS and H. influenzae | 7/10/03 (500 mg levofloxacin); 7/23/03 (500 mg levofloxacin) |
F, female; M, male.
TABLE 3.
MICs (Etest) of quinolones and mutations in gyrA/B and parC/E of QRDR with S. agalactiae
| Strain | Status | MIC (μg/ml)a
|
Different codond (amino acid) in:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LVXb | GEMb | MOXb | GARb | GATb | GEMc | gyrA (codon 81) | gyrA (codon 85) | parC (codon 79) | gyrB (codon 367) | parE | ||
| 0488 | Resistant isolate | >32.0 | 0.75 | 4.0 | 3.0 | 12.0 | 1.0 | TTA (Leu) | GAA (Glu) | TAC (Tyr) | GCA (Ala) | No difference |
| 0493 | Resistant isolate | >32.0 | 0.38 | 3.0 | 1.0 | 6.0 | 1.0 | TCA (Ser) | AAA (Lys) | TTC (Phe) | GCT (Ala) | No difference |
| 0612 | Resistant isolate | >32.0 | 2.0 | >32.0 | 4.0 | >32.0 | 2.0 | TTA (Leu) | GAA (Glu) | TAC (Tyr) | GCA (Ala) | No difference |
| 0823 | Resistant isolate | >32.0 | 1.5 | >32.0 | 3.0 | >32.0 | 1.0 | TTA (Leu) | GAA (Glu) | TAC (Tyr) | GCT (Ala) | No difference |
| 1110 | Resistant isolate | 12.0 | 0.25 | 0.75 | 0.5 | 1.0 | 0.25 | TTA (Leu) | GAA (Glu) | GCC (Ala) | GCA (Ala) | No difference |
| 1286 | Lab strain | 0.5 | 0.023 | 0.125 | 0.047 | 0.25 | 0.032 | TCA (Ser) | GAA (Glu) | TCC (Ser) | GCT (Ala) | No difference |
| NEM316 | GenBank strain | TCA (Ser) | GAA (Glu) | TCC (Ser) | GCT (Ala) | |||||||
LVX, levofloxacin; GEM, gemifloxacin; MOX, moxifloxacin; GAR, garenoxacin; GAT, gatifloxacin.
By Etest measurement.
By microdilution measurement.
Primers, indicated by nucleotide numbers (nt) of the respective genes, were as follows: nt 58 to 79 (forward), nt 674 to 795 (reverse), and nt 644 to 664 (sequence) for gyrA; nt 902 to 923 (forward and sequence) and nt 1512 to 1533 (reverse) for gyrB; nt 12 to 22 (forward and sequence) and nt 577 to 597 (reverse) for parC; and nt 1135 to 1155 (forward), nt 1735 to 1755 (reverse), and nt 1609 to 1628 (sequence) for parE. Abbreviations: Ser, serine; Ala, alanine; Glu, glutamic acid; Lys, lysine; Leu, leucine; Phe, phenylalanine; Tyr, tyrosine.
Nucleotide sequences of quinolone resistance-determining regions (QRDRs) in gyrA, gyrB, parC, and parE were determined for five resistant isolates and one laboratory strain. For laboratory strain 1286, no difference was found in QRDRs of gyrA, gyrB, parC, and parE relative to those in GenBank S. agalactiae strain NEM316. For each resistant isolate, one point mutation in gyrA and one in parC relative to the GenBank entry and laboratory strain 1286 were observed (Table 3). Two mutational positions in gyrA were observed: a Ser-81-to-Leu substitution was found in four resistant isolates, and a Glu-85-to-Lys substitution was found in one (Table 3). All isolates had a substitution at codon 79 in parC. Among five resistant isolates, three mutations were observed at this position: Ser-79 to Tyr (0488, 0612, and 0823), Ser-79 to Phe (0483), and Ser-79 to Ala (1110). A silent mutation in gyrB at codon 367 (GCT to GCA) was observed; in 0488, 0612, and 1110, GCT changed to GCA. No mutation in parE was found.
In summary, we identified quinolone-resistant group B streptococci in Queens, N.Y., among clinical isolates from 1999 to 2002. Of 508 urinary and blood isolates tested for levofloxacin susceptibility, 23 (4.4%) demonstrated resistance to levofloxacin according to NCCLS breakpoints; all LR-GBS isolates were resistant to other quinolones tested. The prevalences of resistance of GBS isolates to erythromycin (34.6%) and to clindamycin (16%) were higher than those in two recent reports (4, 8). Nucleotide sequence analysis of LR-GBS isolates revealed the same changes in the quinolone resistance-determining regions, corresponding to a change from Ser-81 to Leu in GyrA, as reported previously with Japanese isolates (6). A change from Glu-85 to Lys in one isolate was not observed previously (6). All resistant mutants also contained alterations of ParC. Recovery of double mutants may have reflected a high breakpoint definition of resistance. Changes in the GyrB QRDR revealed that an Ala-367 codon was changed from GCT to GCA. The significance of this gyrB change remains to be determined, since this alanine is present in the wild-type strain.
Unintended selection and proliferation of nontargeted microorganisms due to empirical use of antimicrobials is a growing issue facing physicians. For example, Clostridium difficile infection occurs during quinolone therapy, and the risk of intestinal colonization by vancomycin-resistant enterococci increases after the administration of various β-lactams (11). In addition, a levofloxacin-resistant strain of Mycobacterium tuberculosis arose in a patient treated with ofloxacin for gram-negative pneumonia 1 year before tuberculosis diagnosis (12). We now report that seven patients residing in the same LTCF had LR-GBS, quinolone-resistant H. influenzae, and other quinolone-resistant bacteria in sputum, consistent with inadvertent consequences of quinolone use. These findings emphasize the importance of monitoring antibiotic susceptibility to quinolone antibiotics at LTCFs and the need to replace empirical therapy with rapid molecular diagnosis (13).
Acknowledgments
Financial support was from BMA Medical Foundation, the Beatrice Snyder Foundation, Agnes Varis, and National Institutes of Health grant AI35257.
REFERENCES
- 1.Diekema, D. J., J. I. Andrews, H. Huynh, P. R. Rhomberg, S. R. Doktor, J. Beyer, V. D. Shortridge, R. K. Flamm, R. N. Jones, and M. A. Pfaller. 2003. Molecular epidemiology of macrolide resistance in neonatal bloodstream isolates of group B streptococci. J. Clin. Microbiol. 41:2659-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farley, M. M., R. C. Harvey, T. Stull, J. D. Smith, A. Schughat, J. D. Wenger, and D. S. Stephens. 1993. A population based assessment of invasive disease due to group B streptococcus in nonpregnant adults. N. Engl. J. Med. 328:1807-1811. [DOI] [PubMed] [Google Scholar]
- 3.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 4.Heelan, J. S., M. E. Hasenbein, and A. J. McAdam. 2004. Resistance of group B streptococcus to selected antibiotics, including erythromycin and clindamycin. J. Clin. Microbiol. 42:1263-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henning, K. J., E. L. Hall, and D. M. Dwyer. 2001. Invasive group B streptococcal disease in Maryland nursing home residents. J. Infect. Dis. 183:1138-1142. [DOI] [PubMed] [Google Scholar]
- 6.Kawamura, Y., H. Fujiwara, N. Mishima, Y. Tanaka, A. Tanimoto, S. Ikawa, Y. Itoh, and T. Ezaki. 2003. First Streptococcus agalactiae isolates highly resistant to quinolones with point mutations in gyrA and parC. Antimicrob. Agents Chemother. 47:3605-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, X., N. Mariano, J. J. Rahal, C. M. Urban, and K. Drlica. 2004. Quinolone-resistant Haemophilus influenzae in a long-term care facility: nucleotide sequence characterization of alterations in the genes encoding DNA gyrase and DNA topoisomerase. Antimicrob. Agents Chemother. 48:3570-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murdoch, D. R., and L. B. Reller. 2001. Antimicrobial susceptibilities of group B streptococci isolated from patients with invasive disease: 10 year perspective. Antimicrob. Agents Chemother. 45:3623-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazir, J., C. Urban, N. Mariano, J. Burns, B. Tommasulo, C. Rosenberg, S. Segal-Maurer, and J. J. Rahal. 2004. Quinolone resistant Haemophilus influenzae in a long term care facility: clinical and molecular epidemiology. Clin. Infect. Dis. 38:1564-1569. [DOI] [PubMed] [Google Scholar]
- 10.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing: fourteenth informational supplement. NCCLS document M100-14. NCCLS, Wayne, Pa.
- 11.Paterson, D. L. 2004. “Collateral damage” from cephalosporin or quinolone antibiotic therapy. Clin. Infect. Dis. 38(Suppl. 4):S341-S345. [DOI] [PubMed] [Google Scholar]
- 12.Perman, D. C., W. M. El Sadr, L. B. Heifets, E. T. Nelson, J. P. Matts, K. Chirgwin, N. Salomon, E. E. Telzak, O. Klein, B. N. Kreiswirth, J. M. Musser, and R. Hafner. 1997. Susceptibility to levofloxacin of Mycobacterium tuberculosis isolates from patients with HIV-related tuberculosis and characterization of a strain with levofloxacin monoresistance. AIDS 11:1473-1478. [DOI] [PubMed] [Google Scholar]
- 13.Peterson, L. R., and A. Dalhoff. 2004. Towards targeted prescribing: will the cure for antimicrobial resistance be specific, directed through improved diagnostic testing? J. Antimicrob. Chemother. 53:902-905. [DOI] [PubMed] [Google Scholar]
- 14.Sambola, A., J. M. Miro, M. P. Tornos, B. Almirante, A. Moreno-Torrico, M. Gurgui, E. Martinez, A. Del Rio, M. Azqueta, F. Marco, and J. M. Gatell. 2002. Streptococcus agalactiae infective endocarditis: analysis of 30 cases and review of the literature, 1962-1998. Clin. Infect. Dis. 34:1576-1584. [DOI] [PubMed] [Google Scholar]
- 15.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyrrell, G. J., L. D. Senzilet, J. S. Spika, D. A. Kertesz, M. Alagaratnam, M. Lovgren, and J. A. Talbot. 2000. Invasive disease due to group B streptococcal infection in adults: results from a Canadian, population-based, active laboratory surveillance study—1996. J. Infect. Dis. 182:168-173. [DOI] [PubMed] [Google Scholar]
- 17.Wu, J. J., K. Y. Lin, P. R. Hsueh, J. W. Liu, H. I. Pan, and S. M. Sheu. 1997. High incidence of erythromycin-resistant streptococci in Taiwan. Antimicrob. Agents Chemother. 41:844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]