Abstract
In addition to encoding the structural and regulatory proteins, many viruses encode auxiliary proteins, some of which have been shown to play important roles in lytic and latent states of the viruses. The human neurotropic JC virus (JCV) genome encodes an auxiliary protein called Agno whose function remains unknown. Here, we investigated the functional role of JCV Agno protein on transcription and replication of the viral genome in glial cells. Results from transfection of human glial cells showed that Agno protein suppresses both T-antigen-mediated transcription of the viral late gene promoter and T-antigen-induced replication of viral DNA. Affinity chromatography and coimmunoprecipitation assays demonstrated that the Agno protein and T antigen physically interact with each other. Through the use of a series of deletion mutants, we demonstrated that the T-antigen-interacting region of Agno protein is localized to its amino-terminal half and the Agno-interacting domain of T antigen maps to its central portion. Furthermore, utilizing various Agno deletion mutants in functional studies, we confirmed the importance of the Agno-T antigen interaction in the observed down-modulation of T antigen function upon viral gene transcription and DNA replication by Agno protein. Taken together these data suggest that the Agno protein of JCV, which is produced late during the late phase of the lytic cycle, can physically and functionally interact with the viral early protein, T antigen, and downregulate viral gene expression and DNA replication. The importance of these observations in the lytic cycle of JCV is discussed.
Results from a large volume of studies have shown that the auxiliary proteins produced by various eukaryotic viruses may play a critical role in orchestrating the viral lytic cycle and the state of viral latency. This group of proteins has a diverse effect on various stages of infection including transcription (11, 13, 14, 27, 41, 61), translation (19), replication (12, 35, 50), viral assembly (39), release of viral particles (51, 56), and export of viral transcripts from nucleus to cytoplasm (15). In addition, this group of viral proteins may have an impact upon host function and, by deregulating expression of key cellular genes, contribute to the pathogenesis of viral-induced disease.
The human neurotropic polyomavirus, JC virus (JCV), contains an open reading frame for expressing a 71-amino-acid peptide whose function in the viral life cycle remains unknown. Clinically, replication of JCV in glial cells induces the fatal demyelinating disease of the brain, progressive multifocal leukoencephalopathy (PML) (5, 8, 60).
The genome of JCV is composed of a double-stranded covalently linked circular DNA which is composed of three functional regions (17), including viral early and late coding regions and the viral noncoding regulatory region (Fig. 1A). The viral early coding region encodes two regulatory proteins, small and large T antigens. Although little is known about the function of small t antigen, the large T antigen was shown to be a multifunctional phosphoprotein which, by interacting with several cellular proteins including Purα and YB-1 (18, 47, 48), can modulate both the initiation of viral DNA replication and activation of JCV late gene transcription. In addition to small and large T antigens, the viral early genome encodes several spliced variants of early proteins, due to alternative splicing of the early transcripts (57). These variants of T antigen have been shown to differentially interact with the retinoblastoma family of tumor suppressor proteins (7). Additionally, JCV T antigen is oncogenic and its expression can induce development of tumors of neural origin in experimental animals (31, 49, 58), and its genome has been found in several human tumors (32, 33, 45). The viral late coding region is responsible for expression of three structural proteins, VP1, VP2, and VP3, all of which participate in the formation of viral capsids (36). In addition, the leader of the late transcripts contains an open reading frame for the 71-amino-acid Agno protein.
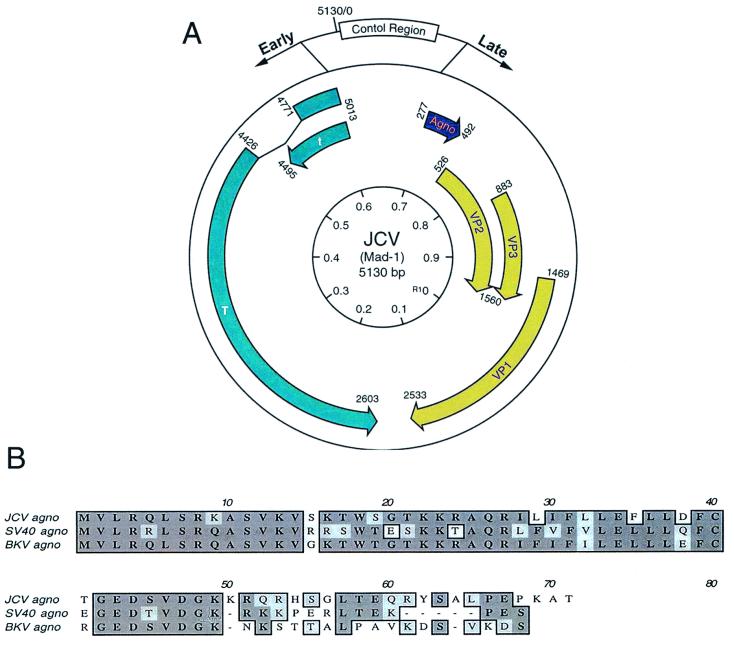
FIG. 1.
Structural organization of the JCV genome and Agno protein. (A) JCV genomic structure, depicting the direction of the genes encoding the early and late proteins with arrows. Early genes encode small and large T antigens. Late genes encode viral capsid proteins, VP-1, -2, and -3, and Agno protein. The control region is located between the early and late genes. (B) Comparison of primary amino acid sequences of JCV Agno protein with its counterparts from SV40 and BKV.
JCV is closely related to the other polyomaviruses, including the simian vacuolating virus (SV40) and the human BK virus (BKV) (16, 17). These viruses share significant sequence homology, particularly in their coding regions (16). JCV and SV40 exhibit approximately 80% homology in their early coding regions, close to 70% homology in their late coding sequences for VP1, VP2, and VP3, and less than 70% homology in the region corresponding to Agno protein. The highest degree of divergence between JCV and SV40 Agno proteins is clustered at the far carboxyl terminal regions (Fig. 1B). JCV and BKV also share a similar degree of homology in those respective regions. The regulatory region of JCV is distinct from those of SV40 and BKV and is responsible for cell-type-specific transcription of the viral genome in central nervous system (CNS) cells (1, 28, 42, 43, 54, 55). Earlier studies on both BKV and SV40 established that the Agno gene is expressed during the late phase of infection (25, 26, 30, 38, 46). Mutational analysis of SV40 Agno protein revealed the importance of this protein in the regulation of the SV40 lytic cycle and virion production (4, 38, 40). While the mechanisms involved in this process remain unknown, it is postulated that SV40 Agno protein may have a functional role at the level of viral assembly (26, 37, 38, 40), maturation (24, 26), transcription, and translation (3, 21, 22).
In this study, we examined the effect of JCV Agno protein on transcription and replication of the viral genome in glial cells, in the absence and presence of the viral early protein T antigen. We demonstrate that functional and structural interaction of Agno protein with T antigen results in suppression of viral gene expression and DNA replication in glial cells.
MATERIALS AND METHODS
Cell lines.
HJC-15b (44) cells were derived from hamster brain tumors induced by JCV (59) and were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum and antibiotics (penicillin-streptomycin, 100 μg/ml). U-87MG, a human glial origin cell line, was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics (penicillin-streptomycin, 100 μg/ml). All cells were maintained at 37°C in a humidified atmosphere with 7% CO2.
Plasmid constructs.
The pBLCAT3-Mad-1L reporter construct containing the regulatory region of JCV Mad-1 strain in the late orientation was described previously (10). The glutathione S-transferase (GST) fusion protein of JCV T antigen (pGEX2T-T-antigen) and its C-terminal [pGEX2T-T-antigen (1-411), pGEX2T-T-antigen (1-265), and pGEX2T-T-antigen (1-82)] and N-terminal [pGEX2T-T-antigen (266-688), pGEX2T-T-antigen (412-688), and pGEX2T-T-antigen (629-688)] deletion mutants were previously described (47). pGEX2T-T-antigen (250-450) was created by PCR amplification of the respective region by forward (5′-ACTACTTCGGATCCGGGGGCCTTAAGGAGCATGACTTT-3′) and reverse (5′-ACTACTTG AATTCAACATTTAATGACTTTCCCCC-3′) primers. The resulting fragment was cloned in frame into BamHI/EcoRI sites of pGEX2T vector. A plasmid containing an intronless JCV T-antigen coding region (6) was used as a template in PCR amplification. PGEX1λT-Agno and its deletion mutants were also created by PCR amplification utilizing the following specific primers. Forward primers (FP) were Agno 5′-BamHI (5′-ACTGACGGATCCGCCACCATGGTTCTTCGCCAGCTGTCA-3′), FP aa 18 (5′-ACTGACGGATCCGCCACCATGAGTGGAACTAAAAAAAGAGCT-3′), FP aa 37 (5′ - AC TGACGGATCCGCCACCATGC TGGAC T T T TGCACAGG TGAA - 3′), and FP aa 55 (5′-ACTGACGGATCCGCCACCATGAGTGGTTTGACTGAGCAGACATAC-3′). Reverse primers (RP) were Agno-3′ EcoRI (5′-ACTGACGAATTCCTACTATGTAGCTTTTGGTTCAGG-3′), RP aa 54 (5′-ACTGACGAATTCCTAGTGTCTCTGTCTTTTTTTCCC-3′), and RP aa 36 (5′-ACTGACGAATTCCTACAAAAATTCTAACAAAAAAAT-3′). A pBR322-based plasmid containing the entire JCV Mad-1 DNA was used as a template in PCR amplification. The PCR products were digested with BamHI/EcoRI and cloned in the pGEX11T vector. Further, full-length Agno and its two deletion mutants (amino acids 1 to 36 and 55 to 71) were also subcloned into pCDNA3 expression vector (Invitrogen) at BamHI/EcoRI sites by utilizing PCR amplification with the respective primers as described above. pEBV-HisA-Agno plasmid was created by PCR amplification with the following primers: Agno-5′ (5′-CCGCTTAGGATCCATGGTTCTTGGCCAGCTGTCA-3′) and Agno-3′ (5′-ACGTCCAAAGCTTCTATGTAGCTTTTGGTTCAGG-3′). The respective PCR product was digested with BamHI and HindIII enzymes and subcloned into BamHI/HindIII sites of the vector. pEBV-HisA-SV40-Agno plasmid was also created by PCR amplification with SV40 5′ (5′-ACACAAAGGATCCCGCCGCCATGGTGCTGCGCCGGCTGTCACGC-3′) and SV40 Agno 3′ (5′-ACACAAAAAGCTTTTAACTTTCTGGTTTTTCAGT-3′) primers, and the resulting PCR products were subcloned into BamHI/HindIII sites of the vector.
Transient transfection assays.
U-87MG cells were transfected by the calcium phosphate precipitation method (20) with reporter constructs alone or in combination with Agno and T-antigen expression plasmids. Plasmid concentrations used in each transfection experiment are indicated below and/or in the figure legends. The total amount of DNA transfected into the cells was normalized with relevant empty vector DNA. A glycerol shock was applied at 4 h posttransfection and cells were harvested after 36 h. Chloramphenicol acetyl transferase (CAT) activity of samples was determined by utilizing 40 μg of protein for each sample. Transfections were repeated at least three times with different plasmid preparations and standard deviations are indicated by error bars.
Expression and purification of recombinant GST fusion proteins.
Fifty-milliliter overnight cultures of Escherichia coli DH5α transformed with either pGEX1λT-Agno or pGEX2T-JCV T antigen or their respective deletion mutant plasmids were diluted 1:10 in fresh Luria-Bertani broth supplemented with ampicillin (100 μg/ml). Cultures were induced with 0.3M isopropyl-β-thiogalactopyranoside (IPTG) at an optical density of 0.5 and incubated for an additional 2 h at 37°C. Cells were collected by centrifugation and resuspended in 10 ml of lysis buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 supplemented with 1 mM phenylmethylsulfonyl fluoride, 2 mM pepstatin A, 0.6 mM leupeptin, and 2 mM benzamidine. After sonication, clear cell lysates were prepared by centrifugation at 12,000 × g. Lysates were then incubated with 150 μl of 50% glutathione-Sepharose beads (Pharmacia, Piscataway, N.J.) overnight at 4°C. GST fusion proteins were purified by three cycles of washing and centrifugation with 5 ml of lysis buffer. Fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining.
GST affinity chromatography assays (GST pull-down).
For GST pull-down assays, 2 μg of either GST alone, GST-Agno, or its deletion mutants immobilized on Sepharose beads was incubated with 0.5 mg of whole-cell extracts prepared from HJC cells constitutively expressing JCV T antigen for 4 h at 4°C in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.5% Nonidet P-40. The protein-complexed beads were washed extensively with lysis buffer and resolved by SDS–10% PAGE followed by Western blot analysis using an antibody (Ab-2 416) directed against JCV T antigen. In a different method, GST or GST-JCV T antigen (2 μg of each) immobilized on Sepharose beads was incubated overnight with 4 μl of [35S]-labeled in vitro-translated Agno in lysis buffer in a 300-μl total volume. The protein-complexed beads were extensively washed with lysis buffer and resolved by SDS–15% PAGE. Proteins were detected by fluorography for the presence of Agno.
Coimmunoprecipitation and Western blot analysis.
pEBV-His-Agno expression plasmid was transfected into HJC-15b (hamster astrocytic cell line constitutively expressing JCV T antigen) cells via the calcium phosphate precipitation method (20). At 36 h posttransfection, cells were lysed in lysis buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.4), and 0.25% Nonidet P-40 supplemented with a cocktail of proteinase inhibitors including 1 mM phenylmethylsulfonyl fluoride, leupeptin (10 μg/ml), aprotinin (1 μg/ml), and 50 mM sodium fluoride. Five hundred micrograms of whole-cell extract in a total volume of 0.5 ml was incubated either with 2 μg of anti-T7 antibody (α-T7) (Novagen, Madison, Wis.) directed against the His-T7-tagged Agno or 2 μg of preimmune antisera (α-pre) overnight at 4°C. Immunocomplexes were precipitated with the addition of protein A-Sepharose beads (Pharmacia) for an additional 2 h and washed extensively with lysis buffer. Immunocomplexes were then resolved by SDS–10% PAGE and analyzed by Western blotting using an anti-T-antigen antibody (Ab-2 416) (Oncogene, Uniondale, N.Y.) and developed with an ECL detection kit (Amersham, Arlington Heights, Ill.) according to the manufacturer's recommendations.
In vitro transcription and translation assay.
Full-length Agno protein was radiolabeled with [35S]methionine using a TNT-coupled in vitro transcription-translation system (Promega, Madison, Wis.) according to the recommendations of the manufacturer.
Replication assay.
Replication assays were carried out as previously described (9). Briefly, a replication-competent plasmid, pBLCAT3-Mad-1L, containing the regulatory region of the Mad-1 strain of JCV, was introduced alone or in combination with expression vectors, CMV-T-antigen and CMV-Agno, into U-87MG (0.4 × 106 cells per plate) cells with the calcium phosphate precipitation method. Plasmid concentrations used in transfections are indicated in the respective figure legends, and the total amount of DNA transfected into the cells was normalized with appropriate empty vectors. A glycerol shock was applied at 4 h posttransfection and the medium was replenished. At 72 h posttransfection, low-molecular-weight DNA containing both input and replicated plasmids was isolated by the Hirt method (23), digested with SacI and DpnI enzymes, resolved on a 0.8% agarose gel and analyzed by using Southern blotting. Probes for Southern blots were prepared from a DNA fragment encompassing the regulatory region of JCV Mad-1 by utilizing a ready-prime random labeling kit (Amersham/Pharmacia Biotechnologies). The bands corresponding to the replicated DNA were quantitated utilizing a densitometer (Bio-Rad Fx PhosphorImager) with Quantity One software. The degree of inhibition of T-antigen-mediated JCV DNA replication by Agno protein was expressed as percent inhibition with respect to the degree of viral DNA replication in the presence of JCV T antigen alone.
RESULTS
Effect of Agno protein on transcriptional activity of the JCV promoters.
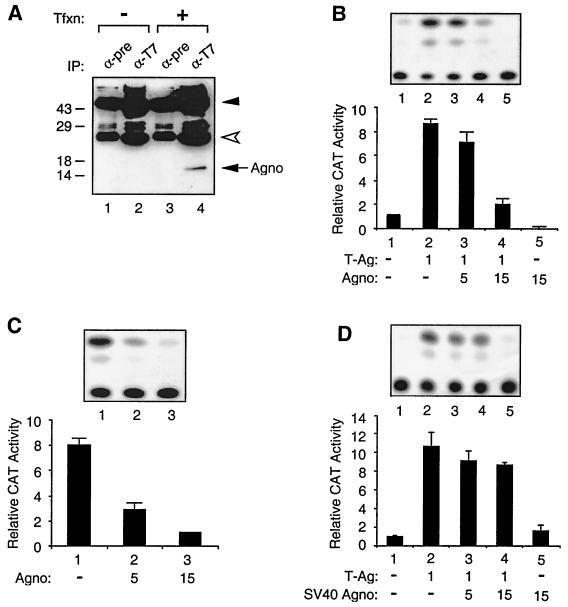
To determine the functional importance of Agno protein on transcriptional activity of the JCV late promoter, a plasmid that permits expression of Agno protein under the control of the Rous sarcoma virus (RSV) promoter was created and the first stable production of Agno protein was determined by Western blot analysis. As shown in Fig. 2A, anti-T7 antibody specifically immunoprecipitated histidine-tagged Agno protein in extracts from U-87MG cells transfected with a His-tagged Agno expression plasmid (compare lane 4 to lane 3). The specificity of this immunoprecipitation was demonstrated by the use of both normal mouse serum (lane 1) and anti-T7 antibody (lane 2), both of which showed no cross-reactivity with the proteins prepared from the untransfected cells. Next, we performed cotransfection experiments in U-87MG cells by using a reporter construct containing the JCV late gene promoter and expression plasmids for Agno and JCV T antigen. As shown in Fig. 2B, in the absence of T antigen a CAT reporter construct containing the JCV late promoter was poorly expressed (lane 1). Cotransfection of the reporter construct with a T-antigen expression plasmid resulted in a substantial increase (ninefold) in the transcriptional activity from the late promoter (compare lanes 1 and 2). In contrast, when increasing amounts of a plasmid expressing Agno protein were cointroduced along with a constant amount of T-antigen expression plasmid into glial cells, we observed a suppressive effect of Agno on the T-antigen-mediated activation of the late promoter (compare lane 2 to lanes 3 and 4). At the highest DNA concentration, expression of Agno protein alone further decreased the basal expression of the late promoter, suggesting that Agno may have a negative regulatory effect on the basal expression of JCV late promoter. To further test this possibility, we carried out transfection experiments using a constant amount of reporter construct containing JCV late promoter alone (Fig. 2C, lane 1) or in combination with an increasing amount of Agno expression plasmid (lanes 2 and 3). As shown in Fig. 2C, production of Agno protein in U-87MG caused a considerable decrease in the basal level of JCV late gene transcription (lanes 2 and 3). In addition, we have also performed transfection experiments to assess the effect of SV40 Agno protein on JCV late promoter expression. As shown in Fig. 2D, SV40 Agno showed only a minor inhibitory effect on the transcription of the JCV late promoter in the presence of T antigen (Fig. 2D), suggesting that the observed negative effect of Agno on JCV gene expression is specific. These observations indicate that Agno protein negatively regulates both basal and T-antigen-induced transcription of the JCV late promoter.
FIG. 2.
Effect of Agno protein on transcriptional activity of the JCV late promoter. (A) Expression of JCV Agno gene. pEBV-His-Agno expression plasmid was introduced into U-87MG cells. Two hundred micrograms of whole-cell extracts prepared from untransfected (lanes 1 and 2) and transfected (lanes 3 and 4) cells was immunoprecipitated with either preimmune (α-pre) (lanes 1 and 3) or anti-T7 (α-T7) (lanes 2 and 4) antibodies. Immunocomplexes were resolved by an SDS–15% PAGE and analyzed by Western blotting using anti-T7 antibody. Tfxn, transfection; IP, immunoprecipitation. Closed and open arrowheads indicate large and small IgG fragments, respectively. The arrow indicates His-tagged Agno. The positions of molecular mass markers (in Kilodaltons) are shown to the left of the panel. (B) Agno suppresses T-antigen-mediated transactivation of JCV late promoter. A reporter plasmid (pBLCAT3-Mad-1L) (7 μg) was introduced into U-87MG cells alone or in combination with plasmids expressing Agno protein and T antigen (T-Ag) as indicated for each lane. At 36 h posttransfection, CAT enzymatic activity was determined. (C) Agno protein suppresses the basal expression of JCV late promoter. pBLCAT3-Mad-1L reporter plasmid (10 μg) was introduced into U-87MG cells alone or together with 5 and 15 μg of Agno protein expression plasmid. Bar graph shows CAT values relative to the basal expression of the reporter construct alone. (D) Effect of SV40 Agno protein on T-antigen-mediated activation of JCV late promoter. Transfections were carried out as described for Fig. 2B. The results of a representative CAT assay for each panel (B, C, and D) are shown on top. Results shown in each panel represent the mean of three independent experiments and bars indicate standard deviation.
A similar set of experiments was carried out to evaluate the effect of Agno protein on transcription of the JCV early genome. The results showed a suppressive effect of Agno protein on basal transcription of the JCV early promoter. Agno protein had no noticeable effect on the T-antigen-mediated transcriptional suppression of the JCV early promoter (data not shown).
Effect of Agno on T-antigen-mediated JCV DNA replication.
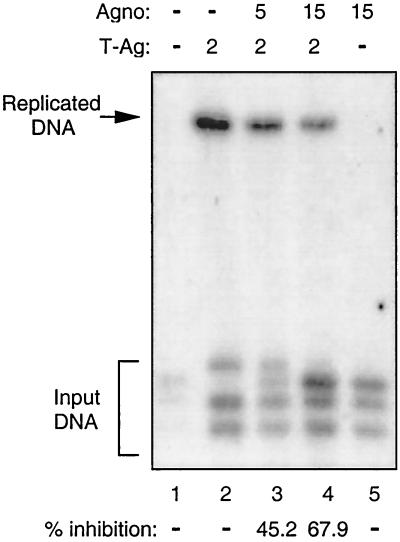
The observed effect of Agno protein on T-antigen-mediated transcription of the JCV late genome suggested that Agno protein may also exert a regulatory effect on T-antigen-mediated viral DNA replication. To investigate this possibility, we performed DpnI replication assays (9). A replication-competent plasmid containing the JCV origin of DNA replication was introduced alone or in combination with an Agno gene expression plasmid into U-87MG cells. At 72 h posttransfection, low-molecular-weight DNA was isolated by the Hirt procedure (23) and newly replicated DNA was analyzed by Southern blot analysis. As shown in Fig. 3, in the presence of T antigen, the plasmid containing viral DNA was efficiently replicated (lane 2). However, in the presence of Agno protein, a substantial decrease in T-antigen-induced viral DNA replication was observed. Agno protein alone showed no ability to induce viral DNA replication (lane 5). Of note, in recent studies it was shown that JCV with a mutation in Agno has a different growth cycle compared to the wild-type virus, suggesting the importance of Agno in the viral lytic cycle (Y. Okada et al., unpublished data). These observations indicate that Agno protein can negatively affect T-antigen-induced replication of JCV DNA, perhaps through its interaction with and inactivation of T antigen.
FIG. 3.
Agno protein inhibits T-antigen-mediated JCV DNA replication. A replication-competent plasmid, pBLCAT3-Mad-1L (5 μg), containing the regulatory region of the Mad-1 strain of JCV was transfected alone or in combination with the expression plasmids CMV-T-antigen and CMV-Agno into U-87MG cells as described in Materials and Methods. Plasmid concentrations used in transfections are indicated on top (in micrograms). The total amount of DNA transfected into the cells was normalized with appropriate empty vectors. At 72 h posttransfection, low-molecular-weight DNA was isolated by the Hirt method (23), digested with SacI and DpnI enzymes, resolved on a 0.8% agarose gel, and analyzed by Southern blotting. The bands corresponding to the replicated DNA bands were quantitated by densitometry (see Materials and Methods) and expressed as percent inhibition with respect to the viral DNA replication in the presence of T antigen alone. The results for percent inhibition are shown at the bottom.
Interaction of Agno protein and T antigen.
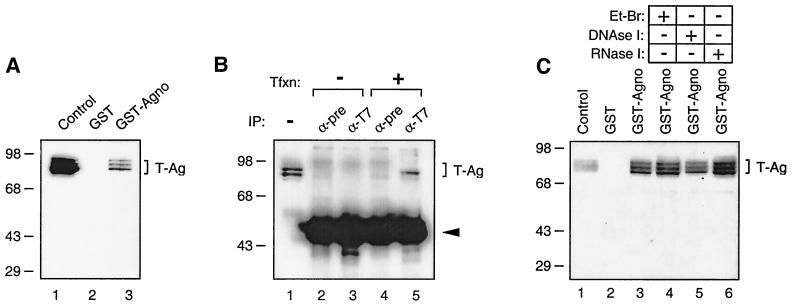
Results from transcription and replication studies suggested that JCV large T antigen may physically interact with the late auxiliary Agno protein. To test this possibility, we performed affinity chromatography (GST pull-down) experiments in which one of the two proteins was prokaryotically expressed as a GST fusion protein and bound to a glutathione resin while the other protein was passed over the resin and analyzed for its ability to be specifically retained by the GST fusion protein. In the first experiment, GST or GST-Agno protein was immobilized on glutathione-Sepharose beads and incubated with whole-cell extracts from HJC-15b cells, which constitutively express T antigen. Proteins bound to beads were extensively washed and were analyzed by Western blot analysis using antibodies specific for T antigen. As demonstrated in Fig. 4A, T antigen was retained on the Sepharose column containing GST-Agno (lane 3). Interestingly, Agno protein is able to interact with three isoforms of JCV T antigen expressed in HJC-15b cells. These three isoforms of T antigen are believed to represent different phosphorylated forms of T antigen (53). This interaction was not observed between T antigen and GST (lane 2).
FIG. 4.
In vitro and in vivo interaction between Agno protein and T antigen. (A) Agno protein interacts with T antigen in GST pull-down assays. Whole-cell extracts prepared from HJC-15b cells, which express JCV T antigen constitutively, were incubated with either GST alone (lane 2) or GST-Agno protein (lane 3) as described in Materials and Methods. Beads were washed extensively and proteins interacting with GST or GST-T-Agno were resolved by SDS-PAGE and analyzed by Western blotting using anti-T-antigen antibody (Ab-2 416). (B) Agno coimmunoprecipitates with T antigen. Five hundred micrograms of whole-cell extracts prepared from HJC-15b cells and HJC-15b cells transfected with an Agno expression plasmid (pEBV-Agno) was immunoprecipitated either with normal serum (α-pre) (lane 4) or anti-T7 (α-T7) (lane 5) antibodies. Immunocomplexes were resolved on an SDS–10% PAGE and analyzed by Western blotting for the presence of T antigen with an anti-T-antigen antibody (Ab-2 416). Tfxn, transfection. The arrowhead indicates the position of immunoglobulin heavy chain detected by the secondary antibody. (C) Whole-cell extracts from HJC-15b cells treated with either ethidium bromide (100 ng/ml, lane 4) (Et-Br), DNase I (0.2 U/μg of protein, lane 5) (47), or RNaseI (0.5 U/μg of protein, lane 6). In lane 1 for each panel, the whole-cell extracts prepared from HJC-15b cells were loaded as a migration control for T antigen.
To further examine the association of Agno protein with T antigen, protein extracts from HJC-15b cells that were either untransfected or transfected with the His-Agno expression plasmid were immunoprecipitated with control preimmune serum and with an anti-T7 antibody that recognizes His-tagged Agno protein. Immunocomplexes were resolved by Western blotting and examined for the presence of T antigen using anti-T-antigen antibody. As shown in Fig. 4B, anti-T7 antibody detected T antigen in extracts from HJC-15b cells transfected with a plasmid expressing His-tagged Agno protein (lane 5). The specificity of this coimmunoprecipitation was verified by the absence of a band corresponding to T antigen when protein extracts from the transfected cells were incubated with normal mouse serum (lanes 2 and 4). Also, anti-T7 antibody showed no cross-reactivity with cellular proteins from untransfected cells (lane 3). To examine the involvement of RNA or DNA in the association of Agno with T antigen, GST-Agno fusion protein and the whole-cell lysate were treated with ethidium bromide, DNase I, or RNase prior to pull-down experiments. As shown in Fig. 4C, treatment of the whole-cell extract with ethidium bromide, DNase I, and RNase had no significant effect on the binding of GST-Agno to T antigen. Altogether, results from both in vitro GST pull-down and coimmunoprecipitation experiments demonstrate that Agno protein physically associates with JCV T antigen.
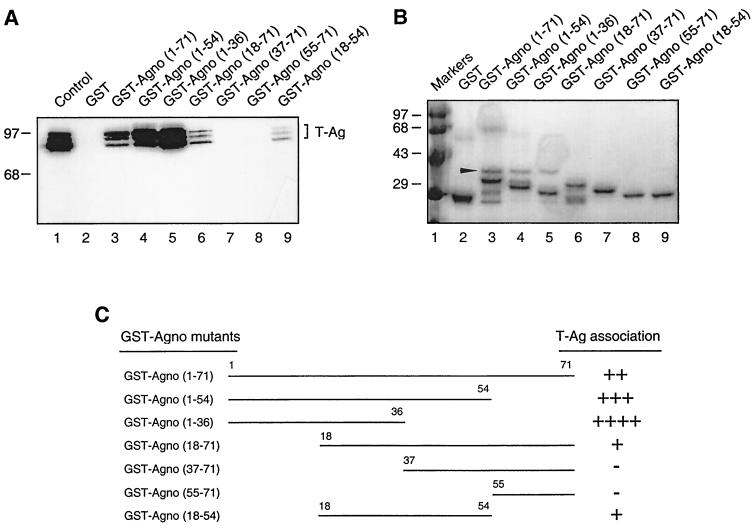
Mapping of the region within Agno protein which interacts with T antigen.
In the next series of experiments, we attempted to map the region of Agno protein which is involved in the interaction with T antigen. A series of deletion mutants in the Agno gene was created and mutant Agno proteins fused to GST were incubated with whole-cell lysates from HJC-15b cells. Bound complexes were resolved by SDS-PAGE and analyzed by Western blotting using anti-T-antigen antibody. As shown in Fig. 5A, removal of the region of the Agno protein which is positioned between residues 55 to 71 and 36 to 71 enhanced the binding ability of Agno protein to T antigen in comparison to that observed with the full-length Agno protein (compare lanes 4 and 5 to lane 3). Deletion of the region spanning residues 1 to 18 showed a moderate effect on the binding ability of Agno protein to T antigen (compare lane 6 to lane 3). Removal of the residues between 37 to 71 and 55 to 71 completely abrogated association of Agno protein and T antigen. These observations suggest that the region positioned between residues 18 to 37 is important for allowing Agno protein to interact with T antigen. In support of this conclusion, results from the binding of a truncated Agno protein encompassing residues 18 to 54 revealed the ability of this protein to interact with T antigen (lane 9). This study, in addition, demonstrates that amino acid residues between 36 and 71 have a negative effect on the interaction between Agno and T antigen. Figure 5B illustrates the Coomassie blue-stained SDS-PAGE of the full-length and mutant Agno proteins which were used in this study and verifies the integrity of the protein preparations. Figure 5C summarizes the results of the GST pull-down assay and depicts the regions of Agno which bind to T antigen.
FIG. 5.
Mapping of the interaction domain of Agno protein with T antigen. Whole-cell extracts from HJC-15b cells were incubated with either GST alone (lane 2), GST-Agno (lane 3), or deletion mutants of GST-Agno fusion proteins (lanes 4 to 9) immobilized on glutathione-Sepharose beads. Bound complexes were washed extensively, resolved by SDS-PAGE, and analyzed by Western blotting using an anti-T-antigen antibody (Ab-2 416). HJC-15b whole-cell extract was loaded as a migration control (lane 1). The bracket indicates the position of bound T antigen. (B) Analysis of the GST and GST-Agno protein and its deletion mutants by SDS-PAGE. The similarly sized nonspecific bacterial protein which was copurified in our protein purification assays is indicated by an arrowhead. (C) Summary of the results obtained from in vitro mapping assays. The ability of Agno protein and its deletion mutants to interact with T antigen is depicted on the right as follows: ++++ or +++, very strong interaction; ++, strong interaction; +, reduced interaction; −, no interaction.
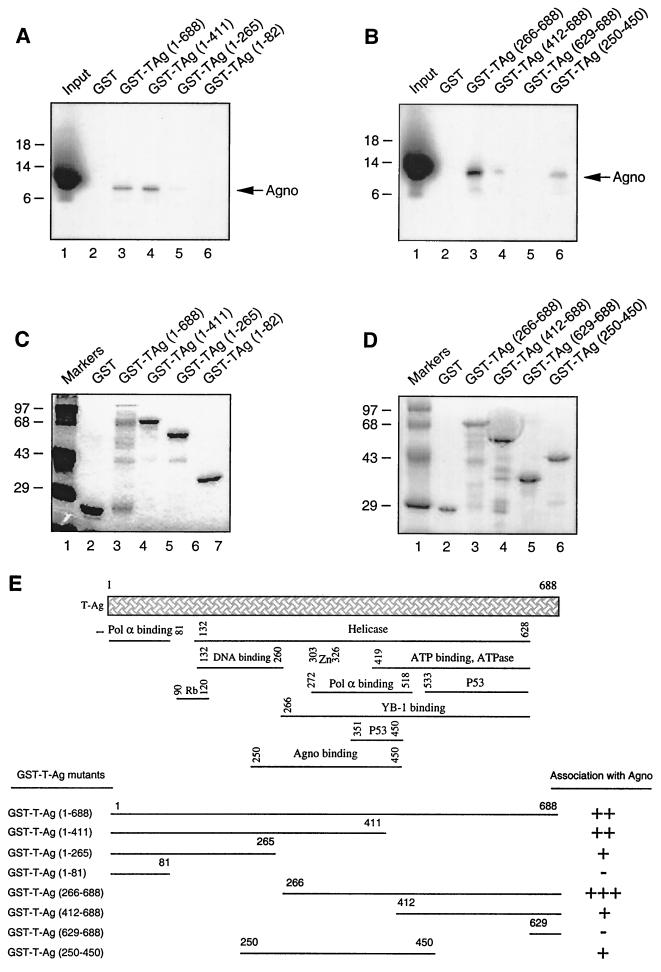
Identification of the region within T antigen which is important for its interaction with Agno protein.
To identify the region(s) of T antigen necessary for its interaction with Agno protein, a series of carboxy-terminal (Fig. 6A) and amino-terminal (Fig. 6B) deletion mutants of T antigen were prokaryotically expressed as GST fusion proteins and incubated with in vitro-translated [35S]methionine-labeled Agno protein. Consistent with previous observations (Fig. 3A), while full-length T antigen fused to GST interacted with Agno protein (lane 3), GST protein alone showed no binding to T antigen (lane 2). The carboxy-terminal deletion mutant of T antigen containing residues 1 to 411 retained its binding ability to Agno protein (compare lanes 3 to 4). The removal of residues 1 to 266 significantly affected the ability of T antigen to interact with Agno protein (lane 5). A further carboxy-terminal deletion completely abolished interaction of Agno protein with T antigen (lane 6).
FIG. 6.
Localization of the interaction domain of T antigen with Agno protein. (A and B) GST C-terminal (A) or N-terminal (B) deletion mutants of T antigen immobilized on glutathione-Sepharose beads were incubated with in vitro-translated [35S]methionine-labeled Agno protein. The Sepharose beads were washed extensively, and bound proteins were resolved by SDS-PAGE and analyzed by autoradiography. One-tenth of the input Agno protein used in each reaction was loaded for migration controls (lane 1 in each panel). The arrows indicate the position of in vitro-translated [35S]methionine-labeled Agno protein. (C and D) Analysis of GST, GST-T-antigen, GST-T-antigen C-terminal (C), and GST-T-antigen N-terminal (D) deletion mutants by SDS-PAGE. (E) Summary of the results obtained from in vitro mapping assays. A schematic representation of T antigen is shown at the top (not shown to scale). The abilities of T antigen and its deletion mutants to interact with Agno protein are shown on the right. +++, very strong interaction; ++, strong interaction; +, minimal interaction; −, no interaction.
A deletion mutant of T antigen lacking the amino-terminal 265 amino acids (lane 3) showed a slight increase in its ability to interact with Agno protein in comparison to that seen with full-length T antigen (compare lane 3 in Panel A with lane 4 in Panel B). A further removal of amino acids from the amino-terminal region up to residue 411 significantly decreased the interaction between these two proteins (lane 4). An amino-terminal deletion mutant retaining residues 626 to 688 failed to interact with Agno protein. Furthermore, an amino- and carboxy-terminal deletion mutant retaining the central portion of the protein encompassing the amino acids 250 to 450 showed reduced interaction with Agno protein (lane 6). Altogether, results from mapping experiments demonstrate that the minimal region of T antigen which is important for interaction with Agno protein lies within the central portion of the protein between amino acids 250 and 450. A summary of these observations is schematized in Fig. 6E.
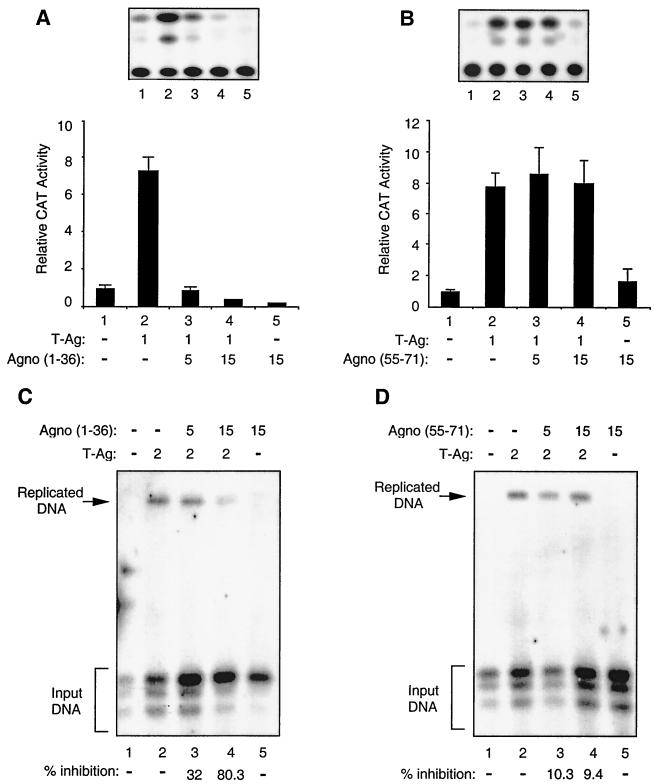
Effect of mutant Agno proteins on T-antigen-induced JCV gene transcription and viral DNA replication.
To further assess the functional interaction between Agno protein and T antigen, we examined the ability of mutant Agno proteins which have retained or lost their binding activity with T antigen upon JCV gene transcription and replication in glial cells. We chose mutant protein Agno (1-36), which showed the ability to interact with T antigen in in vitro GST pull-down assays, and mutant protein Agno(55-71), which showed no binding activity to T antigen. For the transcriptional assays, we performed transfections using a reporter JCV late CAT construct alone or in combination with the T-antigen expression plasmid and plasmids expressing protein mutants Agno(1-36) and Agno(55-s71). As shown in Fig. 7A, Agno(1-36), which strongly binds to T antigen, showed a drastic negative effect on the T-antigen-mediated activation of the JCV late promoter (Fig. 7A) compared to the full-length protein (Fig. 2B), while mutant Agno(55-71) had no effect on the level of transcriptional activation of the JCV promoter by T antigen (Fig. 7B).
FIG. 7.
Functional interaction of two Agno protein deletion mutants with T antigen. (A and B) Effect of Agno protein deletion mutants on T-antigen-mediated transcriptional activation of JCV late promoter. A CAT reporter plasmid (pBLCAT3-Mad-1L) (7 μg) containing the JCV late promoter was transfected into U-87MG cells alone or in combination with Agno protein deletion mutants Agno(1-36) (A) and Agno(55-71) (B) and T-antigen expression plasmids. The concentrations of expression plasmids are indicated at the bottom of the respective panels in micrograms. A representative CAT assay for each panel is shown on the top. The results shown in each panel represent the mean of three independent experiments. (C and D) Effect of Agno protein mutants on T-antigen-mediated JCV DNA replication. A replication-competent plasmid, pBLCAT3-Mad-1L (5 μg), containing the regulatory region of the Mad-1 strain of JCV, was transfected alone or in combination with Agno protein deletion mutants CMV-Agno(1-36) (C) and CMV-Agno(55-71) (D) and CMV-T-antigen expression plasmids into U-87MG cells. Plasmid concentrations used in transfections are indicated on top in micrograms. The total amount of DNA transfected into the cells was normalized with appropriate empty vectors. Replication assays were carried out as described for Fig. 3. The replicated DNA is indicated by an arrow and input DNA is indicated by a bracket. The quantitation of the bands corresponding to the replicated viral DNA was carried out as described for Fig. 3.
We also examined the effects of these two mutants [Agno(1-36) and Agno(55-71)] on JCV DNA replication. In agreement with the above observations from transfection assays, Agno (1-36) strongly inhibited T-antigen-mediated JCV DNA replication (Fig. 7C). The mutant protein Agno(55-71) showed no appreciable inhibitory activity on the replication of viral DNA in glial cells (Fig. 7D). Interestingly, comparison of the results from the studies on the effect of full-length Agno protein on T-antigen-mediated transcription and replication of JCV DNA (Fig. 2 and 3) with those obtained with mutant Agno(1-36) (Fig. 7A and C) suggests that the mutant Agno protein is more potent in modulating T-antigen function than the full-length Agno protein. This notion corroborates with results from protein-protein interaction studies (Fig. 5), where we observed that Agno(1-36) exhibited stronger binding activity for T-antigen than the full-length Agno protein. We have also used an additional Agno mutant [Agno(1-54)] in our transfection and replication studies to further confirm the specificity of the interaction between Agno and T antigen. This mutant also interacts with T antigen more strongly than wild-type Agno in GST pull-down assays (Fig. 5). We observed very similar results for this mutant and the Agno(1-36) (data not shown). These results also corroborate with the results obtained from GST pull-down assays (Fig. 5). Of note, the stable expression of Agno mutants used in transfection assays was investigated utilizing indirect immunostaining and Western blotting assays, and therefore the absence of functional interaction between T antigen and Agno(55-71) may not be attributed to its lack of expression in transfected cells (data not shown). Taken together, the data from transcription and replication studies utilizing mutants of Agno protein, two of which physically and functionally interact with T antigen [Agno(1-36) and Agno(1-54)] and one which lacks such characteristics [Agno(55-71)], further confirm the significance of physical and functional interaction between Agno and T antigen in regulation of JCV gene expression and replication.
DISCUSSION
The 71-amino-acid Agno protein of JCV may play an important role in the viral lytic cycle by modulating the rate of viral gene transcription and DNA replication. To exert its activity, Agno protein may interact with the viral key regulatory protein, T antigen, which is expressed early during infection. This is an interesting observation, as it demonstrates that interaction of Agno protein, which is produced at the late phase of the infection cycle, with the early protein, T antigen, can modulate the activity of this protein during the late stage of lytic infection. While the importance of this regulatory event in the pathogenesis of JCV-induced demyelination of the CNS remains to be elucidated, one may speculate that, by slowing the rate of late gene expression and viral DNA replication, Agno protein may prevent disproportional production of capsid proteins and viral DNA during the course of the infection cycle and optimize the efficiency of virion formation. Furthermore, Agno protein may prolong the course of the lytic cycle in CNS cells, and this may further contribute to the pathogenesis of the disease, which is manifested by gradual demyelination of the CNS.
Similar to SV40 T antigen, JCV T antigen is composed of several interesting domains which interact with several functionally important cellular proteins. For example, the region which is important for Agno protein interaction, i.e., residues 250 to 450, overlaps with the binding sites for p53, YB-1, and polymerase α and has several activities such as helicase and Zn binding. Thus, it is likely that Agno protein, by associating with T antigen, controls several of these activities during the late stages of the infection cycle. In addition, structurally, Agno protein has several interesting features. First, the protein is highly basic and contains several posttranslational modification sites including phosphorylation by protein kinase C and casein kinase II. While the phosphorylation sites for protein kinase C are clustered at the amino terminal of the Agno protein, the carboxyl terminal of the protein is the target for phosphorylation by casein kinase II. Although the phosphorylation of JCV Agno protein remains to be illustrated, earlier studies have shown that BKV Agno protein can be phosphorylated (46). Future studies in our laboratory are aimed to further characterize the structural features of the Agno protein and their importance in regulation of the JCV lytic cycle and host function during the course of infection.
The studies presented in this communication provide evidence, for the first time, that the human papovavirus Agno protein, by interacting with the viral regulatory protein, T antigen, can modulate the activity of this protein upon viral gene expression and DNA replication. With the notion that JCV infection occurs in greater than 70% of the human population during childhood and the virus remains in the latent state throughout life, one may question whether Agno protein plays any role in maintaining the virus at latency. Furthermore, it is interesting to determine the function of Agno protein during the course of immunosuppression, when the virus gains an opportunity to exit from latency and actively replicates in brain cells. Our recent preliminary observations have shown expression of the Agno protein in oligodendrocytes of patients with PML, suggesting that this protein is expressed during the course of the disease. As such, one may hypothesize that the negative regulatory effect of Agno protein on viral replication may alter the course of disease progression, or that a mechanism induced upon immunosuppression prevents the Agno protein from imposing its suppressive function on viral gene transcription and replication. Our future experiments, which are aimed at the identification of the proteins which are associated with Agno protein in the infected oligodendrocytes during the course of infection, should provide some clues regarding the role of Agno protein in the pathogenesis of PML.
ACKNOWLEDGMENTS
We thank past and present members of the Center for Neurovirology and Cancer Biology for insightful discussions, sharing of ideas, and reagents. We thank Cynthia Schriver for editorial assistance.
This work was made possible by grants awarded by NIH to K.K. Y.O. is a research fellow of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Ahmed S, Rappaport J, Tada H, Kerr D, Khalili K. A nuclear protein derived from brain cells stimulates transcription of the human neurotropic virus promoter. J Biol Chem. 1990;265:13899–13905. [PubMed] [Google Scholar]
- 2.Ahmed S, Chowdhury M, Khalili K. Regulation of a human neurotropic virus promoter, JCVE: identification of a novel activator domain located upstream from the 98 bp enhancer promoter region. Nucleic Acids Res. 1990;18:7417. doi: 10.1093/nar/18.24.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alwine J. Evidence for simian virus 40 late transcriptional control: mixed infections of wild-type simian virus 40 and a leader deletion mutant exhibit trans effects on late viral RNA synthesis. J Virol. 1982;37:730–737. doi: 10.1128/jvi.42.3.798-803.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkan A, Mertz J E. DNA sequence analysis of simian virus 40 mutants with deletions mapping in the leader region of the late viral mRNA's: mutants with deletions similar in size and position exhibit varied phenotypes. J Virol. 1981;37:730–737. doi: 10.1128/jvi.37.2.730-737.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger J R, Concha M. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J Neurovirol. 1995;1:5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- 6.Bollag B, Mackeen B C, Frisque R J. Purified JC virus T antigen derived from insect cells preferentially interacts with binding site II of the viral core origin under replication conditions. Virology. 1995;218:81–93. doi: 10.1006/viro.1996.0168. [DOI] [PubMed] [Google Scholar]
- 7.Bollag B, Prins C, Snyder E L, Frisque R. Purified JC T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology. 2000;274:165–178. doi: 10.1006/viro.2000.0451. [DOI] [PubMed] [Google Scholar]
- 8.Brooks B R, Walker D L. Progressive multifocal leukoencephalopathy. Neurol Clin. 1984;2:299–313. [PubMed] [Google Scholar]
- 9.Chang C-F, Gallia G L, Muralidharan V, Chen N N, Zolteck P, Johnson E, Khalili K. Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence-specific single-stranded DNA-binding protein Pura. J Virol. 1996;70:4150–4156. doi: 10.1128/jvi.70.6.4150-4156.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N N, Khalili K. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur α in glial cells. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen E A, Subbramanian R A, Gottlinger H G. Role of auxiliary proteins in retroviral morphogenesis. Curr Top Microbiol Immunol. 1996;214:219–235. doi: 10.1007/978-3-642-80145-7_7. [DOI] [PubMed] [Google Scholar]
- 12.Collette Y, Olive D. The primate lentivirus-encoded Nef protein can regulate several steps of the viral replication cycle. Virology. 1999;265:173–177. doi: 10.1006/viro.1999.0053. [DOI] [PubMed] [Google Scholar]
- 13.Cullen B. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 14.Diaz-Guerra M, Esteban M. Vaccinia virus nucleoside triphosphate phosphohydrolase I controls early and late gene expression by regulating the rate of transcription. J Virol. 1993;67:7561–7572. doi: 10.1128/jvi.67.12.7561-7572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher U, Huber J, Boelens W C, Mattaj I W, Baltimore D. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 16.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frisque R J, White F A., III . The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy. In: Roos E R P, editor. Molecular neurovirology: pathogenesis of viral CNS infections. Totowa, N.J: Humana Press Inc.; 1992. pp. 25–158. [Google Scholar]
- 18.Gallia L G, Safak M, Khalili K. Interaction of the single-stranded DNA-binding protein Pura with the human polyomavirus JC virus early protein T-antigen. J Biol Chem. 1998;273:32662–32669. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 20.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 21.Haggerty S, Walker D L, Frisque R J. JC virus-simian virus 40 genomes containing heterologous regulatory signals and chimeric early regions: identification of regions restricting transformation by JC virus. J Virol. 1989;63:2180–2190. doi: 10.1128/jvi.63.5.2180-2190.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay N, Skolnik D H, Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982;29:183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- 23.Hirt B J. Selective extraction of polyoma DNA from infected mouse cell culture. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 24.Hou-Jong M H, Larsen S H, Roman A. Role of the agnoprotein in regulation of simian virus 40 replication and maturation pathways. J Virol. 1987;61:937–939. doi: 10.1128/jvi.61.3.937-939.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson V, Chalkley R. Use of whole-cell fixation to visualize replicating and maturing simian virus 40: identification of a new viral gene product. Proc Natl Acad Sci USA. 1981;78:6082–6085. doi: 10.1073/pnas.78.10.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jay G, Nomura S, Anderson C W, Khoury G. Identification of the SV40 agnogene product: a DNA binding protein. Nature. 1981;291:346–349. doi: 10.1038/291346a0. [DOI] [PubMed] [Google Scholar]
- 27.Kao S-Y, Calman A F, Luciw P A, Peterlin B M. Antitermination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 28.Kenney S, Natarajan V, Strike D, Khoury G, Salzman N P. JC virus enhancer-promoter active in human brain cells. Science. 1984;226:1337–1339. doi: 10.1126/science.6095453. [DOI] [PubMed] [Google Scholar]
- 29.Khalili K, Feigenbaum L, Khoury G. Evidence for a shift in 5′-termini of early viral RNA during the lytic cycle of JC virus. Virology. 1987;158:469–472. doi: 10.1016/0042-6822(87)90224-8. [DOI] [PubMed] [Google Scholar]
- 30.Khalili K, Brady J, Papas J, Spence S, Sadofsky M, Khoury G. COOH-terminal mutants of large tumor antigen of simian virus 40: a role for the early protein late in the lytic cycle. Proc Natl Acad Sci USA. 1988;85:354–358. doi: 10.1073/pnas.85.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krynska B, Otte J, Franks R, Khalili K, Croul C. Human ubiquitous JCV(CY) T-antigen gene induces brain tumors in experimental animals. Oncogene. 1999;18:39–46. doi: 10.1038/sj.onc.1202278. [DOI] [PubMed] [Google Scholar]
- 32.Krynska B, Del Valle L, Croul S, Gordon J, Katsetos C D, Carbone M, Giordano A, Khalili K. Detection of human neurotropic JC virus DNA sequence and expression of the viral oncogenic protein in pediatric medulloblastomas. Proc Natl Acad Sci USA. 1999;96:11519–11524. doi: 10.1073/pnas.96.20.11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laghi L, Randolph A E, Chauhan D P, Marra G, Major E O, Neel J V, Boland C R. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci USA. 1999;96:7484–7489. doi: 10.1073/pnas.96.13.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lashgari M, Tada H, Amini S, Khalili K. Regulation of JCVL promoter function. Transactivation of JCVL promoter by JCV and SV40 early proteins. Virology. 1989;170:292–295. doi: 10.1016/0042-6822(89)90381-4. [DOI] [PubMed] [Google Scholar]
- 35.Levy N D, Refaeli Y, Weiner D B. Extracellular Vpr protein increases cellular permissiveness to human immunodeficiency virus replication and reactivates virus from latency. J Virol. 1995;68:1243–1252. doi: 10.1128/jvi.69.2.1243-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Major E O, Amemiya K, Tornatore C S, Houff S A, Berger J R. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margolskee R F, Nathans D. Suppression of a VP1 mutant of simian virus 40 by missense mutations in serine codons of the viral agnogene. J Virol. 1983;48:405–409. doi: 10.1128/jvi.48.2.405-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mertz J E, Murphy A, Barkan A. Mutants deleted in the agnogene of simian virus 40 define a new complementation group. J Virol. 1983;45:36–46. doi: 10.1128/jvi.45.1.36-46.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michaud G, Zachary A, Rao V B, Black L W. Membrane-associated assembly of a phage T4 DNA entrance vertex structure studied with expression vectors. J Mol Med. 1989;209:667–681. doi: 10.1016/0022-2836(89)90599-8. [DOI] [PubMed] [Google Scholar]
- 40.Ng S C, Mertz J E, Sanden-Will S, Bina M. Simian virus 40 maturation in cells harboring mutants deleted in the agnogene. J Biol Chem. 1985;260:1127–1132. [PubMed] [Google Scholar]
- 41.Perini G, Wagner S, Green M R. Recognition of bZIP proteins by the human T-cell leukaemia virus transactivator Tax. Nature. 1995;376:602–605. doi: 10.1038/376602a0. [DOI] [PubMed] [Google Scholar]
- 42.Raj G, Khalili K. Transcriptional regulation: lessons from the human neurotropic polyomavirus, JCV. Virology. 1995;213:283–291. doi: 10.1006/viro.1995.0001. [DOI] [PubMed] [Google Scholar]
- 43.Raj G V, Khalili K. Identification and characterization of a novel GGA/C-binding protein, GBP-i, that is rapidly inducible by cytokines. Mol Cell Biol. 1994;14:7770–7781. doi: 10.1128/mcb.14.12.7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raj G V, Gordon J, Logan T J, Hall D J, Deluca A, Giordano A, Khalili K. Characterization of glioma cells derived from polyomavirus-induced brain tumors in hamsters. Int J Oncol. 1995;7:801–808. doi: 10.3892/ijo.7.4.801. [DOI] [PubMed] [Google Scholar]
- 45.Rencic A, Gordon J, Otte J, Curtis M, Kovatich A, Zoltick P, Khalili K, Andrews D. Detection of JC virus DNA sequence and expression of the viral oncoprotein, tumor antigen, in brain of immunocompetent patient with oligoastrocytoma. Proc Natl Acad Sci USA. 1996;93:7352–7357. doi: 10.1073/pnas.93.14.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rinaldo C H, Traavik T, Hey A. The agnogene of the human polyomavirus BK is expressed. J Virol. 1998;72:6233–6236. doi: 10.1128/jvi.72.7.6233-6236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Safak M, Gallia G L, Ansari S A, Khalili K. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J Virol. 1999;73:10146–10157. doi: 10.1128/jvi.73.12.10146-10157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Safak M, Gallia G L, Khalili K. Reciprocal interaction between two cellular proteins, Purα and YB-1, modulates transcriptional activity of JCVCY in glial cells. Mol Cell Biol. 1999;19:2712–2723. doi: 10.1128/mcb.19.4.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Small J A, Khoury G, Jay G, Howley P M, Scangos G M. Early regions of JC virus and BK virus induce distinct and tissue-specific tumors in transgenic mice. Proc Natl Acad Sci USA. 1986;83:8288–8292. doi: 10.1073/pnas.83.21.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strebel K, Klimkait T, Maldarelli F, Martin M A. Molecular and biochemical analysis of human immunodeficiency virus type 1 Vpu protein. J Virol. 1989;68:3784–3791. doi: 10.1128/jvi.63.9.3784-3791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swenson J J, Trowbridge P W, Frisque R J. Replication activity of JC virus large T antigen phosphorylation and zinc finger domain mutants. J Neurovirol. 1996;2:78–86. doi: 10.3109/13550289609146541. [DOI] [PubMed] [Google Scholar]
- 53.Swenson J J, Frisque R J. Biochemical characterization and localization of JC virus large T antigen phosphorylation domains. Virology. 1995;212:295–308. doi: 10.1006/viro.1995.1487. [DOI] [PubMed] [Google Scholar]
- 54.Tada H, Lashgari M, Rappaport J, Khalili K. Cell type-specific expression of JC virus early promoter is determined by positive and negative regulation. J Virol. 1989;63:463–466. doi: 10.1128/jvi.63.1.463-466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tada H, Lashgari M S, Khalili K. Regulation of JCVL promoter function: evidence that a pentanucleotide “silencer” repeat sequence AGGGAAGGGA down-regulates transcription of the JC virus late promoter. Virology. 1991;180:327–338. doi: 10.1016/0042-6822(91)90037-c. [DOI] [PubMed] [Google Scholar]
- 56.Terwilliger E F, Cohen E A, Lu Y C, Sodroski J G, Hasetine W A. Functional role of the human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trowbridge P M, Frisque R. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J Neurovirol. 1995;1:195–206. doi: 10.3109/13550289509113966. [DOI] [PubMed] [Google Scholar]
- 58.Varakis J, Zu Rhein G M, Padgett B L, Walker D L. Induction of peripheral neuroblastomas in Syrian hamsters after injection of neonates with JC virus. Cancer Res. 1978;38:1718–1722. [PubMed] [Google Scholar]
- 59.Walker D L, Padgett B L, ZuRhein G M, Albert A E, Marshal R F. Human papovavirus (JC): induction of brain tumors in hamsters. Science. 1973;17:674–676. doi: 10.1126/science.181.4100.674. [DOI] [PubMed] [Google Scholar]
- 60.Walker D L. Progressive multifocal encephalopathy: an opportunistic viral infection of the central nervous system. In: Vinken P J a B, Bruyn G W., editors. Handbook of clinical neurology. New York, N.Y: Elsevier North Holland; 1978. pp. 307–329. [Google Scholar]
- 61.Wei P, Garber M E, Fang S-M, Fisher W H, Jones K A. A novel CDK-9-associated C-type cyclin interacts directly with HIV-1 tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]