Abstract
Antimicrobial resistance mediated by extended-spectrum beta-lactamase (ESBL)- and plasmid-mediated cephalosporinase (AmpC)-producing Enterobacterales, as well as carbapenemase-producing Enterobacterales have globally increased among companion animals, posing a potential health risk to humans in contact with them. This prospective longitudinal study investigates the transfer of ESBL/AmpC- and carbapenemase-producing Enterobacterales between companion animals and their cohabitant humans in Portugal (PT) and the United Kingdom (UK) during animal infection. Fecal samples and nasal swabs collected from dogs and cats with urinary tract infection (UTI) or skin and soft tissue infection (SSTI), and their cohabitant humans were screened for resistant strains. Relatedness between animal and human strains was established by whole-genome sequencing (WGS). ESBL/AmpC-producing Enterobacterales were detected in companion animals (PT = 55.8%; UK = 36.4%) and humans (PT = 35.9%; UK = 12.5%). Carbapenemase-producing Enterobacterales carriage was observed in one dog from Portugal (2.6%) and another dog from the UK (4.5%). Transmission of index clinical ESBL-producing Escherichia coli and Klebsiella pneumoniae strains to cohabitant humans was observed in three Portuguese households (6.9%, n = 43), with repeated isolation of the index strains on fecal samples from the animals and their cohabiting humans. In addition, longitudinal sharing of E. coli strains carried by companion animals and their owners was observed in other two Portuguese households and two households from the UK. Furthermore, a multidrug-resistant ACT-24-producing Enterobacter hormaechei subsp. hoffmannii strains were also shared within another Portuguese household. These results highlight the importance of the household as an epidemiological unit in the efforts to mitigate the spread of antimicrobial resistance, further emphasizing the need for antimicrobial surveillance in this context, capable of producing data that can inform and evaluate public health actions.
Keywords: one health, ExPEC pathotypes, animal–human sharing, Klebsiella pneumoniae, Enterobacter hormaechei subsp. hoffmannii, CTX-M-15 ESBL, CTX-M-27, CMY-2
1. Introduction
The gastrointestinal tract is an important reservoir for antimicrobial-resistant Enterobacterales that may cause both intestinal and extraintestinal infections (Geurtsen et al., 2022). One critical mechanism driving antimicrobial resistance (AMR) in Enterobacterales involves the enzymatic inactivation of a wide range of beta-lactam antimicrobials, including third- and fourth-generation cephalosporins (3GCs and 4GCs) through the action of extended-spectrum β-lactamases (ESBLs) and plasmid-mediated cephalosporinases (AmpCs), and also carbapenem inactivation by carbapenemase-producing Enterobacterales (CPE) (Eiamphungporn et al., 2018). These medically important antimicrobials are critical in the management of serious infections caused by multidrug-resistant bacteria, and resistance to them poses a significant public health concern (World Health Organization, 2024).
Antimicrobial use can increase the burden of AMR, leading to long-term disturbance of commensal bacterial populations and prolonged carriage of antibiotic-resistant strains in the host environment even in the absence of selective pressure (Jernberg et al., 2010; Schmidt et al., 2018). In the endeavor to attenuate the emergence of resistance within the veterinary settings, the European Medicines Agency (EMA) has categorized 3GCs and 4CGs as ‘Restrict’ (Category B) in veterinary medicine and carbapenems as ‘Avoid’ (Category A) (EMA, 2019), the latter being recently classified as authorized only for use in humans by the World Health Organization (WHO), over concerns regarding the impact of the development of resistance in animals on humans (World Health Organization, 2024). However, the prevalence of ESBL- and AmpC-producing Enterobacterales (ESBL/AmpC-E) as well as CPE strains has been increasing worldwide in companion animals (Grönthal et al., 2018; Hong et al., 2020; Dazio et al., 2021).
ESBL/AmpC-E and CPE strains carried by companion animals are of particular One Health concern as they may act as a reservoir for self-infection and further transmit resistance genes and/or pathogens into other hosts including humans, other animals, and the environment (Damborg et al., 2016; Pomba et al., 2020; Donà et al., 2023). The close contact between humans and companion animals provides ample opportunities for the exchange of microbiota, potentially facilitating the spread of AMR (Pomba et al., 2020). Recent studies have shown that these bacteria can be transmitted between pets and their owners, indicating a bidirectional flow of resistant strains (Grönthal et al., 2018; Toombs-Ruane et al., 2020; Van Den Bunt et al., 2020). However, little is known about the dynamics of transmission in humans and companion animals belonging to the same household, particularly from sick companion animals to their healthy owners.
We hypothesize that companion animals under antibiotic treatment will share resistant bacteria with their cohabitant humans. As such, this study aims to explore the contribution of companion animals under antibiotic treatment in the AMR dissemination to the community setting. Hence, to assess the within-household transmission of resistant bacteria, a prospective and longitudinal study was conducted to describe the molecular epidemiology and evaluate the sharing of ESBL/AmpC-E and CPE strains between companion animals under antimicrobial treatment for skin and soft tissue infections (SSTI) or urinary tract infections (UTIs) and their cohabitant humans in the community. The sharing of ESBL/AmpC- and carbapenemase-producing strains was analyzed with two approaches, after the detection of their carriage among companion animals and their cohabitant humans: (i) the transmission of the animals’ clinical pathogenic strain to their cohabitant household humans and (ii) the transfer of resistant strains between infected companion animals under antibiotic therapy and their cohabitant humans, resulting in carriage and/or colonization. This was a 2-year, multicenter study conducted in Lisbon, Portugal, and the South-East United Kingdom (UK).
2. Materials and methods
2.1. Ethics approval
Ethical clearance for the collection of samples from humans and companion animals was obtained from the Faculty of Veterinary Medicine, University of Lisbon (FMV-ULisboa) Ethics Committee for Research and Education and the Royal Veterinary College (RVC) Ethics and Welfare Committee (ethics reference number: CEBEA 027/2018 and URN 2017 1750–3, respectively). Written consent for sample collection and the use of participants’ anonymized questionnaire-derived data was obtained from owners prior to enrollment in the study. Research was conducted in accordance with the Declaration of Helsinki and national and institutional standards.
2.2. Study design and setting
This study was part of an international prospective longitudinal observational study conducted at the Small Animal Veterinary Teaching Hospital of the Faculty of Veterinary Medicine, University of Lisbon, Portugal, and the RVC Small Animal Veterinary Referral Service, Royal Veterinary College, Hatfield, UK.
Between 2018 and 2021, dogs (n = 60) and cats (n = 5) that were presented to the veterinary hospitals for care for infection consultations were enrolled on a voluntary basis, together with their cohabitant humans (n = 102), if they fulfilled the criteria for diagnosis of the following infections: UTI according to the International Society of Companion Animal Infectious Diseases (ISCAID) guidelines (Weese et al., 2019), SSTI according to results of diagnostic tests (e.g., cytology and/or culture), and typical clinical signs of superficial pyoderma, deep pyoderma, and wound infections. Other inclusion criteria for enrollment of companion animals and humans were as follows: (i) no systemic antimicrobial therapy in the last 3 months; (ii) no topical antimicrobial therapy 2 days before sampling; and (iii) have lived together for at least 3 months (cohabitant). To ensure that participation was anonymous, households, humans, and animals were coded.
Epidemiological questionnaires assessing participants’ general health status, current or previous medical treatments, and exposure to healthcare facilities, were filled out by the human participants. In addition, owners were asked about their animal’s lifestyle, diet, contact with other animals, and closeness of the contact with their human counterparties, such as kissing/licking the owner’s face. For all variables on the questionnaire, the option ‘Prefer not to answer’ was available. The number of answers collected for any specific factor depended on whether the owner decided to disclose the information. Questionnaire variables and responses, by country, are listed in Table 1.
Table 1.
Questionnaire responses on demographic, social, and clinical data of dogs (n = 60), cats (n = 5), and their cohabiting humans (n = 102) by country, 2018–2021.
| Variables | Participants | Portugal | United Kingdom | ||
|---|---|---|---|---|---|
| Positive responses | Beta-lactam resistance carriage | Positives responses | Beta-lactam resistance carriage | ||
| Demographic | |||||
| Female | Humans | 46 (n = 78) | 15 | 6 (n = 11) | 0 |
| Dogs | 20 (n = 38) | 14 | 3 (n = 7) | 1 | |
| Cats | 2 (n = 5) | 0 | N/A | N/A | |
| Male | Humans | 32 (n = 78) | 13 | 5 (n = 11) | 2 |
| Dogs | 18 (n = 38) | 7 | 3 (n = 6) | 2 | |
| Cats | 3 (n = 5) | 3 | N/A | N/A | |
| Median age (years) | Humans | 43.4 (n = 75) | N/A | 47 (n = 13) | N/A |
| Dogs | 8.3 (n = 38) | N/A | 6.5 (n = 7) | N/A | |
| Cats | 8.8 (n = 5) | N/A | N/A | N/A | |
| Clinical | |||||
| Antibiotic treatment 3 to 12 months before sampling | Humans | 31 (n = 75) | 15 | 4 (n = 13) | 3 |
| Dogs | 31 (n = 38) | 17 | 14 (n = 22) | 5 | |
| Cats | 4 (n = 5) | 3 | N/A | N/A | |
| Antibiotic treatment 3 to 6 months before sampling | Humans | 17 (n = 77) | 9 | 3 (n = 12) | 2 |
| Dogs | 15 (n = 38) | 8 | 2 (n = 18) | 2 | |
| Cats | 0 (n = 5) | N/A | N/A | N/A | |
| Hospitalization in the 12 months before sampling | Humans | 6 (n = 77) | 2 | 1 (n = 13) | 0 |
| Dogs | 14 (n = 38) | 10 | 3 (n = 8) | 2 | |
| Cats | 3 (n = 5) | 2 | N/A | N/A | |
| The companion animal had probiotic treatment in the last year | Dogs | 5 (n = 38) | 4 | 2 (n = 8) | 1 |
| Cats | 1 (n = 5) | 1 | N/A | N/A | |
| The companion animal had glucocorticoid treatment in the last year | Dogs | 16 (n = 38) | 6 | N/A | N/A |
| Cats | 0 (n = 5) | N/A | N/A | N/A | |
| The human has a chronic disease | Humans | 29 (n = 74) | 14 | 2 (n = 12) | 0 |
| Social | |||||
| The companion animal has an indoor lifestyle | Dogs | 34 (n = 38) | 20 | 11 (n = 13) | 4 |
| Cats | 5 (n = 5) | 3 | N/A | N/A | |
| The companion animal socializes with other animals outside the household | Dogs | 17 (n = 38) | 8 | 7 (n = 13) | 3 |
| Cats | 0 (n = 5) | N/A | N/A | N/A | |
| The companion animal participates in pet animal shows | Dogs | 0 (n = 38) | N/A | 0 (n = 13) | N/A |
| Cats | 0 (n = 5) | N/A | N/A | N/A | |
| The companion animal is a working animal | Dogs | 0 (n = 38) | N/A | 0 (n = 13) | N/A |
| Cats | 0 (n = 5) | N/A | N/A | N/A | |
| The companion animal stayed in a pet hotel in the last year | Dogs | 6 (n = 38) | 2 | 1 (n = 13) | 0 |
| Cats | 0 (n = 5) | N/A | N/A | N/A | |
| The companion animal sleeps in the human bed | Dogs | 14 (n = 38) | 8 | 3 (n = 13) | 0 |
| Cats | 4 (n = 5) | 3 | N/A | N/A | |
| The human cleans up companion animal waste | Humans | 49 (n = 78) | 27 | 13 (n = 13) | 6 |
| The human feeds companion animal | Humans | 59 (n = 78) | 35 | 13 (n = 13) | 6 |
| The human pets/cuddles with companion animals | Humans | 71 (n = 78) | 42 | 13 (n = 13) | 6 |
| The human allows kissing/ licking by companion animals | Humans | 36 (n = 76) | 20 | 13 (n = 13) | 6 |
| Other | |||||
| The companion animal eats raw meat/raw fish | Dogs | 3 (n = 38) | 2 | 2 (n = 13) | 1 |
| Cats | 0 (n = 5) | N/A | N/A | N/A | |
| The human is a healthcare professional | Humans | 8 (n = 78) | 3 | 1 (n = 13) | 0 |
| The human traveled abroad in the past 12 months | Humans | 35 (n = 76) | 22 | 8 (n = 13) | 2 |
Data are reported as the number of individuals; n indicates the total responses. For carriage results, data are reported as a number of positive participants who presented the characteristic. N/A, not applicable.
2.3. Sample collection
Samples were collected, immediately after enrollment (T0), 1 week after antimicrobial treatment started (T1), 1 month after antimicrobial treatment started (T2), and 2 months after antimicrobial treatment started (T3) (Table 2). At each timepoint, the inclusion criteria were reviewed, and participants were excluded if any change was reported. Epidemiological variables were also monitored to ensure that changes in these factors, such as household environment, that could be possible contamination sources, were accounted for throughout the study. Acquisition of follow-up samples depended on the owner’s willingness to continue to participate in the study with their respective companion animal. For these reasons, at T1, sample collection was performed only for 18 households in Portugal and only in one household in the UK, while for T2, 19 households from Portugal and 16 households from the UK were studied. Some participants did not deliver samples a week after the enrollment and returned to give samples a month later at T2 (Table 2).
Table 2.
Data collection timepoints for the longitudinal study by country, 2018–2021.
| Data collection timepoints | Portugal (n = 43 households) | United Kingdom (n = 22 households) | ||
|---|---|---|---|---|
| Animals (n = 43) | Humans (n = 78) | Animals (n = 22) | Humans (n = 24) | |
| T0: recruitment | 33 | 59 | 22 | 24 |
| T1: antibiotic treatmenta,b | 18 | 41 | 1 | 1 |
| T2: 1 month after T0c | 19 | 37 | 16 | 17 |
| T3: 2 months after T0 | 14 | 27 | 11 | 12 |
aT1 was performed 1 week after antimicrobial treatment started.
bFor 10 households from Portugal (10 animals and 18 humans), recruitment started at this timepoint.
cFor 10 households from Portugal (10 animals and 15 humans) and 15 households from the UK (15 animals and 17 humans), sample collection was not possible at T1 but only at T2.
At each time point, animals were evaluated by the attending veterinarian, and infection samples were collected and analyzed by the hospital’s Microbiology diagnostic laboratory according to standard procedures. In Portugal, the clinical strains were subsequently molecularly characterized for research purposes.
Instructions for at-home fecal sample collection were given to the owners. Human feces were to be collected directly into sterile plastic containers or by using ‘FeCol’ feces collection papers (Alpha Laboratories Ltd., United Kingdom) and then transferred into a sterile plastic container. Infant fecal samples were collected from diapers by their parents. Companion animal’s partial fecal samples (that did not touch the ground) were collected using sterile gloves and placed into a sterile container. Nasal swabs were collected by an attending veterinarian; in brief, a single sterile cotton swab was inserted approximately 1 cm into one nostril and then rotated along the mucous membrane for 5 s. The samples were stored for a maximum of 48 h at 4°C until processing.
2.4. Sample processing
One gram of homogenized fecal sample was added to 10 mL of sterile 0.85% NaCl (Merck, Germany) solution and mixed thoroughly by vortexing. Ten microliters of fecal suspension were plated onto MacConkey agar plates (Biokar Diagnostics, France) with and without 1.5 μg/mL of cefotaxime (Sigma-Aldrich, US) or 1 μg/mL meropenem (Sigma-Aldrich, US) supplementation; MacConkey agar plates containing temocillin 30μg disk (Mast Group, UK) were used as phenotypic indicator of OXA-48-like production (Da Silva et al., 2022). To improve the detection of low numbers of resistant bacteria, 1 g of feces was also diluted in 10 mL of sterile buffered peptone water (Biokar Diagnostics, France) and incubated at 36 ± 1°C for 24 h, followed by inoculation of 10 μL onto selective plates as described above. Nasal swabs were inoculated in 3 mL of NaCl 0.85% (Merck, Germany), and then 100 mL of this were streaked on selective media described above. After overnight incubation at 36 ± 1°C, up to five resistant suspected colonies of each different morphology were isolated and stored in 20% glycerol (Sigma-Aldrich, US) brain heart infusion broth (Biokar Diagnostics, France) at −20°C until processing.
2.5. Molecular characterization of isolates
Bacterial DNA was extracted by heat lysis and centrifugation for all obtained isolates (Féria et al., 2002) and species identification was performed by 16S rRNA gene sequencing, as described elsewhere (Salisbury et al., 1997). An extended PCR scheme described by Doumith et al. (2012) was used to assign the Escherichia coli isolates to one of the four major phylogroups (A, B1, B2, and D).
Genetic relationships between all E. coli isolates were initially determined by repetitive element sequence-based PCR (REP-PCR) typing (Versalovic et al., 1991). Persistence was defined as the isolation of strains with matching molecular profiles (REP-PCR and resistance determinants) from repeated samples of the same subject.
All isolates were tested for the presence of specific ESBLs (blaCTX-M-type, blaTEM, and blaSHV) (Eckert et al., 2006; Pomba et al., 2006), AmpC variants (blaCIT-type, blaFOX-type, blaMOX-type, blaDHA-type, blaACT-type, and blaMIR-type) (Marques et al., 2019), and carbapenemases (blaAIM, blaDIM, blaGIM, blaSIM, blaIMP, blaVIM, blaSPM, blaNDM, blaKPC, blaBIC, and blaOXA-48-like) (Poirel et al., 2011) encoding genes by PCR and sequencing, due to their clinical relevance, prevalence in antimicrobial resistance, and major public health impact (Bevan et al., 2017; Salgado-Caxito et al., 2021; Da Silva et al., 2022; Zamudio et al., 2022; UK Health Security Agency, 2024).
2.6. Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) of a panel of antibiotics were determined by broth microdilution using the MicroScan® Neg MIC Panel Type 44 (Beckman Coulter, US) following guidelines by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints 2024 (The European Committee on Antimicrobial Susceptibility Testing, 2024) and the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2024). Multidrug resistance was defined as non-susceptibility to at least one agent in three or more antimicrobial class categories (Sweeney et al., 2018). For E. coli strains, antimicrobial susceptibility testing was only performed for one representative isolate from each REP-PCR profile detected per participant per timepoint.
2.7. Whole-genome sequencing
ESBL/AmpC-producing Enterobacterales strains (n = 38) shared by companion animals and cohabitant humans based on REP-PCR profiling were further characterized by WGS. For this part of the study, genomic DNA was extracted using the NZY Tissue gDNA Isolation kit (NZYTech, Portugal). Library preparation was performed with the TruSeq DNA PCR-Free preparation kit and sequenced using the Illumina NovaSeq 6000 system with 2 × 150 bp paired-end reads (Illumina, San Diego, California, US) at a commercial company (Macrogen, Seoul, Republic of Korea).
The raw sequence reads were assessed for quality using FastQC v0.11.9 (Andrews, 2010) and filtered for low-quality reads using PRINSEQ v0.20.4 (Schmieder and Edwards, 2011) (mean base quality score of ≥20 and minimum read length of 90 nt), making an average of 9.1×106 high-quality reads per library. De novo assemblies were generated with SPAdes v3.14.1 (Bankevich et al., 2012) followed by two runs of polishing with Pilon v1.24 (Walker et al., 2014) and annotated using Prokka v1.14.6 (Seemann, 2014). Assemblies’ quality was assessed using QUAST v5.0.2 (Gurevich et al., 2013). Genome assemblies presented an average of L50 = 9.6 (range from 6 to 14) and N50 = 1.9×105 (range from 9.1×104 to 3.9×105) and an average depth of 258x; full WGS statistics are listed in Supplementary Table S1.
2.7.1. Genome sequencing analysis
Antimicrobial resistance genes were assessed using AMRFinder (Feldgarden et al., 2021) and ResFinder (Bortolaia et al., 2020); plasmid replicons were obtained using PlasmidFinder (Carattoli et al., 2014), and sequence types were assigned using the MLST 2.0 (Larsen et al., 2012). E. coli virulence factors were obtained using VirulenceFinder 2.0 (Joensen et al., 2014), whereas Klebsiella spp. virulence factors were obtained using the Pathogenwatch web application (Centre for Genomic Pathogen Surveillance, 2018). E. coli strains were classified as extraintestinal pathogenic E. coli (ExPEC), uropathogenic E. coli (UPEC), or avian pathogenic E. coli (APEC) based on established molecular definitions (Menezes et al., 2023).
2.7.2. Clonal relationships
Three datasets were defined for the possible shared 3GC-resistant strains, one including E. coli strains (n = 29), a second comprising K. pneumoniae strains (n = 7), and a final one with Enterobacter hormaechei strains (n = 2). Parsnp v1.2 (Treangen et al., 2014) was used to generate single nucleotide polymorphism (SNP) alignment. The complete genome sequence of E. coli K-12 MG1655 (GenBank accession: GCA_000005845.2) was used as the reference genome for the E. coli dataset, K. pneumoniae subsp. pneumoniae MGH 78578 (GenBank accession: GCA_000016305.1) as reference for K. pneumoniae dataset, and E. hormaechei NCTC 9394 (GenBank accession: GCA_000210775.1) as reference for E. hormaechei dataset. Gubbins (Croucher et al., 2015) and RaxML-NG (Kozlov et al., 2019) were used to provide a maximum likelihood tree with 100 bootstrap repeats based on the recombination core genome alignment. Snp-dists v.0.8.2 (Seemann et al., 2021) was used to extract the number of SNPs between strains. Microreact platform (Argimón et al., 2016) was used to visualize the phylogenetic tree linked to antimicrobial resistance data. Comparison of ORFs from companion animal’s-owner paired strains was performed by using the EasyFig and the BLASTn algorithm (Sullivan et al., 2011).
2.8. Statistical analysis
Statistical analysis was performed using the SAS statistical software package for Windows, version 9.3 (SAS Institute Inc., Cary, United States). For comparative analysis of baseline characteristics, Fisher’s exact test was used for categorical variables, and a p-value<0.05 was considered significant.
To identify risk factors for ESBL/AmpC-E carriage, contingency tables were generated using the collected demographic and clinical data to perform univariable logistic regression analysis. Since no variables with a p-value<0.1 were identified, it was not possible to build multivariable models.
3. Results
3.1. Study population
3.1.1. Dogs and cats
A total of 43 households from Portugal and 22 from the UK were enrolled in this study (Table 2; Supplementary Figure S1), and each household had one companion animal.
In Portuguese households, 43 companion animals were included, consisting of 38 dogs and 5 cats. Among these animals, 21 had UTIs and 22 had SSTIs. In the UK households, 22 companion animals were included, all of which were dogs. Four of these dogs had UTIs, and 18 had SSTIs.
Table 1 presents detailed demographic, social, and clinical data for the study participants. The ages of the companion animals ranged from 1.8 to 15 years, with a median age of 8 years (n = 50). Most of the companion animals in both countries lived indoors. In Portugal, cats and dogs tend to sleep more often in their owners’ beds compared to dogs in the UK. Moreover, in Portugal, 81.6% (n = 31/38) of the dogs and 80% of the cats received antimicrobials within the 12 months before sampling, and for the UK dogs, this percentage was 63.6% (n = 14/22) (Table 1).
3.1.2. Pet owners
In Portugal, a total of 78 humans cohabitating with companion animals with SSTI or UTI provided nasal and fecal samples. In the UK households, 24 humans were enrolled. The households had different compositions, with 1 to 5 humans per household. Human ages ranged from 3 months to 82 years, with a median age of 43.3 years (n = 88).
A high number of humans reported close contact behaviors with their companion animals, such as petting/cuddling and being kissed/licked by them. Regarding antibiotic treatments, among the humans who filled in the questionnaire, 41.3% (n = 31/75) from Portugal and 30.8% (n = 4/13) from the UK took antibiotics within the 12 months before sampling (Table 1).
3.2. Prevalence of ESBL/AmpC- and carbapenemase-producing Enterobacterales carriage
Considering all four sampling timepoints, CPE strains were recovered from only two dogs (3.3, 95%CI, 0–8, n = 2/60), specifically one from Portugal (2.6, 95%CI, 0–7.9, n = 1/38) and the other from the UK (4.6, 95%CI, 0–13.9, n = 1/22).
ESBL/AmpC-E strains were isolated from 30.4% (95%CI, 21.3–39.5, n = 31/102) humans and 49.2% (95%CI, 36.7–61.7, n = 32/65) companion animals (dogs and cats) in at least one timepoint sample. There was no significant difference between the prevalence of ESBL/AmpC-E carriage in humans and companion animals (p = 0.158). In addition, no significant difference was found between carriage in animals with SSTI or UTI (p = 0.060); hence, these infection groups were combined for further analysis.
3.2.1. Comparative analysis by country
When comparing the two countries, no significant difference was found in the prevalence of ESBL/AmpC-E strains in companion animals from Portugal (55.8, 95%CI, 40.3–71.3, n = 24/43) and the United kingdom (36.4, 95%CI, 14.5–58.2, n = 8/22) (p = 0.138).
For humans, a significant difference was detected with a higher prevalence of ESBL/AmpC-E strains in Portugal (35.9, 95%CI, 25–46.8, n = 28/78) than in the United kingdom (12.5, 95%CI, 0–26.8, n = 3/24; p = 0.029). Due to this variation in results according to the country, the risk factor analysis for both companion animals and humans was carried out separately for each country.
Table 1 shows the complete set of variables considered in the risk factor analysis. None of them were significantly associated with ESBL/AmpC-E or CPE carriage in humans or companion animals (p-values>0.1).
3.2.2. Isolate distribution
In total, 254 ESBL/AmpC-E and/or CPE carriage isolates were recovered from 63 participants (31 humans and 32 companion animals). These comprised two Enterobacter hormaechei subsp. hoffmannii isolates, 23 K. pneumoniae, and 229 E. coli isolates. REP-PCR profiling showed that only 115 E. coli strains were non-duplicate (Supplementary Figures S2-S4: dendrograms generated from REP-PCR fingerprinting of PT and UK strains). Among these non-duplicate E. coli isolates, phylogroup A and B1 were most frequently found in Portugal, followed by B2, while in the UK, phylogroup D was the most common (Supplementary Table S2).
3.2.3. Antimicrobial susceptibility
Enterobacterales non-duplicate strains (n = 140) were further characterized through antimicrobial susceptibility testing, and MICs for each strain are displayed in Supplementary Table S2. All the strains were resistant to cefotaxime, a 3GC agent, presenting MIC values for this antibiotic ranging from 4 to >32 mg/L. Among these, two strains from a Portuguese dog were recovered from MacConkey agar plates supplemented with temocillin (30 μg) antibiotic disks and were susceptible to meropenem (MIC values ≤1 mg/L). Other four strains from a UK dog were recovered from meropenem-supplemented MacConkey agar plates, presenting a resistant profile to this antibiotic (MIC values >8 mg/L).
In Portugal, high resistance rates among companion animal strains were also observed for ampicillin (94.8%), the combination of ampicillin/sulbactam (84.5%), fourth-generation cephalosporin, cefepime (75.9%), and ciprofloxacin (63.8%). Human strains showed similar resistance patterns, with all of them being resistant to ampicillin and 88.7% to cefepime. Notably, a large percentage of carriage strains from companion animals were multidrug-resistant (96.2%), while 66% of the human strains presented this profile (Table 3).
Table 3.
Antimicrobial resistance of ESBL/AmpC- and Carbapenemase-producing Enterobacterales strains isolated from feces and nasal swabs of companion animals and their cohabitant humans in Portugal and the United Kingdom.
| Antimicrobial | Portugal | United Kingdom | ||
|---|---|---|---|---|
| Companion animals’ strains (n = 58) |
Humans strains (n = 53) |
Companion animals’ strains (n = 24) |
Humans strains (n = 5) |
|
| %R (No) | %R (No) | %R (No) | %R (No) | |
| Amikacin | 1.7 (1) | 0 (0) | 0 (0) | 0 (0) |
| Amoxicillin/clavulanate | 36.2 (21) | 7.5 (4) | 79.2 (19) | 80 (4) |
| Ampicillin/sulbactam | 84.5 (49) | 71.7 (38) | 95.8 (23) | 100 (5) |
| Ampicillin | 94.8 (55) | 100 (53) | 87.5 (21) | 100 (5) |
| Aztreonam | 81 (47) | 60.4 (32) | 91.7 (22) | 80 (4) |
| Ceftazidime | 77.6 (45) | 73.6 (39) | 95.8 (23) | 100 (5) |
| Cefepime | 75.9 (44) | 88.7 (47) | 50 (12) | 20 (1) |
| Cefotaxime | 100 (58) | 100 (53) | 100 (24) | 100 (5) |
| Cefoxitin | 20.7 (12) | 7.5 (4) | 70.8 (17) | 80 (4) |
| Cephalothin | 100 (58) | 100 (53) | 100 (24) | 100 (5) |
| Ciprofloxacin | 63.8 (37) | 43.4 (23) | 16.7 (4) | 0.2 (1) |
| Chloramphenicol | 37.9 (22) | 15.1 (8) | 29.2 (7) | 0 (0) |
| Ertapenem | 5.2 (3) | 1.9 (1) | 16.7 (4) | 0 (0) |
| Gentamicin | 18.9 (11) | 7.6 (4) | 29.2 (7) | 0 (0) |
| Imipenem | 0 (0) | 0 (0) | 16.7 (4) | 0 (0) |
| Meropenem | 0 (0) | 0 (0) | 16.7 (4) | 0 (0) |
| Piperacillin/tazobactam | 17.2 (10) | 7.6 (4) | 25 (6) | 0 (0) |
| Trimethoprim/sulfamethoxazole | 53.5 (31) | 67.9 (36) | 37.5 (9) | 0 (0) |
| Multidrug-resistant | 86.2 (50) | 66 (35) | 54.2 (13) | 0 (0) |
Minimum inhibitory concentrations (MICs) were interpreted using the criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2024 (The European Committee on Antimicrobial Susceptibility Testing, 2024), except for amoxicillin/clavulanate, ampicillin/sulbactam, and trimethoprim/sulfamethoxazole, for which criteria from the Clinical and Laboratory Standards Institute (CLSI) were used (CLSI, 2024). Data are reported as a number (No) of resistant (R) strains; n, total number of non-duplicate strains tested.
In the United Kingdom, resistance rates for companion animal strains were high for ampicillin/sulbactam (95.8) and aztreonam (91.7%). However, the resistance to ciprofloxacin was lower in companion animal strains (16.7%) compared to Portugal. Human strains showed 100% resistance to ampicillin. None of the human strains presented a multidrug-resistant profile, and only 54.2% of the companion animal strains did (Table 3).
3.2.4. Genes conferring resistance to third-generation cephalosporins and carbapenems
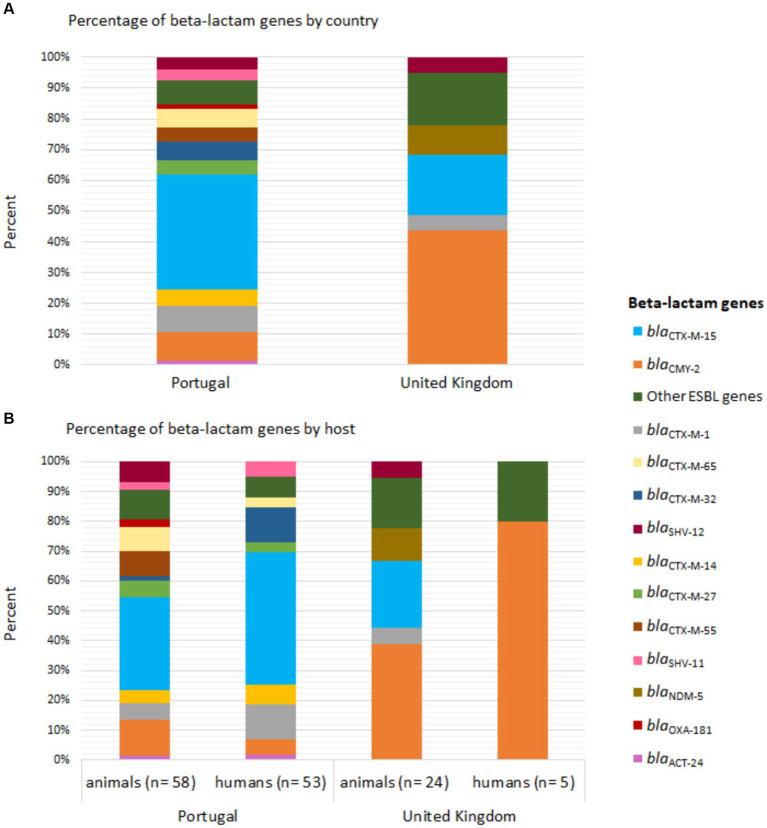
The distribution of beta-lactam resistance genes varied between strains from the two countries. The blaCTX-M-15 gene was the most frequent in strains from Portugal (humans: 49.1%, n = 26/53; animals: 39.7%, n = 23/58), followed by blaCTX-M-1 and blaCTX-M-32 in the humans (13.2%, n = 7/53) and blaCMY-2 in animals (15.5%, n = 9/58) (Figure 1). In the UK, the blaCMY-2 gene was the most common (humans: 80%, n = 4/5; animals: 58.3%, n = 14/24) (Figure 1).
Figure 1.
Distribution of beta-lactam genes in Enterobacterales strains carried by companion animals and their cohabitant humans in Portugal and the United Kingdom. (A) Percentage of beta-lactam genes by country and (B) host. The bars are color-coded according to the gene, as indicated in the legend inset.
Furthermore, carbapenemase-encoding genes were found in E. coli strains recovered from meropenem and temocillin-supplemented MacConkey agar plates. In Portugal, a dog carried the blaOXA-181 gene in two meropenem-susceptible E. coli strains; in the UK, four meropenem-resistant strains harboring the blaNDM-5 gene were isolated from one dog, as has been previously reported (Brilhante et al., 2020; Menezes et al., 2023).
3.3. Longitudinal isolation of beta-lactam-resistant Enterobacterales carriage strains
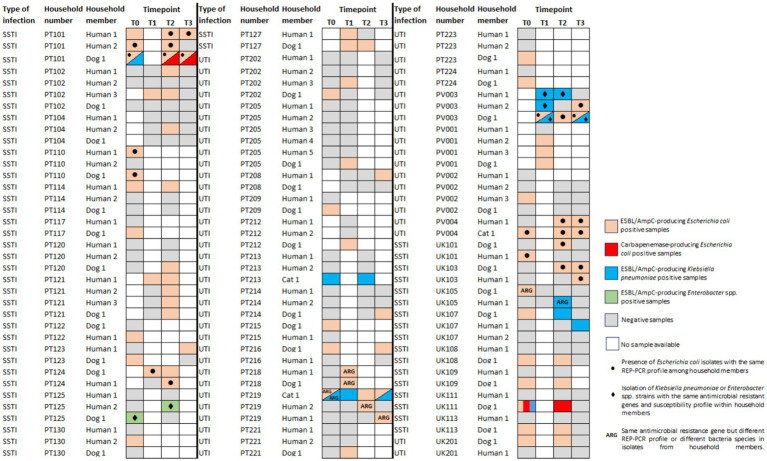
In Portugal, of the 43 households enrolled, 32 had at least one participant who carried an ESBL/AmpC-E and/or CPE strain. As the collection of follow-up samples was dependent on the owner’s willingness to continue to participate in the study, only 22 of the 32 positive households delivered samples at two or more timepoints (Figure 2). Repeated isolation of ESBL/AmpC-E strains was observed for eight humans and eight companion animals from 11 households (Figure 2). Persistence of the same ESBL/AmpC-producing E. coli strains was detected by REP-PCR profiling in three humans (PT101H1, PT101H2, and PV004H1) and four companion animals (PT101D1, PT127D1, PV003D1, and PV004C1) (Figure 2; Supplementary Figure S3).
Figure 2.
Distribution of ESBL/AmpC- or carbapenemase-producing Enterobacterales across fecal/nasal swabs sampling timepoints of 32 positive households in Portugal (PT) and nine households from the United Kingdom (UK). See the color key on the right side of the figure. SSTI, skin and soft tissue infection; UTI, urinary tract infection.
In the UK, nine of the 22 households had at least one participant colonized by an ESBL/AmpC-E and/or CPE strain. All positive households had follow-up samples (Figure 2). Two dogs had the same E. coli strain identified in subsequent samples (UK103D1 and UK111D1) (Supplementary Figure S4); another dog (UK109D1) had K. pneumoniae strains with the same susceptibility profile and resistant genes observed in different timepoints (Figure 2).
The acquisition of ESBL/AmpC-E strains by companion animals that were initially negative (PT: n = 32; UK: n = 15) occurred in five (15.6%) companion animals from Portugal (1 week after treatment for three dogs; 1 month after treatment for another dog; and 2 months after treatment for another dog) and in two dogs from the UK (13.3%), 1 month after treatment (Figure 2).
3.4. Bacterial strains causing infection in companion animals
A variety of bacterial strains were diagnosed as causes of infections in the 65 companion animals enrolled. Most of the SSTIs in both countries were caused by staphylococci, while UTIs were mostly due to E. coli (Supplementary Table S3). In Portugal, a total of 22 Enterobacterales clinical strains were isolated, of which 10 of them (47.6%) were resistant to 3GCs. Molecular analyses showed the presence of ESBL/AmpC encoding genes in all of them (Table 4). Regarding UK clinical strains, seven strains were identified as Enterobacterales, and only two displayed resistance to 3GCs. These resistant strains were not subjected to further investigation.
Table 4.
Genotypic characteristics of third- and fourth-generation cephalosporin-resistant Enterobacterales strains causing infection in companion animals (n = 10).
| Household | Host | Type of infectiona | Strain code | Bacterial species | Pattern of resistanceb | Beta-lactam-resistant genes |
|---|---|---|---|---|---|---|
| PT110 | Dog | SSTI | PT110/0-D1F3E1 | Escherichia coli | AMC, AMP, C, CIP, CFL, CTX, F, FOX, TMS | blaCTX-M-65, blaTEM-1 |
| PT125 | Dog | SSTI | PT125/0-D1I3E1 | Escherichia coli | AK, AMP, CAZ, CFL, CIP, CTX, CPM, F | bla CTX-M-27 |
| PT202 | Dog | UTI | PT202/0-D1I3E1 | Escherichia coli | AMP, CIP, CFL, CTX, TMS | blaCTX-M-1, blaTEM-1 |
| PT209 | Dog | UTI | PT209/0-D1I3E1 | Escherichia coli | AMC, AMP, CAZ, CFL, CN, CTX, F, FOX, TMS | blaCMY-2, blaTEM-1 |
| PT212 | Dog | UTI | PT212/0-D1I4E1 | Escherichia coli | AMP, CAZ, CFL, CIP, CTX, FOX | bla CTX-M-15 |
| PT213 | Cat | UTI | PT213/T0-C1I3K1 | Klebsiella pneumoniae | AMC, AMP, CAZ, CFL, CN, CPM, CTX, F, FOX, TET, TMS | blaCTX-M-15, blaSHV-28, blaTEM-1 |
| PV001 | Dog | UTI | PV001G1D1U1 | Enterobacter spp. | AMC, AMP, CAZ, CFL, C, CIP, CN, CTX, F, FOX, TET, TMS | bla CMY-2 |
| PV002 | Dog | UTI | PV002V1UD1U1 | Klebsiella pneumoniae | AMC, CAZ, CIP, CFL, CN, CTX, CPM, TMS | blaCTX-M-15, blaCMY-2, blaTEM-1 |
| PV003 | Dog | UTI | PV003V1UD1K5 | Klebsiella pneumoniae | AMP, CAZ, CFL, CTX, CPM, F, TET, TMS | blaCTX-M-15, blaSHV-11, blaTEM-1 |
| PV004 | Cat | UTI | PV004V0C1I0E1 | Escherichia coli | AMP, CAZ, CFL, CIP, CTX, CPM, TET, TMS | bla CTX-M-27 |
a UTI, urinary tract infection; SSTI, skin and soft tissue infection.
b Antibiotics tested: AK, amikacin; AMC, amoxicillin/clavulanate; AMP, ampicillin; C, chloramphenicol; CAZ, ceftazidime; CFL, cephalothin; CIP, ciprofloxacin; CN, gentamicin; CTX, cefotaxime; CPM, cefepime; ETP, ertapenem; F, nitrofurantoin; FOX, cefoxitin; IMP, imipenem; MEM, meropenem; TET, tetracycline; TMS, trimethoprim/sulfamethoxazole.
Minimum inhibitory concentrations (MICs) were interpreted using the criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2024 (The European Committee on Antimicrobial Susceptibility Testing, 2024), except for amoxicillin/clavulanate, ampicillin/sulbactam, and trimethoprim/sulfamethoxazole, for which criteria from the Clinical and Laboratory Standards Institute (CLSI) were used (CLSI, 2024).
Among clinical ESBL/AmpC-producing Enterobacterales strains from Portugal, a K. pneumoniae (PV003V1UD1K5) and two E. coli (PT110/0-D1I3E1 and PV004V0C1I0E1) strains from different animals presented the same genetic profile and resistance pattern as carriage strains from the respective companion animal and their cohabiting humans. To further explore the genetic relatedness of these clinical and carriage strains, one representative carriage strain from each household member per timepoint and the clinical strains were analyzed by WGS. In a total, 9 E. coli and 7 K. pneumoniae strains were analyzed by WGS.
3.5. Co-carriage of ESBL/AmpC-producing Enterobacterales strains
The presence of ESBL/AmpC-E carriage strains in companion animals and cohabitant humans harboring the same resistance genes was observed in eight Portuguese households (18.6%, n = 8/43) and two households from the UK (9.1%, n = 2/22) (Figure 2). Of these, five households (PT = 3; UK = 2) included companion animal–human E. coli paired strains with matching REP-PCR that were selected for WGS analysis (one representative strain from each household member per timepoint, comprising a total of 20 E. coli strains).
For another household, E. hormaechei strains were recovered from an animal/owner pair (Figure 2) presenting the same resistant genes and susceptibility profile; these strains (n = 2) were also sequenced.
For one household from the UK (UK105), the blaCMY-2 gene was associated with an E. coli in the dog and a K. pneumoniae in the owner; in a similar way, in a household from Portugal (PT219), the blaCTX-M-15 gene was found in a K. pneumoniae and E. coli from the cat and at E. coli strains from its cohabitant humans. These strains were not sequenced as companion animals’ and owners’ strains belonged to different bacterial species and different E. coli REP-PCR profiles (Figure 2; Supplementary Table S2).
3.6. Core genome relatedness between animals and humans’ strains
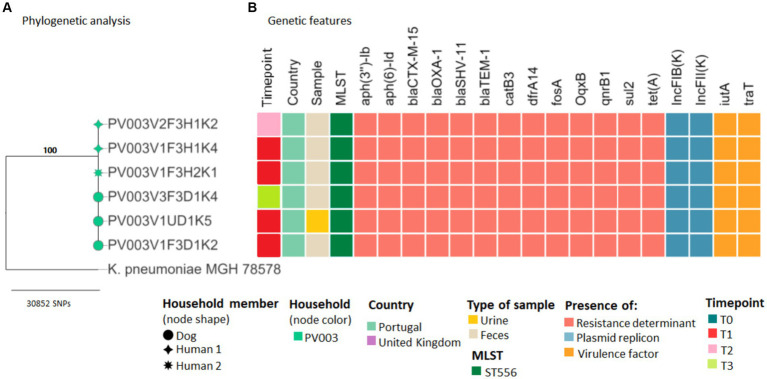
Evidence of sharing was found in several households. In household PV003, K. pneumoniae ST556 strains harboring the blaCTX-M-15 gene, recovered from dog and owners’ fecal samples at different timepoints, had ≤7 SNP differences from animals’ clinical strains, indicating possible transmission of the pathogenic bacteria (Figure 3; Supplementary Table S4).
Figure 3.
Core genome SNP analysis and genetic features of clinical and carriage ESBL/AmpC-producing Klebsiella pneumoniae strains from companion animals and their cohabitant humans. (A) Maximum likelihood phylogeny of the core genome of six K. pneumoniae strains and the K. pneumoniae subsp. pneumoniae MGH 78578 strain. Bootstrap support values are shown in bold on each node. (B) Heatmap shows the sequence types, antimicrobial resistance determinants, plasmid replicons, and virulence factors for each strain (see color key at the bottom of the figure).
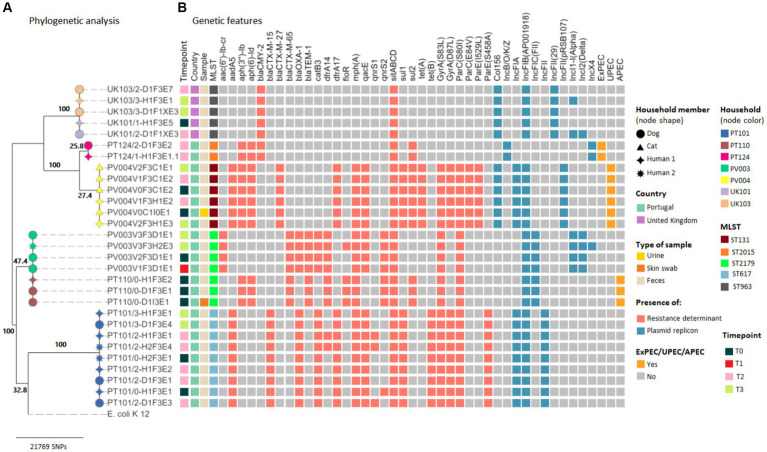
Maximum likelihood phylogenetic tree based on the SNP alignment of 29 E. coli core genomes showed six large clusters corresponding to five different households from Portugal and one cluster comprising strains from two UK households (Figure 4).
Figure 4.
Core genome SNP analysis and genetic features of clinical and carriage ESBL/AmpC-producing Escherichia coli strains from companion animals and their cohabitant humans. (A) Maximum likelihood phylogeny of the core genome of 33 E. coli strains and the E. coli K-12 MG1655 strain. Bootstrap support values are shown in bold on each node. (B) Heatmap shows the E. coli sequence types, antimicrobial resistance determinants, plasmid replicons, and pathotypes based on a repertoire of virulence factors for each strain (see color key on the right side of the figure).
In two of the Portuguese households clusters (PT110 and PV004), the sharing of companion animal’s E. coli clinical strains was confirmed (Figure 4). At household PT110, an E. coli ST2179 strain harboring the blaCTX-M-65 and blaTEM-1 genes causing SSTI in the dog was shared between the pet and owner (2 SNP difference). These strains were classified as APEC due to the presence of hlyF, iutA, iroN, iss, and ompT virulence genes (Supplementary Table 5).
For household PV004, the cat–human pair was colonized by an E. coli ST131 strain harboring the blaCMY-2 gene causing UTI in the cat. Cat’s clinical and colonization strains from T0, T2, and T3, as well as its owner’s strains, recovered at timepoint T2 and T3, displayed ≤3 SNP difference (Figure 3; Supplementary Table S6). The strains from household PV004 displayed virulence genes typically associated with UPEC (chuA, fyuA, and yfcV) (Supplementary Table S5).
Among the remaining three Portuguese household clusters (PV003, PT101, and PT124), analysis of the core genomes confirmed that no distinction could be made between E. coli carriage strains from the three companion animal–human pairs as within-household strains presented ≤4 SNPs differences (Figure 4). For households, PT101 sharing over time of the E. coli ST617 strains harboring blaCTX-M-15 was also observed as strains were isolated from the dog at T2 and T3 and its owners at T0, T2, and T3. Furthermore, clonal strains presented the same MLST result, the same resistance genes, plasmid replicons, and pathotypes (Figure 4; Supplementary Table S5). Strains from household PT124 displayed virulence genes typically associated with pathogenic E. coli strains and were classified as ExPEC (kpsMII, papC, and sfaD) (Figure 4; Supplementary Table S5).
The strains from the two UK households clustered together (UK101 and UK103), presented only 9 SNPs of difference between them (Figure 4; Supplementary Table S6). These results demonstrated that sharing of the same ESBL/AmpC-producing E. coli carriage strains occurred between companion animals and owners and between participants from different households in the UK. These E. coli strains belong to ST963 harboring the blaCMY-2 gene.
In addition to the ESBL/AmpC genes, sequenced strains exhibited a wide variety of genes encoding for narrow-spectrum beta lactamases (blaTEM–1 and blaOXA-1) and resistance against aminoglycosides [aadA5, aph(6)-Id, and aph(3″)-Ib], fluoroquinolones (qnrS and qnrB1), macrolides [mph(A)], phenicols (catB3 and floR), sulphonamides (sul1 and sul2), trimethoprim (dfrA14 and dfrA17), tetracyclines [tet(A) and tet(B)], and point mutations in gyrA, parC, and/or parE genes, which could confer resistance to nalidixic acid and fluoroquinolones (Figures 3, 4; Supplementary Table S5).
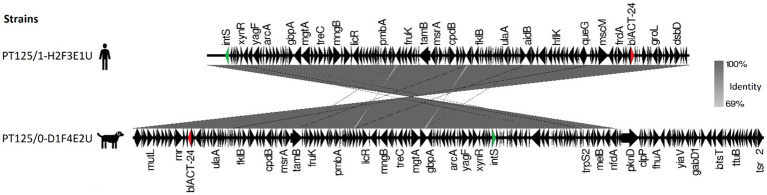
E. hormaechei dog-owner paired strains share high nucleotide similarity presenting 16 SNPs difference (Supplementary Table S7). These strains harbored a blaACT-24 gene, flanked by multiple regulon genes and an integrase, suggesting that it has been integrated into the chromosomal DNA (Figure 5).
Figure 5.
Map of blaACT-24 genetic environment comparison between carriage Enterobacter hormaechei subsp. hoffmannii strains from a dog and their cohabitant human fecal samples. Antibiotic resistance genes are represented by red arrows, integrase by green arrows, and other genes with black arrows.
4. Discussion
The close relationship between pets and humans supports the likelihood of transferring antibiotic-resistant bacteria and resistance-mediating mobile genetic elements, between them (Pomba et al., 2017). Here, we presented insights on the co-carriage of ESBL/AmpC-E and CPE strains between companion animals receiving antimicrobials and their owners in Portugal and the UK, as well as the transmission of animals’ clinical strains within the household. WGS combined with phylogenetic analysis was used to infer bacterial resistance transmission dynamics.
This study used a prospective, longitudinal design to examine the effect of antibiotic treatment on the selection and carriage of ESBL/AmpC-E and CPE strains in a cohort of dogs and cats with UTI or SSTI. The prevalence of ESBL/AmpC-E carriage in companion animals under antibiotic therapy in this study (PT = 55.8%; UK = 36.4%) was higher compared to the prevalence in healthy animals in both countries (12.7 and 8.5% for Portugal and the UK, respectively) during the same timeframe (Menezes et al., 2023). While care must be taken when comparing data from different studies, the similarity in recruitment and methods between these studies allows for a meaningful comparison. The critical distinction that one study focused on healthy animals, while the current study involves companion animals under antibiotic therapy, supports the potential for antibiotic selection pressure being exerted upon the animals in the current study based on the observed prevalence rates. Furthermore, in the present study, 15.6% of the companion animals from Portugal and 13.3% from the UK acquired ESBL/AmpC-E strains after antibiotic treatment. These findings suggest a role for companion animals under antibiotic treatment as reservoirs of AMR that may be potentially transmissible to the environment and to other hosts, including their owners.
Despite the absence of significant differences, the prevalence of ESBL/AmpC-E strains in companion animals was higher than in their owners (PT = 35.9%; UK = 12.5%), following the pattern previously found in other studies (Abbas et al., 2019; Dazio et al., 2021) that suggest companion animals as a source of this type of resistance in a household.
The observed differences in the detection of ESBL/AmpC-E strains between Portuguese and British individuals align with the broader patterns of higher prevalence of 3GC-resistant Enterobacterales isolates in Southern European countries compared to Northern European countries (Marques et al., 2016; WHO Regional Office for Europe/European Centre for Disease Prevention and Control, 2022). This suggests that geographical variation could be associated with the acquisition of resistant bacteria, potentially emerging as a risk factor. Such geographical disparities may reflect underlying regional differences in antimicrobial usage. In Portugal, there is a higher rate of antimicrobial use in the community, especially for systemic treatments with cephalosporins and other beta-lactam agents, compared to England (European Centre for Disease Prevention and Control, 2023; UK Health Security Agency, 2023). However, the substantial discrepancy in sample size between Portugal (n = 78 humans) and the UK (n = 24) might have also contributed to the observed difference. Although statistical adjustments were made to account for these variations, the impact of sample size on the robustness of our comparisons and the generalizability of our findings should be considered.
CPE carriage was only found in companion animals. This finding was somewhat surprising as such isolates are primarily reported in human clinical samples and carbapenems are not authorized for veterinary use (EMA, 2019; World Health Organization, 2024). The lack of positive cases in humans may be attributed to the limited sample size featured in this study. In addition, recent studies have shown that carbapenemase carriage is generally rare among healthy individuals globally (Diebold et al., 2023). Hospital settings, however, are known to harbor antimicrobial resistance genes, making carbapenemase genes more common in inpatient and outpatient cohorts compared to healthy individuals who were the focus of our study. In contrast, the animals in our study were exposed to veterinary hospital environments, where antibiotic use could facilitate the co-selection of resistant strains, potentially explaining their CPE carriage. These findings emphasize the importance of incorporating dogs/cats in the assessment of antimicrobial resistance circulation in the community setting.
We did not identify any risk factors for ESBL/AmpC-E or CPE carriage. In previous studies, raw food diets and prior antimicrobial use have been identified as risk factors for ESBL-E carriage in companion animals (Baede et al., 2017; Schmidt et al., 2018). However, no association between the consumption of raw food and ESBL/AmpC-E or CPE carriage was found (p = 0.6817 in Portugal; p = 0.7175 in the UK) or between antimicrobial treatment 3–12 months prior to sampling and carriage (p = 0.9121 in Portugal and p = 0.9333 in the UK). Yet, the acquisition of resistant bacteria was observed after a single course of antibiotics. Similarly, hospital setting contact has also been strongly associated with AMR carriage (Adler et al., 2016; Nigg et al., 2019); here, being a healthcare professional (p = 0.9206 in Portugal; p = 0.9792 in the UK) or recently hospitalized (≤12 months) (p = 0.9680 in Portugal; p = 0.9792 in the UK) was not established as risk factors among humans participants. This may be explained by the small sample size of participants presenting these characteristics, which was not powered to detect changes. Although given the extensive dissemination of ESBL/AmpC-E strains, there are numerous routes for acquiring these resistant strains, making the identification of risk factors difficult (Salgado-Caxito et al., 2021; WHO Regional Office for Europe/European Centre for Disease Prevention and Control, 2022; Zamudio et al., 2022).
E. coli was the most predominant species identified in 3CG-resistant carriage isolates, suggesting that this bacterial species may act as a reservoir of ESBL/AmpC-E genes. The detection of phylogroup B2 and D strains in high rates of occurrence in the community emphasizes the need for ongoing surveillance. These two phylogroups are commonly associated with strains causing extraintestinal infection, namely, urinary tract infection (Schmiedel et al., 2014).
ESBL/AmpC-producing K. pneumoniae was the second most found among carriage strains. K. pneumoniae stands as a healthcare-associated pathogen known for causing various infections and its ability to develop antibiotic resistance (WHO Regional Office for Europe/European Centre for Disease Prevention and Control, 2022). The prevalence reported here for ESBL/AmpC-producing K. pneumoniae is higher in contrast with previous observations by our group in healthy companion animals and humans in both countries (Menezes et al., 2023) but in agreement with studies in dogs and cats presented to veterinary hospitals in Switzerland (12.3%) (Dazio et al., 2021). Also of notice is the high proportion of multidrug-resistant ESBL/AmpC-E strains pointing toward a rising trend of AMR within the community setting.
Despite the ubiquitous nature of resistance genes, there was a variation in their distribution and frequency between geographical origins. The blaCTX-M-15 gene was more frequent in Portugal compared to the UK, where the blaCMY-2 gene was more common. These observations are consistent with previous resistance determinant patterns observed in both countries (Wedley et al., 2017; Carvalho et al., 2021). On the other hand, the blaACT-24 gene was only identified in shared E. hormaechei strains. Enterobacter spp. strains often harbor an inducible chromosomal ampC gene encoding AmpC beta-lactamase (Meini et al., 2019). The blaACT-24 gene has been identified before as chromosomally integrated in E. hormaechei strains (Lasarte-Monterrubio et al., 2023). In our study, this resistant gene was found surrounded by an integrase indicating that it was chromosomally integrated and highlighting its potential for mobilization and spread within microbial communities.
Transmission of bacteria strains causing infections in companion animals to their household members has been sporadically reported, encompassing bacteria resistant to medically important antimicrobials, such as NDM-5-producing E. coli (Grönthal et al., 2018), as well as dog-derived UTI ExPEC strains resulting in UTI in the owners (Johnson et al., 2008), and the reverse scenario has also been described (Johnson and Clabots, 2006; Johnson et al., 2008; Toombs-Ruane et al., 2020). In the present study, three households from Portugal had owners carrying the ESBL/AmpC-E strains causing infection in their cohabiting companion animal. The shared index E. coli strains belong to UPEC and APEC pathotypes. UPEC is the primary cause of urinary tract infections in humans, while APEC typically causes a range of infections in poultry (Geurtsen et al., 2022). Intriguingly, studies have shown that APEC exhibits genetic similarity with human ExPECs, including the possession of virulence genes associated with UTIs and meningitis (Tivendale et al., 2010; Mellata, 2013), thus emphasizing its potential for zoonotic transmission. The co-occurrence of both variants causing infection in companion animals and their owners’ fecal samples is particularly noteworthy, indicating that companion animals may act as reservoirs and disseminators of these strains. This highlights the importance of infection control measures during animal disease.
Overall, here we documented the sharing of carriage of ESBL/AmpC-E between companion animals and owners within the same households in Portugal (13.9%, n = 6/43) and the UK (9.1%, n = 2/22) based on core genome analysis. When considering only households with positive participants for ESBL/AmpC carriage, the sharing frequency from Portugal increases to 18.8% (n = 6/32) and 22.2% in the UK (n = 2/9). These frequencies are higher compared to the ones found among households with healthy companion animals in both countries (sharing of 4.9% in Portugal and 0% in the UK) (Menezes et al., 2023). These differences could be associated with the animal’s exposure to antibiotic selective pressure, as mentioned above, that increased carriage, making the occurrence of sharing more likely. Nevertheless, in a similar study to ours, no co-carriage was observed between dogs and cats presented to veterinary clinics and hospitals in Switzerland and their owners (Dazio et al., 2021). This could be related to the unique patterns of antimicrobial usage and resistance observed in both animals and humans across different geographical locations, as reported elsewhere (European Centre for Disease Prevention and Control, 2023; European Medicines Agency and European Surveillance of Veterinary Antimicrobial Consumption, 2023; UK Health Security Agency, 2023). Southern European countries, such as Portugal, generally exhibit higher levels of antimicrobial resistance compared to Northern countries (Marques et al., 2016; WHO Regional Office for Europe/European Centre for Disease Prevention and Control, 2022). Consequently, places with higher resistance levels have an increased likelihood of sharing resistant bacteria.
Although a high prevalence of intra-familial co-carriage was observed, underestimation cannot be excluded since data rely on bacterial culture, a methodology that has a low sensitivity (Tofteland et al., 2007).
Shared strains within households belonged to five different E. coli sequence types (ST131, ST2015, ST2179, ST617, and ST963), K. pneumoniae ST556, and E. hormaechei, which include bacterial lineages that have been already reported in the animal and human settings before (Jure et al., 2019; Sepp et al., 2019; Salgado-Caxito et al., 2021; Donà et al., 2023). Although direct transmission cannot be confirmed, the diminutive number of SNP differences suggests very recent acquisition from a common source or recent direct transmission. The last one could have occurred via petting, cuddling, licking the owner’s face, or cleaning the pet’s waste, behaviors that were reported on the questionnaires to occur at least occasionally in over 90% of the households. Determining the direction of transmission or whether both humans and dogs were contaminated by the same source is challenging, given that the study design precluded its assessment. Numerous potential routes of exposure exist, including common food and water, the surrounding household environment, and contaminated hospital environment.
In two households from the UK (UK101 and UK103), dogs and their cohabitant humans carried indistinguishable E. coli ST963 strains, indicating a common source of exposure. Notably, both dogs attended the same veterinary hospital, where they may have acquired the bacteria through a contaminated hospital environment and then transmitted it to their owners. Veterinary hospitals have been implicated in the selection and dissemination of AMR among patients (Nigg et al., 2019; Dazio et al., 2021). The lack of rigorous infection prevention and control (IPC) measures in these settings exacerbates this issue (Willemsen et al., 2019). Implementing robust IPC programs tailored to veterinary clinics is crucial to mitigate the occurrence of transmission events that could further spread to the animal and human population and the environment.
Further exhaustive study into plasmids harboring antimicrobial resistance, coupled with the role of other mobile genetic elements, including transposons, in the transmission and epidemiology of these resistance genes, is needed. This constitutes a significant limitation of the current study as there is the potential for plasmid-mediated gene transfer to occur, for example, as in the case of the blaCMY-2 and blaCTX-M-15 genes, which were associated with two different bacterial species in the UK105 and PT219 households, respectively. Another limitation is the noteworthy proportion of participants failing to conclude the longitudinal study, notwithstanding favorable initial recruitment. Consequently, this study may potentially underestimate the prevalence of persistent carriage of ESBL/AmpC-E and CPE strains.
This study highlights the need for long-term, globally standardized monitoring of AMR among companion animals and cohabiting humans to understand the dynamics and relative significance of the various sources influencing the burden of AMR over time. The potential for inadvertent selection of antibiotic-resistant colonizing bacteria should be considered in prospective antimicrobial stewardship strategies aimed at limiting the development and dissemination of AMR and thus the importance of adopting a One Health approach that integrates both human and animal health.
Acknowledgments
We thank the participating animals and owners. We are grateful for the assistance of the veterinarians, technicians, and staff at the Small Animal Veterinary Teaching Hospital of the Faculty of Veterinary Medicine, University of Lisbon, Portugal, and the RVC Small Animal Veterinary Referral Service, Royal Veterinary College, Hatfield, UK. We acknowledge the PET-Risk Consortium and all its members: Adriana Belas, Cátia Marques, Luís Telo Gama, Rodolfo Leal, Mafalda Lourenço, Hugo Pereira, and Joana Moreira da Silva (Faculty of Veterinary Medicine, University of Lisbon, Portugal) and Claudia Feudi (Freie Universität Berlin, Berlin, Germany).
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Portuguese Foundation for Science and Technology (FCT), under projects UIDB/00276/2020 (CIISA), LA/P/0059/2020 (AL4AnimalS), JPIAMR/0002/2016 (PET-Risk Consortium) and 2022.08669. PTDC (VetCare project), and by the Medical Research Council (MRC), UK (MR/R000042/1); JM was supported by a PhD fellowship (DOI: 10.54499/2020.07562.BD); AA was supported by CEEC 4th edition (2021.02058.CEECIND).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB55525. High resolution E. coli phylogenetic tree linked to molecular data is available at Microreact platform, https://microreact.org/project/6r3o1DHTVwF5qPVK4LbPpM-e-coli-transmission-during-different-types-of-animal-infection; K. pneumoniae phylogenetic tree can also be found at https://microreact.org/project/iHYMJ8K9dZpkzctDeCprdd-klebsiella-pneumoniae-sharing.
Ethics statement
The studies involving humans and animals were approved by the Faculty of Veterinary Medicine, University of Lisbon (FMV-Ulisboa) Ethics Committee for Research and Education and the Royal Veterinary College (RVC) Ethics and Welfare Committee (ethics reference number: CEBEA 027/2018 and URN 2017 1750–3, respectively). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for human participation in this study was provided by the participants’ legal guardians/next of kin. For the animal studies, written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
JM: Writing—original draft, Formal analysis, and Methodology. S-MF: Methodology and Writing—reviewing and editing. SW: Conceptualization and Writing—reviewing and editing. VP: Conceptualization and Writing—reviewing and editing. SS: Conceptualization and writing—reviewing and editing. AA: Formal analysis and Writing—reviewing and editing. AL: Writing—reviewing and editing, Conceptualization, and Project administration. CP: Writing—reviewing and editing, Conceptualization, and Project administration.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1432240/full#supplementary-material
References
- Abbas G., Khan I., Mohsin M., Sajjad-ur-Rahman S. U. R., Younas T., Ali S. (2019). High rates of CTX-M group-1 extended-spectrum β-lactamases producing Escherichia coli from pets and their owners in Faisalabad. Pakistan. Infect. Drug Resist. 12, 571–578. doi: 10.2147/IDR.S189884, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler A., Katz D. E., Marchaim D. (2016). The continuing plague of extended-spectrum β-lactamase–producing Enterobacteriaceae infections. Infect. Dis. Clin. N. Am. 30, 347–375. doi: 10.1016/j.idc.2016.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010) A quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed: 10 January 2021).
- Argimón S., Abudahab K., Goater R. J. E., Fedosejev A., Bhai J., Glasner C., et al. (2016). Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microbial genomics 2:e000093. doi: 10.1099/mgen.0.000093, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baede V. O., Broens E. M., Spaninks M. P., Timmerman A. J., Graveland H., Wagenaar J. A., et al. (2017). Raw pet food as a risk factor for shedding of extended-spectrum beta-lactamase-producing Enterobacteriaceae in household cats. PLoS One 12:e0187239. doi: 10.1371/journal.pone.0187239, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan E. R., Jones A. M., Hawkey P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146, PMID: [DOI] [PubMed] [Google Scholar]
- Bortolaia V., Kaas R. S., Ruppe E., Roberts M. C., Schwarz S., Cattoir V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhante M., Menezes J., Belas A., Feudi C., Schwarz S., Pomba C., et al. (2020). OXA-181-producing extra-intestinal pathogenic Escherichia coli ST410 isolated from a dog in Portugal. Antimicrob. Agents Chemother. 64, e02298–e02219. doi: 10.1128/AAC.02298-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial Agents Chemotherapy 58, 3895–3903. doi: 10.1128/AAC.02412-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho I., Cunha R., Martins C., Martínez-Álvarez S., Safia Chenouf N., Pimenta P., et al. (2021). Antimicrobial resistance genes and diversity of clones among faecal ESBL-producing Escherichia coli isolated from healthy and sick dogs living in Portugal. Antibiotics 10, 1–15. doi: 10.3390/antibiotics10081013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centre for Genomic Pathogen Surveillance (2018) Pathogenwatch. Available at: https://pathogen.watch/ (Accessed March 15, 2023).
- CLSI (2024). “Performance standards for antimicrobial disk and dilution susceptibility tests for Bacteria isolated from animals” in CLSI Supplement VET01S. 7th ed (Wayne, Pennsylvania: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Croucher N. J., Page A. J., Connor T. R., Delaney A. J., Keane J. A., Bentley S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva J. M., Menezes J., Marques C., Pomba C. F. (2022). Companion animals — an overlooked and misdiagnosed reservoir of Carbapenem resistance, Antibiotics, 11:533. doi: 10.3390/antibiotics11040533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damborg P., Broens E. M., Chomel B. B., Guenther S., Pasmans F., Wagenaar J. A., et al. (2016). Bacterial Zoonoses transmitted by household pets: state-of-the-art and future perspectives for targeted research and policy actions. J. Comp. Pathol. 155, S27–S40. doi: 10.1016/j.jcpa.2015.03.004 [DOI] [PubMed] [Google Scholar]
- Dazio V., Nigg A., Schmidt J. S., Brilhante M., Campos-Madueno E. I., Mauri N., et al. (2021). Duration of carriage of multidrug-resistant bacteria in dogs and cats in veterinary care and co-carriage with their owners. One Health 13:100322. doi: 10.1016/j.onehlt.2021.100322, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazio V., Nigg A., Schmidt J. S., Brilhante M., Mauri N., Kuster S. P., et al. (2021). Acquisition and carriage of multidrug-resistant organisms in dogs and cats presented to small animal practices and clinics in Switzerland. J. Vet. Intern. Med. 35, 970–979. doi: 10.1111/jvim.16038, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold P. J., Rhee M. W., Shi Q., Trung N. V., Umrani F., Ahmed S., et al. (2023). ‘Clinically relevant antibiotic resistance genes are linked to a limited set of taxa within gut microbiome worldwide’, nature. Communications 14:7366. doi: 10.1038/s41467-023-42998-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donà V., Nordmann P., Kittl S., Schuller S., Bouvier M., Poirel L., et al. (2023). Emergence of OXA-48-producing Enterobacter hormaechei in a Swiss companion animal clinic and their genetic relationship to clinical human isolates. J. Antimicrob. Chemother. 78, 2950–2960. doi: 10.1093/jac/dkad337, PMID: [DOI] [PubMed] [Google Scholar]
- Doumith M., Day M. J., Hope R., Wain J., Woodford N. (2012). Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J Clin Microbiol. 50, 3108–3001. doi: 10.1128/JCM.01468-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C., Gautier V., Arlet G. (2006). DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57, 14–23. doi: 10.1093/jac/dki398 [DOI] [PubMed] [Google Scholar]
- Eiamphungporn W., Schaduangrat N., Malik A. A., Nantasenamat C. (2018). Tackling the antibiotic resistance caused by class a β-lactamases through the use of β-lactamase inhibitory protein. Int. J. Mol. Sci. 19:2222. doi: 10.3390/ijms19082222, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA (2019) Categorisation of antibiotics in the European Union. Amsterdam: European Medicines Agency, (EMA/CVMP/CHMP/682198/2017). Available at: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific-advice-impact-public-health-and-animal-health-use-antibiotics-animals_en.pdf. [Google Scholar]
- European Centre for Disease Prevention and Control (2023). Antimicrobial consumption in the EU/EEA (ESAC-net) - annual epidemiological report 2022. Stockholm: ECDC. [Google Scholar]
- European Medicines Agency and European Surveillance of Veterinary Antimicrobial Consumption (2023) ‘Sales of veterinary antimicrobial agents in 31 European countries in 2022’. Amsterdam: European Medicines Agency. (EMA/299538/2023). Available at: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2022-trends-2010-2022-thirteenth-esvac-report_en.pdf [Google Scholar]
- Feldgarden M., Brover V., Gonzalez-Escalona N., Frye J. G., Haendiges J., Haft D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep., 11, 1–9. doi: 10.1038/s41598-021-91456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Féria C., Ferreira E., Correia J. D., Gonçalves J., Caniça M. (2002). Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J. Antimicrob. Chemother. 49, 77–85. doi: 10.1093/jac/49.1.77, PMID: [DOI] [PubMed] [Google Scholar]
- Geurtsen J., de Been M., Weerdenburg E., Zomer A., McNally A., Poolman J. (2022). Genomics and pathotypes of the many faces of Escherichia coli. FEMS Microbiol. Rev. 46, 1–30. doi: 10.1093/femsre/fuac031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönthal T., Österblad M., Eklund M., Jalava J., Nykäsenoja S., Pekkanen K., et al. (2018). Sharing more than friendship – transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill. 23:1700497. doi: 10.2807/1560-7917.ES.2018.23.27.1700497, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. S., Song W., Jeong S. H. (2020). Molecular characteristics of NDM-5-producing Escherichia coli from a cat and a dog in South Korea. Microb. Drug Resist. 26, 1005–1008. doi: 10.1089/mdr.2019.0382, PMID: [DOI] [PubMed] [Google Scholar]
- Jernberg C., Löfmark S., Edlund C., Jansson J. K. (2010). Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156, 3216–3223. doi: 10.1099/mic.0.040618-0 [DOI] [PubMed] [Google Scholar]
- Joensen K. G., Scheutz F., Lund O., Hasman H., Kaas R. S., Nielsen E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Clabots C. (2006). Sharing of virulent Escherichia coli clones among household members of a woman with acute cystitis. Clin. Infect. Dis. 43, e101–e108. doi: 10.1086/508541, PMID: [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Clabots C., Kuskowski M. A. (2008). Multiple-host sharing, long-term persistence, and virulence of Escherichia coli clones from human and animal household members. J. Clin. Microbiol. 46, 4078–4082. doi: 10.1128/JCM.00980-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Owens K., Gajewski A., Clabots C. (2008). Escherichia coli colonization patterns among human household members and pets, with attention to acute urinary tract infection. J. Infect. Dis. 197, 218–224. doi: 10.1086/524844, PMID: [DOI] [PubMed] [Google Scholar]
- Jure M. A., Castillo M. E., Musa H. E., López C. G., Cáceres M. A., Mochi S. D., et al. (2019). Novel patterns in the molecular epidemiology of KPC-producing Klebsiella pneumoniae in Tucumán, Argentina. J. Glob. Antimicrob. Res. 19, 183–187. doi: 10.1016/j.jgar.2019.02.015, PMID: [DOI] [PubMed] [Google Scholar]
- Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of Total-genome-sequenced Bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasarte-Monterrubio C., Guijarro-Sánchez P., Vázquez-Ucha J. C., Alonso-Garcia I., Alvarez-Fraga L., Outeda M., et al. (2023). Antimicrobial activity of Cefiderocol against the Carbapenemase-producing Enterobacter cloacae Complex and characterization of reduced susceptibility associated with Metallo-β-lactamase VIM-1. Antimicrob. Agents Chemother. 67:e0150522. doi: 10.1128/aac.01505-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques C., Gama L. T., Belas A., Bergström K., Beurlet S., Briend-Marchal A., et al. (2016). European multicenter study on antimicrobial resistance in bacteria isolated from companion animal urinary tract infections. BMC Vet. Res. 12, 1–17. doi: 10.1186/s12917-016-0840-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques C., Menezes J., Belas A., Aboim C., Cavaco-Silva P., Trigueiro G., et al. (2019). Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: population structure, antimicrobial resistance and virulence genes. J. Antimicrob. Chemother. 74, 594–602. doi: 10.1093/jac/dky499, PMID: [DOI] [PubMed] [Google Scholar]
- Meini S., Tascini C., Cei M., Sozio E., Rossolini G. M. (2019). AmpC β-lactamase-producing Enterobacterales: what a clinician should know. Infection 47, 363–375. doi: 10.1007/s15010-019-01291-9, PMID: [DOI] [PubMed] [Google Scholar]
- Mellata M. (2013). Human and avian Extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10, 916–932. doi: 10.1089/fpd.2013.1533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes J., Frosini S. M., Amaral A. J., Loeffler A., Pomba C. (2023). Dissemination of blaNDM-5-carrying IncX3-type plasmid among non-clonal Escherichia coli strains colonising a dog with a skin infection caused by a carbapenem-resistant Klebsiella pneumoniae, United Kingdom. J. Infect. Pub. Health 16, 190–193. doi: 10.1016/j.jiph.2023.10.048, PMID: [DOI] [PubMed] [Google Scholar]
- Menezes J., Frosini S. M., Belas A., Marques C., da Silva J. M., Amaral A. J., et al. (2023). Longitudinal study of ESBL/AmpC-producing Enterobacterales strains sharing between cohabiting healthy companion animals and humans in Portugal and in the United Kingdom. Eur. J. Clin. Microbiol. Infect. Dis. 42, 1011–1024. doi: 10.1007/s10096-023-04629-2, PMID: [DOI] [PubMed] [Google Scholar]
- Nigg A., Brilhante M., Dazio V., Clément M., Collaud A., Gobeli Brawand S., et al. (2019). ‘Shedding of OXA-181 carbapenemase-producing Escherichia coli from companion animals after hospitalisation in Switzerland: an outbreak in 2018’, Eurosurveillance. Eur. Centre Dis. Control Prevent. 24, 1–12. doi: 10.2807/1560-7917.ES.2019.24.39.1900071, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Walsh T. R., Cuvillier V., Nordmann P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diag. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002, PMID: [DOI] [PubMed] [Google Scholar]
- Pomba C., Belas A., Menezes J., Marques C. (2020). ‘The public health risk of companion animal to human transmission of antimicrobial resistance during different types of animal infection’, in advances in animal health. Med. Prod., 265–278. doi: 10.1007/978-3-030-61981-7_14 [DOI] [Google Scholar]
- Pomba C., Mendonça N., Costa M., Louro D., Baptista B., Ferreira M., et al. (2006). Improved multiplex PCR method for the rapid detection of β-lactamase genes in Escherichia coli of animal origin. Diagn. Microbiol. Infect. Dis. 56, 103–106. doi: 10.1016/j.diagmicrobio.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Pomba C., Rantala M., Greko C., Baptiste K. E., Catry B., van Duijkeren E., et al. (2017). Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 72, dkw481–dkw968. doi: 10.1093/jac/dkw481 [DOI] [PubMed] [Google Scholar]
- Salgado-Caxito M., Benavides J. A., Adell A. D., Paes A. C., Moreno-Switt A. I. (2021). Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats – a scoping review and meta-analysis. One Health 12:100236. doi: 10.1016/j.onehlt.2021.100236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury S. M., Sabatini L. M., Spiegel C. A. (1997). Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction assay. Am. J. Clin. Pathol. 107, 368–373. doi: 10.1093/ajcp/107.3.368 [DOI] [PubMed] [Google Scholar]
- Schmidt V. M., Pinchbeck G., McIntyre K. M., Nuttall T., McEwan N., Dawson S., et al. (2018). Routine antibiotic therapy in dogs increases the detection of antimicrobial-resistant faecal Escherichia coli. J. Antimicrob. Chemother. 73, 3305–3316. doi: 10.1093/jac/dky352, PMID: [DOI] [PubMed] [Google Scholar]
- Schmiedel J., Falgenhauer L., Domann E., Bauerfeind R., Prenger-Berninghoff E., Imirzalioglu C., et al. (2014). Multiresistant extended-spectrum β-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in Central Hesse, Germany. BMC Microbiol. 14, 1–13. doi: 10.1186/1471-2180-14-187, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153, PMID: [DOI] [PubMed] [Google Scholar]
- Seemann T., Klötzl F., Page A. J. (2021) snp-dists. Available at: https://github.com/tseemann/snp-dists (Accessed January 20, 2022).
- Sepp E., Andreson R., Balode A., Bilozor A., Brauer A., Egorova S., et al. (2019). Phenotypic and molecular epidemiology of ESBL-, AmpC-, and Carbapenemase-producing Escherichia coli in northern and Eastern Europe. Front. Microbiol. 10:2465. doi: 10.3389/fmicb.2019.02465, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M. T., Lubbers B. V., Schwarz S., Watts J. L. (2018). Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 73, 1460–1463. doi: 10.1093/jac/dky043, PMID: [DOI] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing (2024) Breakpoint tables for interpretation of MICs and zone diameters European committee on antimicrobial susceptibility testing breakpoint tables for interpretation of MICs and zone diameters version 14.0. Available at: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf (Accessed April 8, 2024).
- Tivendale K. A., Logue C. M., Kariyawasam S., Jordan D., Hussein A., Li G., et al. (2010). Avian-pathogenic Escherichia coli strains are similar to neonatal Meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78, 3412–3419. doi: 10.1128/IAI.00347-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofteland S., Haldorsen B., Dahl K. H., Simonsen G. S., Steinbakk M., Walsh T. R., et al. (2007). Effects of phenotype and genotype on methods for detection of extended-spectrum-β-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J. Clin. Microbiol. 45, 199–205. doi: 10.1128/JCM.01319-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs-Ruane L. J., Benschop J., French N. P., Biggs P. J., Midwinter A. C., Marshall J. C., et al. (2020). Carriage of extended-Spectrum-Beta-lactamase- and AmpC Beta-lactamase-producing Escherichia coli strains from humans and pets in the same households. Appl. Environ. Microbiol. 86, 1–15. doi: 10.1128/AEM.01613-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T. J., Ondov B. D., Koren S., Phillippy A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524–515. doi: 10.1186/s13059-014-0524-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK Health Security Agency (2023). English surveillance programme for antimicrobial utilisation and resistance (ESPAUR) report 2022 to 2023. London: UK Health Security Agency. [Google Scholar]
- UK Health Security Agency (2024) Carbapenemase-producing gram-negative organisms in England since October 2020: quarterly update, Q4 2023. Available at: www.gov.uk/government/publications/carbapenemase-producing-gram-negative-bacteria-laboratory-surveillance/carbapenemase-producing-gram-negative-organisms-in-england-since-october-2020-quarterly-update-q4-2023 (Accessed July 16, 2024).
- Van Den Bunt G., Fluit A. C., Spaninks M. P., Timmerman A. J., Geurts Y., Kant A., et al. (2020). Faecal carriage, risk factors, acquisition and persistence of ESBL-producing Enterobacteriaceae in dogs and cats and co-carriage with humans belonging to the same household. J. Antimicrob. Chemother. 75, 342–350. doi: 10.1093/jac/dkz462, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedley A. L., Dawson S., Maddox T. W., Coyne K. P., Pinchbeck G. L., Clegg P., et al. (2017). Carriage of antimicrobial resistant Escherichia coli in dogs: prevalence, associated risk factors and molecular characteristics. Vet. Microbiol. 199, 23–30. doi: 10.1016/j.vetmic.2016.11.017, PMID: [DOI] [PubMed] [Google Scholar]
- Weese J. S., Blondeau J., Boothe D., Guardabassi L. G., Gumley N., Papich M., et al. (2019). International Society for Companion Animal Infectious Diseases (ISCAID) guidelines for the diagnosis and management of bacterial urinary tract infections in dogs and cats. Vet. J. 247, 8–25. doi: 10.1016/j.tvjl.2019.02.008 [DOI] [PubMed] [Google Scholar]
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control (2022). Antimicrobial resistance surveillance in Europe 2022–2020 data. Copenhagen: WHO Regional Office for Europe. [Google Scholar]
- Willemsen A., Cobbold R., Gibson J., Wilks K., Lawler S., Reid S. (2019). Infection control practices employed within small animal veterinary practices—a systematic review. Zoonoses Public Health 66, 439–457. doi: 10.1111/zph.12589, PMID: [DOI] [PubMed] [Google Scholar]
- World Health Organization (2024). WHO’s list of medically important antimicrobials: A risk management tool for mitigating antimicrobial resistance due to non-human use. Geneva: Licence; CC BY-NC-SA 3.0 IGO. [Google Scholar]
- Zamudio R., Boerlin P., Beyrouthy R., Madec J. Y., Schwarz S., Mulvey M. R., et al. (2022). ‘Dynamics of extended-spectrum cephalosporin resistance genes in Escherichia coli from Europe and North America. Nature Commun. 13:7490. doi: 10.1038/s41467-022-34970-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, PRJEB55525. High resolution E. coli phylogenetic tree linked to molecular data is available at Microreact platform, https://microreact.org/project/6r3o1DHTVwF5qPVK4LbPpM-e-coli-transmission-during-different-types-of-animal-infection; K. pneumoniae phylogenetic tree can also be found at https://microreact.org/project/iHYMJ8K9dZpkzctDeCprdd-klebsiella-pneumoniae-sharing.