Abstract
A 30-residue N-terminally acetylated peptide derived from the N-terminal part of histone H1 was identified as the dominant antimicrobial peptide in skin mucus from Atlantic salmon (Salmo salar). The peptide (termed salmon antimicrobial peptide [SAMP H1]) was purified to homogeneity by a combination of reversed-phase and cation-exchange chromatographies. By Edman degradation of the deacetylated peptide and by sequencing of the PCR-amplified DNA that encodes the peptide, the complete amino acid sequence was determined to be AEVAPAPAAAAPAKAPKKKAAAKPKKAGPS. The theoretical molecular weight of N-terminally acetylated SAMP H1 was calculated to be 2,836, which is the same as that determined by matrix-assisted laser desorption ionization mass spectrometry. The peptide was active against both gram-negative and -positive bacteria. The N-terminal acetyl group was not necessary for activity since deacetylation did not reduce the activity. A synthetic peptide whose sequence was identical to that of the isolated fragment was initially inactive but could be activated by binding it to a cation-exchange column. Treatment of the synthetic peptide when it was bound to the exchange column with peptidylproline cis-trans-isomerase increased the amount of active peptide, indicating that isomerization of the proline peptide bond(s) was necessary for activation of the synthetic peptide. Comparison of the active and inactive forms by circular dichroism and chromatographic analyses suggests that the active form, both the natural and the synthetic forms, is more structured, condensed, and rigid than the inactive form, which has a more nonstructured conformation. This work shows for the first time the importance of proline isomers in the activity of an antimicrobial peptide.
Antimicrobial peptides (AMPs) are widespread in nature, being produced by bacteria, plants, and a wide variety of vertebrates and invertebrates (3, 10, 11, 16, 22). These peptides are thought to be important in the natural defense against invading microorganisms. In animals, AMPs are found in tissues that are likely to be in contact with microorganisms, such as at mucosal surfaces and within immunogenic cells and tissue. Most AMPs are cationic, containing 20 to 50 amino acid residues, and they are often amphipathic and/or hydrophobic, which reflects the fact that many of these peptides interact with and permeabilize target cell membranes (15, 16). Membrane permeabilization is thought to be the mechanism by which most AMPs kill their target cells, although other mechanisms may be involved for some AMPs (15, 25, 29).
Most AMPs are derived from precursor molecules encoded by dedicated genes, but some are the result of proteolysis of larger proteins with other functions. The latter AMPs include the ribosomal protein-derived cecropins (31); pepsinogen-derived bPaAP and bPcAP (21); histone H2A-derived buforin (26), parasin (28), and hipposin (2); and lactoferrin-derived lactoferricin (7). Antimicrobial activity has in some cases also been reported to be associated with the whole proteins from which AMPs are derived (7, 9, 23, 34).
Although fish live in a microbe-rich environment, they have primitive immune systems compared to those of higher vertebrates (8). The innate or unspecific immune system, of which AMPs are a part, seems to be an important component in their defense against infectious agents. Several AMPs have recently been purified from fish, especially from the skin and skin mucus. These include pardaxin (24), pleurocidin (6), moronecidin (18), misgurin (27), bass hepcidin (34), and the histone H2A-derived peptides parasin (28) and hipposin (2). These peptides kill a wide range of gram-negative and -positive bacteria, and some of the peptides are also active against fungi (12, 13).
A 26-amino-acid N-terminal fragment of histone H1 was recently identified in coho salmon (Oncorhynchus kisutch) mucus preparations and was shown to exhibit antimicrobial activity (29). However, a synthetic peptide identical in sequence to the histone fragment was inactive, suggesting that the histone fragment was not an AMP per se. It was suggested that the histone fragment rather acts by potentiating the activities of other antimicrobial polypeptides, such as lysozyme and pleurocidin (29). In this study we have identified the 30-mer N-terminal fragment of histone H1 as the dominant antimicrobial peptide in mucus preparations purified from Atlantic salmon (Salmo salar). A synthetic peptide whose sequence was identical to that of the isolated fragment was initially inactive, but we demonstrate that the synthetic peptide may be transformed into a potent AMP upon exposure to peptidylproline cis-trans-isomerase (PPIase; EC 5.2.1.8). Thus, the N-terminal fragment of histone H1 is indeed an AMP and is referred to as salmon antimicrobial peptide H1 (SAMP H1).
MATERIALS AND METHODS
Bacterial cultures and growth conditions.
The following bacteria were used for testing of antimicrobial activity: Aeromonas salmonicida subsp. salmonicida 1851/97 (V3819) and Vibrio anguillarum 1190/97 (V3767) were obtained from the Section for Fish Health at the National Veterinary Institute in Oslo, Norway; Bacillus subtilis (ATCC 6633) and Listeria ivanovii Li4 were obtained from the Norwegian Food Research Institute, Ås, Norway; and Escherichia coli (ATCC 14763) and Salmonella enterica serovar Typhimurium (LMGT3085) were from our own culture collection. All bacteria were grown at 25°C in Trypticase soy broth (TSB; Becton Dickson Biosciences, Sparks, Md.).
Peptide purification.
Skin mucus was scraped from the surfaces of the Atlantic salmon and stored at −20°C. The frozen skin mucus was mixed with 4 volumes of cold 2% (vol/vol) acetic acid containing 1 mM phenylmethylsulfonyl fluoride and centrifuged at 18,000 × g for 30 min. The centrifuged extract could be stored at 4°C for several months without a loss of activity, but due to the large volumes (typically several liters) used, the extract was subjected to the next step as soon as possible. All further purification steps were performed at room temperature (20 to 25°C).
The supernatant was applied to a SepPak C18 solid-phase extraction cartridge (Waters, Milford, Mass.), which was first activated with methanol and washed with 0.1% (vol/vol) trifluoroacetic acid (TFA). Approximately 100 ml acid extract was applied per g column material. The cartridge was washed with 10 column volumes 0.1% TFA, and bound material was eluted with 5 column volumes 0.1% TFA in water-2-propanol (1:1).
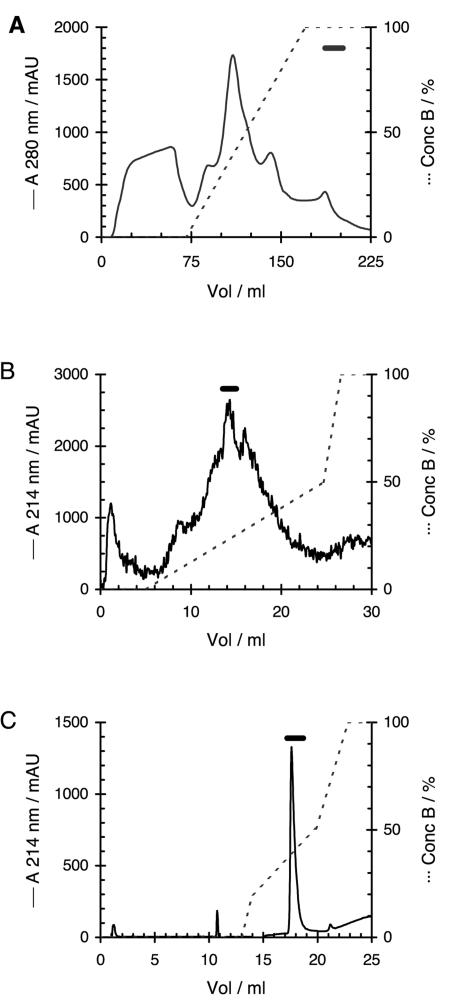
The SepPak eluate was applied to a HiPrep SP 16/10 cation-exchange column (Amersham Biosciences, Uppsala, Sweden) connected to a fast-performance liquid chromatography system (Amersham Biosciences). The column was equilibrated with buffer A (20 mM ammonium acetate, 0.1% [vol/vol] TFA, pH 4.0) before the sample was injected. The column was washed with 2 volumes of buffer A, and the bound material was eluted with a linear gradient of buffer A to B (200 mM ammonium acetate, 0.1% [vol/vol] TFA, pH 8.0) in 100 ml at a flow rate of 5 ml/min (Fig. 1A). The purification was monitored by measuring the absorbance at 280 nm.
FIG. 1.
Purification of SAMP H1. (A) HiPrep SP cation-exchange chromatography of SepPac C18-concentrated skin mucus extract; (B) Mono S cation-exchange chromatography with urea of RPC-concentrated active fractions obtained upon HiPrep SP chromatography; (C) final rechromatography on Mono S without urea. Solid lines, absorbance at 280 nm for panel A and absorbance at 214 nm for panels B and C; dotted lines, concentration (conc) of buffer B. The horizontal bars show the fractions that contained antimicrobial activity. mAU, milli-absorbance units.
All further purification steps were performed with an ÄKTA purifier system (Amersham Biosciences). The purifications were monitored by measuring the absorbance at 214 nm, 254 nm, and 280 nm simultaneously. Active fractions from the HiPrep SP chromatography were pooled and concentrated by binding to a Resource reversed-phase chromatography (RPC) column (Amersham Biosciences) equilibrated with 0.1% TFA. After the bound peptides were washed, they were eluted in 2 to 3 ml of 0.1% TFA in water-2-propanol (1:1) at a flow rate of 1 ml/min.
Urea was added to the concentrated sample to a final concentration of 6 M. The sample was then applied to a Mono S HR5/5 column (Amersham Biosciences) with buffer A containing 6 M urea and eluted with a linear gradient from 0 to 0.5 M sodium chloride in 20 ml followed by a gradient from 0.5 to 1 M sodium chloride in 3 ml at a flow rate of 1 ml/min (Fig. 1B). Final purification was achieved with repeated chromatography of the active fractions on the same column, but with buffers without urea and a gradient from 0.2 to 0.5 M sodium chloride in 6 ml at a flow rate of 1 ml/min (Fig. 1C).
Final desalting was done by adjusting the pH of the pure peptide to approximately 7 with sodium hydroxide, and the peptide preparation was then applied to a Resource RPC column equilibrated with 10 mM ammonium acetate. The column was washed with the same buffer before the bound peptide was recovered with 0.1% TFA in water-2-propanol (1:1) at a flow rate of 1 ml/min.
Antimicrobial assays.
The antimicrobial activities of all fractions were measured during purification by using a double agar diffusion assay (19). Briefly, an aliquot of each fraction was lyophilized and redissolved in a small volume (10 to 100 μl) of 0.01% (vol/vol) acetic acid. Approximately 6 ml underlayer agar containing 10 mM sodium phosphate buffer, pH 6.5, 1% (vol/vol) TSB, 1% (wt/vol) agarose, and approximately 1 × 106 CFU E. coli was poured in a petri dish (diameter, 9 cm). Wells (diameter, 2.5 to 3 mm) were made in the solidified agar, and 5-μl samples were added to each well. Pleurocidin (0.05 mg/ml, 5 μl) and acetic acid (0.01%, 5 μl) were used as positive and negative controls, respectively. The samples were allowed to diffuse into the agar for 2 to 3 h before 6 ml of normal-strength (30 g/liter) TSB containing 1% (wt/vol) agarose was added. After incubation overnight, antimicrobial activity was detected as zones without bacterial growth.
MICs were measured in microtiter plates (36). Twofold dilutions of peptides were made in 50 μl 0.2% (wt/vol) bovine serum albumin, 0.01% (vol/vol) acetic acid in polypropylene microtiter plates (Corning Inc., Corning, N.Y.), and approximately 1 × 105 CFU of bacteria in 50 μl TSB was added. Controls without peptide (growth control) and without bacteria (sterility control) were included. The plates were incubated overnight at room temperature before growth was measured at 620 nm in a Multiscan Ascent microtiter plate reader (Labsystems, Helsinki, Finland). The MIC was taken as the lowest concentration at which greater than 90% growth inhibition was observed. TSB was chosen as a suitable growth medium since it is able to support the growth of all the bacteria tested. The ionic strength of TSB is similar to that of the more commonly used Mueller-Hinton medium, so the results obtained are believed to be of the same order.
Physical characterization.
The molecular weight of the purified peptides was determined on a Voyager-DE RP MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, Calif.) with α-cyano-4-hydroxycinnamic acid as the matrix material. The partial N-terminal amino acid sequence was determined by Edman degradation and amino acid composition analysis, as described previously (20). N-acetyl deblocking was performed by incubating the peptide with TFA-methanol (1:1) at 47°C for 3 days (14).
Circular dichroic (CD) spectra were recorded by using a Jasco J-810 spectropolarimeter (Jasco International Co., Ltd., Tokyo, Japan) calibrated with ammonium d-camphor-10-sulfonate (Icatayama Chemicals, Tokyo, Japan). Measurements were performed at 20°C by using a quartz cuvette (Starna, Essex, United Kingdom) with a path length of 0.1 cm. All the measurements were performed with a protein concentration of 0.3 mg/ml in 0.5 to 1.0 M sodium chloride buffer (pH 4). Samples were scanned five times at 50 nm/min with a bandwidth of 1 nm and a response time of 1 s over a wavelength range of 190 to 260 nm. The data were averaged, and the spectrum of a sample-free control sample was subtracted. The spectra were smoothed (means movement; convolution width, 5), and all measurements were made at least twice.
PCR and DNA sequencing.
Suitable primers for PCR (Table 1) were designed on the basis of comparison of the histone H1 gene sequences available in the EMBL database (http://www.ebi.ac.uk/). The primers were purchased from Invitrogen (Carlsbad, Calif.). PCR was performed with Taq DNA polymerase (QIAGEN, Hilden, Germany) on a Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany). Conditions were 97°C for 3 min and 35 cycles at 94°C for 30 s, 45 to 65°C for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 5 min. The PCR products were purified with QIAquick PCR purification kit (QIAGEN). Direct DNA sequencing of the PCR products was done by following the BigDye protocol of Applied Biosystems, and the reaction was analyzed on an ABI Prism 377 DNA sequencer (Applied Biosystems).
TABLE 1.
Primers used for PCR and DNA sequencing
| Primer | Sequence 5′-3′ |
|---|---|
| Histone H1 N-term 1 | ACACAAGTGAAAGAGCATTTGGCCAGCCC |
| Histone H1 N-term 2 | AAGGCCAGCCCCCTGCACGGT |
| Histone H1 N-term 3 | TGGAGCTTCAATAGCGCAGAGCAG |
| Histone H1 N-term 5 | TTTACCGGACCGAACGACAGACATGGC |
| Histone H1 N-term P | ATGGCAGAAGTCGCACCAGCACC |
| Histone H1 C-term (antisense) | TTCTTGAGCGCGGCCAGGGACA |
Peptide synthesis.
Synthetic peptide (purified to >90% with RPC and analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry [MALDI-TOF MS]) equal to SAMP H1 was obtained from Joe Gray, Molecular Biology Unit of the University of Newcastle Upon Tyne, Newcastle Upon Tyne, United Kingdom. The synthetic peptide was activated by chromatography of small amounts (5 to 10 μg) on a Mono S cation exchanger. This chromatography was performed in the same manner as the last purification step (Fig. 1C). If necessary for further analysis, the active and the inactive forms obtained upon the cation-exchange chromatography were separated by RPC. A Resource RPC column connected to an ÄKTA purifier system was equilibrated with 0.1% TFA, and bound peptide was eluted with a linear gradient of 0 to 50% 2-propanol containing 0.1% TFA in 10 ml at a flow rate of 1 ml/min. The process was monitored by measuring the absorbance at 214 nm, 254 nm, and 280 nm simultaneously.
The different forms of SAMP H1 were compared by cation-exchange chromatography and gel filtration. As described above, the conditions for the cation exchanger were the same as those for the last chromatography during purification. The gel filtration was performed on a Superdex Peptide HR10/30 column (molecular weight separation range, 7,000 to 100; Amersham Biosciences) with 20 mM ammonium acetate, 0.1% TFA, 0.3 M sodium chloride, pH 4.0. The analysis was run on the ÄKTA purifier system at a flow rate of 0.5 ml/min; and the process was monitored by reading the absorbance at 214 nm, 254 nm, and 280 nm simultaneously.
PPIase treatment.
Synthetic SAMP H1 (0.125 to 0.5 mg/ml) was mixed with Brugia malayi cyclophilin-1 PPIase (0.5 to 2 μg/ml; New England Biolabs, Beverly, Mass.) in either 20 mM ammonium acetate, 0.1% (vol/vol) TFA, 0.3 M sodium chloride, pH 4.0, or 50 mM Tris HCl, pH 7.5, with or without 0.6 M urea (total volume, 0.1 ml); and the mixture was incubated at 25 or 37°C for 1 to 48 h.
Alternatively, 50 μg SAMP H1 in 1 ml buffer A was applied to a Mono S column, immediately followed by the addition of 1 μg PPIase in 1 ml buffer A. After 1 to 16 h incubation at room temperature, the peptide was eluted with the same elution buffers and gradient used in the final cation-exchange purification step. The amount of peptide activated was calculated by reading of the absorbance at 214 nm after separation of the active and inactive forms on a Resource RPC column.
Nucleotide and amino acid sequence accession numbers.
The sequences of the partial salmon histone H1 gene and the complete SAMP H1 peptide are deposited in the EMBL database as accession numbers AJ877042 and P84408, respectively.
RESULTS
Peptide purification.
Antimicrobial activity was detected in acetic acid extracts from the blood, whole eyes, and skin mucus of Atlantic salmon. Skin mucus was chosen as a suitable starting material for isolation of the antimicrobial substance, since mucus could easily be obtained in large amounts. Active SAMP H1 was purified to homogeneity by a combination of reversed-phase and cation-exchange chromatographies.
No activity was lost after the SepPak C18 solid-phase extraction. Antimicrobial activity eluted at the end of the gradient upon HiPrep SP cation-exchange chromatography (Fig. 1A). The presence of 0.1% TFA in the chromatography buffers increased the stability of the antimicrobial activity at this stage. In the absence of TFA, all activity was lost within a few days after chromatography (at 4°C). Most contaminating material was removed after the SepPak extraction and HiPrep SP cation-exchange chromatography. The active fractions from the HiPrep SP step were concentrated on a Resource RPC column. Elution with a gradient did not increase the specific activity but led to the additional loss of sample since the antimicrobial activity was spread out over the whole gradient. Likewise, omission of urea in the buffers used in the subsequent Mono S chromatography step (Fig. 1B) resulted in scattering of the antimicrobial activity along the whole gradient. There were also large batch-to-batch variations in the elution profile of the antimicrobial activity in the absence of urea. The final purification steps of the antimicrobial substance were carried out by repetitious (two to five times) Mono S chromatography until a single, well-defined peak was obtained (Fig. 1C). In these subsequent Mono S chromatography steps, urea was not included in the buffers, since a homogeneous elution peak of antimicrobial activity was more easily obtained without urea. These subsequent chromatography steps were, however, strongly affected by the amount of peptide applied on the Mono S column. Only relatively small amounts (5 to 10 μg) could be applied, as larger amounts resulted in marked broadening of the activity peak. The final recovery (compared to that of the crude acetic acid mucus extract) was about 20%. Desalting of the sample obtained after Mono S chromatography was done by using a Resource RPC column under basic-neutral conditions, but it resulted in an approximately 50% loss of the antimicrobial substance applied to the column. Unlike the earlier concentration step, the antimicrobial substance did not bind at this final stage to the Resource RPC column under acidic (0.1% TFA) conditions. The purified peptide could be stored for several weeks without a loss of activity in the absence of TFA.
Determination of the primary structure of the antimicrobial substance.
Initial Edman N-terminal amino acid sequencing gave no result; however, after N-acetyl deblocking with TFA-methanol, repeated steps of sequencing revealed 15 of the first 16 amino acids, thus indicating that the peptide was acetylated at the N terminus. The identified amino acid residues are highlighted in boldface in Fig. 2. A FASTA search (30) with the amino acid sequence obtained against the sequences in the EMBL database showed that this fragment was identical to the N-terminal part of histone H1 from rainbow trout (Oncorhynchus mykiss) and brown trout (Salmo trutta). This prompted us to obtain the corresponding gene sequence by PCR. Since the gene sequence was unknown at that time, the PCR was run in a gradient thermocycler with 12 different annealing temperatures in the range of 45 to 65°C. Most PCR primer pairs, however, gave products of the expected size in the entire range, with the exceptions being primers N-term 1 and C-term (Table 1), which yielded two products at the lowest temperature, and primers N-term 5 and C-term, which provided a very low yield at the highest temperature (results not shown). For sequencing, the two largest fragments, which were obtained with primers N-term 1 and C-term and primers N-term 2 and C-term and which were obtained at the middle annealing temperature (approximately 55°C), were directly sequenced with all primers. The complete sequence is presented in Fig. 2, together with the sequence of the rainbow trout histone H1 and the derived protein sequence.
FIG. 2.
Histone H1 sequences. Histone H1 gene sequences of rainbow trout (OMHIS1; sequence from the EMBL database) and Atlantic salmon (SSHIS1) and the corresponding protein sequences are shown. Differences in the salmon and the trout gene sequences are shown in boldface, and primer sequences are underlined (the dotted underline signals overlapping primer sequences). The protein sequence obtained by Edman sequencing is shown in boldface, while the histone H1 sequence outside the antimicrobial fragment is shown in grey italics.
The molecular weight of the purified peptide was determined to be 2,836 by MALDI-TOF MS (result not shown). This is the theoretical molecular weight of the 30 first residues of the histone H1 sequence obtained, including the N-terminal acetylated residue. Analysis of the amino acid composition showed that it was in accordance with the deduced sequence of the peptide (results not shown).
Activation of synthetic peptide with PPIase.
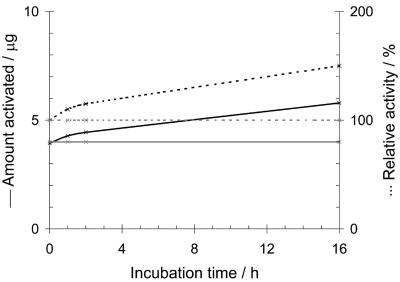
N-terminal acetylation was not necessary for antimicrobial activity since the deacetylation did not reduce the activity. The acetyl group was therefore not included in the synthesis of SAMP H1. The synthetic peptide was inactive, as determined by the standard radial diffusion assay. However, the synthetic peptide was partly activated upon chromatography on the Mono S cation-exchange column (Fig. 3) by use of the same chromatography conditions used in the final purification step. The absolute amount of SAMP H1 activated did not increase by applying larger amounts of peptide to the column: approximately 5 μg of the peptide was activated, irrespective of the amounts applied. This observation led us to further studies on the activation of the synthetic peptide. Treatment of SAMP H1 bound to the chromatographic column with PPIase increased the amount of active peptide. Moreover, prolonged exposure to PPIase increased the amount of active peptide (Fig. 3). PPIase treatment of the inactive peptide in buffer (not bound to the column) did not activate the peptide, nor did PPIase treatment of the active peptide in buffer inactivate the peptide (data not shown). In all cases the integrity of the peptides was controlled by MALDI-TOF MS (data not shown).
FIG. 3.
Activation of synthetic SAMP H1. SAMP H1 (50 μg) was applied to a Mono S column with (black lines) and without (grey lines) addition of PPIase. Solid lines, number of μg of peptide activated; dotted lines, relative activity. The amount of activated peptide was measured by determination of the A214 after separation of the active and inactive forms by Resource RPC. Antimicrobial activity was measured by the radial diffusion assay, with 100% relative activity being equal to the activity obtained without PPIase at 0 h.
Characterization of the active and inactive forms of SAMP H1.
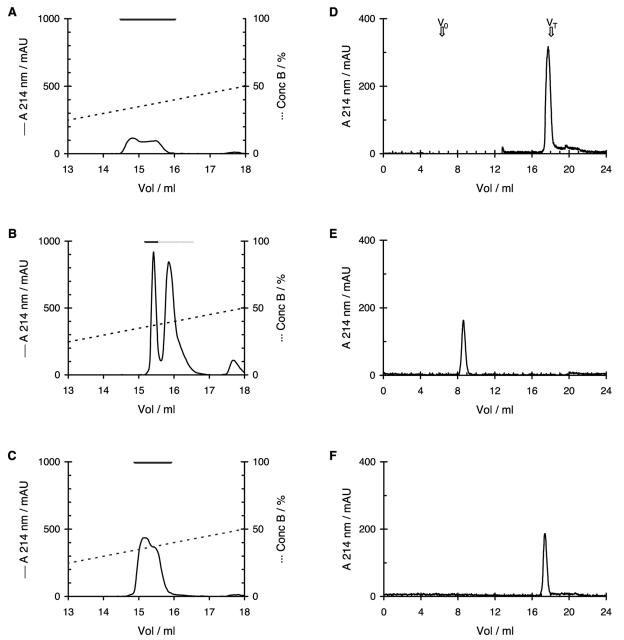
The active and inactive forms of SAMP H1 had identical molecular weights, as determined by MS (data not shown). Their chromatographic patterns, however, were different. Upon Mono S chromatography, the purified peptide (Fig. 4A) and the active form of the synthetic peptide (Fig. 4C) eluted slightly ahead of the inactive synthetic form (Fig. 4B), although the two forms could not be entirely separated. Upon gel filtration the inactive form of the synthetic peptide eluted as expected for a peptide of this size (Fig. 4E), whereas the purified peptide (Fig. 4D) and the active form of the synthetic peptide (Fig. 4F) lagged behind and eluted as if they had a molecular weight similar to that of sodium chloride. Most striking were the differences obtained upon chromatography on the Resource RPC column, in which the inactive form of the synthetic peptide bound to the column in the presence of 0.1% TFA, whereas the purified peptide and the active form of the synthetic peptide passed through in the exclusion volume of the column (data not shown).
FIG. 4.
Chromatographic properties of the different forms of SAMP H1. Chromatographic properties of natural (A and D), inactive synthetic (B and E), and activated synthetic (C and F) SAMP H1 on Mono S (A to C) and Superdex Peptide (D to F) columns. The black horizontal bars in panels A to C show the fractions that contained antimicrobial activity, while the dotted bar in panel B shows the fractions that contained inactive SAMP H1. The void volume (V0) and total volume (VT) of the Superdex Peptide column are indicated by the arrows in panel D. Solid lines, absorbance at 214 nm; dotted lines, concentration of buffer B; mAU, milli-absorption units.
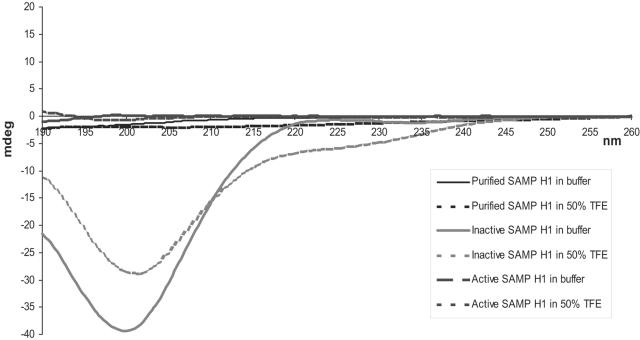
Structural analysis by CD confirmed that the active and inactive forms of SAMP H1 have different conformations. In buffer, inactive SAMP H1 had a CD spectrum characteristic of that of a nonstructured, random-coil conformation (Fig. 5). In the presence of 50% TFE (trifluoroethanol), which induces and stabilizes α-helical structures in peptides, part of the peptide seems to form an α helix, as witnessed by the negative value of about 220 to 230 nm (Fig. 5). Active SAMP H1, however, was optically inactive in both buffer and 50% TFE (Fig. 5), probably due to a condensed structure. One should note that the purified, biological peptide gave the same CD spectra as the active form of the synthetic peptide. The integrity of the peptides was controlled by MALDI-TOF MS after each chromatography run and CD experiment (data not shown).
FIG. 5.
CD spectra of SAMP H1. The CD spectra of natural, inactive synthetic, and active synthetic SAMP H1 in buffer and 50% TFE are shown. The experiments were performed as described in Materials and Methods, and values are given in mean relative ellipticity.
Antimicrobial activity.
The MICs of SAMP H1 against the microorganisms examined are shown in Table 2 and are compared to those of pleurocidin. SAMP H1 was active against both gram-negative and -positive bacteria and had a slightly lower potency than pleurocidin against all organisms except the fish pathogen A. salmonicida. The natural, purified peptide and the synthetic, activated peptide had the same specific activities in all cases. The activities of AMPs are often reduced in the presence of salt. In this study antimicrobial assays were often performed with samples containing 0.3 M NaCl, without a measurable effect on activity. However, due to a limited amount of material, no systematic evaluation of the salt sensitivity was performed.
TABLE 2.
MICs for SAMP H1 and pleurocidin against some microorganismsa
| Microorganism | MIC (μM)
|
||
|---|---|---|---|
| SAMP H1
|
Pleuro- cidin | ||
| Purified | Activated | ||
| Escherichia coli | 2 | 2 | 0.5 |
| Aeromonas salmonicida subsp. salmonicida | 4 | 4 | 32 |
| Vibrio anguillarum | 1 | 1 | 0.3 |
| Salmonella enterica serovar Typhimurium | 4 | 4 | 2 |
| Bacillus subtilis | 2 | 2 | 0.5 |
| Listeria ivanovii | 4 | 4 | 4 |
The values were determined by using TSB, as described in Materials and Methods. All tests were performed in duplicate; the same values were obtained both times.
DISCUSSION
We were able to detect antimicrobial activity in the skin mucus of Atlantic salmon, unlike Richards et al. (33), who identified antimicrobial activity in extracts from the liver, intestine, and stomach but not in extracts from the skin. This may have been because their extraction method was not harsh enough. Our experience shows that acidic extraction, preferably with acetic acid, is necessary to release antimicrobial peptides from the mucus of fish. Richards et al. (33) assigned the antimicrobial activity to the entire histone H1 protein; but the presence of specific fragments, like the one reported here, cannot be ruled out.
The purification of SAMP H1 was problematic due to large batch-to-batch differences in the chromatographic properties of the antimicrobial activity, especially in the step in which the Mono S column was used. This may have been due to unfavorable interactions with other compounds from the mucus, and the presence of urea eliminated the problem. The presence of urea in the subsequent Mono S runs was not necessary, however, probably because the interfering compounds were removed.
The sequence of the 30-mer peptide was quite unique in that it contained high amounts of alanine (43%), lysine (23%), and proline (20%); and a sequence homology search identified the peptide as a fragment of histone H1. Surprisingly, unlike other synthetic AMPs, the synthetic version of the peptide initially had no antimicrobial activity. A small fraction of it, however, became activated upon binding to the Mono S cation-exchange column. The active form apparently had an altered three-dimensional structure, since it eluted ahead of the inactive form upon chromatography on the Mono S column; it was (in contrast to the inactive form) not retained on the reversed-phase column, and it eluted with an apparent molecular weight of about 100 upon gel filtration, in contrast to the inactive form, which eluted at the expected molecular weight of about 2,800. Moreover, the CD spectra of the active form were entirely different from those of the inactive form, whose CD spectra were those of a nonstructured peptide. The activity cannot be due to a degraded product, since the active form had the expected molecular weight, as determined by MALDI-TOF MS. These observations demonstrate that the activated synthetic peptide has the same apparent physiochemical properties as the biological isolated SAMP H1. A peculiar finding was that the same amount of peptide was activated, irrespective of the amount applied to the column or the incubation time (Fig. 3). Thus, the activation probably occurs at the solvent-matrix interface and is dependent only on the column cross section.
Comparison of the active and inactive forms of SAMP H1 by CD and chromatographic analyses revealed that the active form, both the natural and the synthetic forms, has a structure different from that of the inactive synthetic form. The active form is more structured and is probably not oligomerized but is more condensed, since it is retarded upon gel filtration and it passed through a filtration membrane with a nominal molecular weight cutoff of 3,000. The active form apparently exposes its hydrophilic residues and buries the hydrophobic ones to a greater extent than the inactive form, since the active form did not bind to the RPC column. The slightly earlier elution of the active peptide upon cation-exchange chromatography might be due to a more rigid structure that limits the number of positively charged groups that may interact with the column material at the same time. The inactive form, being nonstructured and presumably more flexible, may permit most of its positive groups to interact with the column.
It is known that the peptide bond on the N-terminal side of proline residues can exist in both the trans and the cis conformations, in contrast to the peptide bond adjacent to other residues, which is in the trans conformation (35). This prompted us to consider the possibility that the synthetic peptide did not contain the proper proline peptide bond conformation necessary for the peptide to be active. When the synthetic peptide was free in buffer, treatment with PPIase did not produce the active form of SAMP H1. The CD spectra suggested an unstructured random coil, in which the all-trans form might be most stable. It has previously been shown that PPIase may be more effective in forming the correct structure of a protein when it is bound to a chromatographic support than when it is in solution and that even higher yields of correctly folded protein were achieved when the PPIase was supplemented with a minichaperone and a protein disulfide isomerase (1). For SAMP H1 to obtain the necessary isomerization upon PPIase treatment, the peptide must probably be structurally stabilized, such that the cis form becomes energetically favorable, and such stabilization apparently occurs upon binding of the peptide to the cation-exchange column.
An inducible mechanism for the proteolytic production of AMPs from histones has been found for the histone H2A-derived peptides buforin I and parasin I. Buforin I is produced from cytoplasmic histone H2A by the action of pepsin in the gastric gland cells of the Asian toad (17). Parasin I is produced in a similar way in the skin mucus of catfish by the action of cathepsin D (4), which itself is activated by matrix metalloproteinase 2 (5). A similar mechanism may exist for the formation of SAMP H1 by a specific proteolytic degradation of the histone H1 during proliferation/degradation of the epidermal tissue of the fish. The antimicrobial active structure of the peptide is already formed by the folding during the biosynthesis of the histone and, consequently, is present in the antimicrobial entity. On the other hand, synthesized SAMP H1 is not active, since the conditions for chemical synthesis of the peptide do not support such a specific isomeric formation of the correct proline isomeric forms that are needed for an active peptide.
It is not possible at present to deduce which of the prolyl peptide bonds are responsible for the high degrees of stability of the two forms of SAMP H1. Even though extensive studies have been performed with model peptides (32), the results cannot be directly transformed to natural peptides and proteins. Of the six proline residues in SAMP H1, four are preceded by alanine residues, and the remaining two are preceded by lysine and glycine, respectively. Neither of these peptide pairs showed a propensity to yield cis amide bonds in the aforementioned study. It is likely that several possible isomeric structures of SAMP H1 possess antimicrobial activity.
Patrzykat et al. (29) reported on the presence of a shorter form (26-mer) of SAMP H1 in coho salmon in which the four C-terminal amino acid residues were missing but the synthetic form of the peptide was inactive. Judging from our results, this was probably because the correct proline peptide bond conformation was not present in their synthetic peptide. Their inactive peptide was, however, reported to potentiate the antimicrobial activity of pleurocidin and lysozyme, whereas our inactive SAMP H1 showed no synergy with pleurocidin (data not shown).
This work shows for the first time the importance of proline isomers in the activity of a peptide and that it can be enzymatically converted from an inactive form to an active form and also confirms that the structures of such small antimicrobial peptides can be essential for the antimicrobial activity.
Acknowledgments
We are grateful to AKVAFORSK Averøy Research Station, Bremnes Fryseri, and the Research Aquaculture Facility, Department of Agricultural Engineering, Norwegian University of Life Sciences, for supplying fish material and to Dimitris Mantzilas, Program of Biochemistry and Molecular Biology, Department of Molecular Biosciences, University of Oslo, for assistance with the MS and CD studies.
This work was supported by grants from the Research Council of Norway and EU (project QLK2-CT-2000-00422).
REFERENCES
- 1.Altamirano, M. M., C. García, L. D. Possani, and A. R. Fersht. 1999. Oxidative refolding chromatography: folding of the scorpion toxin Cn5. Nat. Biotechnol. 17:187-191. [DOI] [PubMed] [Google Scholar]
- 2.Birkemo, G. A., T. Lüders, Ø. Andersen, I. F. Nes, and J. Nissen-Meyer. 2003. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta 1646:207-215. [DOI] [PubMed] [Google Scholar]
- 3.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61-92. [DOI] [PubMed] [Google Scholar]
- 4.Cho, J. H., I. Y. Park, H. S. Kim, W. T. Lee, M. S. Kim, and S. C. Kim. 2002. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 16:429-431. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J. H., I. Y. Park, M. S. Kim, and S. C. Kim. 2002. Matrix metalloproteinase 2 is involved in the regulation of the antimicrobial peptide parasin I production in catfish skin mucosa. FEBS Lett. 531:459-463. [DOI] [PubMed] [Google Scholar]
- 6.Cole, A. M., P. Weis, and G. Diamond. 1997. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272:12008-12013. [DOI] [PubMed] [Google Scholar]
- 7.Dionysius, D. A., and J. M. Milne. 1997. Antibacterial peptides of bovine lactoferrin: purification and characterization. J. Dairy Sci. 80:667-674. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, A. 1992. Differences between the immune mechanisms of fish and higher vertebrates in microbial diseases of fishes, p. 1-30. In R. J. Roberts (ed.), Microbial diseases of fish. Academic Press, London, United Kingdom.
- 9.Faye, T., D. A. Brede, T. Langsrud, I. F. Nes, and H. Holo. 2002. An antimicrobial peptide is produced by extracellular processing of a protein from Propionibacterium jensenii. J. Bacteriol. 184:3649-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganz, T. 1994. Biosynthesis of defensins and other antimicrobial peptides. Ciba Found. Symp. 186:62-71. [DOI] [PubMed] [Google Scholar]
- 11.Ganz, T., and R. I. Lehrer. 1999. Antibiotic peptides from higher eukaryotes: biology and applications. Mol. Med. Today 5:292-297. [DOI] [PubMed] [Google Scholar]
- 12.Ganz, T., M. E. Selsted, D. Szklarek, S. S. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, A. G., S. M. Hakimi, C. A. Mittanck, Y. Wu, B. M. Woerner, D. M. Stark, D. M. Shah, J. Liang, and C. M. Rommens. 2000. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18:1307-1310. [DOI] [PubMed] [Google Scholar]
- 14.Gheorghe, M. T., H. Jörnvall, and T. Bergman. 1997. Optimized alcoholytic deacetylation of N-acetyl-blocked polypeptides for subsequent Edman degradation. Anal. Biochem. 254:119-125. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E. 2001. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 1:156-164. [DOI] [PubMed] [Google Scholar]
- 16.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. S., H. Yoon, I. Minn, C. B. Park, W. T. Lee, M. Zasloff, and S. C. Kim. 2000. Pepsin-mediated processing of the cytoplasmic histone H2A to strong antimicrobial peptide buforin I. J. Immunol. 165:3268-3274. [DOI] [PubMed] [Google Scholar]
- 18.Lauth, X., H. Shike, J. C. Burns, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. Van Olst, V. Nizet, S. W. Taylor, C. Shimizu, and P. Bulet. 2002. Discovery and characterization of two isoforms of moronecidin, a novel antimicrobial peptide from hybrid striped bass. J. Biol. Chem. 277:5030-5039. [DOI] [PubMed] [Google Scholar]
- 19.Lehrer, R. I., M. Rosenman, S. S. Harwig, R. Jackson, and P. Eisenhauer. 1991. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137:167-173. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Cuesta, M. C., G. Buist, J. Kok, H. H. Hauge, J. Nissen-Meyer, C. Peláez, and T. Requena. 2000. Biological and molecular characterization of a two-peptide lantibiotic produced by Lactococcus lactis IFPL105. J. Appl. Microbiol. 89:249-260. [DOI] [PubMed] [Google Scholar]
- 21.Minn, I., H. S. Kim, and S. C. Kim. 1998. Antimicrobial peptides derived from pepsinogens in the stomach of the bullfrog, Rana catesbeiana. Biochim. Biophys. Acta 1407:31-39. [DOI] [PubMed] [Google Scholar]
- 22.Nissen-Meyer, J., and I. F. Nes. 1997. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch. Microbiol. 167:67-77. [PubMed] [Google Scholar]
- 23.Noga, E. J., Z. Fan, and U. Silphaduang. 2001. Histone-like proteins from fish are lethal to the parasitic dinoflagellate Amyloodinium ocellatum. Parasitology 123:57-65. [DOI] [PubMed] [Google Scholar]
- 24.Oren, Z., and Y. Shai. 1996. A class of highly potent antibacterial peptides derived from pardaxin, a pore-forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur. J. Biochem. 237:303-310. [DOI] [PubMed] [Google Scholar]
- 25.Oren, Z., and Y. Shai. 1998. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers 47:451-463. [DOI] [PubMed] [Google Scholar]
- 26.Park, C. B., M. S. Kim, and S. C. Kim. 1996. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem. Biophys. Res. Commun. 218:408-413. [DOI] [PubMed] [Google Scholar]
- 27.Park, C. B., J. H. Lee, I. Y. Park, M. S. Kim, and S. C. Kim. 1997. A novel antimicrobial peptide from the loach, Misgurnus anguillicaudatus. FEBS Lett. 411:173-178. [DOI] [PubMed] [Google Scholar]
- 28.Park, I. Y., C. B. Park, M. S. Kim, and S. C. Kim. 1998. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 437:258-262. [DOI] [PubMed] [Google Scholar]
- 29.Patrzykat, A., L. Zhang, V. Mendoza, G. K. Iwama, and R. E. Hancock. 2001. Synergy of histone-derived peptides of coho salmon with lysozyme and flounder pleurocidin. Antimicrob. Agents Chemother. 45:1337-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pütsep, K., S. Normark, and H. G. Boman. 1999. The origin of cecropins; implications from synthetic peptides derived from ribosomal protein L1. FEBS Lett. 451:249-252. [DOI] [PubMed] [Google Scholar]
- 32.Reimer, U., G. Scherer, M. Drewello, S. Kruber, M. Schutkowski, and G. Fischer. 1998. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 279:449-460. [DOI] [PubMed] [Google Scholar]
- 33.Richards, R. C., D. B. O'Neil, P. Thibault, and K. V. Ewart. 2001. Histone H1: an antimicrobial protein of Atlantic salmon (Salmo salar). Biochem. Biophys. Res. Commun. 284:549-555. [DOI] [PubMed] [Google Scholar]
- 34.Shike, H., X. Lauth, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. Van Olst, C. Shimizu, P. Bulet, and J. C. Burns. 2002. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur. J. Biochem. 269:2232-2237. [DOI] [PubMed] [Google Scholar]
- 35.Stewart, D. E., A. Sarkar, and J. E. Wampler. 1990. Occurrence and role of cis peptide bonds in protein structures. J. Mol. Biol. 214:253-260. [DOI] [PubMed] [Google Scholar]
- 36.Wu, M., and R. E. Hancock. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274:29-35. [DOI] [PubMed] [Google Scholar]