Abstract
Mu et al. (Mu, J., M. T. Ferdig, X. Feng, D. A. Joy, J. Duan, T. Furuya, G. Subramanian, L. Aravind, R. A. Cooper, J. C. Wootton, M. Xiong, and X. Z. Su, Mol. Microbiol. 49:977-989, 2003) recently reported exciting associations between nine new candidate transporter genes and in vitro resistance to chloroquine (CQ) and quinine (QN), with six of these loci showing association with CQ or QN in a southeast Asian population sample. We replicated and extended this work by examining polymorphisms in these genes and in vitro resistance to eight drugs in parasites collected from the Thailand-Burma border. To minimize problems of multiple testing, we used a two-phase study design, while to minimize problems caused by population structure, we analyzed parasite isolates collected from a single clinic. We first examined associations between genotype and drug response in 108 unique single-clone parasite isolates. We found strong associations between single nucleotide polymorphisms in pfmdr and mefloquine (MFQ), artesunate (AS), and lumefantrine (LUM) response. We also observed associations between an ABC transporter (G7) and response to QN and AS and between another ABC transporter (G49) and response to dihydro-artemisinin (DHA). We reexamined significant associations in an independent sample of 199 unique single-clone infections from the same location. The significant associations with pfmdr-1042 detected in the first survey remained. However, with the exception of the G7-artesunate association, all other associations observed with the nine new candidate transporters disappeared. We also examined linkage disequilibrium (LD) between markers and phenotypic correlations between drug responses. We found minimal LD between genes. Furthermore, we found no correlation between chloroquine and quinine responses, although we did find expected strong correlations between MFQ, QN, AS, DHA, and LUM. To conclude, we found no evidence for an association between 8/9 candidate genes and response to eight different antimalarial drugs. However, the consistent association observed between a 3-bp indel in G7 and AS response merits further investigation.
Transporter loci are attractive candidates for antimalarial drug resistance genes. Two of four known drug resistance loci in Plasmodium falciparum are transporters, and transporters play major roles in drug resistance in both bacteria and cancer cells (1, 6, 13). A previous study (14) located 49 genes encoding transporters in the P. falciparum genome and sequenced these genes in a panel of 97 isolates from worldwide locations. Polymorphisms in 11/49 genes showed significant association with in vitro drug resistance to chloroquine (CQ) and/or quinine (QN). Two of these genes, pfcrt and pfmdr, are already known to play a role in resistance (9, 12, 26, 28), while the remaining nine candidates are new. Of these nine candidates, six were associated with either a CQ response or a QN response in a southeast Asian population sample. There was also strong linkage disequilibrium (LD) between loci on different chromosomes, consistent with coselection or epistasis (8), and strong correlations between 50% inhibitory concentrations (IC50) with CQ and QN, suggesting that common genes may underlie resistance to both drugs.
While these results are exciting, some caution is needed in interpreting such candidate gene studies. There are two particular problems. First, when multiple tests are carried out, some significant results may occur by chance (false positives or type I errors). This problem may be overcome by using more-stringent criteria for determining significance by procedures such as the Bonferroni correction (4). However, this may result in discounting truly significant results and inflating numbers of false negatives (type II errors) and so is not entirely satisfactory (19). A second major problem with association studies is population structure (23-25). If samples are collected from two or more populations that differ in both allele frequency and phenotype, then spurious association may occur with genetic loci that have no real relationship with the phenotype of interest. For example, suppose parasites are sampled from two populations, one with 90% and the other with 10% of parasites resistant to a drug. Suppose also that allele frequencies have diverged by genetic drift in these two populations at a “neutral” locus that has no influence on resistance status, so that frequencies of an allele A at this locus are 40% in one population and 60% in the other. Assuming equal numbers of parasites from each population are analyzed, the population sample will contain 50% resistant parasites and 50% will carry allele A. However, resistant parasites will carry allele A more often than allele B, generating a false positive association between this locus and drug resistance. For the same reasons, sampling from multiple populations results in elevated LD and can generate correlations between drug phenotypes even when the different sets of genes underlie resistance to each drug.
Study replication using samples collected in a single region over a limited time frame remains the most convincing test of whether genotype-phenotype associations are real or spurious (11). To minimize the potential problem of geographical population structure, we reexamine associations between the 11 candidate genes and resistance to antimalarial drugs in parasites collected from a single clinic on the Thailand-Burma border. Furthermore, to minimize type I errors due to multiple statistical comparisons, we used a two-phase study design, in which significant associations were reexamined in an independent population sample. We investigated genotypic associations with response to mefloquine (MFQ), artesunate (AS), dihydro-artemisinin (DHA), lumefantrine (LUM), doxycycline (DOX), and atovaquone (AQ) in addition to QN and CQ. We found no evidence for association between polymorphisms in eight of the new transporters and drug response. However, we did observe significant association between an ABC transporter (G7) on chromosome 13 (Chr 13) and artesunate response in both population samples examined.
MATERIALS AND METHODS
Study design.
We used a two-phase design. In phase I, we genotyped polymorphisms in all 11 genes (9 new transporters, pfmdr, and pfcrt) for comparison with in vitro resistance to 8 different antimalarial drugs. In phase II, we retyped only those polymorphisms that showed significant association in phase I in an independent population sample collected from the same location.
Parasite sampling.
We collected 5 ml of P. falciparum-infected blood samples with >0.5% parasitemia from patients visiting the malaria clinic at Mawker-Thai on the Thai-Burma border. This site is ∼100 miles from Maela, Thailand, where recent studies of in vitro resistance have been conducted (5, 22). We collected samples from patients that had not visited the malaria clinic within the past 60 days and had no history of prior malaria treatment. The clinic serves people on both sides of the border; 90% of patients travel from within a 10-km radius around the clinic (François Nosten, personal communication). Collection protocols were approved by the Ethical Committee of the Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, and by the Institutional Review board at the University of Texas Health Science Center at San Antonio. Parasite isolates used in phase I were collected between December 1998 and July 2001, while parasites used in phase II were collected between November 2001 and September 2003. Patients in this region have been treated with mefloquine-artesunate combination therapy since 1994 (18).
Measurement of in vitro resistance.
Infected blood samples were transported to the laboratory in Mae Sot, Thailand, within 4 h of collection. We measured in vitro responses to CQ, QN, MFQ, LUM, AS, DHA, AQ, and DOX using a 48-h [3H]hypoxanthine incorporation assay (5, 7). Tests were performed using a starting parasitemia of between 0.5 and 1%, and duplicate assays were performed at each of 7 or 11 doubling drug concentrations. Dose-response curves, IC50s, and coefficients of variation were calculated by fitting the data to an inhibitory E-max pharmacokinetic model using WINNONLIN version 3.1 (Pharsight Corporation). To ensure data quality, we rejected all IC50s with coefficients of variation (standard error × 100)/mean of estimated IC50s of >20% and those in which the incorporation in control wells (parasites, no drug) was <5 times the background level (red blood cells only). A single outlier was removed in some cases. IC50s were retained if removal of an outlier reduced the coefficient of variation (CV) to less than 20% and did not alter the IC50 value by greater than 10%. For curves from highly resistant samples, the range of dilutions used in the drug tests was insufficiently wide to obtain accurate measures of IC50. In these cases, the curves were “forced” by adding an extra data point (zero, indicating no growth) at the next doubling concentration. This procedure results in conservative IC50s, while allowing us to retain data from interesting parasite isolates with unusually high IC50s.
Microsatellite genotyping of infections.
Infections containing multiple clones provide ambiguous information for both genotype and drug response data. We genotyped seven microsatellite loci to identify infections containing multiple clones. We also used the genotype information to identify multilocus haplotypes that were represented more than once in the data set. Inclusion of “clonally identical” parasites can bias association studies, just as inclusion of relatives can bias association studies in human populations. Parasite DNA was prepared by phenol/chloroform extraction of whole blood, following removal of buffy coats as described previously (15). The microsatellite loci amplified were ARA2 (Chr 11), POLYA (Chr 4), TA1 (Chr 6), C2M1 (Chr 2), C3M54 (Chr 3), TA60 (Chr 13), C3M85 (Chr 3), and C4M30 (Chr 4). Primers and protocols for amplifying these loci are described on the NIH website (http://www.ncbi.nlm.nih.gov/projects/Malaria/Mapsmarkers/PfGMap/pfgmap.html). We amplified microsatellites using fluorescent end-labeled oligonucleotides, ran products on an ABI 3100 capillary sequencer, and scored alleles using GENESCAN and GENOTYPER software. Multiple-clone infections were defined as those in which one or more of the seven loci showed multiple alleles. Minor alleles were scored if they were >33% of the height of predominant alleles (2, 3).
Genotyping of transporter polymorphisms.
We used a combination of primer extension (“SNaPshot” genotyping), sequencing, and microsatellite genotyping to score polymorphisms in the 11 candidate transporter genes (Table 1). Primer extension was performed using the ABI PRISM SNaPshot Multiplex kit (Applied Biosystems) and scored on an ABI 3100 capillary sequencer using GENESCAN and GENOTYPER software. In this technique, mutations are detected by fluorescent single-base extension of primers designed adjacent to polymorphic sites. Multiple mutations can be genotyped in a single reaction and scored in a single lane of a gel or a single capillary on an automated sequencer. The methods used were similar to protocols described previously (15). Table 1 lists primer sequences and conditions used for the different loci. For pfcrt we used a combination of primer extension and direct sequencing. We genotyped five codons (codons 220, 271, 326, 356, and 371) in the 3′ region of pfcrt by primer extension, while mutations in the 5′ region of pfcrt (amino acids 72, 74, 75, and 76) were scored by direct sequencing using ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems). We used sequencing, rather than primer extension, for the 5′ region of pfcrt, since the cluster of mutations from amino acids 72 to 76 makes primer design problematic for primer extension or other single nucleotide polymorphism (SNP) typing methods. We used PCR spin columns (QIAGEN) to remove excess nucleotides and oligonucleotides from the PCRs prior to the sequencing reactions. As controls, we genotyped a variety of laboratory parasite lines (3D7, HB3, Dd2, VS/1, and FCR3) with known pfmdr and pfcrt sequences. These were obtained from MR4/American Type Culture Collection (Manassas, Virginia).
TABLE 1.
Methods and oligonucleotide sequences used for genotyping transporter loci
| Genea | Position | AA changeb | Methodc | Snapshot oligod | Template oligod |
|---|---|---|---|---|---|
| pfcrt | 72 | C-S | DS | ||
| 74 | M-I | DS | pfcrt-F:GGTGGAGGTTCTTGTCTTGG | ||
| 75 | N-E | DS | pfcrt-R:GAATTTCCCTTTTTATTTCCA | ||
| 76 | K-T | DS | |||
| 220 | A-S | SS | ctgactCATATTTAATCTTGTCTTAATTAGT | ||
| 271 | F-Q | SS | ctgacCTTTCCTAATTAATTCTTACGTT | ||
| 326 | N-D/S | SS | gactgacGGTTATTAAATTATCACAAATGY | ||
| gactgactgTGGTTATTAAATTATCACAAATG | pfcrt2F:TCTTTTGAAACACAAGAAGAAA | ||||
| 356 | I-T/L | SS | gacTGTATACAAGGTCCAGCA | pfcrt-2R:AATTTCACACTTACCAAAGTTAC | |
| actgactTGTATACAAGGTCCAGCA(AT) | |||||
| 371 | R-I | SS | gactgactgactgAATTTTATAGGGTGATGTTGTAA | ||
| pfmdr | 86 | N-Y | SS | gactgactgactgactAGGATTAATATCATCACCTAAAT | pfmdr5′-F TGGGTAAAGAGCAGAAAGAG |
| 184 | Y-F | SS | gactgactgactCATTTTTTAATGACCAAATA | pfmdr5′-R ACGGAAAAACGCAAGTAATA | |
| 1034 | S-C | SS | ATGCAGCTTTATGGGGATTC | pfmdr3′-F TTTCTGTAATTTGATAGAAAAAGC | |
| 1042 | N-D | SS | gactATCCAAACCAATAGGCAAAACTAT | pfmdr3′-R TTGCATCTTCTCTTCCAAAT | |
| gactgactGGGTTCTTGACTAACTATTGAAAATAAGT | |||||
| 1246 | D-Y | SS | TTCTAAGAT | ||
| G2 (PFA0590w) | 191 | Y-H | SS | GTTTTTTGAAGCTCTTTTG | G2-F TAGTGGTAGTGATGATAATGTTTTT |
| 437 | A-S | SS | GACTGAGTAACATAGGAGTTTTTATAAGT | G2-R GATATGGAAGGATCTAAAGATGTAA | |
| G7 (PF13-0271) | 1390 | &1 | MS | G7-F AAACAAATCCAAATATTACGAAAA | |
| G7-R CAAACTACTTTTTCCTGAACCA | |||||
| G25 (PF14-0679) | Intron | G-A | SS | CTTGTAACAGTTTTATGTGTAAGTA | C25-F ATGGGATGCCTTCATCATAG |
| C25-R TTCCAGTTAACACACCATTTG | |||||
| G30 (PF14-0292) | Intron | C-G | SS | GACTGGAAGGCATACTTAAATTACTT | G30-F AAAGGGAAGGAAGGCATAC |
| G30-R GCCATATTCACAACAATTCA | |||||
| G47 (PFE0775c) | 241 | L-V | SS | GTAATTATTCTATATATTATGAGGATAAAAAATTA | G47-F TGCCAAAGAAAAAGAACGAT |
| G47-R TTTTCAAATACACTCGCCATA | |||||
| G49 (PF08-0078) | 146 | Q-E | SS | tATTCATCTTCTTCTAATTCATCTT | G49-1F GGAAGGCATTATAGCGAAACC |
| G49-1R TGTTCATCCAGGTCATCTTCA | |||||
| 1046 | L-I | SS | ctGTTTAAGTTCATGTTTTTAAATATATAT | G49-2F GGTGAAAAATACGGAACAAA | |
| 1116 | L-I | SS | TGGATACAAATTTTTCGAAC | G49-2R TCACCTCCACTTTTGTCTTT | |
| G54 (PF14-0260) | 141 | Y-Y | SS | TAGTGTAAAAAATGATAGTGATTA | G54-F TTTTAAAAGAATCGTTTGTG |
| 144 | T-T | SS | gactATACTCATATCATCTATTAATTTTGT | G54-R TCTTTGTAATTTGTTTGGAT | |
| G55 (PF14-0133) | Intron | &2 | MS | G55-F GAGTTACAGTTTCCCATTG | |
| G55-R TGAACAAAATAAAATGAACA | |||||
| G70 (PFL0620c) | 105 | E-K | SS | gactgactgactGGTATTGGTCATATATGTTGGAAT | G70-F AAAATTGGCTAGTGTTATTG |
| G70-R TTTTAACAACTGGAAACATT |
PlasmoDB (version 4) gene identifiers are shown in brackets.
Italicized letters indicate nucleotide changes in noncoding regions; &1 and &2 are length variants resulting from indels or microsatellites. AA, amino acid.
SS, snapshot; DS, direct sequencing; MS, microsatellite genotyping.
Oligo, oligonucleotide. Tails (lowercase) were added to some oligos to adjust length of products and allow multiplexing of multiple SNP assays.
Statistical analysis.
Prior to analysis, in vitro drug response data were normalized by log transformation. We investigated associations between polymorphisms (SNPs or microsatellites) and in vitro resistance using t tests. To examine genotype-genotype associations, we measured LD between all possible pairwise combinations of polymorphic SNPs, indels, or microsatellites. To assess the significance of LD, we permuted the data 184,000 times and counted the number of times that LD statistics from permuted data sets were greater than those observed. This analysis was performed using FSTAT (version 2.9.3.2) (10). We measured correlations between log-transformed IC50s for all pairwise combinations of the eight drugs analyzed in both phase I and II population samples.
RESULTS
Genotype-phenotype associations.
Parasite isolates used in the phase I survey were collected between December 1998 and July 2001. Following removal of multiple infections, 121 samples that contained a single predominant clone and in vitro data (IC50 CV of <20%) for ≥2 drugs remained. We found 99 genotypes that were represented a single time, while 9 genotypes (defined by microsatellite loci and transporter polymorphisms) were represented two to four times in the data set. To minimize bias, we analyzed unique genotypes only, including only one representative of each genotype and using mean values for the phenotype data from multiply represented clones. This reduced the sample size to 108 for phase I. The IC50s for the eight drugs are summarized in Table 2. All 108 of the pfcrt sequences examined contained 76T, critical for CQ resistance. All were identical to the published Dd2 sequence (CIETSESSTTI at positions 72, 74, 75, 76, 220, 271, 326, 356, and 371, respectively) except for one parasite that had I rather than T at residue 356. pfcrt was therefore excluded from subsequent analyses. Two SNPs in pfmdr (codons 1034 and 1246) and 1 SNP in G2 (codon 151) were monomorphic, leaving 12 polymorphisms (11 SNPs and 1 3-bp indel) in coding regions and 1 microsatellite and 2 SNPs within introns in the 10 genes (pfcrt excluded) for analysis. Both in vitro drug response and polymorphism data for phase I are provided in the supplemental material.
TABLE 2.
Summary statistics for IC50 data from phases I and IIa
| Study phase or parameter | Value for drug
|
|||||||
|---|---|---|---|---|---|---|---|---|
| CQ | QN | AQ | AS | LUM | DHA | DOX | MFQ | |
| Phase I | ||||||||
| n | 85 | 100 | 68 | 83 | 71 | 86 | 62 | 98 |
| Mean | 231.8 | 758 | 6.2 | 3.4 | 56.7 | 4.6 | 15,947 | 75.5 |
| Median | 187.4 | 567 | 5.6 | 2.5 | 41.9 | 3.9 | 15,708 | 56.5 |
| Min | 51.11 | 73 | 0.7 | 0.2 | 4.5 | 0.3 | 4574 | 5.4 |
| Max | 753.6 | 2,325 | 22.9 | 18.6 | 279.6 | 28.8 | 29,848 | 476.8 |
| Phase II | ||||||||
| n | 190 | 168 | 183 | 158 | 163 | 171 | 167 | 194 |
| Mean | 248.5 | 771 | 3.7 | 2.3 | 53.2 | 3.3 | 11,771 | 87.4 |
| Median | 218.1 | 704 | 2.5 | 1.7 | 44.2 | 2.7 | 11,498 | 74.3 |
| Min | 56.48 | 104 | 0.3 | 0.4 | 4.0 | 0.3 | 1406 | 6.1 |
| Max | 707.5 | 1,878 | 22.2 | 8.8 | 176.0 | 13.5 | 36,722 | 353.1 |
| Significance | NSb | NS | *** | * | NS | NS | *** | *** |
Units are nM, and IC50s are shown. The significance of differences in mean IC50 between phase I and II population samples was evaluated using t tests of log-transformed data and is indicated by asterisks. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
NS, not significant.
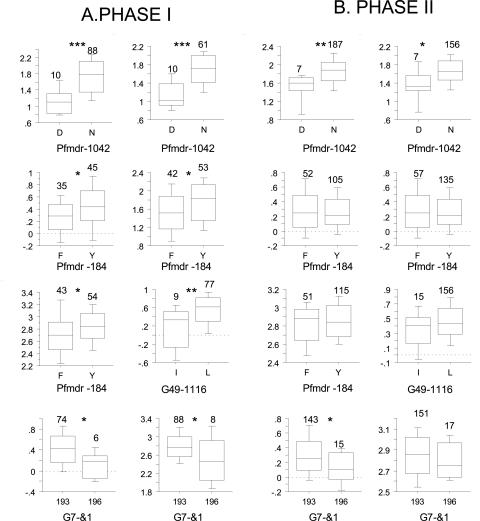
Table 3 shows the relationship between drug response and SNP genotype. Four SNPs were either invariant or had minor alleles at low frequency and were uninformative. Five of 120 t tests were significant at the 5% level, one at the 1% level, and two at the 0.1% level (Fig. 1). Five of eight significant associations involved pfmdr. We found strong associations (P < 0.001) between pfmdr-1042 and MFQ and LUM, while weaker associations were observed between pfmdr-184 and QN, AS, and MFQ. The remaining three associations involved the nine new candidate transporters. We found an association between a 3-bp indel in the coding region of an ABC transporter (G7) and in vitro response to QN and AS and between another ABC transporter (G49 codon 1116) and DHA.
TABLE 3.
Associations between SNPs or indels in 10 putative transporter genes and IC50s for eight antimalarial drugsa
| Gene | Predicted product | Position | Amino acid changeb | Nucleotide changec | N | Freq. common variantd |
P value
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CQ | QN | AQ | AS | LUM | DHA | DOX | MFQ | |||||||
| pfmdr | ABC transporter | 86 | N-Y | AAT-TAT | 108 | 104 | 0.357 | 0.646 | 0.992 | 0.576 | 0.809 | 0.843 | 0.717 | 0.079 |
| 184 | Y-F | TAT-TTT | 105 | 61 | 0.405 | 0.040 | 0.374 | 0.025 | 0.134 | 0.970 | 0.568 | 0.020 | ||
| 1034 | S-C | AGT-TGT | 107 | 107 | NI | NI | NI | NI | NI | NI | NI | NI | ||
| 1042 | N-D | AAT-GAT | 108 | 98 | 0.778 | 0.063 | 0.635 | 0.144 | 0.000 | 0.270 | 0.763 | 0.000 | ||
| 1246 | D-Y | GAT-TAT | 106 | 105 | NI | NI | NI | NI | NI | NI | NI | NI | ||
| G2 | ABC transporter | 191 | Y-H | TAT-CAT | 107 | 107 | NI | NI | NI | NI | NI | NI | NI | NI |
| 437 | A-S | GCA-TCA | 105 | 90 | 0.149 | 0.795 | 0.540 | 0.995 | 0.304 | 0.093 | 0.426 | 0.768 | ||
| G7 | ABC transporter | 1390 | &1 | NA | 104 | 96 | 0.061 | 0.015 | 0.622 | 0.042 | 0.450 | 0.258 | 0.712 | 0.246 |
| G25 | Sulfate transporter | Intron | G-A | G-A | 101 | 77 | 0.771 | 0.838 | 0.224 | 0.702 | 0.138 | 0.624 | 0.071 | 0.579 |
| G30 | GTPase | Intron | C-G | C-G | 108 | 104 | 0.289 | 0.870 | 0.485 | 0.106 | 0.638 | 0.091 | 0.977 | 0.272 |
| G47 | Glycine transporter | 241 | L-V | TTA-GTA | 105 | 94 | 0.386 | 0.085 | 0.315 | 0.389 | 0.284 | 0.702 | 0.995 | 0.799 |
| G49 | ABC/ATPase | 146 | Q-E | CAA-GAA | 108 | 66 | 0.696 | 0.937 | 0.711 | 0.606 | 0.299 | 0.117 | 0.145 | 0.988 |
| 1046 | K-I | AAA-ATA | 108 | 78 | 0.260 | 0.349 | 0.638 | 0.316 | 0.401 | 0.831 | 0.457 | 0.817 | ||
| 1116 | L-I | TTA-ATA | 108 | 96 | 0.063 | 0.714 | 0.435 | 0.293 | 0.514 | 0.002 | 0.187 | 0.867 | ||
| G54 | Membrane protein | 141 | Y-Y | TAC-TAT | 102 | 54 | 0.209 | 0.904 | 0.194 | 0.848 | 0.165 | 0.751 | 0.443 | 0.965 |
| 144 | T-T | ACG-ACA | 102 | 54 | 0.209 | 0.904 | 0.194 | 0.848 | 0.165 | 0.751 | 0.443 | 0.965 | ||
| G55 | ABC transporter | Intron | &2 | NA | 107 | 60 | 0.227 | 0.212 | 0.475 | 0.414 | 0.161 | 0.608 | 0.429 | 0.938 |
| G70 | Choline transporter | 105 | E-K | GAA-AAA | 102 | 52 | 0.535 | 0.292 | 0.866 | 0.225 | 0.497 | 0.865 | 0.195 | 0.557 |
Pfcrt is not included in the table because it was monomorphic in all but one isolate examined. Drug abbreviations are as described in the text. We used t tests to compare IC50s in parasites with polymorphisms, and significant results (P < 0.05) are shown in bold. Both polymorphism data and IC50s are listed in the supplemental material online. Comparisons that were significant in Mu et al.'s southeast Asian data set (14) are shown underlined (CQ and QN columns only).
&1 is a 3-bp indel. &2 is a dinucleotide microsatellite in an intron. We found four alleles. For this analysis, alleles were grouped into those that were ≤194 and ≥196. Some loci showed insufficient variation (<2 alleles with one allelic state); these are labeled NI.
NA, not applicable.
Frequency of common variant.
FIG. 1.
(A) Box-and-whisker plots showing associations between polymorphisms and drug response in phase I samples. (B) The same associations examined in phase II samples. Asterisks above graphs indicate significance level (*, P < 0.05; **, P < 0.01; ***, P < 0.001). The boxes indicate the 25th and 75th percentiles, the whiskers indicate the 10th and 90th percentiles, and the lines show the median values. The numbers at the top of each graph indicate the sample sizes for each of the alternate states at the polymorphism examined. Only the pfmdr-1042 and G7 polymorphisms remain significantly associated with drug response in both phase I and II population samples examined.
To investigate whether the associations found in the phase I data set were real or type 1 errors, we reexamined these polymorphisms (pfmdr-184/1042, G7-&1, G49-1116) in an independent population sample comprising 256 single-clone infections collected from Mawker-Thai between November 2001 and September 2003 (phase II samples). Thirty-six multilocus haplotypes were represented between two and six times in the phase II data set. We analyzed unique genotypes only, including only one representative of each genotype and mean values for the phenotype data from multiply represented clones. This reduced the sample size to 199 for phase II samples. The IC50s for the eight drugs are summarized in Table 2. We found significant associations once again between pfmdr-1042 and MFQ (P = 0.0099) and LUM (P = 0.019) response but not between pfmdr-184 and MFQ (P = 0.070), QN (P = 0.189), or AS (P = 0.718) response. The associations between G49-1116 and DHA and between G7 and response QN observed in phase I samples disappeared in the phase II analysis (Fig. 1). However, significant association between G7 and AS response remained in both phase I and phase II samples (P < 0.042, phase I; P < 0.029, phase II) (Fig. 1).
Phenotype-phenotype associations.
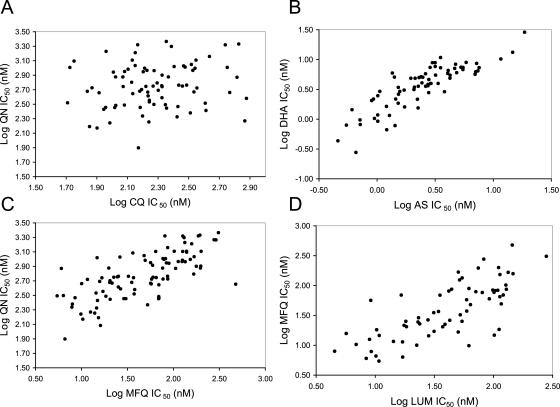
We examined correlations between IC50s for the eight drugs in both phase I (108 isolates) and phase II (199 isolates) population samples (Fig. 2 and Table 4). We found strong consistent correlations (P < 0.0001) in comparisons between AS, DHA, LUM, MFQ, and QN in both population samples. Correlations between AS and DHA and between MFQ and LUM/QN were particularly strong, with r2 values of >0.3 in both phase I and phase II samples (Fig. 2). Phenotypic correlations between the responses to these drugs have been observed in previous southeast Asian studies (5, 21). We also observed weaker correlations (r2 < 0.1) between CQ and AQ/LUM and between DOX and DHA in phase II samples. We found no significant phenotypic correlation between CQ and QN responses in either phase I or phase II samples (Fig. 2). In comparison, there was a strong correlation between CQ and QN in Mu et al.'s previous southeast Asian data set (r2 = 0.478; P < 0.001) (14). This correlation became weaker but remained significant (r2 = 0.145; P < 0.018) when the four isolates bearing wild-type pfcrt alleles were excluded.
FIG. 2.
Scatter plots showing phenotypic correlations between log (IC50) values for different drugs. (A) CQ and QN; (B) DHA and AS; (C) MFQ and QN; (D) LUM and MFQ. Correlation coefficients (r2), sample sizes, and significance for all pairwise comparisons are shown in Table 4.
TABLE 4.
Matrix of correlation coefficients (r2) between drug responsesa
| Study phase | Drug |
r2 value for drug responses
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CQ | DOX | AQ | AS | DHA | LUM | MFQ | QN | ||
| Phase I | CQ | 0.003 | 0.016 | 0.001 | 0.014 | 0.014 | 0.005 | 0.027 | |
| DOX | 55 | 0.007 | 0.006 | 0 | 0.008 | 0.011 | 0.016 | ||
| AQ | 57 | 49 | 0.097 | 0.1 | 0.079 | 0.047 | 0.02 | ||
| AS | 67 | 54 | 56 | 0.728 | 0.27 | 0.267 | 0.219 | ||
| DHA | 69 | 56 | 57 | 74 | 0.297 | 0.22 | 0.16 | ||
| LUM | 63 | 48 | 50 | 63 | 63 | 0.559 | 0.355 | ||
| MFQ | 80 | 55 | 65 | 73 | 76 | 66 | 0.506 | ||
| QN | 80 | 59 | 65 | 76 | 78 | 66 | 94 | ||
| Phase II | CQ | 0.03 | 0.075 | 0.002 | 0.035 | 0.048 | 0.013 | 0.015 | |
| DOX | 161 | 0.033 | 0.001 | 0.051 | 0.037 | 0 | 0 | ||
| AQ | 175 | 158 | 0.008 | 0.018 | 0.023 | 0.006 | 0 | ||
| AS | 151 | 135 | 152 | 0.383 | 0.16 | 0.334 | 0.152 | ||
| DHA | 164 | 145 | 163 | 152 | 0.227 | 0.216 | 0.169 | ||
| LUM | 157 | 139 | 149 | 141 | 147 | 0.51 | 0.318 | ||
| MFQ | 186 | 162 | 178 | 156 | 169 | 163 | 0.554 | ||
| QN | 161 | 164 | 154 | 133 | 145 | 140 | 164 | ||
Significance of correlations is indicated: bold, P < 0.0001; italic bold, P < 0.001; underline, P < 0.01. Correlations were performed on log-transformed IC50 data.
Genotype-genotype associations.
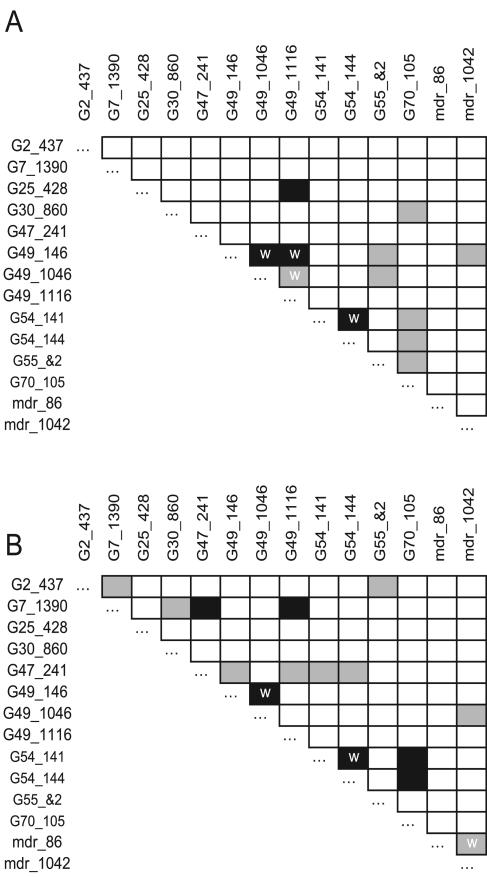
We examined the significance of LD between different polymorphisms by permutation (Fig. 3). To allow comparison between this data set and our previous data set (14), we report LD using those loci that were polymorphic in both these studies. In both data sets, we observed strong LD between markers within genes, as expected. In the current study, we observed limited amounts of LD in comparisons between markers on different genes. In 86 pairwise comparisons between genes, we found one interlocus comparison significant at the 1% level (G25-428/G49-117 [P < 0.0098]) and six at the 5% level (G49-146/pfmdr-1042; [P < 0.014]; G49-146/G55&2 [P < 0.015]; G1046/G55 [P < 0.025]; G54-141 and G54-144/G70-105 [P < 0.039]; G55&2/G70-105 [P < 0.043]). For comparison, four comparisons were significant at the 1% level and eight at the 5% level in the previous southeast Asian data set (Fig. 3B) (14). Two of the significant between-gene comparisons were significant in both studies (G70-105 with G54-141/G54-144). The numbers of comparisons where the P value is <0.05 do not differ significantly between the two data sets (Fisher's exact test, P = 0.47). However, when the sizes of the population samples were equalized by random removal of parasites from the Mawker-Thai data set, the difference in LD between the two data sets was considerable, with an average of 2.3/86 comparisons significant in 10 resampled datasets. LD was not examined in phase II, since only five polymorphisms were genotyped in this population sample.
FIG. 3.
Linkage disequilibrium between 15 polymorphic markers (SNPs or microsatellites) in 10 transporter genes. Panel A shows results from this study. Panel B shows results from Mu et al.'s Southeast Asian data set for comparison (14). Significance was assessed by permutation as described in the text (black shading, P < 0.01; grey shading, P < 0.05). Significant comparisons between markers located within the same gene are marked with a white “w” to distinguish them from between-gene comparisons.
DISCUSSION
We found a single association between an ABC transporter (G7) on chromosome 13 and AS response that remained significant in both population samples. In both, a trinucleotide insertion was associated with increased IC50s. To date no clinical resistance to AS has been reported: this association reflects quantitative differences in drug response measured in the laboratory and is not likely to be of clinical significance. Nevertheless, should this association be supported by future work, identification of the gene and mutations involved will provide a useful tool for dissecting the mechanism of action of artemisinin derivatives. The gene contains 1,049 amino acids and has 37% identity to an ABC transporter in the Arabidopsis genome. The association between G7 and AS is of borderline significance in both populations (P < 0.042 and P < 0.029) and should be treated with some suspicion until it has been substantiated.
Our current results differed in a number of respects from the previous southeast Asian data set (14). We observed no significant associations between CQ response and the polymorphisms in the nine novel transporters and just a single association with QN response in the phase I samples examined. Furthermore, we observed just two additional associations between responses to other drugs examined and the nine novel candidate transporter loci (G7 with AS and G49-1116 with DHA). With the exception of comparisons involving pfmdr-1042 and between G7 and AS, these associations disappeared when retested in a large independent sample of parasites, suggesting that they were type I errors. We observed lower levels of LD after correction for sample size differences. Finally, we observed no correlation between CQ and QN drug responses in the phase I samples or phase II samples (Fig. 2 and 3). This study and the previous association analysis (14) differ in two key respects: in the methods used for measuring in vitro resistance and in the sampling regime and study design. Examination of these aspects of the two studies' designs may indicate why our results differ.
The ability to detect associations between genotype and phenotype is strongly dependent on the quality of the phenotype data. In the original study (14), drug response was measured in isolates adapted to laboratory culture using a minimum of five replicate runs per parasite. As a further control, Dd2 assays were run concurrently, and the data were standardized to Dd2. These procedures were adopted to maximize the quality of the in vitro data. In the present study, we measured in vitro resistance using isolates freshly isolated from patients, and we used two replicates at each drug concentration. Can additional noise in the in vitro data reported here explain the difference between the two studies? We believe that there is a strong signal in the in vitro data reported here. (i) We used stringent criteria to remove low-quality in vitro data (see Materials and Methods). In particular, IC50 data were used only if standard errors were less than 20% of the mean (CV < 20%). (ii) The data show expected strong phenotypic correlations between AS and DHA and between MFQ and QN and LUM (Fig. 2). Similar strong correlations have been observed in previous southeast Asian studies (5, 17, 30), suggesting that the data are reliable. (iii) We also see expected strong associations between pfmdr-1042 and in vitro resistance to MFQ and LUM (21). Unlike the other associations detected in the first phase of this study, these associations remained on retesting in a larger parasite sample. Since pfmdr is a known modulator of resistance to a variety of antimalarial compounds (20-22, 26), this provides an important positive control for this data set. (iv) Real-time PCR measurement of pfmdr copy number showed highly significant associations with drug response. An increased pfmdr copy number was significantly associated with higher IC50s for MFQ (P < 0.0001), LUM (P < 0.0001), QN (P = 0.0001), DHA (P < 0.05), and AS (P < 0.05) (T. J. C. Anderson et al., unpublished data), confirming results from a number of recent studies (20-22) and providing further strong validation for this data set. (v) Finally, lower precision in our IC50 measures is likely to have been offset by larger sample sizes. Our sample comprised 108 unique parasite isolates, of which 85 and 100 gave reliable in vitro data for CQ and QN. This is more than two times larger than the southeast Asian sample collection examined previously (14).
On the other hand, three features of the in vitro data reported here do suggest some measurement error. First, we found significant differences between IC50s for strain K1 parasites in the different assay periods for a number of drugs. We are unsure of the reasons for this. Second, we observed significant differences in drug responses measured during the phase I and phase II samplings (Table 2). Some of these differences may reflect real temporal changes in allele frequencies in loci encoding drug resistance genes. However, decreases in DOX and AQ IC50s are more difficult to explain. Third, we observed elevated levels of resistance to some drugs within particular batches of plates used in the drug resistance assays, suggesting some error in preparation of assay plates. The consistent associations with pfmdr polymorphisms (Table 3; Fig. 1) and existence of expected correlations between drug responses (Fig. 2 and Table 4) demonstrate a strong signal-to-noise ratio in the in vitro data and make it unlikely that associations with genes of large effect would be missed. However, we cannot exclude the possibility that noise in our phenotype data precluded us from detecting subtle associations and resulted in Type II errors in the present study.
Association studies are notoriously prone to type I errors due to multiple testing and population structure (23-25). Here we used a two-phase design to minimize type I errors, while in the previous study (14), a single population sample was used and statistical approaches were employed to minimize such errors. Furthermore, we used parasite isolates collected from a single location over a short time period, while the previous study used cultured parasite isolates collected from disparate locations over a longer period. The two-phase design used here clearly helped us to avoid type I errors, since of eight positive associations detected in the first phase, five were rejected during the phase II survey.
To minimize type I errors due to population structure in the previous data set (14), two procedures were used. First, SNPs in housekeeping genes were typed to provide “genomic controls” for spurious associations. None of these housekeeping genes showed positive associations. However, unfortunately, only 2/39 genes examined were sufficiently polymorphic for inclusion. Second, population samples were grouped into three different geographical regions (Africa, southeast Asia, and South America), and samples from each region were analyzed independently. However, within-region population structure could still generate bias and lead to type I errors. If population structure is an important concern in southeast Asia, then we would expect to see greater LD in the southeast Asian population analyzed previously than in the data set presented here from a local P. falciparum population. Consistent with this, we found significantly less LD in the parasite population from Mawker-Thai than in our previous data set (14) after equalizing the sample sizes in the two studies. The fact that there is less LD in a local southeast Asian parasite population suggests that population structure could have generated some of the borderline associations reported previously (14).
Mixed population samples are likely to be a problem only if population allele frequencies and drug responses vary regionally or temporally within southeast Asia. There is independent evidence for weak population structure in southeast Asia. We have documented weak but significant population structure in allele frequencies at 9/11 putatively neutral SNPs on chromosomes 2 and 3, with much of the variation being attributable to populations from Laos and Bangladesh (Anderson et al., unpublished). There is also good evidence that levels of in vitro resistance vary both within and between countries within southeast Asia. These data come from measurement of in vitro resistance (17, 30), from drug efficacy rates (27), and from genotyping of known drug resistance markers (16).
In conclusion, we found suggestive evidence that G7 is associated with AS response, and we encourage others to try to replicate this result. However, we did not find strong evidence supporting a role for the remaining eight new candidate genes and resistance to CQ or QN or to six other drugs. Our failure to replicate these associations (14) may stem from differences in methods used for measuring resistance phenotypes and/or differences in design used in this and the previous study (14). Sampling from a single location and the two-phase study design provides an effective approach to minimizing problems with population structure and type I errors resulting from multiple testing. On the other hand, replicate measurement of IC50s in cultured parasites, as used in the initial study (14), provide high quality in vitro data and can help avoid type II errors (false negatives). We suggest that these approaches should be combined in future candidate gene association studies for drug resistance genes. These largely negative results should not be used as grounds to discard this approach to resistance gene location. Transporter genes remain extremely promising candidates. While eight of nine loci examined do not appear to show associations in southeast Asia, it is possible that some of these loci play a role in drug resistance in other regions of the world. In particular, the associations observed between polymorphisms in G7, G30, and G55 with CQ and/or QN response in African parasites in the work of Mu et al. (14) urgently require reexamination. African parasite populations show less geographical variation in allele frequencies than Asian or South American populations (2). Associations between SNPs and in vitro response are therefore less susceptible to bias due to population structure in African samples. A further caveat is worth mentioning. There is increasing evidence that pfmdr copy number, rather than point mutations, plays the key role in drug resistance in southeast Asia (20-22, 29). If copy number rather than point mutations underlies resistance in other transporters, then screens for association with SNPs may fail to incriminate candidate transporter loci. Examination of copy number and/or expression levels would provide a valuable complementary approach to SNP association for screening candidate transporter loci.
Supplementary Material
Acknowledgments
We thank Jianbing Mu and Xin-Zhuan Su for providing details of SNPs within transporter genes prior to publication, for encouraging us to examine these SNPs in our Thai samples, and for assistance throughout this project. We thank the staff of the SMRU for assistance with field collection of blood samples and the patients visiting the field clinic at Mawker-Thai. Comments from Ric Price improved the manuscript.
The SMRU is part of the Wellcome-Trust Mahidol University-Oxford Tropical Medicine Research Program supported by the Wellcome Trust of Great Britain. F.N. is a Wellcome Trust Senior Clinical Fellow. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program grant number C06 RR-98001 from the National Center for Research Resources at NIH. This work was funded by NIH grant AI48071 to T.J.C.A.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ambudkar, S. V., C. Kimchi-Sarfaty, Z. E. Sauna, and M. M. Gottesman. 2003. P-glycoprotein: from genomics to mechanism. Oncogene 22:7468-7485. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, T. J., B. Haubold, J. T. Williams, J. G. Estrada-Franco, L. Richardson, R. Mollinedo, M. Bockarie, J. Mokili, S. Mharakurwa, N. French, J. Whitworth, I. D. Velez, A. H. Brockman, F. Nosten, M. U. Ferreira, and K. P. Day. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467-1482. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, T. J., X. Z. Su, M. Bockarie, M. Lagog, and K. P. Day. 1999. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 119:113-125. [DOI] [PubMed] [Google Scholar]
- 4.Bland, J. M., and D. G. Altman. 1995. Multiple significance tests: the Bonferroni method. Br. Med. J 310:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockman, A., R. N. Price, M. van Vugt, D. G. Heppner, D. Walsh, P. Sookto, T. Wimonwattrawatee, S. Looareesuwan, N. J. White, and F. Nosten. 2000. Plasmodium falciparum antimalarial drug susceptibility on the north-western border of Thailand during five years of extensive use of artesunate-mefloquine. Trans. R. Soc. Trop. Med. Hyg. 94:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, G. 2003. Multidrug resistance ABC transporters. FEBS Lett. 555:102-105. [DOI] [PubMed] [Google Scholar]
- 7.Desjardins, R. E., C. J. Canfield, J. D. Haynes, and J. D. Chulay. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duraisingh, M. T., L. V. von Seidlein, A. Jepson, P. Jones, I. Sambou, M. Pinder, and D. C. Warhurst. 2000. Linkage disequilibrium between two chromosomally distinct loci associated with increased resistance to chloroquine in Plasmodium falciparum. Parasitology 121:1-7. [DOI] [PubMed] [Google Scholar]
- 9.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. Ursos, A. B. Sidhu, B. Naude, K. W. Deitsch, X. Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goudet, J. 2000. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.1).
- 11.Hirschhorn, J. N., and D. Altshuler. 2002. Once and again—issues surrounding replication in genetic association studies. J. Clin. Endocrinol. Metab. 87:4438-4441. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, D. J., D. A. Fidock, M. Mungthin, V. Lakshmanan, A. B. Sidhu, P. G. Bray, and S. A. Ward. 2004. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol. Cell 15:867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, J. H., and M. Yamazaki. 2003. Clinical relevance of P-glycoprotein in drug therapy. Drug Metab. Rev. 35:417-454. [DOI] [PubMed] [Google Scholar]
- 14.Mu, J., M. T. Ferdig, X. Feng, D. A. Joy, J. Duan, T. Furuya, G. Subramanian, L. Aravind, R. A. Cooper, J. C. Wootton, M. Xiong, and X. Z. Su. 2003. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol. Microbiol. 49:977-989. [DOI] [PubMed] [Google Scholar]
- 15.Nair, S., A. Brockman, L. Paiphun, F. Nosten, and T. J. Anderson. 2002. Rapid genotyping of loci involved in antifolate drug resistance in Plasmodium falciparum by primer extension. Int. J. Parasitol. 32:852-858. [DOI] [PubMed] [Google Scholar]
- 16.Nair, S., J. T. Williams, A. Brockman, L. Paiphun, M. Mayxay, P. N. Newton, J. P. Guthmann, F. M. Smithuis, T. T. Hien, N. J. White, F. Nosten, and T. J. Anderson. 2003. A selective sweep driven by pyrimethamine treatment in SE Asian malaria parasites. Mol. Biol. Evol. 20:1526-1536. [DOI] [PubMed] [Google Scholar]
- 17.Noedl, H., T. Allmendinger, S. Prajakwong, G. Wernsdorfer, and W. H. Wernsdorfer. 2001. Desbutyl-benflumetol, a novel antimalarial compound: in vitro activity in fresh isolates of Plasmodium falciparum from Thailand. Antimicrob. Agents Chemother. 45:2106-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nosten, F., M. van Vugt, R. Price, C. Luxemburger, K. L. Thway, A. Brockman, R. McGready, F. ter Kuile, S. Looareesuwan, and N. J. White. 2000. Effects of artesunate-mefloquine combination on incidence of Plasmodium falciparum malaria and mefloquine resistance in western Thailand: a prospective study. Lancet 356:297-302. [DOI] [PubMed] [Google Scholar]
- 19.Perneger, T. V. 1998. What's wrong with Bonferroni adjustments. Br. Med. J 316:1236-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickard, A. L., C. Wongsrichanalai, A. Purfield, D. Kamwendo, K. Emery, C. Zalewski, F. Kawamoto, R. S. Miller, and S. R. Meshnick. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price, R. N., C. Cassar, A. Brockman, M. Duraisingh, M. van Vugt, N. J. White, F. Nosten, and S. Krishna. 1999. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 43:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price, R. N., A. C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard, J. K., and P. Donnelly. 2001. Case-control studies of association in structured or admixed populations. Theor. Popul. Biol. 60:227-237. [DOI] [PubMed] [Google Scholar]
- 24.Pritchard, J. K., and N. A. Rosenberg. 1999. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 65:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pritchard, J. K., M. Stephens, N. A. Rosenberg, and P. Donnelly. 2000. Association mapping in structured populations. Am. J. Hum. Genet. 67:170-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 27.Rojanawatsirivej, C., S. Vijaykadga, I. Amklad, P. Wilairatna, and S. Looareesuwan. 2003. Monitoring the therapeutic efficacy of antimalarials against uncomplicated falciparum malaria in Thailand. Southeast Asian J. Trop. Med. Public Health 34:536-541. [PubMed] [Google Scholar]
- 28.Sidhu, A. B., D. Verdier-Pinard, and D. A. Fidock. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, C. M., S. K. Volkman, S. Thaithong, R. K. Martin, D. E. Kyle, W. K. Milhous, and D. F. Wirth. 1993. Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol. Biochem. Parasitol. 57:151-160. [DOI] [PubMed] [Google Scholar]
- 30.Wongsrichanalai, C., T. Wimonwattrawatee, P. Sookto, A. Laoboonchai, D. G. Heppner, D. E. Kyle, and W. H. Wernsdorfer. 1999. In vitro sensitivity of Plasmodium falciparum to artesunate in Thailand. Bull. W. H. O. 77:392-398. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.