Abstract
Human papillomaviruses (HPVs) and herpesviruses are detected in patients with epithelial ovarian cancer (EOC). We sought to analyze the prevalence of HPV’s 16 and 18, cytomegalovirus (CMV), and Epstein-Barr virus (EBV) DNA in peripheral blood, ovarian, and fallopian tube (FT) tissue samples collected from 97 EOC patients, including 71 cases of high-grade serous ovarian carcinoma (HGSOC), and from 60 women with other tumors or non-neoplastic gynecological diseases. DNA isolates were analyzed by PCR methods, including droplet digital PCR. The results demonstrate that (1) HPV16 DNA has been detected in one-third of the FT and tumor samples from EOCs; (2) the prevalence and quantity of HPV16 DNA were significantly higher in FT samples from HGSOCs, non-HGSOCs, and ovarian metastases than in those from non-neoplastic diseases; (3) CMV and EBV have been detected in approximately one-seventh of EOC samples. The results suggest that HPV16 might be a potential risk factor for EOC development.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-72814-0.
Keywords: Epithelial ovarian cancer, Fallopian tube, Papillomavirus, Herpesvirus, High-grade serous ovarian carcinoma
Subject terms: Cancer, Ovarian cancer, Human papilloma virus
Introduction
Ovarian cancer (OC) affected almost 314 thousand women and was a reason for at least 207 thousand deaths in 2020 worldwide1. In 2023, approximately 19.7 thousand new cases of OC and 13.2 thousand deaths were likely to be reported in the United States2. This cancer is currently the eighth most common women’s cancer and the eighth cause of cancer death in women. The high mortality rate is caused by asymptomatic tumor growth and the lack of appropriate screening tests that allow for early diagnosis of the disease. Epithelial OC (EOC) is the most common type of OC, occurring in more than 90% of cases. EOC consists of the four most common subtypes, including serous, endometrioid, clear cell, and mucinous carcinoma3,4. They can affect the ovaries, fallopian tubes (FT), and the primary peritoneal cavity. However, the most fatal epithelial cancer is that of the serous histology. More than 90% of serous carcinomas are very aggressive high-grade serous ovarian carcinomas (HGSOCs), while 10% are low-grade serous ovarian carcinomas (LGSOCs), which generally leading to a better prognosis5. HGSOC is the most lethal gynecological cancer with a 5-year survival rate of only approximately 30% in patients diagnosed at an advanced stage6. The fallopian tube epithelium (FTE) and ovarian surface epithelium (OSE) are considered to be the HGSOC cell of origin7. In FTE, the presence of proliferative lesions known as serous tubal intraepithelial carcinoma (STIC) is observed8,9. Approximately 80% of ovarian HGSOCs are associated with the involvement of FT fimbria, including the form of STIC10. As demonstrated using mouse models as well as FTE- and OSE-derived organoids, HGSOC can originate from both cell lineages11. The cell of origin of HGSOC plays an important role in progression, metastasis, and drug response. Ovarian tumors possess a unique tumor microenvironment (TME) whose immunosuppressive features are associated with shorter overall survival rates. HGSOC is characterized by TP53 mutations in almost all tumors (96%)12. Other common factors for ovarian carcinogenesis include amplification of Cyclin E (CCNE1, 20%), germline and somatic mutations of BRCA1/2 (20–40%), and other aberrations in pathways of DNA damage response12–14. The other risk factors include family history of the disease, older age at menopause, hormone replacement therapy, chronic inflammation, environmental and lifestyle factors, and persistent infections15. The p53 and STICs signature lesions are precursors of serous ovarian cancer with a progression difference from STICs to ovarian cancer with subsequent metastasis of 6.5 years9. Some data suggest that the cervicovaginal microbiome may also be a risk factor for the development of ovarian cancer16. Low levels of Lactobacillus species and vaginal dysbiosis, as well as viral infections, can lead to pelvic inflammatory disease (PID), vaginosis, and the development of cancer.
Human papillomaviruses (HPVs) can infect and replicate in the nucleus of cells in the basal layer of the epithelium. This virus has immune evasion strategies that allow it the unique ability to persist in the host’s epithelium for a long time17. HPV types 16 and 18 are the most carcinogenic of the high-risk types (HR-HPV) and are associated with 70% and 60% of all cervical cancer (CC) cases and cervical intraepithelial neoplasia (CIN), respectively18. Detection of HR-HPV DNA appears to be a useful prognostic marker of malignant transformation and the risk of CIN progression, even at low copy numbers19. HPV-positive lesions increase the risk of other cancers, including cancer of the vulva, vagina, anus, penis, and oropharynx. The relationship between HPV infection and the development of cancers of the upper genital tract, including ovarian and endometrial cancer, remains controversial. Both HPV16 and HPV18 were found to be the predominant types detected in cancerous ovarian tissues20–31. The prevalence and distribution of HPV types in ovarian tissue samples varied among countries worldwide32–34. Some studies have confirmed the presence of the HPV DNA and/or proteins in malignant ovarian tissues20–31,35 and FT specimens20,21, while others have not confirmed this observation36–38. A positive correlation was detected between HPV detection and p53 expression in EOC tissue samples22. In addition, a higher prevalence of herpesviruses, including Epstein-Barr virus (EBV)39,40 and human cytomegalovirus (CMV)21,41,42 DNA, was found in ovarian tissue samples from patients with EOC compared to controls. Both herpesviruses have two replication cycles, lytic and latent, and are known as oncomodulatory viruses that can alter TME.
In this study, we determined the prevalence of HPV16, HPV18, CMV, and EBV DNA in peripheral blood, tumor, and FT tissue samples collected from 121 women with suspected EOC. The control group consisted of 36 women with benign ovarian tumors or non-neoplastic diseases. Because EOC has previously been found to be complicated by the low number of viral DNA copies per cell21, sensitive and specific molecular techniques were used for the detection of viral genome sequences, including quantitative droplet digital PCR (ddPCR) and real-time PCR (qPCR), as well as qualitative nested PCR (nPCR).
Results
Study population
One hundred and twenty-one women with suspected EOC were enrolled in the study. The demographic and clinical characteristics of the patients are shown in Table 1. Tissue material from 97 women with confirmed EOC (median age: 64, range: 40–87 years) consisted of 71 HGSOCs (approximately 73.2%; median age: 65, range: 42–85) and 26 other ovarian neoplasms (non-HGSOCs), including clear cell ovarian cancer (CCOC), mucinous ovarian cancer (MOC), endometroid ovarian cancer (ENOC), and others (median age: 60.5, range: 40–87). Furthermore, nine cases of serous or mucinous borderline ovarian tumor (BOT) (median age: 61, range: 26–76 years) and fifteen women with ovarian metastases from other organs e.g. colon, breast, and gastric cancers (median age: 65, range: 39–82 years) were identified. For comparison, patients with metastatic tumors and BOTs were included in separate groups. The control group included 36 women with benign ovarian tumors or other non-cancerous diseases of the genital tract (median age: 61, range: 26–88 years). Ovarian tissue and whole blood samples were collected from all patients, while FT specimens were obtained from 67/97 (69.1%) women with primary EOC, 12/15 (80.0%) women with ovarian metastases, 7/9 (77.8%) patients with BOT, and 21/36 (56.3%) of individuals in the control group. Our initial expectation that viral infection is related solely to ovarian tumors resulted in fewer FT specimens collected. When we embarked on collecting both tumors and FT specimens from all patients, the only cases in which the FT specimens were missing were due to medical risks involved in such a procedure. There was a trend towards a higher proportion of the International Federation of Gynecology and Obstetrics (FIGO) stage IV in women with HGSOC (15/71, 21.1%) compared to women with non-HGSOC types of EOC (1/26, 3.8%) (p = 0.061, Fisher’s exact test). Only 7% (5/71) of HGSOCs were diagnosed as stage I, whereas this stage was observed in approximately 30% (8/26) of non-HGSOC cases (p = 0.005, Fisher’s exact test).
Table 1.
Demographic and clinical characteristics of female patients.
| Patient group | Number of patients |
|---|---|
| I. Epithelial ovarian cancer | 97 |
| Median age, years (range) | 64 (40–87) |
| Tumor histology | n (%) |
| HGSOC | 71 (73.2) |
| Other types | 26 (26.8) |
| CCOC | 10 |
| MOC | 6 |
| ENOC | 6 |
| Othera | 4 |
| FIGO stage | n (%) |
| I | 13 (13.4) |
| II | 10 (10.3) |
| III | 56 (57.7) |
| IV | 16 (16.5) |
| No data | 2 (2.1) |
| II. Metastatic ovarian cancer | 15 |
| Median age, years (range) | 65 (39–82) |
| Primary tumor | |
| Colon cancer | 9 |
| Breast cancer | 3 |
| Gastric cancer | 2 |
| Undetermined primary site | 1 |
| III. BOT | 9 |
| Median age, years (range) | 61 (26–76) |
| FIGO stage | n (%) |
| I | 7 (77.8) |
| II | 1 (11.1) |
| III | 1 (11.1) |
| IV. Benign ovarian tumor or other disease | 36 |
| Median age, years (range) | 61 (26–88) |
HGSOC high-grade serous ovarian cancer, CCOC clear cell ovarian cancer, BOT serous borderline ovarian tumor, MOC mucinous ovarian cancer, ENOC endometroid ovarian cancer, FIGO International Federation of Gynecology and Obstetrics (Fédération Internationale de Gynécologie et d’Obstétrique). aOther epithelial ovarian cancer types, including low-grade serous papillary adenocarcinoma, carcinosarcoma, and undifferentiated carcinoma.
HPV DNA is detected in one-third of the fallopian tube samples from patients with ovarian cancer
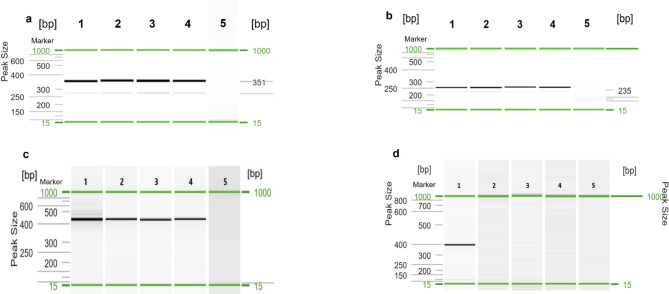
HPV16 and/or HPV18 DNA was detected by nPCR (Fig. 1), ddPCR (Fig. 2) or qPCR in tubal fimbriae and/or ovarian samples from 50/97 (51.5%) of patients with EOC (Table 2), including 36/71 (50.7%) of HGSOCs and 15/26 (57.7%) of non-HGSOC cases (Table 3). Analysis of FT specimens revealed that HPV DNA was present in 24/67 (35.8%) of EOC patients, 6/12 (50.0%) of patients with ovarian metastasis, 2/7 (28.6%) of BOT cases, and 4/21 (19.0%) of control patients. HPV16 was the most frequent viral type in single infections and was detected more frequently in FT isolates from patients with EOC and women with ovarian metastases than in samples from control individuals i.e. 31.3% and 41.7% vs. 4.8%; p = 0.019 and p = 0.016, respectively (Fisher’s exact test). HPV16 DNA detection was associated with a higher risk of EOC and metastasis compared with the controls (odds ratio, OR 9.1; 95% confidence interval, 95% CI, 1.7–169.2; p = 0.037 and OR 14.3; 95% CI, 1.9–300.2; p = 0.024, respectively). A trend of increased risk of HPV16 infection in FT samples collected from women with EOC diagnosed as stage I/II in comparison to stage IV could be observed (p = 0.058; Fisher’s exact test). Mixed infection with two HR-HPV types, HPV16 and HPV18, was found in only FT samples collected from only two EOC patients. EBV and CMV sequences were detected in approximately one-seventh of FT samples from women with EOC, while HPV18 infection occurred in these cases less frequently. Using qualitative nPCR alone, HPV16 DNA was also detected more frequently in FT samples from patients with EOC than in samples from control individuals (26.9% vs. 4.8%; p = 0.036, Fisher’s exact test). All amplified PCR products were then purified and analyzed using the Sanger sequencing, and no discrepancies were found. The EBV EBNA-2 gene was not detected in any of the DNA isolates tested by nPCR (Fig. 1d).
Fig. 1.
Visualization of nPCR products for HPV16 (a), HPV18 (b), CMV (c), and EBV (d) DNA. Gel image: (1) Positive control (DNA isolated from Ca Ski cells for HPV16, HeLa cells for HPV18, MRC-5 cells infected with CMV AD169 or Davis for CMV, and Namalwa cells for EBV); (2) Cancerous ovarian tissue; (3) Fallopian tube; (4) Peripheral blood; (5) Negative control. Alignment markers (15 bp, 1 kbp).
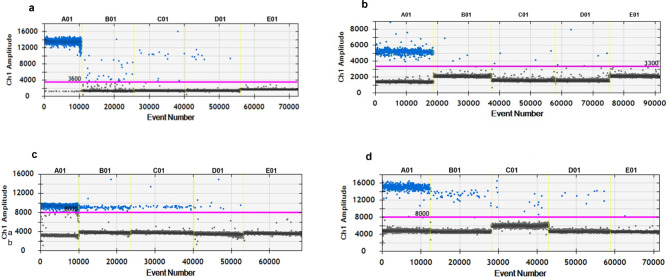
Fig. 2.
Detection of the fragments of the HPV16 E6 (a), HPV18 E7 (b), CMV UL55 (c), and EBV EBNA1 (d) genes by ddPCR in tissue samples collected from patients with epithelial ovarian cancer. A01—positive control; DNA isolated from Ca Ski, HeLa, MCR-5 infected with CMV, and Namalwa cells, respectively. B01, C01, D01—DNA isolated from clinical materials, including fallopian tube, ovarian tumor, and whole blood samples. E01—no-template control (NTC). Channel 1 (Ch1)—positive droplets containing the virus gene amplicon are colored blue, while negative drops are marked in black.
Table 2.
Distribution of HPV16, HPV18, CMV, and EBV DNA in tissue materials from patients with primary epithelial ovarian cancer, ovarian metastases, borderline ovarian tumor, and controls, including benign tumors.
| Virus | Tissue type | Prevalence; n (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| EOC | Metastasis | pa-value | Control | pb-value | pc-value | BOT | pd-value | ||
| HPV16 | Tumor or FT | 42/97 (43.3) | 7/15 (46.7) | 1.000 | 9/36 (25) | 0.07 | 0.190 | 4/9 (44.4) | 1.000 |
| Tumor | 33/97 (34.0) | 6/15 (40.0) | 0.772 | 9/36 (25) | 0.402 | 0.324 | 3/9 (33.3) | 1.000 | |
| FT | 21/67 (31.3) | 5/12 (41.7) | 0.516 | 1/21 (4.8) | 0.019 e | 0.016 f | 2/7 (28.6) | 1.000 | |
| Blood | 22/97 (22.7) | 8/15 (53.3.) | 0.061 | 9/36 (25.0) | 0.820 | 0.190 | 4/9 (44.4) | 0.218 | |
| HPV18 | Tumor or FT | 14/97 (14.4) | 2/15 (13.3) | 1.000 | 5/36 (13.9) | 1.000 | 1.000 | 0/9 (0) | 0.603 |
| Tumor | 9/97 (9.3) | 2/15 (13.3) | 0.641 | 3/36 (8.3) | 1.000 | 0.624 | 0/9 (0) | 1.000 | |
| FT | 5/67 (7.5) | 1/12 (8.3) | 1.000 | 4/21 (19.0) | 0.208 | 0.630 | 0/7 (0) | 1.000 | |
| Blood | 9/97 (9.3) | 3/15 (20.0) | 0.202 | 6/36 (16.7) | 0.233 | 1.000 | 1/9 (11.1) | 1.000 | |
| HPV16/18 | Tumor or FT | 6/97 (6.2) | 0/15 (0) | 1.000 | 2/36 (5.6) | 1.000 | 1.000 | 0/9 (0) | 1.000 |
| Tumor | 4/97 (4.1) | 0/15 (0) | 1.000 | 2/36 (5.6) | 0.661 | 1.000 | 0/9 (0) | 1.000 | |
| FT | 2/67 (3.0) | 0/12 (0) | 1.000 | 1/21 (4.8) | 0.564 | 1.000 | 0/7 (0) | 1.000 | |
| Blood | 3/97 (3.1) | 1/15 (6.7) | 0.442 | 3/36 (8.3) | 0.343 | 1.000 | 1/9 (11.1) | 0.303 | |
| CMV | Tumor or FT | 27/97 (27.8) | 7/15 (46.7) | 0.225 | 10/36 (27.8) | 1.000 | 0.212 | 2/9 (22.2) | 1.000 |
| Tumor | 20/97 (20.6) | 6/15 (40.0) | 0.110 | 8/36 (22.2) | 0.815 | 0.301 | 2/9 (22.2) | 1.000 | |
| FT | 12/67 (17.9) | 2/12 (16.7) | 1.000 | 6/21 (28.6) | 0.354 | 0.680 | 1/7 (14.3) | 1.000 | |
| Blood | 12/97 (12.4) | 5/15 (33.3) | 0.051 | 5/36 (13.9) | 0.777 | 0.135 | 2/9 (22.2) | 0.339 | |
| EBV | Tumor or FT | 21/97 (21.6) | 2/15 (13.3) | 0.732 | 4/36 (11.1) | 0.216 | 1.000 | 0/9 (0) | 0.200 |
| Tumor | 14/97 (14.4) | 2/15 (13.3) | 1.000 | 3/36 (8.3) | 0.559 | 0.624 | 0/9 (0) | 0.603 | |
| FT | 10/67 (14.9) | 0/12 (0) | 0.345 | 1/21 (4.8) | 0.448 | 1.000 | 0/7 (0) | 0.583 | |
| Blood | 5/97 (5.2) | 1/15 (6.7) | 0.587 | 2/36 (5.6) | 1.000 | 1.000 | 1/9 (11.1) | 0.421 | |
n number of cases with the virus genotype, EOC epithelial ovarian cancer, BOT borderline ovarian tumor, p two-tailed Fisher’s exact test was used to compare virus distribution between the EOC group and patients with metastasis (pa), EOC and control groups (pb), patients with metastasis and control group (pc), as well as between the EOC and BOT cases (pd), FT fallopian tubes, HPV16/18 coinfection with HPV16 and HPV18, eodds ratio (OR) 9.1, 95% confidence interval (CI) 1.7–169.2, p = 0.037, fOR 14.3, 95% CI 1.9–300.2, p = 0.024.
Significant values are in bold.
Table 3.
Distribution of viral DNA in tissue materials from patients with HGSOC, other types of EOC types, and control, including benign tumors.
| Virus | Tissue type | Prevalence; n (%) | |||||
|---|---|---|---|---|---|---|---|
| HGSOC | Non-HGSOC | pa-value | Control | pb-value | pc-value | ||
| HPV16 | Tumor or FT | 29/71 (40.8) | 14/26 (53.8) | 0.370 | 9/36 (25) | 0.136 | 0.033 e |
| Tumor | 25/71 (35.2) | 9/26 (34.6) | 1.000 | 9/36 (25) | 0.380 | 0.572 | |
| FT | 14/47 (29.8) | 7/20 (35.0) | 0.775 | 1/21 (4.8) | 0.026 d | 0.021 f | |
| Blood | 18/71 (25.3) | 4/26 (15.4) | 0.414 | 9/36 (25.0) | 1.000 | 0.529 | |
| HPV18 | Tumor or FT | 10/71 (14.1) | 4/26 (15.4) | 1.000 | 5/36 (13.9) | 1.000 | 1.000 |
| Tumor | 7/71 (9.9) | 2/26 (7.7) | 1.000 | 3/36 (8.3) | 1.000 | 1.000 | |
| FT | 3/47 (6.4) | 2/20 (10.0) | 0.631 | 4/21 (19.0) | 0.190 | 0.663 | |
| Blood | 6/71 (8.5) | 3/26 (11.5) | 0.698 | 6/36 (16.7) | 0.213 | 0.722 | |
| HPV16 | Tumor or FT | 3/71 (4.2) | 3/26 (11.5) | 0.338 | 2/36 (5.6) | 1.000 | 0.641 |
| Tumor | 3/71 (4.2) | 1/26 (3.8) | 1.000 | 2/36 (5.6) | 1.000 | 1.000 | |
| FT | 0/47 (0) | 2/20 (10.0) | 0.086 | 1/21 (4.8) | 0.309 | 0.606 | |
| Blood | 3/71 (4.2) | 0/26 (0) | 0.562 | 3/36 (8.3) | 0.402 | 0.258 | |
| CMV | Tumor or FT | 19/71 (26.8) | 7/26 (26.9) | 1.000 | 10/36 (27.8) | 1.000 | 1.000 |
| Tumor | 14/71 (19.7) | 5/26 (19.2) | 1.000 | 8/36 (22.2) | 0.803 | 1.000 | |
| FT | 6/47 (12.8) | 3/20 (15.0) | 1.000 | 6/21 (28.6) | 0.168 | 0.454 | |
| Blood | 10/71 (14.1) | 2/26 (7.7) | 0.505 | 5/36 (13.9) | 1.000 | 0.689 | |
| EBV | Tumor or FT | 17/71 (23.9) | 4/26 (15.4) | 0.420 | 4/36 (11.1) | 0.131 | 0.710 |
| Tumor | 11/71 (15.5) | 3/26 (11.5) | 0.753 | 3/36 (8.3) | 0.375 | 0.689 | |
| FT | 8/47 (17.0) | 2/20 (10.0) | 0.711 | 1/21 (4.8) | 0.256 | 0.606 | |
| Blood | 3/71 (4.2) | 2/26 (7.7) | 0.608 | 2/36 (5.6) | 1.000 | 1.000 | |
n number of cases, HGSOC high-grade serous ovarian cancer, p two-tailed Fisher’s exact test was used to compare virus distribution between the HGSOC and non-HGSOC groups (pa), HGSOC group and controls, including patients with benign tumors (pb), as well as between the non-HGSOC group and controls (pc), FT fallopian tubes, dodds ratio (OR) 8.5, 95% confidence interval (CI) 1.5–159.6, p = 0.046; eOR 3.5, 95% CI 1.2–10.7, p = 0.023; fOR 10.8, 95% CI 1.6–213.7, p = 0.035.
Significant values are in bold.
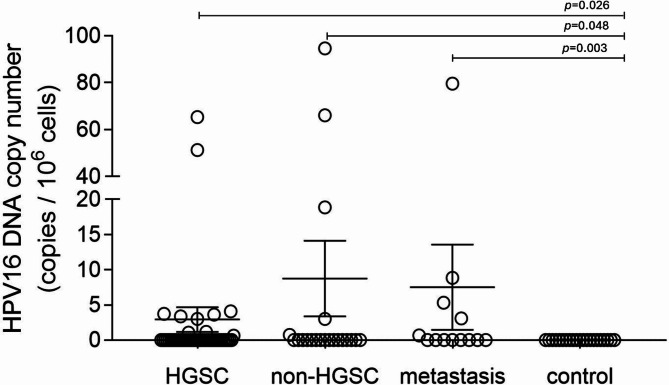
To determine the number of viral DNA copies in the tested DNA isolates, ddPCR and qPCR were used. The HPV16 ddPCR assay was optimized by comparing the published set of primers/probes and thermal profiles. A set of HPV16 primers/probe generated a positive population with a higher amplitude, suggesting high target DNA affinity43,44. The HPV1845 and EBV46 primer/probe sets were adapted from published sequences, while the CMV primer/probe set was used from the qPCR assay47,48, and optimized to match ddPCR-specific parameters. Low amounts of viral DNA were detected in tissue isolates in all patient groups (Supplementary Table 1). The concentration of HPV16 DNA in FT specimens from patients with metastases or EOC was significantly higher compared to the control group (p = 0.003 and p = 0.034, respectively; Fig. 3). The mean HPV16 DNA concentration in FT samples from all EOC patients was almost 2-fold lower (47.4 copies/105 cells) in comparison to that observed in women with metastases (88.7 copies/105 cells; Supplementary Tables 1 and 2). Moreover, significantly higher concentrations of HPV16 than HPV18 DNA were observed in FT samples from EOC patients (p = 0.012, Mann-Whitney U test; Supplementary Table 1). In these samples, mean CMV and EBV DNA concentrations were 36.8 copies/105 cells and 6.9 copies/105 cells, respectively. No other significant differences in viral DNA in FT tissues were observed between patient groups.
Fig. 3.
HPV16 copy numbers in fallopian tube samples from HGSOCs, non-HGSOC cases, patients with ovarian metastases, BOTs, and control group, including benign tumors. The virus copy number was quantified by ddPCR or qPCR. The bars in the scatter plot represent the mean viral load and the whiskers represent the SEM values. The Mann–Whitney U test was used to assess statistically significant differences.
HPV16 DNA is present in ovarian tumor samples in higher amounts than in patients with benign tumors
HPV16 or HPV18 sequences were found in approximately 39.2% (38/97) of tumor samples collected from women with EOC. CMV and EBV DNA sequences were found at lower frequencies in EOC tumor samples, including 20.6% and 14.4%, respectively (Table 2). Surprisingly, HPV16 DNA sequences were detected in EOC tumor samples with a similar frequency as in patients with ovarian metastases, BOT, and controls (p ≥ 0.05). Mixed HPV16 and HPV18 infections were detected in tumor samples from four patients with EOC (4.1% of cases). The amounts of HPV16 and CMV DNA among EOC patients in tumor isolates were 2-fold lower than in FT-specimens (Supplementary Table 1 The mean HPV16 DNA concentration in all malignant ovarian samples was 29.0 copies/105 cells in the EOC group and 32.6 copies/105 cells in metastatic patients, whereas in BOTs and controls it was < 20 copies/105 cells (p = 0.41 and p = 0.45, respectively; Mann-Whitney U test). In EOC cases, the mean CMV, HPV18, and EBV DNA concentrations in ovarian tumor samples were lower than 20 copies/105 cells (Supplementary Table 1). Furthermore, HPV16 DNA levels in tumors from EOC cases were significantly higher compared to HPV18 DNA levels (p < 0.0001, Mann-Whitney U test). Differences in CMV, HPV18, and EBV DNA concentrations in pathological tissues between different groups of patients were not significant (p ≥ 0.05).
The concentration of HPV16 DNA is higher in the fallopian tube and tumor samples from patients with HGSOC than in patients with non-malignant disease
Overall, HPV sequences were found in 45/71 (63.4%) of patients with HGSOC and in 18/26 (69.2%) of non-HGSOC cases. The frequency of HPV16 DNA detection in FT and tumor specimens was similar in HGSOC and non-HGSOC cases (p ≥ 0.05, Table 3). The detection rate of HPV16 DNA in FT samples was higher in both HGSOC and non-HGSOC groups than in the control group (p = 0.026 and p = 0.021, respectively). The frequency of HPV16 DNA detection in FT samples was significantly higher in HGSOC than in controls (OR 8.5; 95% CI, 1.5–159.6; p = 0.046). HPV16 DNA concentration was significantly higher in FT samples from women with HGSOC and non-HGSOC compared to controls (p = 0.026 and p = 0.048, respectively; Mann-Whitney U test; Fig. 3, Supplementary Table 1). HPV16 DNA levels were also higher in FT from non-HGSOC cases compared to controls (p = 0.048, Fig. 3). Furthermore, the concentration of HPV16 DNA in tumor and FT isolates from women with HGSOC was significantly higher than that of HPV18 DNA (p = 0.0006 and p = 0.02, respectively; Mann-Whitney U test; Supplementary Table 1). HPV18, CMV, and EBV DNA was distributed with similar frequency in tissue samples from HGSOC and non-HGSOC cases (p ≥ 0.05).
The number of HPV16 and CMV DNA copies in the blood of patients with ovarian metastases was higher than in patients in the control group
The mean HPV16 DNA concentration in peripheral blood samples was 10.7 copies/106 cells, 44.8 copies/105 cells, and < 10 copies/105 cells for EOCs, ovarian metastases, and BOTs or controls, respectively (Supplementary Tables 1 and 2). The prevalence of HPV16 and CMV DNAemia in blood samples from patients with ovarian metastases was nonsignificantly higher than in women with EOC (p = 0.061 and p = 0.051, respectively; Table 2). Although HPV16 DNA was detected in up to 25% of blood samples in the control group, the level of DNAemia in patients with ovarian metastases was significantly higher than in the control group (p = 0.009; Mann-Whitney U test). The highest mean CMV DNAemia was observed in women with metastasis (36.0 copies/105 cells), whereas 12.5 copies/105 cells in BOTs, and < 10 copies/105 cells in EOCs and controls, respectively (Supplementary Tables 1 and 2). The mean HPV16 DNAemia was 2-fold higher in EOCs compared to ovarian metastases, while the mean CMV DNA concentration was almost 6-fold higher in patients with metastases than in women with EOC.
Discussion
In the present study, HPV DNA was found in at least one-third of FT and cancerous ovarian tissues. Infection with HPV16 was common in patients with EOC and a similar frequency of infection was observed for HGSOC and non-HGSOC cases. The frequency of HPV16 DNA detection was significantly higher in FT of HGSOCs, non-HGSOCs, and ovarian metastases compared to samples from non-malignant tissues. Although a similar prevalence of viral infection was observed for the HGSOC and non-HGSOC groups, the concentration of HPV16 DNA in the FT was significantly higher in the HGSOC and non-HGSOC cases compared to the controls. Moreover, the concentrations of HPV16 DNA in malignant ovarian and FT samples from HGSOCs were significantly higher than that of HPV18 DNA. The presence of genetic material of HPV and herpesviruses, even in a low amount of copies, indicates that these viruses exist in the fallopian tube and ovarian cells during the development of ovarian cancer. HPV16 as an oncovirus, present in higher titers in tubal fimbriae, may contribute to cancer initiation. We hypothesize that HPV16 might contribute to the transformation of the fallopian tube and ovarian cells into cancer cells, while herpesviruses may create a unique inflammatory microenvironment that favors cancer development. Hence, we propose that HR-HPV infection of the tubal fimbriae and ovary may cause some cases of virus-dependent ovarian cancer.
The presence of HR-HPV genotypes in the upper genital tract, including fallopian tubes and ovaries, suggests that the virus may be involved in the development of some EOC cases, including the HGSOC type. HR-HPV is likely to be responsible for the lower expression of p53 in ovarian cancer, especially the type HGSOC. Our results show a reduction in TP53 gene expression in ovarian tissues from EOC and HGSOC cases compared to those from benign tumors (p < 0.05; data not shown). The virus replicates in the nucleus of the basal layer cells of epithelia which probably express the viral E6 and E7 gene products. Oncoproteins can bind to the tumor suppressor proteins, which disrupts cell cycle regulation. The HR-HPV E6 oncoprotein can inactivate p53, while E7 can inhibit pRB, leading to a complementary and synergistic effect that induces cell immortalization, stimulates cell proliferation, and the likelihood of malignant transformation34. The inhibition of p53 seems to be more present in ovarian tumors with worse prognoses49,50. The p53 signature is a potent precursor in the transformation of fallopian tube secretory cells to STICs, which leads to the development of HGSOC. However, the mechanisms that can induce the development of disease remain still unknown. HPV replicates in the nucleus of the basal layer cells of epithelia and viral DNA is maintained in an episomal form with a low-copy number. The integration of HPV DNA within the host genome promotes changes in gene expression and carcinogenesis51. Therefore, detection even of a low number of copies of HPV DNA may be a promotor of tumor transformation. It was reported that the HPV16 and HPV18 genomes were integrated with the host genome in approximately 65% and 54.5% of HPV-positive ovarian tumor tissues, respectively24. The analysis performed by Cherif et al. revealed the highest prevalence of HPV in ovarian tumors in Asia and Eastern Europe (30.9% and 29.3%, respectively)32. The overall pooled prevalence of HPV in women with OC was found as 15.9%32 and was in accordance with previous meta-analyses52,53. Previous studies have shown that HPV DNA occurs in the fallopian tubes of EOC patients20,21. It should be noted that in the analyzed studies, mainly PCR, immunohistochemistry (IHC), as well as combined techniques e.g. PCR/in situ hybridization (ISH) were used. These findings are generally consistent with previous results in a smaller number of subjects with low levels of viral DNA21. The higher rate of HPV frequency in the present study is probably due to the use of a more sensitive method, such as ddPCR. It should be noted that PCR-based methods allow the detection of only the part of the viral genome and not the live infectious virus.
We have shown that the fallopian tubes can also be infected with CMV and EBV, although the viral DNA copy number is low. Both herpesviruses were detected in FT samples from EOC patients, including HGSOC cases, while CMV was found in all groups of patients studied. Overall, CMV or EBV DNA was detected in approximately 15% of FT and tumors of patients with EOC. Previous studies on the detection of CMV DNA and proteins in patients with EOC have focused on tumor tissue21,35,39,41,42,54. CMV immediate-early protein (IE) and CMV tegument protein pp65 proteins were detected in 82% and 97% of Chinese women with EOC and in 36% and 63% of benign tumors, respectively54. A similar frequency of CMV proteins was observed in EOC tumor tissues in a Swedish population41. CMV-IE protein expression was detected in 75% of ovarian cancers and 26% of benign tumors, while pp65 protein was found in 67% of ovarian cancers and 14% of benign tumors. Moreover, high CMV expression in OC biopsies was associated with shorter survival outcomes compared to those without the virus42. In another study, 29% of patients with OC were CMV-positive at diagnosis, while after three cycles of chemotherapy, the CMV was reactivated in 60% of patients55. Our results indicate that CMV and HPV16 viremia was most common in women with ovarian metastases and occurred in at least one-third of the patients studied. Higher CMV- and HPV16 DNAemia that was observed in patients with ovarian metastases is probably a result of long-term effects of cancer. Both viruses promote tumor growth and induce a pro-inflammatory environment. It is known that CMV infects many cell types and establishes chronic latency, while active CMV infection in the tumor creates an immunosuppressive TME that suppress tumor-specific immune responses56. BOTs are primary epithelial ovarian tumors with a low malignant potential. The extensive expression of CMV IE and pp65 proteins were detected in 87% and 40% of BOT tissue sections, respectively57. Although we studied a few BOT cases only, our results confirmed the presence of CMV and HPV16 DNA in 22% and 33% of tumor samples, respectively. HPV18 and EBV DNA were not detected in the tumor and FT samples of BOT. It was previously found that PID is associated with an increased risk of developing serous BOT58. This inflammatory environment can activate monocytes/macrophages and T cells, as well as trigger CMV reactivation57 and HPV replication. All these observations confirm that CMV and EBV can occur at low activity or in a latent phase in FT and ovarian cells. Both herpesviruses have been suggested as biomarkers of immune suppression, but it is possible that these pathogens also play an oncomodulatory role in ovarian cancer.
This study has several strengths and some limitations. This is the first study to focus on HR-HPVs and herpesviruses detection in FT specimens collected from different patient groups and to confirm that the concentrations of HPV16 DNA in FT from EOC patients are higher compared to non-malignant samples. We have found a significant association of HPV16 infection in FT samples with HGSOC and non-HGSOC cases. The main strength of this study is the clinical evaluation of patients, the analysis of valuable clinical materials, and important clinical implications. Because EOC is complicated by the low amounts of viral DNA copies per cell, sensitive qualitative and quantitative PCR methods were developed and used for DNA detection and load quantification in the studied groups of patients. ddPCR is a specific and sensitive technology, which provides an alternative method for nucleic acid quantification and enables its quantification in an absolute manner, without using calibration curves59. Therefore, ddPCR is capable of detecting a few copies of viral DNA with high precision and accuracy. These results indicate that ddPCR provides an alternative method for quantifying and monitoring viral infection in tumor and FT tissues. The few copies of viral DNA detected may suggest that the virus might be present in a persistent or latent phase. It should be noted that our results are not representative of the entire population and that the detected virus distribution is only valid for patients studied. Because the sample population of patients in some groups was small, the significance of these results must be handled with caution. Further studies using larger patient groups from different geographical regions are needed to confirm our findings.
In summary, this report demonstrates that HR-HPV, CMV, and EBV are present in fallopian tubes and ovarian tumors. HPV infections of the genital tract and associated pathologies are still an important burden in different regions, where HPV-related cancers are on the rise. Chronic infection of the fallopian tube and ovarian epithelium by HR-HPV may be a factor associated with the onset and progression of ovarian cancer, while viral oncoproteins are the main drivers of carcinogenesis. It is suggested that persistent HPV infection and chronic inflammation of the fallopian tube and/or ovary may contribute to the development of EOC.
Methods
Ethical statement
The study was approved by the Ethics Committee of the Medical University of Lodz, Poland under resolution numbers RNN/346/17/KE and KE/1147/20. The study was conducted in accordance with the Declaration of Helsinki and the good clinical practice guidelines. Written informed consent was obtained from all participants prior to participation in the study.
Sample collection
Human ovarian and FT tissues, and peripheral blood samples were obtained from patients at the Department of Surgical, Endoscopic and Oncological Gynecology, Polish Mother’s Health Center Research Institute, Lodz, at the Tomaszow Health Center, and the Department of Surgical and Oncological Gynecology, Medical University of Lodz, Poland. The inclusion criterion was referral for surgery to a specialized center on suspicion of EOC. Women were considered ineligible to participate in the study if they met any of the following criteria: synchronous cancer other than EOC and ovarian cancer of non-epithelial origin. Following WHO criteria, the EOC group was limited to patients diagnosed with serous, mucinous, endometrioid, and clear cell tumors. Samples of both tumor tissue and the distal part of the fallopian tube (including tubal fimbriae) unilateral to the tumor site were obtained, as previously described21, and immediately secured for laboratory tests. The control group consisted of 36 women who underwent surgery for uterine fibroids, benign ovarian tumors or other non-cancerous diseases of the reproductive tract. Finally, the tissue material consisted of 71 HGSOCs, 26 ovarian cancers of other histological types, 9 cases of BOT, and 15 metastatic ovarian cancers (Table 1). Because BOT and metastatic ovarian cancer are histologically different from primary ovarian cancer, these patients were excluded from the study group and included in separate groups for comparison. All women were Caucasian with a history of sexual activity and underwent primary cytoreductive surgery or diagnostic laparoscopy between 2018 and 2023. Tissue samples were collected at the time of surgery in ice-cold Dulbecco’s phosphate buffered saline (DPBS; Sigma-Aldrich Co. Ltd., Ayrshire, UK) and processed within 1 h or stored at − 80 °C. Additional pieces of fresh tissue were placed in ice-cold RNAlater solution (Invitrogen by Thermo Fisher Scientific, Vilnius, Lithuania), and snap frozen. Among the examined samples, biospecimens from 24 women with EOC and 4 women with metastatic tumors were retrospectively analyzed. These specimens were included in previous studies using other detection assays21.
DNA extraction
Total DNA was isolated from cell lines and clinical samples using the DNeasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. Total DNA was extracted from 200 µL of blood, and approximately 25 mg of tissue eluted in 100 µL of elution buffer. DNA concentration and purity were assessed using a NanoDrop 2000c UV-Vis spectrophotometer (Thermo Scientific, Waltham, MA, USA) and stored at − 20 °C until use.
Nested polymerase chain reaction
The sequences of the outer pair of primers used in the nPCR method, targeting the E6 gene of the HPV16 and HPV18 were designed in the Laboratory of Virology, the Institute of Medical Biology of the Polish Academy of Sciences in Lodz, Poland using Primer-BLAST (US. National Library of Medicine, National Center for Biotechnology Information). The following primers were used: HPV16 E6 F 5′-TGCACAGAGCTGCAAACAAC-3′ and R 5′-GAGAACAGATGGGGCACACA-3′ (amplicon size 699 bp); HPV18 E6 F 5′-GCGACCCTACAAGCTACCTG-3′ and R 5′-GGAATGCTCGAAGGTCGTCT-3′ (718 bp). The nested primer pair sequences for nPCR have been described previously60. Amplification conditions with all primer sets were as follows: 95 °C for 5 min, 35 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 60 s, with an extension step at 72 °C for 5 min. HPV-positive DNA isolates from Ca Ski (American Type Culture Collection, Rockville, USA; ATCC CRL-1550) and HeLa cells (ATCC CCL-2) were used as positive controls,. Primer sequences for detection of the CMV UL55 gene and the thermal profiles for amplifications have been previously described61. DNA isolates from MCR-5 cells (ATCC CCL-171) infected with CMV strain AD169 (ATCC VR-538) were used as positive controls. The first PCR amplified the EBV EBNA-2 gene region, followed by nested reactions that amplified distinct regions62. The DNA isolate from Namalwa cells (ATCC CRL-1432) was used as a positive control. Nuclease-free water was used as a non-template control in each amplification. Primers targeting a 452 bp region of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene were used to confirm the presence of human DNA63. Samples negative for the GAPDH amplicon were considered unsatisfactory. PCR was performed on a Biometra TAdvanced programmable thermal cycler (Analytik Jena, Göttingen, Germany). Amplicons were separated using the QIAxcel DNA Screening Kit on the QIAxcel capillary electrophoresis system (Qiagen). Sanger sequencing was performed to confirm the detected viral genomes. The sequences were aligned using the Basic Local Alignment Search Tool (blast.ncbi.nlm.nih.gov/) and compared with reference sequences available in the National Center for Biotechnology Information data bank.
Droplet digital polymerase chain reaction
Thw ddPCR was performed using the QX200 Droplet Digital PCR System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer’s instructions. Specific primers and TaqMan probe sets were used to detect the HPV16 E643,44 and HPV18 E745 oncogenes, as well as EBV EBNA1/246 and CMV UL5547,48 genes (Supplementary Table 3). Endogenous control RPP30 assay64 was included in each ddPCR as a copy number reference. Both 6-FAM and HEX fluorescent dye reporters were used to target viral genes and the human RPP30 gene, respectively, whereas black hole quencher type 1 (BHQ-1) was used as the non-fluorescent chromophore. The ddPCR master mix included 2×ddPCR SuperMix for probes (Bio-Rad Laboratories, Inc.), 0.2 µL of specific primers (100 µM), 0.055 µL of specific probes (100 µM), 5 µL of DNA eluate (0.5–5.0 µg) and 1 µL of restriction digest mixture for DNA cleavage, supplemented with nuclease-free water to a final volume of 22 µL. The restriction digest mix consisted of 4 U of EcoRI and HindIII restriction enzymes (Thermo Fisher, Vilnius, Lithuania) per well and was incubated for 30 min at room temperature. Subsequent amplification was performed using a T100 96-well thermal cycler (Bio-Rad Laboratories, Inc.) at a ramp rate of 2 °C/s. Quantitative analysis of droplet numbers was performed using a QX200 Droplet Reader (Bio-Rad Laboratories, Inc.). All reaction runs were performed in triplicate and included negative and positive controls. The ddPCR runs were considered technically successful if the number of accepted droplets per individual sample replicate was > 10,000. DNA isolates from Ca Ski, HeLa, CMV-infected MRC-5, and Namalwa cell lines for the HPV16, HPV18, CMV, and EBV runs, respectively, were used as positive controls. The ddPCR data were analyzed using Quantasoft Version 1.7.4 (Bio-Rad Laboratories, Inc.), which expressed the results in copies per µL. Manual thresholds were applied to both viral and human control RPP30 genes. The cut-offs for positive droplets were determined regarding the non-template and negative controls. Samples were considered positive if there were at least three droplets with an amplitude above the threshold baseline. The final results were expressed as the number of viral DNA copies per 105 cells and are the values reported here.
Real-time polymerase chain reaction
HPV16/HPV18 DNA copy numbers were quantified using multiplex qPCR with the AmpliSens HPV 16/18-FRT PCR kit (Ecoli s.r.o., Prague, Czech Republic), whereas CMV and EBV DNA was quantified with genesig Standard Kits (Primerdesign Ltd., Chandler’s Ford, UK), according to the manufacturers instructions. QuantStudio 5 Real-Time PCR System (Applied Biosystems by Thermo Fisher Scientific) was used for real-time analysis.
Statistical analysis
The normality of the data distribution was verified using the Shapiro–Wilk test using Statistica version 13.3 software (StatSoft, Cracow, Poland). Statistical comparisons were evaluated using GraphPad Prism version 10.1.2 software (Graphpad Software Inc., San Diego, CA, USA). Differences between groups were tested using a two-tailed Fisher’s exact probability test according to the characteristics of data distribution. The two-tailed Mann–Whitney U test was performed for comparisons of viral DNA titers between groups. Univariable logistic regression analysis was used to calculate the ORs and corresponding 95% CIs by comparing the case group to the control group. A p-value less than 0.05 was considered statistically significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Agnieszka Jabłońska and Mirosława Studzińska for preparing materials that were subjected to retrospective analysis, and Katarzyna Bednarska-Szczepaniak for assistance with statistical analysis.
Author contributions
E.P. and J.R.W. Study conception and design. J.R.W., M.W., A.M., and M.N., Patient-related data and tissue collection. E.P., D.A.H., K.D.K., D.J., Experimental plan. J.R.W., M.W., A.M., E.P., D.A.H., K.D.K., D.J., M.N., M.K., Data collection. E.P., D.A.H., J.R.W., K.D.K., D.J., Data analysis and interpretation of results. E.P., Draft manuscript preparation. All authors have reviewed the results and approved the final version of the manuscript.
Funding
This research was funded by the National Science Centre of Poland (Grant no. 2019/33/B/NZ7/02872).
Data availability
Data supporting the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Edyta Paradowska, Daria A. Haręża.
References
- 1.Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin.73, 17–48 (2023). 10.3322/caac.21763 [DOI] [PubMed] [Google Scholar]
- 3.Oliveira, D. V. N. P. et al. Gene expression profile association with poor prognosis in epithelial ovarian cancer patients. Sci. Rep.11, 5438 (2021). 10.1038/s41598-021-84953-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCluggage, W. G. Morphological subtypes of ovarian carcinoma: A review with emphasis on new developments and pathogenesis. Pathology43, 420–432 (2011). 10.1097/PAT.0b013e328348a6e7 [DOI] [PubMed] [Google Scholar]
- 5.Kurman, R. J. & Shih, I. M. Pathogenesis of ovarian Cancer: Lessons from morphology and Molecular Biology and their clinical implications. Int. J. Gynecol. Pathol. PAP (2008). [DOI] [PMC free article] [PubMed]
- 6.Yousefi, M. et al. Current insights into the metastasis of epithelial ovarian cancer—Hopes and hurdles. Cell. Oncol.43, 515–538 (2020). 10.1007/s13402-020-00513-9 [DOI] [PubMed] [Google Scholar]
- 7.Colvin, E. K. & Howell, V. M. Why the dual origins of high grade serous ovarian cancer matter. Nat. Commun.11, 1200 (2020). 10.1038/s41467-020-15089-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarboe, E. et al. Serous carcinogenesis in the fallopian tube: A descriptive classification. Int. J. Gynecol. Pathol.27, 1–9 (2008). 10.1097/pgp.0b013e31814b191f [DOI] [PubMed] [Google Scholar]
- 9.Labidi-Galy, S. I. et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun.8, 1093 (2017). 10.1038/s41467-017-00962-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilks, C. B. et al. Incidental Nonuterine High-grade Serous carcinomas arise in the fallopian tube in most cases: Further evidence for the Tubal origin of high-grade Serous Carcinomas. Am. J. Surg. Pathol.39, 357–364 (2015). 10.1097/PAS.0000000000000353 [DOI] [PubMed] [Google Scholar]
- 11.Zhang, S. et al. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nat. Commun.10, 5367 (2019). 10.1038/s41467-019-13116-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature474, 609–615 (2011). 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuchenbaecker, K. B. et al. Risks of breast, ovarian, and contralateral breast Cancer for BRCA1 and BRCA2 mutation carriers. JAMA317, 2402 (2017). 10.1001/jama.2017.7112 [DOI] [PubMed] [Google Scholar]
- 14.Uboveja, A. & Aird, K. M. Interplay between altered metabolism and DNA damage and repair in ovarian cancer. BioEssays46, 2300166 (2024). 10.1002/bies.202300166 [DOI] [PubMed] [Google Scholar]
- 15.Webb, P. M. & Jordan, S. J. Global epidemiology of epithelial ovarian cancer. Nat. Rev. Clin. Oncol.21, 389–400 (2024). 10.1038/s41571-024-00881-3 [DOI] [PubMed] [Google Scholar]
- 16.Nené, N. R. et al. Association between the cervicovaginal microbiome, BRCA1 mutation status, and risk of ovarian cancer: A case-control study. Lancet Oncol.20, 1171–1182 (2019). 10.1016/S1470-2045(19)30340-7 [DOI] [PubMed] [Google Scholar]
- 17.Steinbach, A. & Riemer, A. B. Immune evasion mechanisms of human papillomavirus: An update. Int. J. Cancer142, 224–229 (2018). 10.1002/ijc.31027 [DOI] [PubMed] [Google Scholar]
- 18.Wang, X., Zeng, Y., Huang, X. & Zhang, Y. Prevalence and genotype distribution of human papillomavirus in Invasive Cervical Cancer, Cervical Intraepithelial Neoplasia, and Asymptomatic women in Southeast China. Biomed. Res. Int.2018, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundström, K. et al. Prospective study of HPV16 viral load and risk of in situ and invasive squamous cervical Cancer. Cancer Epidemiol. Biomarkers Prev.22, 150–158 (2013). 10.1158/1055-9965.EPI-12-0953-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bilyk, O. O., Pande, N. T., Pejovic, T. & Buchinska, L. G. The frequency of human papilloma virus types 16, 18 in upper genital tract of women at high risk of developing ovarian cancer. Exp. Oncol.36, 121–124 (2014). [PubMed] [Google Scholar]
- 21.Paradowska, E., Jabłońska, A., Studzińska, M., Wilczyński, M. & Wilczyński, J. R. Detection and genotyping of CMV and HPV in tumors and fallopian tubes from epithelial ovarian cancer patients. Sci. Rep.9, 19935 (2019). 10.1038/s41598-019-56448-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuscu, E., Ozdemir, B. H., Erkanli, S. & Haberal, A. HPV and p53 expression in epithelial ovarian carcinoma. Eur. J. Gynaecol. Oncol.26, 642–645 (2005). [PubMed] [Google Scholar]
- 23.Atalay, F. et al. Detection of human papillomavirus DNA and genotyping in patients with epithelial ovarian carcinoma. J. Obstet. Gynaecol. Res.33, 823–828 (2007). 10.1111/j.1447-0756.2007.00663.x [DOI] [PubMed] [Google Scholar]
- 24.Al-Shabanah, O. A. et al. Human papillomavirus genotyping and integration in ovarian cancer Saudi patients. Virol. J.10, 343 (2013). 10.1186/1743-422X-10-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, C. H. et al. Human papillomavirus in Benign and malignant ovarian and endometrial tissues. Int. J. Gynecol. Pathol.11, 210–215 (1992). 10.1097/00004347-199207000-00007 [DOI] [PubMed] [Google Scholar]
- 26.Wu, Q. J. et al. Detection of human papillomavirus-16 in ovarian malignancy. Br. J. Cancer89, 672–675 (2003). 10.1038/sj.bjc.6601172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan, Z., Hafez, M. M., Kamel, M. M. & Zekri, N. A. R. Human Papillomavirus Genotypes and Methylation of CADM1, PAX1, MAL and ADCYAP1 Genes in Epithelial Ovarian Cancer Patients. APJCP18 (2017). [DOI] [PMC free article] [PubMed]
- 28.Malisic, E., Jankovic, R. & Jakovljevic, K. Detection and genotyping of human papillomaviruses and their role in the development of ovarian carcinomas. Arch. Gynecol. Obstet.286, 723–728 (2012). 10.1007/s00404-012-2367-6 [DOI] [PubMed] [Google Scholar]
- 29.Dadashi, M. et al. Detection of human papilloma virus type 16 in epithelial ovarian tumors samples. Arch. Clin. Infect. Dis.12, (2016).
- 30.Zhang, P. P. et al. Possible epithelial ovarian cancer association with HPV18 or HPV33 infection. Asian Pac. J. Cancer Prev.17, 2959–2964 (2016). [PubMed] [Google Scholar]
- 31.Roos, P., Orlando, P. A., Fagerstrom, R. M. & Pepper, J. W. In North America, some ovarian cancers express the oncogenes of preventable human papillomavirus HPV-18. Sci. Rep.5, 8645 (2015). 10.1038/srep08645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherif, S. et al. Prevalence of human papillomavirus detection in ovarian cancer: A meta-analysis. Eur. J. Clin. Microbiol. Infect. Dis.40, 1791–1802 (2021). 10.1007/s10096-021-04282-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathak, S., Wilczyński, J. R. & Paradowska, E. Factors in Oncogenesis: Viral infections in Ovarian Cancer. Cancers12, 561 (2020). 10.3390/cancers12030561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haręża, D. A., Wilczyński, J. R. & Paradowska, E. Human papillomaviruses as Infectious agents in Gynecological cancers. Oncogenic properties of viral proteins. IJMS23, 1818 (2022). 10.3390/ijms23031818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanmughapriya, S. et al. Viral and bacterial aetiologies of epithelial ovarian cancer. Eur. J. Clin. Microbiol. Infect. Dis.31, 2311–2317 (2012). 10.1007/s10096-012-1570-5 [DOI] [PubMed] [Google Scholar]
- 36.Idahl, A. et al. Chlamydia trachomatis, Mycoplasma genitalium, Neisseria gonorrhoeae, human papillomavirus, and polyomavirus are not detectable in human tissue with epithelial ovarian cancer, borderline tumor, or benign conditions. Am. J. Obstet. Gynecol.202, 71e1–71e6 (2010). 10.1016/j.ajog.2009.07.042 [DOI] [PubMed] [Google Scholar]
- 37.Ingerslev, K. et al. High-risk HPV is not associated with epithelial ovarian cancer in a caucasian population. Infect. Agents Cancer11, 39 (2016). 10.1186/s13027-016-0087-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anttila, M., Syrjänen, S., Ji, H., Saarikoski, S. & Syrjänen, K. Failure to demonstrate human papillomavirus DNA in epithelial ovarian Cancer by general primer PCR. Gynecol. Oncol.72, 337–341 (1999). 10.1006/gyno.1998.5264 [DOI] [PubMed] [Google Scholar]
- 39.Ingerslev, K. et al. The prevalence of EBV and CMV DNA in epithelial ovarian cancer. Infect. Agents Cancer14, 7 (2019). 10.1186/s13027-019-0223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabarek, B. O. et al. Detection and genotyping of human papillomavirus (HPV16/18), Epstein–Barr Virus (EBV), and human cytomegalovirus (HCMV) in endometrial endometroid and ovarian cancers. Pathogens12, 397 (2023). 10.3390/pathogens12030397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rådestad, A. F. et al. Impact of human cytomegalovirus infection and its Immune response on survival of patients with ovarian Cancer. Transl. Oncol.11, 1292–1300 (2018). 10.1016/j.tranon.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlson, J. W., Rådestad, A. F., Söderberg-Naucler, C. & Rahbar, A. Human cytomegalovirus in high grade serous ovarian cancer possible implications for patients survival. Medicine97, e9685 (2018). 10.1097/MD.0000000000009685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson, A. et al. Droplet digital PCR quantification suggests that higher viral load correlates with improved survival in HPV-positive oropharyngeal tumours. J. Clin. Virol.129, 104505 (2020). 10.1016/j.jcv.2020.104505 [DOI] [PubMed] [Google Scholar]
- 44.McFarlane, M., MacDonald, A. I., Stevenson, A. & Graham, S. V. Human papillomavirus 16 Oncoprotein expression is controlled by the Cellular splicing factor SRSF2 (SC35). J. Virol.89, 5276–5287 (2015). 10.1128/JVI.03434-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanna, G. J. et al. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann. Oncol.29, 1980–1986 (2018). 10.1093/annonc/mdy251 [DOI] [PubMed] [Google Scholar]
- 46.Shuto, T. et al. Establishment of a screening method for Epstein–Barr Virus-Associated gastric carcinoma by Droplet Digital PCR. Microorganisms7, 628 (2019). 10.3390/microorganisms7120628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hassan-Walker, A. F., Mattes, F. M., Griffiths, P. D. & Emery, V. C. Quantity of cytomegalovirus DNA in different leukocyte populations during active infection in vivo and the presence of gB and UL18 transcripts. J. Med. Virol.64, 283–289 (2001). 10.1002/jmv.1048 [DOI] [PubMed] [Google Scholar]
- 48.Paradowska, E. et al. Detection of cytomegalovirus in human placental cells by polymerase chain reaction. Apmis114, 764–771 (2006). 10.1111/j.1600-0463.2006.apm_31.x [DOI] [PubMed] [Google Scholar]
- 49.Nakamura, M., Obata, T., Daikoku, T. & Fujiwara, H. The Association and significance of p53 in gynecologic cancers: The potential of targeted therapy. IJMS20, 5482 (2019). 10.3390/ijms20215482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corney, D. C., Flesken-Nikitin, A., Choi, J. & Nikitin, A. Y. Role of p53 and rb in Ovarian Cancer. In Ovarian Cancer, Vol. 622 (eds Coukos, G., Berchuck, A. & Ozols, R.) 99–117 (Springer New York, New York, 2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rotondo, J. C. et al. Simultaneous detection and viral DNA load quantification of different human papillomavirus types in clinical specimens by the high analytical droplet digital PCR method. Front. Microbiol.11, 591452 (2020). 10.3389/fmicb.2020.591452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosa, M. I. et al. The prevalence of human papillomavirus in ovarian Cancer: A systematic review. Int. J. Gynecol. Cancer23, 437–441 (2013). 10.1097/IGC.0b013e318280f3e0 [DOI] [PubMed] [Google Scholar]
- 53.Svahn, M. F., Faber, M. T., Christensen, J., Norrild, B. & Kjaer, S. K. Prevalence of human papillomavirus in epithelial ovarian cancer tissue. A meta-analysis of observational studies. Acta Obstet. Gynecol. Scand.93, 6–19 (2014). 10.1111/aogs.12254 [DOI] [PubMed] [Google Scholar]
- 54.Yin, M. et al. Detection of human cytomegalovirus in patients with epithelial ovarian cancer and its impacts on survival. Infect. Agents Cancer15, 23 (2020). 10.1186/s13027-020-00289-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogel, R. I. et al. Prevalence of active cytomegalovirus infection at diagnosis of ovarian cancer and during chemotherapy and subsequent changes in cognitive functioning. BMC Cancer23, 1057 (2023). 10.1186/s12885-023-11566-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox, M., Kartikasari, A. E. R., Gorry, P. R., Flanagan, K. L. & Plebanski, M. Potential impact of human cytomegalovirus infection on immunity to ovarian tumours and cancer progression. Biomedicines9, 351 (2021). 10.3390/biomedicines9040351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rahbar, A., Pantalone, M. R., Religa, P. & Rådestad, A. F. Söderberg-Naucler, C. evidence of human cytomegalovirus infection and expression of 5‐lipoxygenase in borderline ovarian tumors. J. Med. Virol.93, 4023–4027 (2021). 10.1002/jmv.26664 [DOI] [PubMed] [Google Scholar]
- 58.Rasmussen, C. B., Jensen, A., Albieri, V., Andersen, K. K. & Kjaer, S. K. Increased risk of borderline ovarian tumors in women with a history of pelvic inflammatory disease: A nationwide population-based cohort study. Gynecol. Oncol.143, 346–351 (2016). 10.1016/j.ygyno.2016.08.318 [DOI] [PubMed] [Google Scholar]
- 59.Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem.83, 8604–8610 (2011). 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okodo, M., Okayama, K., Teruya, K. & Sasagawa, T. Uniplex E6/E7 PCR method detecting E6 or E7 genes in 39 human papillomavirus types. J. Med. Virol.90, 981–988 (2018). 10.1002/jmv.25017 [DOI] [PubMed] [Google Scholar]
- 61.Paradowska, E. et al. Distribution of UL144, US28 and UL55 genotypes in Polish newborns with congenital cytomegalovirus infections. Eur. J. Clin. Microbiol. Infect. Dis.31, 1335–1345 (2012). 10.1007/s10096-011-1447-z [DOI] [PubMed] [Google Scholar]
- 62.Hassan, R. et al. Epstein-Barr Virus (EBV) detection and typing by PCR: A contribution to diagnostic screening of EBV-positive Burkitt’s lymphoma. Diagn. Pathol.1, 17 (2006). 10.1186/1746-1596-1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Z. et al. Effect of advanced glycosylation end products on apoptosis in human adipose tissue-derived stem cells in vitro. Cell. Biosci.5, 3 (2015). 10.1186/2045-3701-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Heetvelde, M. et al. Evaluation of relative quantification of alternatively spliced transcripts using droplet digital PCR. Biomol. Detect. Quantif.13, 40–48 (2017). 10.1016/j.bdq.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author upon reasonable request.