Abstract
The phenylpropanoid pathway is one of the plant metabolic pathways most prominently linked to the transition to terrestrial life, but its evolution and early functions remain elusive. Here, we show that activity of the t-cinnamic acid 4-hydroxylase (C4H), the first plant-specific step in the pathway, emerged concomitantly with the CYP73 gene family in a common ancestor of embryophytes. Through structural studies, we identify conserved CYP73 residues, including a crucial arginine, that have supported C4H activity since the early stages of its evolution. We further demonstrate that impairing C4H function via CYP73 gene inactivation or inhibitor treatment in three bryophyte species—the moss Physcomitrium patens, the liverwort Marchantia polymorpha and the hornwort Anthoceros agrestis—consistently resulted in a shortage of phenylpropanoids and abnormal plant development. The latter could be rescued in the moss by exogenous supply of p-coumaric acid, the product of C4H. Our findings establish the emergence of the CYP73 gene family as a foundational event in the development of the plant phenylpropanoid pathway, and underscore the deep-rooted function of the C4H enzyme in embryophyte biology.
Keywords: Plant Evolution, Bryophytes, Cytochromes P450, Biopolymers, Cinnamic Acid 4-Hydroxylase
Subject terms: Evolution & Ecology, Plant Biology
Synopsis
The phenylpropanoid pathway is assumed to represent a key plant metabolic adaptation associated with terrestrial life, and processes about 30% of all photosynthetic carbon on Earth. This study highlights the importance of the evolutionary co-emergence of the CYP73 gene family, which encodes t-cinnamic acid 4-hydroxylase (C4H) enzymes, in establishing this pathway in a land plant’s ancestor.
The CYP73 gene family emerged in an ancestor of land plants and has subsequently been conserved.
The CYP73 gene family encodes C4H, the first plant-specific enzyme in the phenylpropanoid pathway, from the earliest stages of its evolution.
Alteration of C4H function via CYP73 gene inactivation or inhibitor treatment consistently disrupts phenylpropanoid synthesis and development in land plants.
This paper shows that the t-cinnamic acid 4-hydroxylase activity emerged with the CYP73 gene family in a common ancestor of all land plants.
Introduction
Green plants (i.e., Viridiplantae) have evolved over approximately one billion years and have thrived wherever light was available thanks to their photosynthetic capabilities. The successful evolutionary history of Viridiplantae have yielded an astonishing diversity of forms—from unicellular marine algae to giant redwood trees—and propelled them as the most prevalent living group on Earth from a biomass standpoint (Bar-On et al, 2018).
Most of this biomass is found on land (Bar-On et al, 2018), reflecting the cornerstone functions performed by plants in terrestrial ecosystems, primarily as an entry point for solar energy. Recent phylogenetic evidences indicate that modern land plants, also known as embryophytes, have a unique origin and emerged from freshwater algae about half a billion years ago (Cheng et al, 2019; Morris et al, 2018). The radical change in habitat experienced by early embryophytes, an evolutionary milestone called terrestrialization, involved profound morpho-physiological adaptations. In particular, the ability to synthesize a wide range of metabolites was instrumental by mitigating effects of terrestrial constraints (e.g., UV, lack of buoyancy, drought) and by providing chemical mediators for fast-expanding ecological interactions.
One of the most iconic plant metabolisms associated with terrestrialization is the phenylpropanoid pathway, which generates a suite of phenolic compounds that effectively address terrestrial challenges (Vogt, 2010; Weng and Chapple, 2010). This pathway leads to the synthesis of widespread polyphenolic molecules (e.g., flavonoids) and phenolic esters/amides (e.g., chlorogenic acids), which act as powerful UV screens and antioxidants. The phenylpropanoid pathway also supplies precursors of the four hydrophobic polymers cutin, suberin, sporopollenin, and lignin that strengthen and waterproof the cell wall. These polymers form the framework of various apoplastic diffusion barriers (e.g., cuticle, pollen coat, Casparian strip) and thus help plants to shield their tissues from external aggressions, and to manage water and solutes efficiently. In accordance to these essential functions, hydrophobic biopolymers make up a significant fraction of embryophyte biomass. Lignin for instance is regarded as the second most abundant biopolymer on the planet after cellulose, accounting for ca. 30% of biosphere organic carbon (Boerjan et al, 2003).
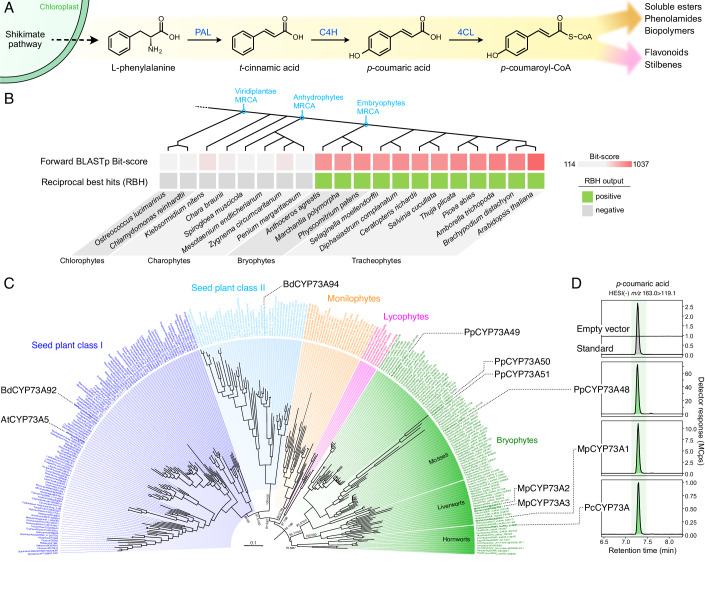
In order to support high biomass production and multiple physiological functions, the phenylpropanoid pathway is meticulously organized and regulated (Mizutani et al, 1997; Bassard et al, 2012). The first three steps of this pathway, collectively known as the general phenylpropanoid pathway (Fig. 1A), are obligatory and hence process the entire metabolic flux. The initial step involves the deamination of phenylalanine by the phenylalanine ammonia-lyase (PAL) to produce trans-cinnamic acid, which is the first bona fide phenylpropanoid molecule. The phenolic ring of t-cinnamic acid is then hydroxylated at position 4 (para position) by the cinnamic acid 4-hydroxylase (C4H) to generate para-coumaric acid. Next, p-coumaric acid is activated with coenzyme A by 4-coumarate:CoA ligase (4CL), which leads to the formation of p-coumaroyl-CoA, a branch-point molecule that feeds into the various downstream pathways (Fig. 1A).
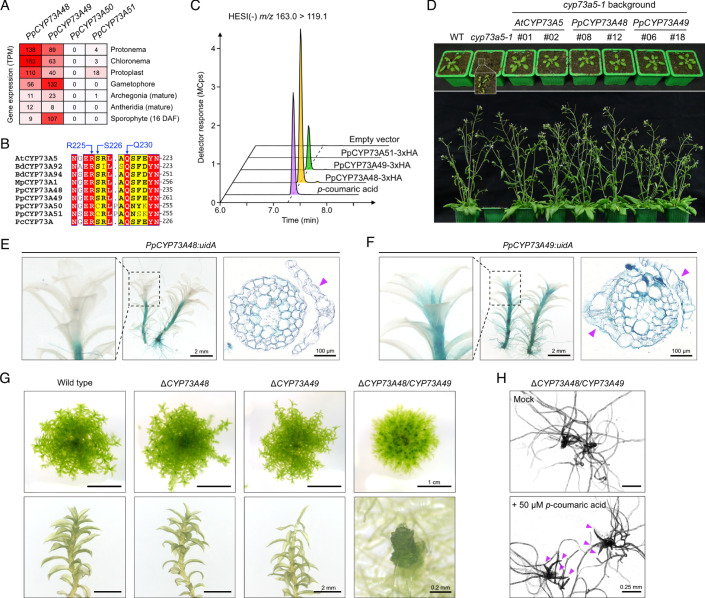
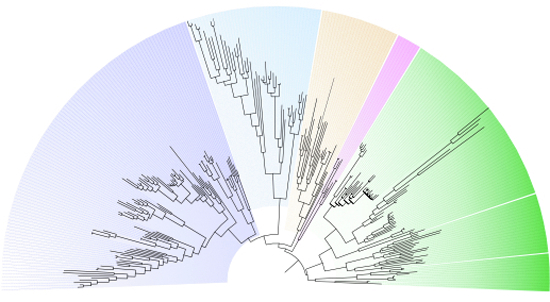
Figure 1. Evolutionary history of the CYP73 family encoding t-cinnamic acid 4-hydroxylase.
(A) Schematic representation of the three steps of the general phenylpropanoid pathway. PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate:CoA ligase. (B) Search for AtCYP73A5 homologs in Viridiplantae by a reciprocal best hit (RBH) strategy. MRCA, most recent common ancestor. (C) Maximum-likelihood nucleotide tree (IQ-TREE2, GTR + F + I + R7) describing the phylogenetic relationships between 275 CYP73 homologous sequences. SH-aLRT test and ultrafast bootstrap (1000 pseudo-replicates) supports are annotated on main branches. CYP73 homologs relevant to the present study are indicated. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Mp, Marchantia polymorpha; Pc, Phaeoceros carolinianus. Scale bar represents the number of nucleotide substitutions per site. Tree was rooted according to the midpoint method. (D) Representative UHPLC-MS/MS chromatograms showing the production in vitro of p-coumaric acid from t-cinnamic acid (C4H activity) by recombinant CYP73 proteins representative of the three major bryophyte groups: mosses (PpCYP73A48), liverworts (MpCYP73A1) and hornworts (PcCYP73A). Assays performed with microsomes derived from yeasts transformed with an empty vector were used as negative controls. Source data are available online for this figure.
PAL activity is not exclusive to plants and is also found in cyanobacteria and fungi (Barros and Dixon, 2019), thus making C4H the first plant-specific step in the phenylpropanoid pathway (Renault et al, 2017b). C4H enzymes belong to the family 73 of cytochrome P450 monooxygenases (CYP73). Cytochromes P450 are highly diversified enzymes that catalyze irreversible and often rate-determining reactions and, as such, are considered to be key drivers of plant metabolism evolution and diversification, and control points in metabolic pathways (Hamberger and Bak, 2013; Liu et al, 2016; Hansen et al, 2021). Accordingly, inactivation of the single CYP73 gene from the tracheophyte model Arabidopsis thaliana led to dramatic developmental defects and phenylpropanoid deficiency (Schilmiller et al, 2009), pointing to the critical role of C4H in tracheophyte physiology. No such in planta investigation has been performed to date in a bryophyte model although these data appear key to improve our understanding of the evolution and early functions of C4H and, by extension, of the phenylpropanoid pathway as a whole.
Here we report a multidisciplinary study of C4H-encoding CYP73 genes. We show that C4H activity emerged with the rise of the CYP73 family in an embryophyte ancestor and identify key residues supporting its catalytic activity. We further show that impairing C4H function via CYP73 gene inactivation or inhibitor treatment compromises phenylpropanoid biosynthesis and development in both tracheophyte and bryophyte species, pointing to a pivotal and conserved role of CYP73 in embryophyte physiology.
Results
CYP73-catalyzed C4H activity originated in an embryophyte progenitor
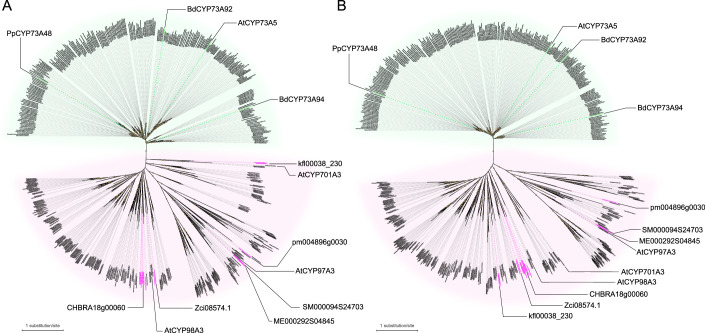
Thus far, the only known enzymes capable of catalyzing the 4-hydroxylation of t-cinnamic acid (C4H activity) are the cytochrome P450 monooxygenases from the 73 family (CYP73). To investigate the evolution of C4H, a survey of CYP73 genes was conducted using a reciprocal best hits (RBH) approach in 20 Viridiplantae genomes, including recent charophyte genomes. The results revealed that potential CYP73 homologs were only found in bryophytes and tracheophytes, indicating that the origin of this CYP family can be traced back to a progenitor of embryophytes (Fig. 1B, Appendix Tab. S1). To bring further support to this evolutionary scenario, we performed a phylogenetic analysis of 275 CYP73 sequences. We included 271 additional sequences from A. thaliana, P. patens, and charophytes to serve as outgroups. Both amino acid- and nucleotide-based phylogenies confirmed that embryophyte CYP73 sequences formed a well-defined monophyletic group (Fig. EV1). Charophyte sequences were not associated with the CYP73 group, but instead grouped with other clan 71 CYP families, such as CYP97, CYP98, or CYP701 (Appendix Tab. S1, Fig. EV1). Phylogenetic analyses failed to identify a precise origin of the CYP73 gene family, or a clearly defined sister family, but suggested that the family arose within the clan 71 (Fig. EV1). To explore the possibility that charophyte proteins might have already a cryptic C4H function, we tested the catalytic activity of the Klebsormidium nitens kfl00038_0230 protein that was uncovered as the best hit among non-embryophyte plants (Appendix Tab. S1). In vitro enzyme assay with K. nitens recombinant proteins did not result in detectable C4H activity under the tested conditions (Appendix Fig. S1), a result that further supported an emergence of bona fide C4H function in a common ancestor of embryophytes.
Figure EV1. Phylogenetic analysis of CYP73 and clan 71 CYP sequences.
Maximum-likelihood trees of 275 CYP73 sequences and 271 additional outgroup sequences derived from A. thaliana, P. patens and charophytes (AtCYP73A5 tBLASTn bit-score >100). (A) Unrooted ML nucleotide tree (IQ-TREE2, SYM + I + R10). (B) Unrooted ML amino acid tree (IQ-TREE2, Q.insect+I + R8). Relevant CYP73 homologs, A. thaliana clan 71 CYP members and charophyte sequences corresponding to BLASTp best hits (see Appendix Tab. S1) are shown. Ultrafast bootstrap support values equal or superior to 80 are annotated on branch as yellow dots. Trees are drawn to scale; scale bars represent the number of substitutions per site.
To better resolve the phylogenetic relationships within the CYP73 family, we performed a focused analysis on the 275 CYP73 sequences only. Apart from the previously reported duplication that occurred in a seed plant common ancestor (Renault et al, 2017b), the topology of the CYP73 tree reflected embryophyte systematics (Fig. 1C). CYP73 family was found fairly diversified in bryophytes with for instance four paralogs in the moss Physcomitrium patens (PpCYP73A48-51) and three in the liverwort Marchantia polymorpha (MpCYP73A1-3). In mosses, this diversity could be at least in part explained by an early duplication that occurred after the Takakia and Sphagnum genera branched off 350 million years ago (Hu et al, 2023) (Fig. 1C). Conservation of C4H function in bryophyte CYP73 proteins was explored through in vitro assays with recombinant proteins representative of the three main groups: PpCYP73A48 (P. patens, moss), MpCYP73A1 (M. polymorpha, liverwort) and PcCYP73A (Phaeoceros carolinianus, hornwort). PpCYP73A48 and MpCYP73A1 proteins were chosen because they were concurrently investigated in planta in the present study; PcCYP73A protein was selected because it had best full-length RNA-seq scaffold quality in the 1kP database (One Thousand Plant Transcriptomes Initiative, 2019). All three bryophyte CYP73 proteins catalyzed the production of p-coumaric acid in vitro, whereas microsomes derived from yeast transformed with the empty vector did not (Fig. 1D). In combination with biochemical data previously obtained in tracheophytes (Mizutani et al, 1997; Renault et al, 2017b), these results indicate that CYP73-catalyzed C4H activity is conserved in embryophytes.
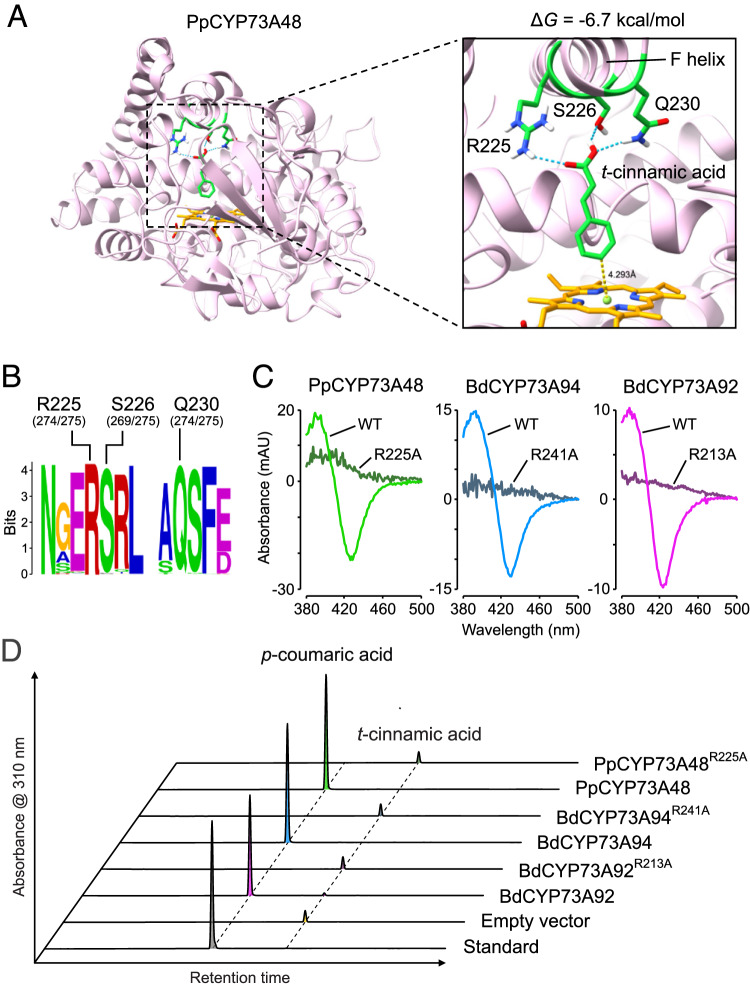
C4H activity is supported by a conserved arginine residue
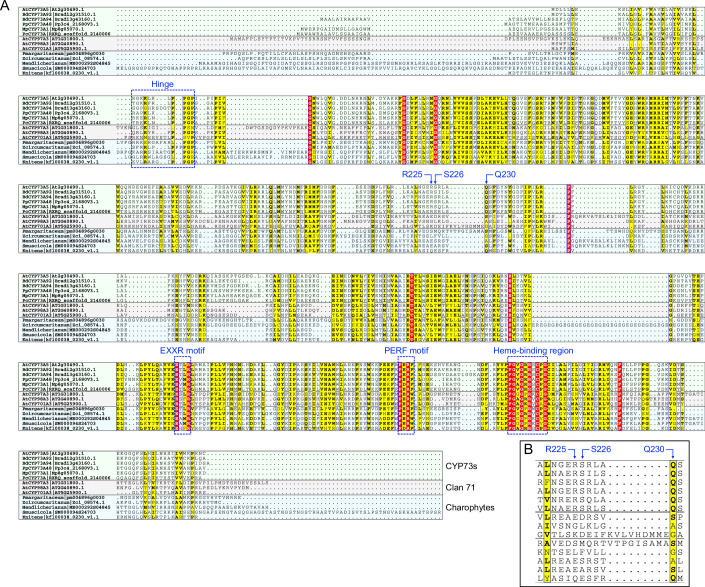
To bring further insights into the origin of C4H function in the CYP73 family, we looked for critical residues that support this catalytic activity and that could have underpinned C4H emergence. We used homology modeling to construct the three-dimensional structure of the P. patens CYP73A48 enzyme, which had the ability to catalyze C4H activity in vitro (Fig. 1D), based on the crystal structure of Sorghum bicolor C4H (Zhang et al, 2020). After docking the heme prosthetic group into the PpCYP73A48 structure, we next docked t-cinnamic acid into its active site. The lowest Gibbs free energy pose (−6.7 kcal/mol) predicted that position 4 of the t-cinnamic acid phenolic ring faced the heme at a distance of 4.3 Å (Fig. 2A), which was consistent with C4H activity. The docking experiment also identified three residues in the F helix of PpCYP73A48—arginine 225 (R225), serine 226 (S226), and glutamine 230 (Q230)—presumably able to form hydrogen bonds with the carboxylic function of t-cinnamic acid and to hold it in the proper orientation with respect to the heme reaction center (Fig. 2A). Multiple sequence alignment and corresponding WebLogo visualization showed that these three residues were highly conserved across the 275 CYP73 proteins used for phylogenetic analysis (Fig. 2B). On the contrary, R225, S226, and Q230 residues were not present in charophyte proteins identified in the RBH analysis and in representative members of A. thaliana clan 71 (Fig. EV2), confirming the specificity of these residues for the CYP73 family.
Figure 2. Structural determinants of t-cinnamic acid 4-hydroxylase activity in CYP73.
(A) Docking of t-cinnamic acid in the active site of the P. patens CYP73A48 enzyme. Pose corresponding to the lowest Gibbs free energy (ΔG) predicts that R225, S226, and Q230 residues can establish hydrogen bonds with the carboxyl group of t-cinnamic acid. In this configuration, para position (or position 4) of the phenolic ring faces the heme prosthetic group at a distance of 4.3 Å. (B) WebLogo showing the conservation of R225, S226, and Q230 residues across the 275 CYP73 sequences used for the phylogenetic analyses. For each residue, absolute count is provided between brackets. (C) Representative type I difference spectra showing the loss of t-cinnamic acid binding ability of P. patens CYP73A48, B. distachyon CYP73A94, and B. distachyon CYP73A92 enzymes mutated in the R225 residue, or its equivalent. Positively charged arginine residue was replaced with neutral alanine. (D) Representative HPLC-UV chromatograms of in vitro enzyme assays indicating loss of C4H activity in CYP73 enzymes mutated in the R225 residue, or its equivalent. Source data are available online for this figure.
Figure EV2. Multiple sequence alignment of CYP73 proteins with clan 71 CYP members.
(A) Full-length protein sequences of biochemically characterized CYP73 proteins from both tracheophytes (AtCYP73A5, BdCYP73A92, BdCYP73A94) and bryophytes (PpCYP73A48, MpCYP73A1, PcCYP73A) were aligned with A. thaliana clan 71 CYP members (AtCYP97A3, AtCYP98A3, AtCYP701A3). Protein sequence corresponding to BLAST best hits in the charophytes P. margaritaceum, Z. circumcaritanum, M. endlicherianum, and K. nitens (Fig. 1B) were included in the alignment. These charophyte proteins were found closely associated to A. thaliana clan 71 CYP members (Appendix Tab. S1, Fig. EV1). Alignment was performed with MUSCLE and visualized using ESPript 3.0. Positions that are identical are highlighted with a red background; positions with >70% similarity are highlighted with a yellow background. Cytochrome P450 conserved regions are highlighted. (B) Focus on the protein region encompassing the three residues critical for t-cinnamic acid binding in CYP73 active site.
To scrutinize the predictions of docking experiments, we focused on the R225 residue of PpCYP73A48, which has a positive charge and can thus strongly interact with the negatively charged carboxyl group of t-cinnamic acid. We expanded the evolutionary scope of the experiment by including the previously characterized CYP73A94 and CYP73A92 proteins from the tracheophyte Brachypodium distachyon (Renault et al, 2017b) (Fig. 1C). We replaced the R225 residue, or its equivalent in BdCYP73A94 (R241) and BdCYP73A92 (R213), with a non-polar and uncharged alanine residue. Carbon monoxide-induced difference spectra showed that the R > A substitution did not compromise the structural stability of CYP73 proteins (Appendix Fig. S2). In contrast, substrate-induced type I difference spectrum revealed that all three R > A mutated CYP73 proteins lost their ability to bind t-cinnamic acid compared to their wild-type counterparts (Fig. 2C). As a result, the R > A CYP73 mutant proteins did not produce p-coumaric acid from t-cinnamic acid in in vitro assays (Fig. 2D), demonstrating that the conserved R225 residue was a critical determinant of CYP73-catalyzed C4H activity.
CYP73 deficiency strongly alters moss development
We performed in planta investigation of the CYP73 genes identified in the moss P. patens (Fig. 1C). Scrutiny of public RNA-seq data revealed that only three paralogs were expressed in at least one condition across P. patens tissues, with PpCYP73A48 and PpCYP73A49 being the most prominent (Fig. 3A). We therefore decided to disregard the PpCYP73A50 paralog, as it displayed no expression and also exhibited substantial variation among the protein region containing the three residues interacting with t-cinnamic acid (Fig. 3B). We then performed in vitro assays with PpCYP73A48, PpCYP73A49, and PpCYP73A51 recombinant proteins tagged with 3xHA. While all three proteins were confirmed to be present in yeast microsomes through Western blot analysis (Appendix Fig. S3), only PpCYP73A48-3xHA and PpCYP73A49-3xHA demonstrated the ability to convert t-cinnamic acid into p-coumaric acid (Fig. 3C). This observation was further supported in planta by the fact that PpCYP73A48 and PpCYP73A49 fully complemented the stunted phenotype of the Arabidopsis cyp73a5-1 mutant (Fig. 3D), whereas no mutant plant rescued with PpCYP73A51 was ever found.
Figure 3. Functional analysis of CYP73 genes in the moss Physcomitrium patens.
(A) Expression profiles of the four P. patens CYP73 paralogs in various tissues. Data are derived from the CoNekT database (https://evorepro.sbs.ntu.edu.sg; Proost and Mutwil, 2018). DAF, days after fertilization; TPM, transcripts per kilobase million. (B) Angiosperm and bryophyte CYP73 multiple sequence alignment centered on the protein region encompassing t-cinnamic acid binding residues identified by docking. At, Arabidopsis thaliana, Bd, Brachypodium distachyon; Mp, Marchantia polymorpha; Pc, Phaeoceros carolinianus; Pp, Physcomitrium patens. (C) Representative UHPLC-MS/MS chromatograms of in vitro C4H assays performed with recombinant PpCYP73-3xHA tagged proteins. Assays performed with microsomes derived from yeasts transformed with an empty vector were used as negative control. (D) Phenotype of A. thaliana 3-week-old (upper panel) and 6-week-old (lower panel) wild type, cyp73a5-1 mutant and cyp73a5-1 complemented with AtCYP73A5, PpCYP73A48, and PpCYP73A49 coding sequences. Two independent complemented lines are depicted for each gene. (E, F) Representative GUS staining pattern in 2-month-old gametophores of PpCYP73A48:uidA (E) and PpCYP73A49:uidA (F) reporter lines. For each gene, the central, left, and right pictures show whole gametophores, the apex of a gametophore and a gametophore cross-section, respectively. Magenta arrowheads point to phyllids. (G) Pictures of 2-month-old colonies of wild type, ΔPpCYP73A48 and ΔPpCYP73A49 single mutants, and ΔPpCYP73A48/CYP73A49 double mutant. Close-ups on individual gametophores from each genotype are visible in the lower part. (H) Pictures of 5-week-old ΔPpCYP73A48/CYP73A49 gametophores grown in low-melting point agarose Knop medium supplemented, or not (control), with 50 µM p-coumaric acid. Magenta arrowheads point to developed phyllids. Source data are available online for this figure.
To refine the analysis of PpCYP73A48 and PpCYP73A49 expression patterns, we generated knock-in lines where the STOP codon was replaced with the uidA reporter gene. These lines revealed that PpCYP73A48 and PpCYP73A49 had overlapping expression domains in gametophores, particularly in the stem (Fig. 3E,F). It was noted, however, that PpCYP73A49 expression extended further to the apex and in the basal parts of phyllids. We then disrupted both genes individually or concurrently by inserting a selection cassette through homologous recombination, resulting in a loss of corresponding transcripts (Appendix Fig. S4). No strong difference was visible between single mutants and the wild type, except in the ΔPpCYP73A49 lines where the phyllids at the apex appeared thinner and more fragile (Fig. 3G; Appendix Fig. S5). This latter observation was in accordance with PpCYP73A49 expression pattern (Fig. 3F). In contrast, the ΔPpCYP73A48/CYP73A49 double mutant consistently exhibited a very strong developmental phenotype characterized by restricted gametophore growth, phyllid fusion and an overproduction of protonema (Fig. 3G; Appendix Fig. S5). No major changes in the expression of PpCYP73A48 and PpCYP73A49 were observed in single mutant backgrounds (Appendix Fig. S6). We then attempted to chemically rescue the double mutant by providing exogenous p-coumaric acid. This treatment successfully restored phyllid expansion compared to mock treatment (Fig. 3H; Appendix Fig. S7), indicating that the ΔPpCYP73A48/CYP73A49 phenotype was, at least in part, caused by a shortage in p-coumaric acid, the product of C4H activity.
CYP73 deficiency leads to phenylpropanoid shortage and cuticle defect in moss
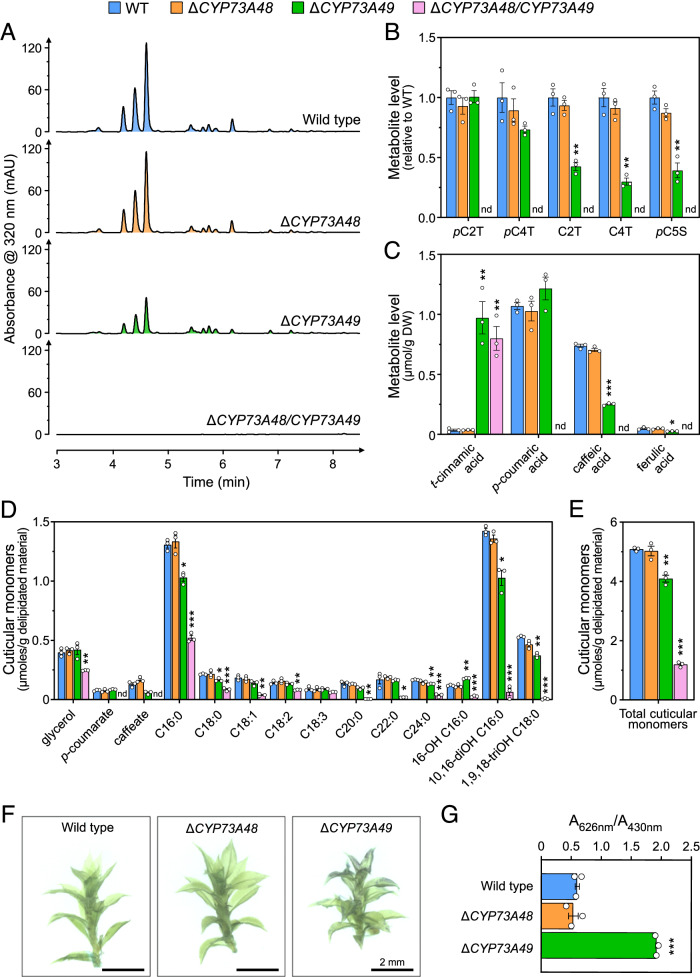
Next, we examined the metabolic consequences of CYP73 deficiency in the moss. UHPLC-UV fingerprints of ΔPpCYP73 mutant crude extracts revealed the complete loss of UV-absorbing molecules in double mutants (Fig. 4A), the major ones being hydroxycinnamate esters as reported before (Renault et al, 2017a; Kriegshauser et al, 2021). We also noticed a moderate decrease in the main peaks in ΔPpCYP73A49 mutants as compared to ΔPpCYP73A48 and the wild-type plants (Fig. 4A). These findings were confirmed through targeted UHPLC-MS/MS analysis of diagnostic phenylpropanoids in both crude and saponified extracts (Fig. 4B,C). None of the targeted phenylpropanoids downstream of the C4H enzymatic step was detected in metabolic extracts from ΔPpCYP73A48/CYP73A49 double mutants, whereas we observed concomitant accumulation of t-cinnamic acid, the C4H substrate, in saponified extracts (Fig. 4B,C). These results indicated that simultaneous disruption of PpCYP73A48 and PpCYP73A49 was sufficient to comprehensively abolish C4H function. Although no notable changes in metabolic profile of ΔPpCYP73A48 mutants were observed, a significant decrease in p-coumaroyl-5-O-shikimate, caffeoyl-2-O-threonate and caffeoyl-4-O-threonate level was evident in ΔPpCYP73A49 crude extracts. Corresponding saponified samples showed the accumulation of t-cinnamic acid and a corollary reduction in the amount of caffeic acid, while p-coumaric acid level remained unchanged (Fig. 4C). Since cis-cinnamic acid, a t-cinnamic acid stereoisomer, was suggested to play a role in the developmental abnormalities linked to C4H deficiency in Arabidopsis (Houari et al, 2021), we conducted a targeted analysis of these two compounds in crude extracts. Neither free t-cinnamic acid nor c-cinnamic acid accumulated in P. patens mutants (Appendix Fig. S8).
Figure 4. Metabolic and physiological characterization of P. patens ΔCYP73 mutants.
(A) Representative UHPLC-UV chromatograms of 2-month-old wild-type, ΔPpCYP73A48, ΔPpCYP73A49, and ΔPpCYP73A48/CYP73A49 crude extracts showing the loss of UV-absorbing molecules in the double mutants. (B) Relative levels of threonate and shikimate phenolic esters in 2-month-old gametophores. pC2T, p-coumaroyl-2-O-threonate; pC4T, p-coumaroyl-4-O-threonate; C2T, caffeoyl-2-O-threonate; C4T, caffeoyl-4-O-threonate; pC5S, p-coumaroyl-5-O-shikimate. (C) Quantification of hydroxycinnamic acids from saponified extracts of 2-month-old gametophores. (D) Compositional analysis of the cuticular polymer of 2-month-old gametophores. (E) Total amount of cuticular monomers from gametophores. (F) Representative pictures of 2-month-old wild-type, ΔPpCYP73A48 and ΔPpCYP73A49 gametophores after toluidine blue permeability assay. (G) Quantification of permeability to toluidine blue in 2-month-old gametophores. Results in panels (B), (C), (D), (E), and (F) are the mean ± SEM of three independent WT biological replicates and three independent mutant lines. WT versus mutant unpaired t test adjusted P-value: *P < 0.05; **P < 0.01; ***P < 0.001. nd, not detected. Source data are available online for this figure.
We then examined how impairing C4H function affected the composition of the P. patens gametophore cuticular polymer, as its formation relies on phenylpropanoid precursors (Renault et al, 2017a; Kriegshauser et al, 2021). The analysis of polymer monomers revealed a significant decrease in all aliphatic components, except C18:3, in the ΔPpCYP73A48/CYP73A49 double mutants compared to wild type (Fig. 4D). This decrease was accompanied by a complete loss of the two phenolic monomers, p-coumarate and caffeate. The overall quantity of the cuticular polymer, represented by the sum of individual monomers, dropped by 75% in the double mutants compared to the wild-type level (Fig. 4E). There were no changes in cuticular polymer composition of ΔPpCYP73A48 single mutants, consistent with the soluble phenylpropanoid analysis (Fig. 4A–E). In contrast, in ΔPpCYP73A49 single mutants, there were moderate yet statistically significant variations in cuticular polymer composition (Fig. 4D), resulting in a ca. 20% decrease in total monomer level compared to wild type (Fig. 4E). As for phenolic monomers, p-coumarate remained unchanged in the cuticular polymer of ΔPpCYP73A49 mutants, while caffeate level was reduced by 50%, although this finding was not statistically supported (adjusted P-value = 0.084). To understand the consequences of polymer composition changes on cuticle properties, we performed toluidine blue assays on gametophores of single ΔPpCYP73 mutants; the stunted growth of double ΔPpCYP73A48/CYP73A49 gametophores prevented their inclusion in the experiment. As shown in Fig. 4F,G, we observed a significant increase in ΔPpCYP73A49 gametophore permeability to toluidine blue, indicating a strong defect in cuticle diffusion barrier function despite the moderate cuticle compositional changes (Fig. 4D). This function appeared unaltered in the ΔPpCYP73A48 mutant as compared to wild type (Fig. 4F,G).
CYP73 function is conserved in the liverwort Marchantia polymorpha
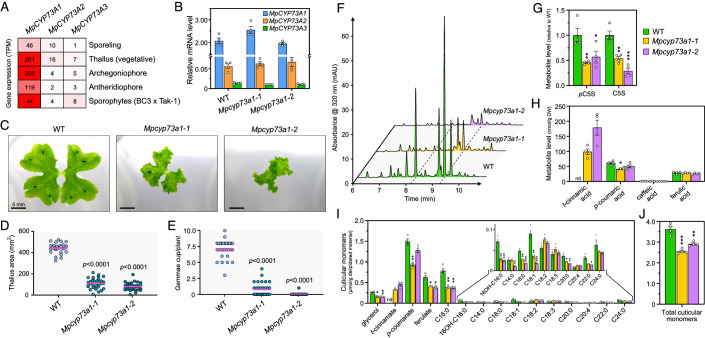
To bring additional evolutionary perspective to the functional analysis of the CYP73 gene family, we expanded our investigations to a second bryophyte species, the liverwort M. polymorpha. We identified the MpCYP73A1 gene as the primary paralog based on its expression level (Fig. 5A,B). In addition, we previously confirmed that the corresponding protein catalyzes C4H activity in vitro (Fig. 1D). We therefore produced two independent CRISPR mutants of the MpCYP73A1 gene, using two distinct protospacers located in the first exon (Appendix Fig. S9). The resulting Mpcyp73a1-1 and Mpcyp73a1-2 alleles featured 12 and 37 nucleotide deletions, respectively, leading to four amino acid loss (residues 94–97) and a premature STOP codon (position 150) at the protein level. The impact of these mutations on M. polymorpha development was significant, leading to a dramatic reduction in both the thallus area and number of gemmae cups per plant compared to the wild type (Fig. 5C–E). Noteworthy, disrupting MpCYP73A1 function did not affect the expression of the two other MpCYP73 paralogs, which remained 28- to 243-times lower in the thallus (Fig. 5B).
Figure 5. Developmental and metabolic consequences of CYP73A1 deficiency in M. polymorpha.
(A) Expression profiles of the three M. polymorpha CYP73 paralogs in various tissues. Data are derived from the CoNekT database (https://evorepro.sbs.ntu.edu.sg; Proost and Mutwil, 2018), except sporophytes for which data derive from the MarpolBase database (https://marchantia.info/mbex/). (B) RT-qPCR expression analysis of the three MpCYP73 genes in 3-week-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 plants. Results are the mean ± SEM of four biological replicates. (C) Representative pictures of 3-week-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 plants. (D, E) Quantification of thallus area (D) and gemmae cup number (E) in 3-week-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 plants. Results are the mean (pink trait) of measurements made on 29–32 plants. WT versus mutant one-way ANOVA adjusted P-value is displayed. (F) Representative UHPLC-UV chromatograms of 1-month-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 crude extracts. (G) Relative quantification of shikimate phenolic esters in 1-month-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 plants. C5S, caffeoyl-5-O-shikimate; pC5S, p-coumaroyl-5-O-shikimate. (H) Quantification of hydroxycinnamic acids from saponified extracts of 1-month-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 plants. (I) Compositional analysis of the cuticular polymer in 1-month-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 plants. (J) Total amount of cuticular monomers in 1-month-old wild-type, Mpcyp73a1-1 and Mpcyp73a1-2 plants. Results in panels (G), (H), (I), and (J) are the mean ± SEM of four independent biological replicates for each genotype. WT versus mutant unpaired t test adjusted P-value: *P < 0.05; **P < 0.01; ***P < 0.001. nd, not detected. Source data are available online for this figure.
We went on with the metabolic analysis of Mpcyp73a1 mutants. Through UHPLC-UV fingerprints of crude metabolic extracts, we observed a decrease in main peaks in mutants as compared to wild type (Fig. 5F). Targeted UHPLC-MS/MS analysis demonstrated ca. 50% reduction in the two essential intermediates of the phenylpropanoid pathway, p-coumaroyl-5-O-shikimate and caffeoyl-5-O-shikimate (Kriegshauser et al, 2021), in Mpcyp73a1 CRISPR lines (Fig. 5G). As with P. patens, we found no evidence of free c-cinnamic acid accumulation in Mpcyp73a1 mutant lines, although t-cinnamic acid was present (Appendix Fig. S8). Saponification of the crude extracts followed by targeted analysis revealed no significant changes in hydroxycinnamic acids, except t-cinnamic acid that appeared in the Mpcyp73a1 mutants (Fig. 5H). These results suggest that the major UV-absorbing peaks in M. polymorpha crude extracts, and which decrease in mutant lines (Fig. 5F), were not hydroxycinnamic acid derivatives but rather belonged to other phenylpropanoid classes (e.g., auronidins) (Berland et al, 2019). Phenylpropanoid biosynthesis was not completely abolished in Mpcyp73a1 mutants, probably due to genetic redundancy since two additional, lowly expressed MpCYP73A paralogs occur in this species (Fig. 5A,B).
We extended the metabolic characterization of Mpcyp73a1 mutants to the compositional analysis of their cuticular polymer. This analysis uncovered phenolics as the most abundant monomers in M. polymorpha (Fig. 5I). Both mutants consistently accumulated t-cinnamate in their polymer and featured a ca. 35% reduction in ferulate compared to the wild type. Impairing MpCYP73A1 function also caused alterations in the aliphatic monomer composition of the cuticular polymer, including decrease in glycerol, palmitate (C16:0), and ω-hydroxypalmitate (16OH-C16:0) (Fig. 5I). Overall, the cuticular polymer quantity was significantly reduced, by up to 25%, in Mpcyp73a1 mutant lines (Fig. 5J).
A selective C4H inhibitor consistently impairs development and phenylpropanoids in embryophytes
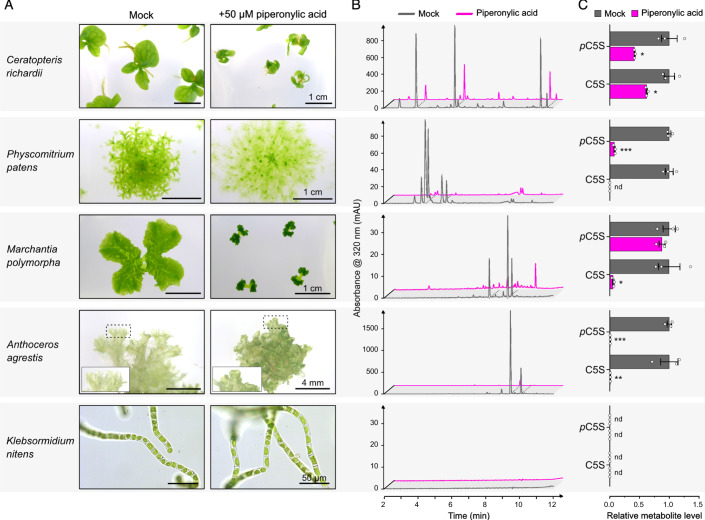
Finally, we performed pharmacological experiments using the selective inhibitor piperonylic acid (PA) to disrupt C4H function in planta (Schalk et al, 1998; Naseer et al, 2012). PA treatment accurately replicated in P. patens wild-type plants the developmental phenotype observed in ΔPpCYP73A48/CYP73A49 double mutants, displaying stunted gametophores and protonema outgrowth (Figs. 3G and 6A). Similarly, we observed a dramatic inhibition by PA of M. polymorpha thallus expansion, which resembled an aggravation of Mpcyp73a1 single mutant phenotypes (Figs. 5C and 6A). Based on these findings, we reasoned that PA treatment could serve as a substitute for CYP73 gene inactivation, allowing to investigate C4H function in a wider range of streptophyte species. We therefore subjected the fern Ceratopteris richardii (tracheophyte), the hornwort Anthoceros agrestis (bryophyte) and the charophyte Klebsormidium nitens to PA treatment. K. nitens has no CYP73 gene and thus virtually lacks PA target enzyme (Fig. 1B; Appendix Tab. S1, Fig. EV1). PA treatment led to developmental changes in C. richardii, where sporophyte growth was retarded, and A. agrestis, where thallus expansion was inhibited and associated with rounded and thickened edges (Fig. 6A). In contrast, no visible deviation in K. nitens development was observed after two weeks in PA-supplemented medium (Fig. 6A). We then investigated the metabolic consequences of PA treatment in the five streptophyte species. UHPLC-UV fingerprints of crude metabolic extracts revealed a consistent decrease in the major UV-absorbing peaks in embryophyte chromatograms (i.e., C. richardii, P. patens, M. polymorpha, and A. agrestis) (Fig. 6B), indicating that phenylpropanoid biosynthesis was impaired. K. nitens was notably devoid of molecules absorbing at 320 nm under the tested conditions, supporting the absence of a phenylpropanoid pathway in this charophyte algae (Fig. 6B). We confirmed UV fingerprint data by the targeted analysis of the two diagnostic phenylpropanoid compounds p-coumaroyl-5-O-shikimate and caffeoyl-5-O-shikimate. This analysis revealed a significant decrease in both molecules in embryophytes after PA treatment, except in M. polymorpha where p-coumaroyl-5-O-shikimate level was unchanged (Fig. 6C). The latter observation may result from an aspecific signal since p-coumaric acid, the corresponding free hydroxycinnamic acid, was significantly decreased in saponified extracts of PA-treated M. polymorpha plants (Appendix Tab. S2). Neither p-coumaroyl-5-O-shikimate nor caffeoyl-5-O-shikimate was detected in the metabolic extracts of K. nitens (Fig. 6C).
Figure 6. Developmental and metabolic effects of the C4H inhibitor piperonylic acid in streptophytes.
Unless otherwise stated, wild-type plants were treated with 50 µM piperonylic acid (PA) or corresponding mock (0.05% DMSO) on agar plates. C. richardii (fern) was treated for three weeks after transfer of 2-week-old sporophytes. P. patens was treated for one month after transfer of individual gametophores. M. polymorpha was treated for three weeks after transfer of individual gemmae. A. agrestis was treated for two months after transfer of thallus pieces. K. nitens (charophyte, Klebsormidiophyceae) was treated in liquid cultures for 2 weeks after subculturing (1:10 ratio). (A) Pictures of plants after PA or mock treatment. (B) Representative UHPLC-UV chromatograms of crude metabolic extracts of mock- and PA-treated plants. P. patens metabolic analysis was performed on plants grown in liquid culture to facilitate tissue collection. (C) Relative quantification of shikimate phenolic esters in mock- and PA-treated plants. C5S, caffeoyl-5-O-shikimate; pC5S, p-coumaroyl-5-O-shikimate. Results are the mean ± SEM of three independent biological replicates for each condition. Mock versus PA unpaired t test adjusted P-value: *P < 0.05; **P < 0.01; ***P < 0.001. nd, not detected. Source data are available online for this figure.
Discussion
Based on both computational and experimental data, our updated evolutionary analysis provides evidence that C4H activity emerged in an embryophyte progenitor together with the CYP73 gene family. These findings resonate with previous studies showing that potential CYP73 orthologs are exclusive to embryophytes (Renault et al, 2017b; de Vries et al, 2017, 2021). Whether this evolutionary scenario will stand valid in light of upcoming, new genomic data from streptophyte algae remains an open question. Nonetheless, the evolutionary pattern of the CYP73 family diverges from that of other genes within the phenylpropanoid pathway, ranging from PAL to CYP98, for which distant homology is evident in charophytes (de Vries et al, 2017, 2021; Renault et al, 2019). The seemingly sudden and delayed emergence of the CYP73 gene family thus raises questions about the evolutionary path and mechanisms that gave birth to this gene family. It also suggests that CYP73 emergence was instrumental in founding the canonical phenylpropanoid pathway in an embryophyte ancestor, in line with the often observed contribution of new CYP families to creating novel metabolic pathways (Mizutani and Ohta, 2010; Hansen et al, 2021). Moreover, previous molecular evolution analysis underscored the consistent trend of strong purifying selection acting on CYP73 genes across both tracheophytes and bryophytes, hinting at the fact they fulfilled essential function from the early stages of land plant evolution (Renault et al, 2017b).
Together with data from the study of the Arabidopsis CYP73A5 gene (Schilmiller et al, 2009), our functional analysis of bryophyte CYP73 genes establishes the C4H step as a critical and evolutionarily conserved element of the canonical phenylpropanoid pathway. The recruitment and fixation of C4H within embryophytes delineates an intriguing facet, particularly when considering the potential of some plant enzymes to establish an alternative route to p-coumaric acid. A prominent example are the bifunctional phenylalanine/tyrosine ammonia-lyases (PTAL), which derive from to the ubiquitous PAL enzyme family. In the monocot Brachypodium distachyon, PTAL bypasses C4H and contributes to nearly half of lignin production through L-tyrosine (Barros et al, 2016). However, PTAL are mainly restricted to grasses, and B. distachyon retained three CYP73 genes that encode functional C4H enzymes (Renault et al, 2017b). The precise benefits of positioning and conserving C4H as a central module in the phenylpropanoid pathway is elusive. Exploration of catalytic properties revealed the high affinity (Km < 10 µM) and relatively slow turnover rates of C4H enzymes across various species (Renault et al, 2017b; Nedelkina et al, 1999; Pierrel et al, 1994; Wu et al, 2018). Furthermore, it was observed that C4H substrate preference is constrained, being capable of using only t-cinnamic acid mimics as substrates (Schalk et al, 1998, 1997; Chen et al, 2007; Pierrel et al, 1994). Thus, the catalytic attributes of C4H potentially prevent metabolic derailment that could result from enzyme promiscuity, while concurrently offering the potential for efficient control over metabolic flux. This perspective is further supported by the documented role of the ER membrane-bound C4H as a nucleation point for a phenylpropanoid metabolon (Bassard et al, 2012; Achnine et al, 2004), facilitating the recruitment of other CYP and soluble enzymes within close proximity for the effective and dynamic channeling of metabolic intermediates.
Our work demonstrates that perturbing C4H function, either via CYP73 gene inactivation or inhibitor treatment, had a significant impact on bryophyte development. This aligns with the findings obtained in the tracheophyte Arabidopsis, where interfering with the five first steps of the phenylpropanoid pathway consistently led to lignin alteration and growth reduction (Schilmiller et al, 2009; Franke et al, 2002; Hoffmann et al, 2004; Huang et al, 2010; Li et al, 2015). Four hypotheses were proposed to explain this phenomenon, ranging from impaired water transport due to xylem collapse, negative feedback through a cell wall surveillance system, and over- or under-accumulation of phenylpropanoid derivatives (reviewed in Muro-Villanueva et al, 2019). Whether the developmental alterations we observed in bryophytes, which lack lignified vascular tissues, falls under some of these hypotheses is an interesting question. It’s worth noting that the impact of impeding C4H function in bryophytes extends beyond mere growth reduction. It profoundly alters the developmental plan, leading for instance in P. patens to phyllid expansion failure and organ fusion symptoms, as observed in mutants of downstream steps (Renault et al, 2017a; Kriegshauser et al, 2021). Interestingly, this strong developmental phenotype could be mitigated by supplying exogenous p-coumaric acid. This suggests that the observed phenotypes were unlikely due to hyperaccumulation of t-cinnamic acid derivatives. As proposed for other P. patens phenylpropanoid mutants (Renault et al, 2017a; Kriegshauser et al, 2021), phenylpropanoid shortage might account for the developmental anomalies in C4H-impaired bryophytes. This hypothesis could be linked to cuticle defects, which might function, in absence of lignin and suberin, as a structural element in bryophytes, contributing to tissue scaffolding. This perspective does not necessarily contradict the two remaining hypotheses—feedback control through cell wall monitoring system or impaired water transport. However, the latter hypothesis appears less substantiated, as neither growing P. patens ΔPpCYP73 mutants in liquid culture nor embedding them in agarose medium could rescue their stunted phenotype. Further investigations will be necessary to elucidate the factors driving the developmental consequences of C4H deficiency in bryophytes and to determine whether these mechanisms are conserved across embryophytes.
Methods
Homolog search and phylogenetic reconstruction
Search for CYP73 homologs involved a reciprocal best hits (RBH) method with AtCYP73A5 protein sequence as the initial query (see Appendix Tab. S1 for RBH output). Briefly, we conducted a forward BLASTp search across 20 Viridiplantae genomes. Next, for each species, the top hit based on the bit-score was used to perform a reverse BLASTp search against the Arabidopsis thaliana genome. The presence of a potential AtCYP73A5 homolog was considered positive when AtCYP73A5 was identified as the best hit in the reverse search. We updated our previously published CYP73 phylogeny (Renault et al, 2017b) with 72 new sequences retrieved from recently released genomes or through an extended search in the 1kP transcriptome dataset (One Thousand Plant Transcriptomes Initiative, 2019), resulting in a total of 275 CYP73 sequences. Additional sequences were retrieved from P. patens, A. thaliana and charophyte genomes by tBLASTn search using AtCYP73A5 protein as query. Applying a bit-score cut-off >100, this search identified 271 CYP sequences, which served as outgroups for subsequent phylogenies. Multiple sequence alignment were performed on protein sequences using the MUSCLE algorithm (Edgar, 2004). Ambiguous regions in obtained alignments were masked using Gblocks (Castresana, 2000) implemented via Seaview (Gouy et al, 2010). For nucleotide phylogenies, protein alignments were reverse-translated to their initial nucleotide sequence prior to phylogenetic analysis (full alignments available in Dataset EV1 and Dataset EV2). Maximum-likelihood phylogenies were reconstructed from the Gblocks alignments using IQ-TREE2 v2.2.2.6 (Minh et al, 2020). The reconstruction employed the following command line: “./iqtree2 -s Gblock_alignment.fst --alrt 1000 -B 1000”, which implements ModelFinder for model selection and computes SH-aLRT tests and ultrafast bootstrap with 1000 pseudo-replicates to determine branch support (tree files available in Datasets EV3, EV4 and EV5). Phylogenetic trees were visualized and edited in FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) or iTOL v6 (https://itol.embl.de), and finally annotated with Affinity Designer.
Homology modeling and docking experiments
Three-dimensional structure of transmembrane segment-free PpCYP73A48 protein (residues 46–517) was built using the SWISS-MODEL server homology modeling pipeline (Waterhouse et al, 2018). Both BLAST and HHblits were used to search for the best template in SWISS-MODEL library. Models were built based on the target-template alignment using ProMod3 v3.1.1 and Sorghum bicolor C4H1 structure (pdb: 6VBY) as template. The resulting structure of PpCYP73A48 exhibited a good structural alignment with SbC4H1, with a global QMEANDisCo score of 0.87 ± 0.05. Heme prosthetic group was first docked into reconstructed PpCYP73A48 3D model using Autodock Vina (Trott and Olson, 2010) in flexible mode, allowing geometry of cysteine 459 to change. Resulting heme-containing PpCYP73A48 3D model (pdb file available in Dataset EV6) was subsequently used for t-cinnamic acid docking via Autodock Vina in rigid mode. A 40 Å × 40 Å × 40 Å search box positioned above the heme was used. Ligand and receptor files required for docking experiments were prepared with AutodockTools 1.5.7 (https://ccsb.scripps.edu/mgltools/). Docking results were visualized with ChimeraX software (Pettersen et al, 2021).
Generation of yeast expression plasmids
Coding sequences (CDS) of Physcomitrium patens CYP73A48 (Pp3c4_21680V3.1), CYP73A49 (Pp3c25_10190V3.1) and CYP73A51 (Pp3c3_17840V3.1) were PCR-amplified from P. patens Gransden cDNA and cloned into the yeast expression plasmid pYeDP60 by restriction/ligation using BamHI/KpnI (CYP73A48 and CYP73A51) or SmaI/KpnI (CYP73A49) restriction sites. Marchantia polymorpha CYP73A1 (Mp6g00020.1) coding sequence was PCR-amplified from M. polymorpha Tak-1 cDNA using Gateway-compatible primers. Yeast codon-optimized Phaeoceros carolinianus CYP73A (PcCYP73A; RXRQ_scaffold_2140006) and native Klebsormidium nitens kfl00038_0230 coding sequences were synthesized as double-strand DNA fragments (gBlock, Integrated DNA Technologies) containing Gateway attB1 and attB2 extensions (primer sequences available in Appendix Tab. S3). After generation of pENTRY plasmids by BP recombination, MpCYP73A1, PcCYP73A, and kfl00038_0230 CDS were shuttled to a Gateway version of pYeDP60 by LR recombination. For 3xHA tagging of CYP73 proteins, STOP codon-free CDS were PCR amplified using Gateway-compatible primers and cloned into pDONR207 by BP recombination. CDS were transferred to a modified pAG425GAL-ccdB-3xHA yeast expression plasmid (Alberti et al, 2007), in which the LEU2 auxotrophic marker was replaced by ADE2. Plasmids containing yeast-optimized CDS of Brachypodium distachyon CYP73A92 and CYP73A94 were described previously (Renault et al, 2017b). For expression of CYP73 mutated proteins, site-directed mutagenesis was performed on pYeDP60 plasmids by overlapping PCR, using primers containing the desired mutations. To reduce background originating from methylated template plasmids, PCR reactions were treated with DpnI prior to E. coli transformation.
Production of recombinant CYP73 proteins in yeast
Yeast expression plasmids harboring CYP73 CDS were introduced into the Saccharomyces cerevisiae WAT11 strain (Urban et al, 1997). Procedures for recombinant CYP expression and microsome purification were the same as in Liu et al (2016). Total microsomal protein concentration was determined according to the Qubit™ protein assay kit (ThermoFisher Scientific). Recombinant CYP proteins were quantified by type II differential spectrophotometry using reduced cytochrome P450 and carbon monoxide (Guengerich et al, 2009). Expression of CYP73-3xHA tagged proteins was analyzed by Western blot. To this end, microsomal proteins were denatured for 5 min at 65 °C in Laemmli buffer containing DTT as reducing agent. Proteins were quantified by the amido black protein assay with bovine serum albumin as reference. Ten micrograms of denatured microsomal proteins were separated by SDS-PAGE using 10% polyacrylamide gel (Mini-PROTEAN TGX precast gel, Biorad) and tris/glycine buffer. After electrophoresis, proteins were transferred on Immobilon-P PVDF membrane. HA-tagged proteins were detected using an anti-HA antibody (1/10,000 dilution; Sigma #H9658) followed by incubation with a peroxidase-conjugated goat anti-mouse IgG antibody (1/5000 dilution; Invitrogen # G21234) and finally revealed by chemiluminescence (Clarity Western ECL, Biorad) on a Fusion-FX imager (Vilber).
In vitro enzyme assays
Standard assay for cinnamic acid 4-hydroxylase activity was performed in a 100 µL reaction containing 50 mM potassium phosphate buffer (pH 7.4), 150 µg microsomal proteins (~5 µl microsomal preparations), 100 µM trans-cinnamic acid (from a 10 mM stock solution prepared in DMSO) and 500 µM NADPH. The reaction was initiated by addition of NADPH and incubated at 28 °C in the dark for 30 min. Reaction was stopped by the addition of 100 µl methanol followed by thorough agitation. Samples were centrifuged for 10 min at 15,000 × g, 4 °C to pellet microsomes. Supernatant was recovered and used for UHPLC-MS/MS analysis. For in vitro assay with mutated CYP73 enzymes, same procedures were followed except that 5 pmoles P450 enzyme per assay were used, and samples were analyzed by HPLC-UV. The t-cinnamic acid binding properties of CYP73 recombinant proteins were assayed spectrophotometrically by monitoring the low-spin to high-spin transition upon substrate binding in the active site (i.e., type I spectra). To this end, absorbance difference in the 380–500 nm range was measured with 150 nM recombinant CYP73 proteins and 100 µM t-cinnamic acid.
Plant material and growth conditions
We used in this study Physcomitrium patens Gransden (Rensing et al, 2008), Marchantia polymorpha Tak-1 (Bowman et al, 2017), Anthoceros agrestis Bonn (Li et al, 2020), Ceratopteris richardii Hn-n (Marchant et al, 2022) and Klebsormidium nitens NIES-2285 (Hori et al, 2014) strains. The Arabidopsis cyp73a5-1 mutant was previously characterized (Renault et al, 2017b). P. patens and A. agrestis were grown axenically in Knop medium as described before (Renault et al, 2017a). M. polymorpha was grown axenically in half-strength Gamborg B5 medium (Duchefa, #G0209) containing 0.5 g/L MES as pH buffering agent. Gamborg B5 medium was adjusted to pH 5.5 with KOH and solidified with 12 g/L agar (#4807, Roth). C. richardii plants were grown axenically from spores to sporophytes on C-medium agar plates as previously described (Hickok and Warne, 1998). K. nitens was grown in half-strength Gamborg B5 liquid medium containing 0.5 g/L HEPES (pH adjusted to 7.0 with KOH). Agar plate cultures were performed in 93 × 21 mm petri plates filled with 30 mL of medium. Liquid cultures were performed in 500 mL Erlenmeyer flasks filled with 200 mL liquid medium and sealed with C-type Silicosen® stoppers. Liquid cultures were constantly agitated at 130 rpm on an orbital shaker. Arabidopsis was grown on soil in 7 cm square pots. Plants were kept under a 22/18 °C, 16 h/8 h light/dark regime. Light was provided by 20W/840 white cool LED tubes (Philips) at an intensity of 50 µmol/m2/s for bryophyte and fern species, and 100 µmol/m2/s for Arabidopsis.
Generation of P. patens transgenic lines
ΔPpCYP73s knock-out mutants were generated via homologous recombination-mediated gene disruption as described previously (Kriegshauser et al, 2021). CYP73A48 and CYP73A49 disruption constructs were excised from vector backbone by BamHI and KpnI digestion, respectively. The ΔPpCYP73A48/CYP73A49 double mutants were produced by disrupting the PpCYP73A49 gene in the ΔPpCYP73A48 #23 mutant background. For uidA reporter lines, two genomic regions for homologous recombination framing the STOP codon were PCR-amplified from genomic DNA and assembled with the uidA reporter gene following the same procedures as in Kriegshauser et al (2021). The CYP73A48:uidA and CYP73A49:uidA constructs were excised from vector backbone by NheI digestion. 25 µg of excised fragment were used for protoplasts transfection. Since CYP73:uidA constructs do not contain a selection marker, it was co-transfected with the pRT101 plasmid containing the NPTII selection cassette (Girke et al, 1998). Transformants were selected on Knop plates supplemented with 25 mg/L geneticin (ΔPpCYP73A48 mutants and PpCYP73A:uidA lines) or 10 mg/L Hygromycin B (ΔPpCYP73A49 mutants). Transgenic lines were molecularly characterized as described previously (Kriegshauser et al, 2021).
Arabidopsis cyp73a5-1 mutant trans-complementation
Coding sequences of CYP73A48, CYP73A49, CYP73A51, and CYP73A5 were transferred by LR recombination into the Gateway pCC1061 binary vector, which contains a 2977 bp promoter fragment from Arabidopsis CYP73A5 gene (Weng et al, 2011). Obtained expression plasmids were introduced into Agrobacterium tumefaciens C58c1 strain and used to transform heterozygous cyp73a5-1 plants by the floral dip method (Clough and Bent, 1998). Transformants were selected based on their resistance to kanamycin. T1 plants homozygous for the cyp73a5-1 allele were identified by PCR (primer sequences available in Appendix Tab. S3) and further confirmed by the full resistance of T2 progeny to sulfadiazine. Experiments were performed with T3 plants homozygous for both the mutant allele and the trans-complementation construct.
GUS staining
Plant tissues were vacuum infiltrated during 10 min with a GUS solution containing 50 mM potassium phosphate buffer pH 7.0, 0.5 mM ferrocyanide, 0.5 mM ferricyanide, 0.1% Triton X-100 and 0.5 mg/mL X-Gluc, and incubated at 37 °C for 4.5 h. Chlorophyll was removed by washing tissues three times in 70% ethanol. For gametophore stem cross-section, cleared gametophores in 70% ethanol were embedded in Paraplast embedding medium (Electron Microscope Sciences). 25 µm sections were prepared with a Leica RM2155 microtome and were imaged on a Leica DMRB microscope.
Chemical complementation with p-coumaric acid
Protoplasts from P. patens wild type and ΔPpCYP73A48/CYP73A49 mutant were embedded in low-melting point agarose as described before (Wiedemann et al, 2018). After three days, regeneration solution that overlaid the solidified film was changed to Knop medium supplemented 50 µM p-coumaric acid. Mock treatment was performed by supplementing Knop medium with 0.1% ethanol. Regeneration of protoplasts was performed in standard growth conditions and monitored over five weeks.
Soluble metabolite extraction and analysis
Tissue collection, lyophilization and grinding were performed as described before (Kriegshauser et al, 2021). Metabolites were extracted from lyophilized plant material using a methanol:chloroform:water protocol as described previously (Kriegshauser et al, 2021). Briefly, 500 µl methanol were added to 10 mg lyophilized plant material. Samples were agitated for 1 h at 1500 rpm at room temperature prior to addition of 250 µl chloroform. After agitation for 5 min, phase separation was induced by addition of 500 µl water followed by vigorous agitation and centrifugation (15,000 × g, 4 °C, 15 min). Supernatants were recovered and constituted the crude metabolic extracts. To release free hydroxycinnamic acid (HCAA) from corresponding soluble esters, crude metabolic extracts were saponified. To this end, 200 µl of extract were dried in vacuo, resuspended in 200 µl 1 M NaOH and incubated for 2 h at 30 °C under 1000 rpm agitation. NaOH was neutralized with 33.3 µl 6 M HCl and HCAA were extracted twice with 1 mL ethyl acetate. Pooled organic phases were dried in vacuo and resuspended in 200 µl 50% methanol prior to analysis. Unless otherwise stated, metabolites were separated and detected on a Dionex UltiMate 3000 UHPLC (ThermoFisher Scientific) system coupled to an EvoQ Elite LC-TQ (Bruker) mass spectrometer as reported before (Kriegshauser et al, 2021). Molecules were ionized in positive or negative mode via a heated electrospray ionization source (HESI, Bruker) and detected by specific multiple reaction monitoring (MRM) methods (Appendix Tab. S4). Concurrently to MS/MS data acquisition, UV-absorbance profiles were recorded with an Ultimate 3000 photodiode array detector operated in the 200–400 nm range (ThermoFisher Scientific). UV data were analyzed with Compass Data Analysis software (Bruker). In the case of mutated CYP73 enzyme assays, metabolites were analyzed by HPLC-UV on a Alliance 2695 chromatographic system (Waters) coupled to a photodiode array detector (PDA 2996; Waters) as described before (Kriegshauser et al, 2021).
Determination of cuticular polymer composition
P. patens cuticular polymer composition, including glycerol, was determined from 2-month-old, lyophilized gametophores grown in liquid culture as previously reported (Philippe et al, 2016). For M. polymorpha, same procedures were followed except that heptadecanoic acid (C17:0) and ribitol were used as internal standards. M. polymorpha samples were analyzed using gas chromatography (GC; Agilent GC8890) coupled with a time-of-flight mass spectrometer (TOFMS; Leco Pegasus BT2). One microliter of derivatized monomers was injected on a HP-5MS Ultra Inert column (30 m × 0.25 mm × 0.25 µm; Agilent) in 100:1 split mode. The temperature gradient was set as follows: 120 °C for 1 min, 120 °C to 340 °C at 10 °C/min, and 340 °C for 3 min. Helium was used as the carrier gas at a flow rate of 1 mL/min. Transfer line and source temperatures were maintained at 250 °C. Analytes were ionized and fragmented by electronic impact at 70 eV. Data were acquired over a m/z 45–500 mass range with a frequency of 8 spectra/s. Compounds were identified according to two orthogonal criteria: spectral similarity score (>800) and retention index (±20). After identification and peak area integration, compound quantification was carried out using the internal standards (ribitol, 5TMS and C17:0, 1TMS) and response factors obtained from the analysis of authentic standards (Appendix Tab. S5). When standards were unavailable, response factors from structurally-related molecules were used.
Tissue permeability assay
P. patens tissue permeability was probed by immersing gametophores for 30 s in a 0.05% toluidine blue solution containing 2% tween20. Care was taken to remove rhizoids prior to staining assay and not to immerse the cut area. Gametophores were then abundantly rinsed with distilled water. For each genotype or replicate, ten gametophores were individually processed and subsequently pooled. Sample pigments were extracted in 400 µl of buffer (200 mM Tris-HCl, 250 mM NaCl, 25 mM EDTA) with two 3 mm steel balls operated at 30 Hz for 5 min. Following addition of 800 µl ethanol, samples were vortexed and centrifuged for 15 min at 18,000 × g. Absorbance of supernatants at 626 nm (A626) and 430 nm (A430) were recorded; A626/A430 ratio was used to quantify toluidine blue levels in plant tissue.
Gene expression analysis by RT-qPCR
Total RNA was extracted from 5-week-old, lyophilized P. patens gametophores and M. polymorpha thalli using TriReagent (Sigma) and was subsequently treated with RQ1 DNAse (Promega). 150 ng (P. patens) or 1 µg (M. polymorpha) of DNase-treated total RNA was retro-transcribed with the SuperScript IV enzyme (ThermoFisher Scientific) and an oligo(dT)18, following the manufacturers’ instructions. qPCR was performed in a 384-well plate, in a reaction volume of 10 µL containing 1 µl (P. patens) or 0.2 µl (M. polymorpha) RT reaction, 0.5 µM of each primer, and 5 µL of Master mix SYBR Green I (Roche). Reactions were run in triplicates using a LightCycler® 480 II (Roche) with the following program: 95 °C for 10 min, followed by 40 amplification cycles [95 °C for 10 s (denaturation) – 60 °C for 15 s (hybridization) – 72 °C for 15 s (extension)], and a melting curve analysis from 55 to 95 °C to verify primer specificity. Crossing points (Cp) were determined using manufacturer’s software and corrected with primer pair efficiency. The expression levels of P. patens CYP73 genes were normalized to the expression levels of PpTRX1 (Pp3c19_1800) and PpSEC15 (Pp3c27_3270); the expression levels of M. polymorpha CYP73 genes were normalized to the expression levels of MpACT7 (Mp6g11010) and MpEF1 (Mp3g2340).
M. polymorpha CRISPR/Cas9-mediated genome editing
Mutant alleles of M. polymorpha CYP73A1 were generated by CRISPR/Cas9 following the procedures described in Sugano et al (2018). Briefly, two independent protospacer sequences in CYP73A1 first exon were identified using the CRISPOR tool (Concordet and Haeussler, 2018). Selected protospacers started with a G for proper U6 promoter-driven expression and had a specificity score of 100. Double-stranded protospacer fragments were reconstituted by hybridization of two complementary oligonucleotides and were BsaI-cloned into the Gateway pMpGE-En03 vector that contains MpU6-1 promoter and gRNA scaffold (Sugano et al, 2018). MpU6pro-gRNA expression cassettes were subsequently transferred by LR recombination into the binary pMpGE011 vector, which contains a SpCas9 expression cassette and allows chlorsulfuron-based selection of plant transformants (Sugano et al, 2018). Recombined pMpGE011 vectors were introduced into A. tumefaciens GV3101 strain and served for the transformation of M. polymorpha Tak-1 accession according to the thallus method (Kubota et al, 2013). Transformants were selected on half-strength Gamborg B5 medium supplemented with 0.5 µM chlorsulfuron. Genome editing was checked in transformants by Sanger sequencing of PCR amplicons, using primers designed by the CRISPOR tool. Edited lines were further propagated from a single gemma to avoid mosaicism and confirm mutant allele.
Piperonylic acid treatment
Standard agar and liquid growth media were supplemented with 50 µM piperonylic acid (PA) using 100 mM stock solution prepared in DMSO. Mock medium was prepared by supplementing growth media with 0.05% DMSO. PA treatment was initiated by transferring wild-type C. richardii sporophytes, P. patens gametophores, M. polymorpha gemmae and A. agrestis thallus pieces in PA and mock media. PA treatment of K. nitens was initiated by inoculating 180 mL PA-supplemented liquid culture with 20 mL standard pre-culture (1:10 final ratio).
Statistical analysis
All statistical analyses were performed with the GraphPad v10 software. Unless otherwise stated, mean comparisons of wild type vs mutants or control vs treatment were performed using unpaired Student t-tests with P-values correction for multiple comparisons according to the Holm-Šídák method. Data for M. polymorpha thallus area and gemmae cup number were analyzed via one-way ANOVA with P-values correction for multiple comparisons according to the Dunnett method.
Supplementary information
Acknowledgements
HR received support from the initiative of excellence IDEX Unistra (ANR-10-IDEX-0002-02) and the Agence Nationale de la Recherche (ANR-19-CE20-0017). HR is grateful to the “Région Grand Est - Fonds Régional de Coopération pour la Recherche 2019” (VitEst project) for co-funding the GC-TOFMS equipment. SK and LK were supported by PhD fellowships from the Ministère de l’Enseignement supérieur et de la Recherche. KT was supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science. RR acknowledges support from the German Research Foundation DFG under Germany’s Excellence Strategy (EXC-2189, CIBSS). HR and RR acknowledge the support of the Freiburg Institute of Advanced Studies FRIAS and the University of Strasbourg Institute of Advanced Study USIAS to the METABEVO project. Anthoceros agrestis Bonn and Ceratopteris richardii Hn.n strains were kindly provided by Péter Szövényi (University of Zurich) and Andrew Plackett (University of Birmingham), respectively. We acknowledge the IBMP Gene Expression Analysis, Plant Imaging and Mass Spectrometry and Plant Production core facilities for their technical assistance.
Expanded view
Author contributions
Samuel Knosp: Conceptualization; Formal analysis; Investigation; Writing—review and editing. Lucie Kriegshauser: Investigation; Writing—review and editing. Kanade Tatsumi: Formal analysis; Investigation; Writing—review and editing. Ludivine Malherbe: Investigation. Mathieu Erhardt: Investigation. Gertrud Wiedemann: Formal analysis; Investigation; Writing—review and editing. Bénédicte Bakan: Formal analysis; Investigation; Writing—review and editing. Takayuki Kohchi: Supervision; Funding acquisition; Methodology; Writing—review and editing. Ralf Reski: Supervision; Funding acquisition; Methodology; Writing—review and editing. Hugues Renault: Conceptualization; Data curation; Formal analysis; Supervision; Funding acquisition; Investigation; Visualization; Writing—original draft; Project administration.
Source data underlying figure panels in this paper may have individual authorship assigned. Where available, figure panel/source data authorship is listed in the following database record: biostudies:S-SCDT-10_1038-S44318-024-00181-7.
Data availability
All the data produced in this study are available in supplementary materials as Source Data and Dataset files. Materials described in this study are available upon request. This study includes no data deposited in external repositories.
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44318-024-00181-7.
Disclosure and competing interests statement
The authors declare no competing interests.
Supplementary information
Expanded view data, supplementary information, appendices are available for this paper at 10.1038/s44318-024-00181-7.
References
- Achnine L, Blancaflor EB, Rasmussen S, Dixon RA (2004) Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16:3098–3109 10.1105/tpc.104.024406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Gitler AD, Lindquist S (2007) A suite of Gateway® cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24:913–919 10.1002/yea.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On YM, Phillips R, Milo R (2018) The biomass distribution on Earth. Proc Natl Acad Sci USA 115:6506–6511 10.1073/pnas.1711842115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros J, Dixon RA (2019) Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci 25:66–79 10.1016/j.tplants.2019.09.011 [DOI] [PubMed] [Google Scholar]
- Barros J, Serrani-Yarce JC, Chen F, Baxter D, Venables BJ, Dixon RA (2016) Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat Plants 2:16050 10.1038/nplants.2016.50 [DOI] [PubMed] [Google Scholar]
- Bassard J-E, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W et al (2012) Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24:4465–4482 10.1105/tpc.112.102566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berland H, Albert NW, Stavland A, Jordheim M, McGhie TK, Zhou Y, Zhang H, Deroles SC, Schwinn KE, Jordan BR et al (2019) Auronidins are a previously unreported class of flavonoid pigments that challenges when anthocyanin biosynthesis evolved in plants. Proc Natl Acad Sci USA 116:20232–20239 10.1073/pnas.1912741116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546 10.1146/annurev.arplant.54.031902.134938 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F et al (2017) Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171:287–304 10.1016/j.cell.2017.09.030 [DOI] [PubMed] [Google Scholar]
- Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chen H, Jiang H, Morgan JA (2007) Non-natural cinnamic acid derivatives as substrates of cinnamate 4-hydroxylase. Phytochemistry 68:306–311 10.1016/j.phytochem.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Cheng S, Xian W, Fu Y, Marin B, Keller J, Wu T, Sun W, Li X, Xu Y, Zhang Y et al (2019) Genomes of subaerial zygnematophyceae provide insights into land plant evolution. Cell 179:1057–1067.e14 10.1016/j.cell.2019.10.019 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Concordet J-P, Haeussler M (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46:W242–W245 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, de Vries S, Slamovits CH, Rose LE, Archibald JM (2017) How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae? Plant Cell Physiol 58:934–945 10.1093/pcp/pcx037 [DOI] [PubMed] [Google Scholar]
- de Vries S, Fürst-Jansen JMR, Irisarri I, Dhabalia Ashok A, Ischebeck T, Feussner K, Abreu IN, Petersen M, Feussner I, de Vries J (2021) The evolution of the phenylpropanoid pathway entailed pronounced radiations and divergences of enzyme families. Plant J 107:975–1002 10.1111/tpj.15387 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30:47–59 10.1046/j.1365-313X.2002.01267.x [DOI] [PubMed] [Google Scholar]
- Girke T, Schmidt H, Zahringer U, Reski R, Heinz E (1998) Identification of a novel delta 6-acyl-group desaturase by targeted gene disruption in Physcomitrella patens. Plant J 15:39–48 10.1046/j.1365-313X.1998.00178.x [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Martin MV, Sohl CD, Cheng Q (2009) Measurement of cytochrome P450 and NADPH–cytochrome P450 reductase. Nat Protoc 4:1245–1251 10.1038/nprot.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Bak S (2013) Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci 368:20120426 10.1098/rstb.2012.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CC, Nelson DR, Møller BL, Werck-Reichhart D (2021) Plant cytochrome P450 plasticity and evolution. Mol Plant 14:1244–1265 10.1016/j.molp.2021.06.028 [DOI] [PubMed] [Google Scholar]
- Hickok LG, Warne TR (1998) C-Fern Manual. Carolina Biological Supply Company, Burlington, VT
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxycinnamoy-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16:1446–1465 10.1105/tpc.020297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N et al (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5:3978 10.1038/ncomms4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houari IE, Beirs CV, Arents HE, Han H, Chanoca A, Opdenacker D, Pollier J, Storme V, Steenackers W, Quareshy M et al (2021) Seedling developmental defects upon blocking CINNAMATE-4-HYDROXYLASE are caused by perturbations in auxin transport. New Phytol 230:2275–2291 10.1111/nph.17349 [DOI] [PubMed] [Google Scholar]
- Hu R, Li X, Hu Y, Zhang R, Lv Q, Zhang M, Sheng X, Zhao F, Chen Z, Ding Y et al (2023) Adaptive evolution of the enigmatic Takakia now facing climate change in Tibet. Cell 186:3558–3576 10.1016/j.cell.2023.07.003 [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y-H, Yu J-Q, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153:1526–1538 10.1104/pp.110.157370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegshauser L, Knosp S, Grienenberger E, Tatsumi K, Gütle DD, Sørensen I, Herrgott L, Zumsteg J, Rose JKC, Reski R et al (2021) Function of the HYDROXYCINNAMOYL-CoA:SHIKIMATE HYDROXYCINNAMOYL TRANSFERASE is evolutionarily conserved in embryophytes. Plant Cell 33:1472–1491 10.1093/plcell/koab044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A, Ishizaki K, Hosaka M, Kohchi T (2013) Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci Biotechnol Biochem 77:167–172 10.1271/bbb.120700 [DOI] [PubMed] [Google Scholar]
- Li FW, Nishiyama T, Waller M, Frangedakis E, Keller J, Li Z, Fernandez-Pozo N, Barker MS, Bennett T, Blázquez MA et al (2020) Anthoceros genomes illuminate the origin of land plants and the unique biology of hornworts. Nat Plants 6:259–272 10.1038/s41477-020-0618-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kim JI, Pysh L, Chapple C (2015) Four isoforms of Arabidopsis 4-Coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol 169:2409–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tavares R, Forsythe ES, Andre F, Lugan R, Jonasson G, Boutet-Mercey S, Tohge T, Beilstein MA, Werck-Reichhart D et al (2016) Evolutionary interplay between sister cytochrome P450 genes shapes plasticity in plant metabolism. Nat Commun 7:13026 10.1038/ncomms13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant DB, Chen G, Cai S, Chen F, Schafran P, Jenkins J, Shu S, Plott C, Webber J, Lovell JT et al (2022) Dynamic genome evolution in a model fern. Nat Plants 8:1038–1051 10.1038/s41477-022-01226-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61:291–315 10.1146/annurev-arplant-042809-112305 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ohta D, Sato R (1997) Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol 113:755–763 10.1104/pp.113.3.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, Wellman CH, Yang Z, Schneider H, Donoghue PCJ (2018) The timescale of early land plant evolution. Proc Natl Acad Sci USA 115:201719588 10.1073/pnas.1719588115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Villanueva F, Mao X, Chapple C (2019) Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr Opin Biotechnol 56:202–208 10.1016/j.copbio.2018.12.008 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109:10101–10106 10.1073/pnas.1205726109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelkina S, Jupe SC, Blee KA, Schalk M, Werck-Reichhart D, Bolwell GP (1999) Novel characteristics and regulation of a divergent cinnamate 4-hydroxylase (CYP73A15) from French bean: engineering expression in yeast. Plant Mol Biol 39:1079–1090 10.1023/A:1006156216654 [DOI] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679–685 10.1038/s41586-019-1693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE (2021) UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe G, Gaillard C, Petit J, Geneix N, Dalgalarrondo M, Bres C, Mauxion J-P, Franke R, Rothan C, Schreiber L et al (2016) Ester cross-link profiling of the cutin polymer of wild-type and cutin synthase tomato mutants highlights different mechanisms of polymerization. Plant Physiol 170:807–20 10.1104/pp.15.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel MA, Batard Y, Kazmaier M, Mignotte-Vieux C, Durst F, Werck-Reichhart D (1994) Catalytic properties of the plant cytochrome P450 CYP73 expressed in yeast. Substrate specificity of a cinnamate hydroxylase. Eur J Biochem 224:835–844 10.1111/j.1432-1033.1994.00835.x [DOI] [PubMed] [Google Scholar]
- Proost S, Mutwil M (2018) CoNekT: an open-source framework for comparative genomic and transcriptomic network analyses. Nucleic Acids Res 46:W133–W140 10.1093/nar/gky336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, Alber AV, Horst NA, Basilio Lopes A, Fich EA, Kriegshauser L, Wiedemann G, Ullmann P, Herrgott L, Erhardt M et al (2017a) A phenol-enriched cuticle is ancestral to lignin evolution in land plants. Nat Commun 8:14713 10.1038/ncomms14713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, De Marothy M, Jonasson G, Lara P, Nelson DR, Nilsson I, Andre F, von Heijne G, Werck-Reichhart D (2017b) Gene duplication leads to altered membrane topology of a cytochrome P450 enzyme in seed plants. Mol Biol Evol 34:2041–2056 10.1093/molbev/msx160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, Werck-Reichhart D, Weng J-K (2019) Harnessing lignin evolution for biotechnological applications. Curr Opin Biotechnol 56:105–111 10.1016/j.copbio.2018.10.011 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319:64–69 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- Schalk M, Batard Y, Seyer A, Nedelkina S, Durst F, Werck-Reichhart D (1997) Design of fluorescent substrates and potent inhibitors of CYP73As, P450s that catalyze 4-hydroxylation of cinnamic acid in higher plants. Biochemistry 36:15253–15261 10.1021/bi971575k [DOI] [PubMed] [Google Scholar]
- Schalk M, Cabello-Hurtado F, Pierrel MA, Atanossova R, Saindrenan P, Werck-Reichhart D (1998) Piperonylic acid, a selective, mechanism-based inactivator of the trans-cinnamate 4-hydroxylase: a new tool to control the flux of metabolites in the phenylpropanoid pathway. Plant Physiol 118:209–218 10.1104/pp.118.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60:771–782 10.1111/j.1365-313X.2009.03996.x [DOI] [PubMed] [Google Scholar]
- Sugano SS, Nishihama R, Shirakawa M, Takagi J, Matsuda Y, Ishida S, Shimada T, Hara-Nishimura I, Osakabe K, Kohchi T (2018) Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS ONE 13:e0205117 10.1371/journal.pone.0205117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272:19176–19186 10.1074/jbc.272.31.19176 [DOI] [PubMed] [Google Scholar]
- Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20 10.1093/mp/ssp106 [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Akiyama T, Ralph J, Chapple C (2011) Independent recruitment of an O-methyltransferase for syringyl lignin biosynthesis in Selaginella moellendorffii. Plant Cell 23:2708–2724 10.1105/tpc.110.081547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Chapple C (2010) The origin and evolution of lignin biosynthesis. New Phytol 187:273–285 10.1111/j.1469-8137.2010.03327.x [DOI] [PubMed] [Google Scholar]
- Wiedemann G, van Gessel N, Köchl F, Hunn L, Schulze K, Maloukh L, Nogué F, Decker EL, Hartung F, Reski R (2018) RecQ helicases function in development, DNA repair, and gene targeting in Physcomitrella patens. Plant Cell 30:717–736 10.1105/tpc.17.00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y-F, Zhao Y, Liu X-Y, Gao S, Cheng A-X, Lou H-X (2018) Isolation and functional characterization of hydroxycinnamoyltransferases from the liverworts Plagiochasma appendiculatum and Marchantia paleacea. Plant Physiol Biochem 129:400–410 10.1016/j.plaphy.2018.06.019 [DOI] [PubMed] [Google Scholar]
- Zhang B, Lewis KM, Abril A, Davydov DR, Vermerris W, Sattler SE, Kang C (2020) Structure and function of the cytochrome P450 monooxygenase cinnamate 4-hydroxylase from Sorghum bicolor. Plant Physiol 183:957–973 10.1104/pp.20.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data produced in this study are available in supplementary materials as Source Data and Dataset files. Materials described in this study are available upon request. This study includes no data deposited in external repositories.
The source data of this paper are collected in the following database record: biostudies:S-SCDT-10_1038-S44318-024-00181-7.