Abstract
Murine bone marrow stromal cells differentiate not only into mesodermal derivatives, such as osteocytes, chondrocytes, adipocytes, skeletal myocytes, and cardiomyocytes, but also into neuroectodermal cells in vitro. Human bone marrow stromal cells are easy to isolate but difficult to study because of their limited life span. To overcome this problem, we attempted to prolong the life span of bone marrow stromal cells and investigated whether bone marrow stromal cells modified with bmi-1, hTERT, E6, and E7 retained their differentiated capability, or multipotency. In this study, we demonstrated that the life span of bone marrow stromal cells derived from a 91-year-old donor could be extended and that the stromal cells with an extended life span differentiated into neuronal cells in vitro. We examined the neuronally differentiated cells morphologically, physiologically, and biologically and compared the gene profiles of undifferentiated and differentiated cells. The neuronally differentiated cells exhibited characteristics similar to those of midbrain neuronal progenitors. Thus, the results of this study support the possible use of autologous-cell graft systems to treat central nervous system diseases in geriatric patients.
Murine and human bone marrow stromal cells differentiate into osteoblasts (2), chondrocytes (13), skeletal myocytes, adipocytes, and cardiomyocytes (24) in vitro and thus are a useful cell source for bone regeneration (26) and in vivo cardiovasculogenesis (11). However, recent studies suggest that bone marrow stromal cells can also differentiate into a neuronal lineage (22), and murine bone marrow-derived multipotent adult progenitor cells differentiate into dopaminergic neuronal cells (16). Since the use of bone marrow stromal cells entails no ethical or immunological problems, and bone marrow aspiration is an established routine procedure, they may be a useful source of cells for transplantation.
Large numbers of cells may be necessary for repairing damaged human tissues to restore function. However, there have been no reports of a sufficient number of differentiated neurons ever having been obtained from human marrow stromal cells. One reason is that normal human cells undergo a limited number of divisions in culture and then enter a nondividing state referred to as “senescence.” Senescence is classified into two categories: “stress-induced premature senescence,” or “telomere-independent senescence,” and “replicative senescence,” or “telomere-dependent senescence” (3, 5, 38). p16Ink4a (p16), a cyclin-dependent kinase (CDK) inhibitor, is induced by certain oncogenes and other damage or stress signals and is required for “premature senescence” in human mammary epithelial cells and keratinocytes. p16 inhibits dephosphorylation of pRb by Cdk4/6-cyclin D, and hypophosphorylated pRb actively represses the genes required for the S phase by sequestering the E2F transcription factors. “Replicative senescence” is caused by telomere size reduction during successive cell divisions because of the chromosome end replication problem. Ectopic expression of telomerase alone bypasses replicative senescence in certain cell types, such as human foreskin fibroblasts.
To obtain enough human cells to restore the function of failing organs and to establish a model of cell therapy, the life span of human marrow stromal cells was extended by infecting them with retrovirus encoding human telomerase reverse transcriptase (hTERT) and the human papillomavirus E6 and E7 genes. Both p16/Rb inactivation by E7 and telomerase activation by E6 are required to extend the life span of human mammary epithelial cells (21) and umbilical cord blood-derived cells (36). E6 also accelerates the degradation of p53, which induces the CDK inhibitor p21 (35). In contrast to foreskin fibroblasts (5), however, the increase in telomerase activity as a result of the introduction of hTERT is insufficient to prolong the life span of marrow stromal cells (27).
The protocols for in vitro differentiation into neuronal cells include the use of a demethylating agent and/or the neurotrophic cytokines, such as nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), and neurotrophin 3. Basic fibroblast growth factor (bFGF) activates essential neuronal transcription factors, such as Hes-1 (10), in neural precursor cells and embryonic stem cells (19, 31). The Notch-Hes1 pathway plays an essential role in inhibition of neuronal differentiation (17), and the B27 supplement is effective in achieving long-term viability of primary neurons in culture (7). Taking these findings into consideration, a simple protocol for neural transdifferentiation was developed in this study.
The first purpose of this study was to determine whether prolonging the cell life span with cell cycle-associated molecules would inhibit neurogenic differentiation of marrow stromal cells in vitro. The second purpose was to determine if transdifferentiation of marrow cells of mesodermal origin to neurogenic cells was accompanied by global changes in gene expression or only leaky expression of some neurogenic markers. The in vitro differentiation process appears to be highly specific, and the life span of marrow-derived stromal cells can be extended by retrovirus-mediated transfer of the bmi-1 gene, which reduces expression of p16 (14, 15, 37), stimulates cell proliferation (9), and is required for maintenance of self-renewing hematopoietic stem cells (29).
MATERIALS AND METHODS
Isolation and cell culture.
After signed informed consent was obtained, bone marrow cells were harvested from a 91-year-old human female donor with the approval (approval numbers 13-1 and 12-1) of the Ethics Committee of Keio University School of Medicine. The cells were resuspended in growth medium (MSCGM, PT-3238, and PT-4105; Cambrex Bio Science Walkersville, Inc., Walkersville, MD) and cultured as previously described (1, 13). Several bone marrow stromal cell strains, designated H4-1, H4-2, and H4-3, were generated from primary or first-passage cells using the limiting dilution method. The cells were cultured for further experiments under the approval (approval numbers 25 and 49) of the Ethics Committee of the National Research Institute for Child Health and Development, Tokyo, Japan.
Infection with recombinant retroviruses.
The cells were prepared for infection with recombinant retroviruses carrying the bmi-1, E6, E7, and hTERT genes, as previously described (1). Stably transduced cells with an expanded life span were designated UBT-5, UBE6T-6, UBE6T-7, UE7T-9, UE6E7T-11, UE6E7T-12, UE7T-13, UBT-15, and UE6E7-16 cells.
Neuronal differentiation of bone marrow stromal cells.
Cells removed from the flask bottom were replated onto a coverslip coated with laminin-polylysine (no. 354455; Becton Dickinson BioScience) in MSCGM. One day after passage, the medium was replaced with B27-supplemented Dulbecco's modified Eagle's medium-F12 (Gibco, BRL) containing 20 ng/ml of BDNF (R&D), 10 ng/ml of bFGF (R&D), and 50 ng/ml of NGF (Invitrogen) for neuroectodermal differentiation. Bone marrow stem cells were processed for immunocytochemistry and reverse transcription (RT)-PCR 7 to 21 days after induction.
RT-PCR.
Total RNA was prepared from cultured cells with Isogen (Nippon Gene, Tokyo, Japan). Human neuronal RNA was purchased (human total brain RNA; lot 2110667; Becton Dickinson BioScience). RNA for RT-PCR was converted to cDNA with a First-strand cDNA Synthesis kit (Amersham Pharmacia Biotech) according to the manufacturer's recommendations. The following PCR primer sets were used for neuron-associated genes: nestin mRNA, sense (5′-AGAGGGGAATTCCTGGAG-3′) and antisense (5′-CTGAGGACCAGGACTCTCTA-3′); NF-M mRNA, sense (5′-TGAGCTACACGTTGGACTCG-3′) and antisense (5′-TCTCCGCCTCAATCTCCTTA-3′); notch-I mRNA, sense (5′-TCACGCTGACGGAGTACAAG-3′) and antisense (5′-CCACACTCGTTGACATCCTG-3′); Sox-2 mRNA, sense (5′-CACAACTCGGAGATCAGCAA-3′) and antisense (5′-GTTCATGTGCGCGTAACTGT-3′); neuronal cell adhesive molecule (N-CAM) mRNA, sense (5′-TCCATCACCTGGAGGACTTC-3′) and antisense (5′-CTCCAGATAGCTGGCAGAGG-3′); MAP-2 mRNA, sense (5′-GGATTCTGGCAGCAGTTCTC-3′) and antisense (5′-TCCTTGCAGACACCTCCTCT-3′); tubulin-beta III mRNA, sense (5′-ACCTCAACCACCTGGTATCG-3′) and antisense (5′-TGCTGTTCTTGCTCTGGATG-3′); and Nur-related factor-1 (Nurr1) mRNA, sense (5′-TTTCTGCCTTCTCCTGCATT-3′) and antisense (5′-GTGGCACCAAGTCTTCCAA-3′); 18S mRNA sense (5′-GTGGAGCGATTTGTCTGGTT-3′) and antisense (5′-CGCTGAGCCAGTCAGTGTAG-3′) were used as a positive control. PCR was performed with TaKaRa Z-Taq (TAKARA SHUZO Co., Ltd.) for 30 cycles, with each cycle consisting of 98°C for 5 s, 68°C or 60°C for 1 s, and 72°C for 10 s, with an additional 30-s incubation at 72°C after completion of the final cycle.
Western blot analysis.
To detect p16, p53, p21, p27, Rb, Bmi-1, and actin, immunoblotting was performed as previously described (30) with antibodies against p16 (G3-245; BD Pharmingen, San Diego, CA), Rb (G174-405; BD Pharmingen), p53 (DO-1), p21 (Ab-1; Oncogene Science, Boston, MA), Bmi-1 (monoclonal antibody generated by T. Kiyono), p27 (BD Pharmingen), and actin (Santa Cruz Biotechnology Inc., Santa Cruz, CA).
Telomere length assay.
Genomic DNA was extracted from cultured cells. Restriction enzyme digestion of genomic DNA was carried out with HinfI and RsaI. The fragments obtained were resolved on 0.7% agarose gels, transferred to a Hybond N membrane (Amersham, United Kingdom), and hybridized with digoxigenin-labeled (TTAGGG)3 probe. The membrane was then incubated with anti-digoxigenin alkaline phosphatase, and detection was performed with a chemiluminescence solution. The size range and intensity were determined with X-ray film.
Telomerase activity analysis.
Telomerase activity was determined with a telomere repeat amplification protocol (TRAP) assay kit, Telo TAGGG telomerase PCR ELISA plus (Roche, Indianapolis, IN), according to the manufacturer's instructions.
G-banding karyotypic analysis.
Metaphase spreads were prepared from cells treated with Colcemid (Karyo Max; Gibco BRL; 100 ng/ml for 6 h). We performed a standard G-banding karyotypic analysis on at least 50 metaphase spreads for each population.
SKY analysis.
Spectral karyotyping (SKY) was performed on metaphase-transduced cells (UBE6T-7, UE6E7T12, and UE7T13) in 80 population doublings (PDs) according to the kit manufacturer's instructions (ASI, Carlsbad, CA) and a previously published method (34).
Immunocytochemical analysis.
Immunocytochemical analysis was performed as previously described (30) with antibodies to MAP-2 (Zymed, San Francisco, CA), tubulin 3 (Sigma, St. Louis, Missouri), GFAP (DAKO, Denmark), Nurr1 (N-20; Santa Cruz), and nestin (Biogenesis, United Kingdom) in phosphate-buffered saline containing 1% bovine serum albumin. As a methodological control, the primary antibody was omitted. After being washed in phosphate-buffered saline, the slides were incubated with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin antibody or phycoerythrin (PE)-conjugated anti-rabbit immunoglobulin antibody (DAKO, Denmark).
GeneChip expression analysis.
Human genomewide gene expression was examined with the Human Genome U133A Probe array (GeneChip; Affymetrix), which contains the oligonucleotide probe set for approximately 23,000 full-length genes and expressed sequence tags, according to the manufacturer's protocol (Expression Analysis technical manual and GeneChip Small Sample Target Labeling Assay version 2 technical note [http://www.affymetrix.com/support/technical/index.affx]). Total RNA was isolated with an RNeasy minikit (QIAGEN, Chatsworth, CA). Double-stranded cDNA was synthesized, and the cDNA was subjected to in vitro transcription in the presence of biotinylated nucleoside triphosphates. The biotinylated cRNA was hybridized with a probe array for 16 h at 45°C, and the hybridized biotinylated cRNA was stained with streptavidin-PE and scanned with a Hewlett-Packard Gene Array Scanner. The fluorescence intensity of each probe was quantified by using the GeneChip Analysis Suite 5.0 computer program (Affymetrix). The expression level of a single mRNA was determined as the average fluorescence intensity among the intensities obtained with 11 paired (perfectly matched and single-nucleotide-mismatched) probes consisting of 25-mer oligonucleotides. If the intensities of mismatched probes were very high, gene expression was judged to be absent, even if high average fluorescence was obtained with the GeneChip Analysis Suite 5.0 program. The level of gene expression was determined with the GeneChip software as the average difference (AD). Specific AD levels were then calculated as percentages of the mean AD level of six probe sets for housekeeping genes (actin and GAPDH [glyceraldehyde-3-phosphate dehydrogenase] genes). Further data analysis was performed with Genespring software version 5 (Silicon Genetics, San Carlos, CA). To normalize the staining intensity variations among chips, the AD values for all genes on a given chip were divided by the median of all measurements on that chip. To eliminate changes within the range of background noise and to select the most differentially expressed genes, data were used only if the raw data values were less than 100 AD and gene expression was judged to be present by the Affymetrix data analysis. Hierarchical clustering analysis with standard correlation was used to identify gene clusters. The separation ratio was set at 0.5. Normalization values were considered significant when (i) expression changed by at least twofold (activation program) and (ii) increased gene expression included at least one present absolute call (Affymetrix algorithm). Normalized values were averaged for two donors and used for the data analysis, and the normalized values were used to classify the genes into up-regulated or down-regulated. There was a statistically significant correlation between the expression levels of genes of the same cells analyzed twice (r = 0.997). These criteria confirmed the reproducibility of the differences that were seen between different cells under different conditions.
Calcium imaging.
The intracellular Ca2+ concentration was monitored by means of the fluorescent Ca2+ indicator Fluo-3. The cultured cells were exposed to 10 μmol/liter Fluo-3 acetoxymethylester (Molecular Probes, Eugene, Oregon) at 37°C for 30 min and then washed in Tyrode's solution containing (in mmol/liter) NaCl, 140; KCl, 4; MgCl2, 0.5; CaCl2, 1.8; HEPES, 5; and d-glucose, 55 (pH adjusted to 7.4 with NaOH). The signal from the cell was monitored with a fluorescence microscope (BX50WI; Olympus, Tokyo, Japan) and a high-resolution optical mapping system (MiCAM-01; SciMedia Ltd. Tokyo, Japan) at a wavelength of 530 nm and an excitation wavelength of 488 nm. The cells were challenged with a depolarization stimulus in the form of high-potassium (144 mmol/liter) Tyrode's solution (KCl substituted for NaCl) applied with a rapid solution changer (RSC-160; Molecular Kinetics, Indianapolis, Indiana) at a flow rate of 100 μl/s at a distance of 1 mm from the cell at a room temperature of 25°C. The image analysis was performed with Igor Pro software (Wavemetrics, Lake Oswego, OR) and by customized procedures. The background fluorescence intensity was subtracted from the data, and the areas were normalized to the control response to normal Tyrode's solution and shown on a color scale.
Quantitative RT-PCR.
RNA was extracted from cells using the RNeasy kit (QIAGEN, Valencia, CA). Contaminating DNA was eliminated by two sequential DNase (Invitrogen) treatments. An aliquot (1 μg) of total RNA was reverse transcribed by using an oligo(dT) primer. For the thermal cycle reactions, cDNA was amplified (ABI PRISM 7000; Perkin-Elmer Applied Biosystems) using the SYBR RT-PCR kit (Takara Bio, Japan) under the following reaction conditions: 40 cycles of PCR (95°C for 15 seconds and 60°C for 1 min) after an initial denaturation (95°C for 10 min). The controls consisted of amplifications without reverse transcription and reactions without the addition of a cDNA template. The authenticity and sizes of the PCR products were confirmed using a melting curve analysis (using software provided by Perkin-Elmer) and a gel analysis. mRNA levels were normalized using the GAPDH gene as a housekeeping gene. The following primer sets were used: hTERT mRNA, sense (5′-CGGTGTGCACCAACATCTACAAG-3′) and antisense (5′-TCAGAGATGACGCGCAGGA-3′); Bmi-1 mRNA, sense (5′-CGCTTGGCTCGCATTC-3′) and antisense (5′-AGCTCAGTGATCTTGATTCTCGTTG-3′); E6 mRNA, sense (5′-GCACAGAGCTGCAAACAACT-3′) and antisense (5′-CTCACGTCGCAGTAACTGTTG-3′); E7 mRNA, sense (5′-ATGACAGCTCAGAGGAGGAG-3′) and antisense (5′-TCCTAGTGTGCCCATTAACAG-3′); NR4A2/Nurr1 mRNA, sense (5′-TTCGGCAGAGTTGAATGAATG-3′) and antisense (5′-GAAATTAAAGGTGGACAGTGTCGTA-3′); and GAPDH mRNA, sense (5′-CCAGCCGAGCCACATCGCTC-3′) and antisense (5′-ATGAGCCCCAGCCTTCTCCAT-3′).
Nucleotide sequence accession numbers.
The gene chip datasets have been deposited in the GEO database with accession number GSE2110 (GSM38114 to GSM38116).
RESULTS
Human marrow stromal cells with an extended life span.
Bone marrow stromal cells were obtained from a 91-year-old human donor and subcloned by limiting dilution. One subclone of the cells isolated was designated H4-1 cells (Fig. 1). To extend the life span of the H4-1 cells and obtain a large number of cells, five different types of cells were obtained by transducing them with combinations of bmi-1, E6, E7, and/or TERT genes (http://1985.jukuin.keio.ac.jp/umezawa/cells/name.html). We referred to the cells transduced with bmi-1 and TERT as UBT-5 and UBT-15 cells, the cells transduced with bmi-1, E6, and TERT as UBE6T-6 and UBE6T-7 cells, the cells transduced with E7 and TERT as UE7T-9 and UE7T-13 cells, the cells transduced with E6, E7, and TERT as UE6E7T-11 and UE6E7T-12 cells, and the cells transduced with E6 and E7 as UE6E7-16 cells. Random amplified polymorphic DNA analysis revealed that the five different types of stable transduced cells (UBT-5, UBE6T-7, UE6E7T-12, UE7T-13, and UE6E7-16) were definitely derived from H4-1 cells (see Fig. S1 in the supplemental material).
FIG. 1.
In vitro growth of parental and transduced strains of bone marrow stromal cells. (A) The population doublings of UE6E7-16, UE7T-13, UE6E7T-12, UBE6T-7, UBT-5, and H4-1 cells are shown. UE7T-13, UE6E7T-12, UBE6T-7, and UBT-5 cells proliferated for over 60 PDs and for more than 400 days. By contrast, parental H4-1 cells and UE6E7-16 cells stopped growing and entered senescence or the growth arrest stage, which is indicated by crosses. (B) H4-1 and UE6E7-16 cells entered senescence at 44 PDs and 70 PDs, respectively, approximately 200 days after the start of culture. (C) Phase-contrast photomicrographs of the bone marrow stromal cells at the semiconfluent stage in a 40-PD culture. (a) H4-1 cells; (b) UBT-5 cells; (c) UBE6T-7 cells; (d) UE6E7T-12 cells; (e) UE7T-13 cells; (f) UE6E7-16 cells.
The UE6E7T-12, UE6E7T-11, UE7T-9, UE7T-13, UBE6T-6, and UBE6T-7 cells were successfully grown and were shown to have extended life spans (Fig. 1A). The life spans of the UE6E7T-12 and UE7T-13 cells were extended to 200 PDs. The life spans of the UBT-5, UBE6T-7, UE6E7T-11, UE7T-9, and UBE6T-6 cells were extended to 60 PDs. The UBT-5 cells also proliferated for more than 60 PDs and were shown to have an extended life span. However, hTERT alone did not extend the life span (data not shown). Nontransduced parental H4-1 cells reached “senescence” in culture at 44 PDs, and UE6E7-16 cells entered a period of “crisis” in culture at 70 PDs (Fig. 1A and B), implying that E6 and E7 are able to prolong the cell life span but that their prolonging effect is limited. The H4-1 cells exhibited large, flat morphology at 40 PDs, while the other cells were small and spindle shaped (Fig. 1C).
None of the cells exhibited malignant transformation activity: they did not form a focus after confluence in vitro; cells grafted into subcutaneous tissue of immunodeficient mice (nonobese diabetic [NOD]-SCID-interleukin 2 receptor knockout mice) did not form tumors, at least during the observation period (more than 30 days); and the morphologies of all clones remained unchanged for 40 to 200 PDs.
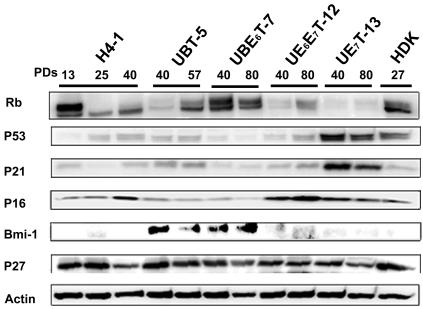
Expression of Bmi-1, Rb, p53, p27, and p21 proteins in transduced cells.
The expression of cell cycle-associated proteins was analyzed in cells transduced with the hTERT and bmi-1, E6, or E7 genes and in parental H4-1 cells (Fig. 2). Human dermal keratinocytes at senescence (PD 27) served as a control for expression of p16Ink4a, hypophosphorylated RB, and p53 proteins at a high level.
FIG. 2.
Time course analysis of cell cycle-associated protein levels in cells with an extended life span. Cell cycle-associated proteins, i.e., Rb, p53, p21, p16, Bmi-1, and p27, in UBT-5, UBE6T-7, UE6E7T-12, UE7T-13, and H4-1 cells were analyzed by Western blotting. Cells cultured for the PDs indicated were assayed. The expression pattern was reproducibly observed in four separate experiments. Expression of actin protein was monitored as a loading control.
Bmi-1 proteins were expressed in the UBT-5 and UBE6T-7 cells, but not in the H4-1, UE6E7-12, and UE7T-13 cells. p16 proteins were down-regulated in the bmi-1-overexpressing cells, i.e., UBT-5 and UBE6T-7 cells. p53 and p21 proteins were down-regulated in E6-overexpressing UBE6T-7 and UE6E7T-12 cells. These results are consistent with the hypothesis that p53 is proteolysed via ubiquitination by E6 (33). Hypophosphorylated Rb was down-regulated in E7-overexpressing cells, probably as a result of proteolysis by E7. There was no significant difference in p27 protein levels between any of the cells tested.
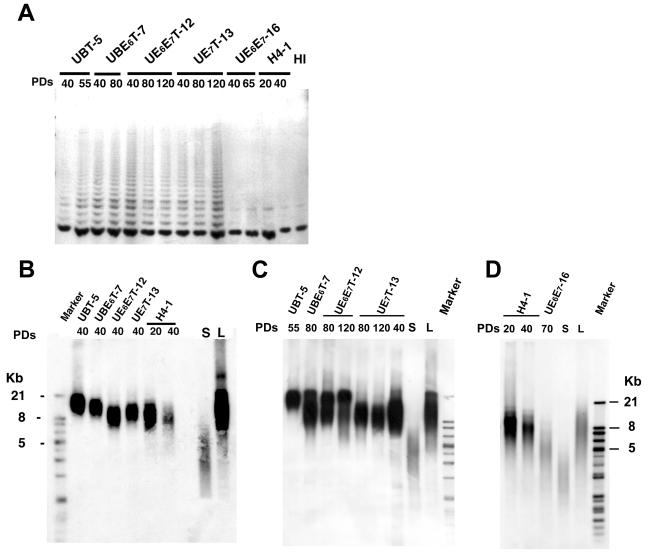
Increase in telomerase activity and maintenance of telomere length in cells transduced with the hTERT gene.
No telomerase activity was detected by the TRAP assay in the parental H4-1 cells at PDs 20 and 40, but telomerase activity was detected in UBT-5, UBE6T-7, UE6E7T-12, and UE7T-13 cells transduced with the hTERT gene at all of the PDs tested. UE6E7-16 cells not transduced with the hTERT gene did not exhibit any telomerase activity at PDs 40 to 120 (Fig. 3A). Likewise, the telomere length of the parental H4-1 cells decreased with the number of PDs, whereas the telomere length of UBT-5, UBE6T-7, UE6E7T-12, and UE7T-13 cells was maintained irrespective of the PD number (Fig. 3B, C, and D).
FIG. 3.
Telomere activities and telomere lengths of the marrow stromal cells transduced with the hTERT and Bmi-1, E6, or E7 genes. (A) Analysis of telomerase activity by PCR assay in UBT-5, UBE6T-7, UE6E7T-12, UE7T-13, UE6E7-16, and H4-1 cells. Telomerase activity is revealed by the characteristic 6-bp ladder of bands. Heat-inactivated cell lysate (HI) was used as a negative control. (B, C, and D) The mean telomere length of the parental and transduced cells was determined at an early stage (B: 20 to 40 PDs) in UBT-5, UBE6T-7, UE6E7T-12, UE7T-13, and H4-1 cells; a late stage (C: 55 to 120 PDs) in UBT-5, UBE6T-7, UE6E7T-12, and UE7T-13 cells; and a senescent stage (D) in H4-1 and UE6E7-16 cells.
Karyotypic analysis of parental and transduced cells with extended life spans.
We performed a karyotypic analysis (G banding and SKY) of both the parental stromal cells (H4-1) and the cells transduced with hTERT and bmi-1, E6, or E7 (see Materials and Methods). SKY permits a detailed analysis of all complex markers and provides insights into their involvement in subsequent rearrangements (34). Our studies raised some interesting points concerning chromosomal abnormality. No genomic abnormalities were found in the parental cells, but an increase in chromosomal number appeared in the other strains, as shown by G-banding and SKY analyses (Table 1 and Fig. 4), except for in the UE7T-13 cells. These abnormalities included translocations, a deletion, other rearrangements, and polyploidy. Surprisingly, we found no genomic changes in the UE7T-13 cells at over 120 PDs.
TABLE 1.
Karyotypic analysis (G banding) in parental cells (H4-1) and clones
| Cells | No. of PDs | No. of cells with indicated no. of chromosomes
|
||||||
|---|---|---|---|---|---|---|---|---|
| 44 | 45 | 46 | 47 | 48 | 49 | 50 | ||
| H4-1 | 40 | 0 | 0 | 50 | 0 | 0 | 0 | 0 |
| UBT-5 | 40 | 2 | 2 | 14 | 11 | 2 | 0 | 0 |
| UBE6T-7 | 40 | 0 | 0 | 32 | 15 | 3 | 0 | 0 |
| UE6E7T-12 | 40 | 0 | 1 | 45 | 2 | 0 | 0 | 0 |
| UE7T-13 | 40 | 0 | 0 | 50 | 0 | 0 | 0 | 0 |
| UE7T-13 | 80 | 0 | 0 | 50 | 0 | 0 | 0 | 0 |
FIG. 4.
Karyotypic analysis of parental and transduced cells with extended life spans. (A, B, C, and D) G-banded karyotyping. (E, F, and G) Spectral karyotyping. Metaphase spreads with structural chromosomal abnormalities were determined after the indicated number of PDs. Normal diploidy was seen in representative parental cells (A: H4-1) and UE7T-13 cells (B and G), but abnormalities were seen in UE6E7T-12 (C and F), UBE6T-7 (E), and UE7T-13 cells (D). Deletion and translocation are indicated by arrowheads and arrows, respectively.
Establishment of a novel protocol for neuroectodermal differentiation of human marrow stromal cells.
Murine stromal cells have been reported to have been transdifferentiated into neuronal lineages (6, 16, 22) by several different protocols. In this study, we devised a simple protocol for neuroectodermal differentiation from mesoderm-derived cells (Fig. 5A). Briefly, growing, adherent stromal cells were trypsinized and pelleted down by centrifugation, and after as much growth medium as possible was aspirated, the pelleted cell aggregates were pipetted with a 20-μl microtip. Each cell aggregate was then replated onto a laminin-coated coverslip in a 7-mm-diameter 24-well culture dish (Fig. 5A), and the growth medium was replaced with B27-supplemented growth medium containing NGF, BDNF, and bFGF. Cells not subjected to this protocol have fibroblast morphology (Fig. 5B, a). The shape of the stromal cells began to change on day 7 after replating, and the cells did not adhere to adjacent cells. The cells then started to exhibit neuron-like morphology that included a refractile round cell body, a neurite-like process(es), and a triangular axon terminal(s) (Fig. 5B, b to d). By days 14 to 21, more than 80% of the bone marrow cells had formed neurite-like processes and exhibited neuron-like morphology. All experiments were repeated at least 10 times using different population doublings (20 to 100 PDs) before using the cells for neuroectoderm differentiation.
FIG. 5.
Neuroectodermal differentiation of bone marrow stromal cells. (A) Scheme of the neuronal differentiation system. (B) Four phase-contrast microscopic images of UE7T-13 cells are shown. (a) Undifferentiated state. (b) Neuronally differentiated cells that had spread out to clusters. (c and d) Higher magnification of neuronally differentiated cells. (C) Fluorescence immunohistochemical analysis of undifferentiated UE7T-13 cells (b, g, l, and p) and neuronally differentiated cells (a, c to f, h to k, n to o, q, and r). (a, b, k, and l) MAP-2; (c) tubulin 3; (d and n) Nurr1. PE-conjugated anti-rabbit immunoglobulin G (IgG) (e) and fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin antibody (m) were used as negative controls. (f, g, h, i, j, o, p, q, and r) Phase-contrast microscopic findings. All images were obtained with a laser scanning confocal microscope. (a to j) UE7T-13 cells; (k to r) H4-1 cells.
Expression of neuron-specific proteins during neuroectodermal differentiation of human marrow stromal cells.
After induction, the UBT-5, UBE6T-7, UE6E7T-12, and UE7T-13 cells became positive for the neuron-specific markers microtuble-associated protein 2 (MAP-2), tubulin 3, and Nurr1 (Fig. 5C, a, c, d, k, and n), implying that they could be induced into neuronal lineages. More than 80% of all the tested cells were positive for MAP-2 and Nurr1, but no staining was detected with antibodies against GFAP (an astrocyte-specific marker) or O4 (an oligodendrocyte-specific marker) (data not shown). Undifferentiated UE7T-13 and H4-1 cells did not exhibit neuroectodermal characteristics (Fig. 5C, b and l).
Gene chip analysis during neuroectodermal differentiation.
To clarify the specific gene expression profile of human marrow stromal cells, we compared the expression levels of approximately 23,000 genes by neuronally differentiated UE7T-13 cells, undifferentiated UE7T-13 cells, and parental H4-1 cells (GEO accession number GSE2110 [http://1954.jukuin.keio.ac.jp/umezawa/chip/mori]) using the Affymetrix gene chip oligonucleotide arrays. Of the approximately 23,000 genes represented on the gene chip, 2,123 genes were up-regulated to more than twice their expression level in undifferentiated cells. Genes whose expression increased or decreased are shown in Table 2. The Nurr1, kynureninase, apolipoprotein, RARRES1, ABC1, rab27B, STC1, WISP2, rhoGAP6, SLC2A5, and palmderphin genes were among the top 0.1% in terms of increase in expression. Surprisingly, expression of Nurr1/NR4A2, which is essential for differentiation of the nigral dopaminergic neurons (23, 32), was up-regulated 26.1-fold. Wnt-5a promotes the acquisition of dopaminergic neurons (8), and its expression was increased 7.1-fold. These findings imply that the characteristics of these cells are similar to those of midbrain neurons. The dendrogram analysis clearly showed that the change in gene expression pattern is dramatic and neuron specific during neuroectodermal differentiation rather than being leaky, subtle, and nonspecific (Fig. 6).
TABLE 2.
Genes regulated under neuronally differentiated conditions
| Probe set | Description | Flag (N/C)a | Normalized valueb | Fold change |
|---|---|---|---|---|
| Increased | ||||
| 204621_s_at | Nurr1 (Nuclear receptor subfamily 4 group A member 2) | P/(A) | 26.1 | 26.1 |
| 210663_s_at | Kynurenidase (l-kyurenine hydrolase) | P/(A) | 60.7 | 9.6 |
| 204430_s_at | Solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | P/(A) | 9.3 | 9.3 |
| 205921_s_at | Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | P/(A) | 6.2 | 6.2 |
| 205156_s_at | ACCN2 (amiloride-sensitive cation channel 2, neuronal) | P/(A) | 4.1 | 3.4 |
| 204846_at | Ceruloplasmin (ferroxidase) | P/(A) | 12.2 | 2.6 |
| 202507_s_at | Synaptosome-associated protein, 25 kDa | P/(A) | 1.5 | 1.5 |
| 205792_at | WNT1-inducible signaling pathway protein 2 | P/(P) | 12.4 | 12.4 |
| 213425_s_at | WNT5A (wingless-type MMTVc integration site family, member 5A) | P/(P) | 4.3 | 7.1 |
| 207594_s_at | Synaptojanin 1 | P/(P) | 3.3 | 6.1 |
| 211806_s_at | Potassium inwardly rectifying channel | P/(A) | 2.5 | 2.5 |
| 211592_s_at | Calcium channel, voltage-dependent, L type | P/(A) | 0.9 | 2.3 |
| 214933_at | Calcium channel, voltage-dependent, P/Q type | P/(A) | 1.0 | 1.4 |
| Decreased | ||||
| 214081_at | Tumor endothelial marker 7 | A/(P) | 0.2 | −19.5 |
| 209287_s_at | Cdc42 effector protein | A/(P) | 0.1 | −11.0 |
| 206898_at | Cadoherin 19, type 2 | A/(P) | 0.9 | −7.1 |
| 205132_at | Actin alpha, cardiac muscle | A/(P) | 0.1 | −7.0 |
| 204736_s_at | Chondroitin 4-sulfotransferase | A/(P) | 0.3 | −5.0 |
| 209652_s_at | Placental growth factor, VEGF-related protein | A/(P) | 1.0 | −3.2 |
| 215177_s_at | Integrin alpha 6 | A/(P) | 0.5 | −2.6 |
N, neuronally differentiated UE7T-13 cells; C, undifferentiated UE7T-13 cells; P, judged to be “present” (expressed) in neuronally differentiated UE7T-13 cells; A, judged to be “absent” (not expressed) in undifferentiated UE7T-13 cells.
The normalized values were calculated from data obtained in UE7T-13 cells as described in Material and Methods.
MMTV, mouse mammary tumor virus.
FIG. 6.
Comparison between the gene profiles of neuroectodermal differentiated cells and undifferentiated cells. Gene expression levels in undifferentiated cells (−) and neuronally differentiated cells (+) are shown by the rows of colored bars that represent one gene. The color bars reflect the magnitude of the response for each gene according to the scale shown on the right. Differentially expressed genes are indicated by a vertical line on the right. The raw data from the gene chip analysis are available at the GEO database with accession number GSE2110 or our laboratory's website (http://1954.jukuin.keio.ac.jp/umezawa/chip/mori).
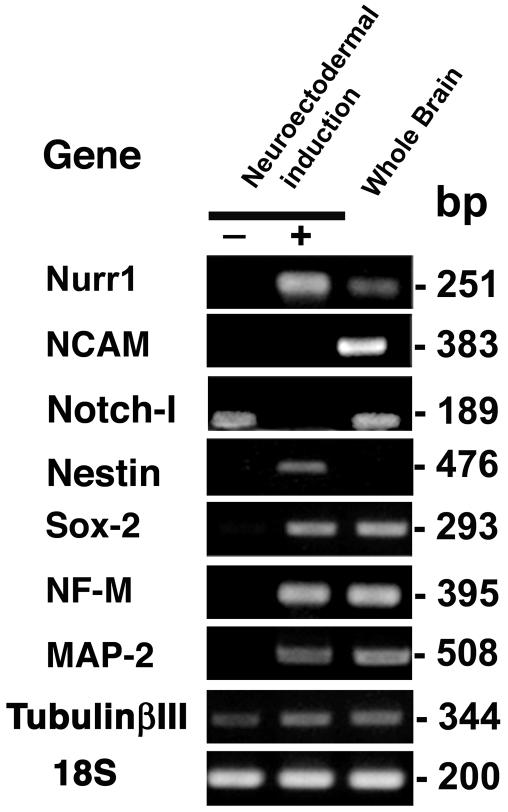
Expression of neuron-specific genes in differentiated UE7T-13 cells.
To confirm the global change in gene expression during neuronal differentiation, RT-PCR was performed with primers that specifically react with human neuronal genes. Differentiated UBT-5, UBE6T-7, UE6E7T-12, UE7T-13, UE6E7-16, and H4-1 cells expressed Nurr1, MAP2, tubulin 3, medium-size neurofilament (NF-M), Sox-2, and nestin, whereas undifferentiated UE7T-13 and H4-1 cells did not express mRNAs for Nurr1, NCAM, nestin, Sox-2, NF-M, or MAP-2 (Fig. 7), implying that these neuron-specific genes began to be expressed in response to the induction protocol (Fig. 5A). Sequence analysis confirmed that the PCR products matched the sequences of the human Nurr1, MAP2, tubulin 3, NF-M, NCAM, Notch-1, Sox-2, and nestin genes. Total brain mRNA was used as a positive control, and expression of all of the genes examined except the nestin gene was observed. The Notch-1 gene, a stem cell-related gene (18), is expressed only in undifferentiated cells and is absent in differentiated cells. Tubulin 3 was detected at low levels in both differentiated and undifferentiated UE7T-13 cells.
FIG. 7.
RT-PCR analysis of expression of neuron-associated genes confirmed the gene chip data. RT-PCR analyses of RNAs from undifferentiated UE7T-13 cells, neuronally differentiated UE7T-13 cells, and human total brain were performed with primers that react with the human genes encoding MAP-2, tubulin 3, NF-M, Notch-1, NCAM, Sox-2, Nestin, and Nurr1. The signals of the 18S gene are approximately the same, indicating an equivalent input for all samples. Most of the neuron-associated genes were expressed by neuronally differentiated UE7T-13 cells and total brain cells.
Rapid and reversible calcium uptake by UE7T-13 cells in response to depolarizing stimuli.
To determine if the neuronal cells derived from UE7T-13 cells are functional, we exposed the cells to depolarizing stimuli and performed calcium imaging analysis. Cells that had a dendrite component and a smooth-surfaced bright round soma body were selected as the targets for calcium-imaging analysis. Rapid and reversible calcium uptake was observed in neuronally differentiated UE7T-13 cells in response to the change in extracellular potassium concentration (Fig. 8), suggesting that the approximately 20% of the cells that showed uptake have both deep resting membrane potentials and voltage-gated Ca2+ channels. The rapid change in extracellular fluid did not cause any stretch-activated channels.
FIG. 8.
Calcium imaging of neuronally differentiated stromal cells. (A) Phase-contrast photomicrograph of neuronally differentiated UE7T-13 cells. (B) The cell indicated by the asterisk in panel A showed rapid and reversible calcium uptake in response to high-potassium stimulation (red line; 115%), but simply changing the extracellular solution to the normal Tyrode's solution did not result in any calcium uptake (control; black line). Images obtained at 10 seconds after the high-potassium stimulation (D) and in the control (C). Rapid and reversible calcium uptake in response to high-potassium stimulation was observed in the four cells within this field.
Quantitative RT-PCR analysis of the bmi-1, TERT, E6, E7, and Nurr1 genes in UBT-5, UBE6T-7, UBE6E7T-12, and UE7T-13 cells with neuroectodermal induction at early and late PDs.
We also performed an additional quantitative RT-PCR experiment to monitor the expression levels of the hTERT, bmi-1, E6, and E7 genes as well as the Nurr1 gene, during the neuronal differentiation of UBT-5, UBE6T-7, UE6E7T-12, and UE7T-13 cells (Table 3). The expression of the hTERT, E6, and E7 genes did not decrease and remained at a high level in neuronally differentiated cells. The bmi-1 gene, on the other hand, was down-regulated. The Nurr-1 gene, a neuron marker, was significantly increased by neural induction, as expected. Although the hTERT, E6, and E7 genes were continuously strongly expressed during differentiation, the phenotype of the cells did not revert to an undifferentiated state and continued to exhibit neuronal phenotypes, even when bFGF, NGF, and BDNF were removed from the culture media.
TABLE 3.
Quantitative RT-PCR analyses of the bmi-1, TERT, E6, E7, and Nurr1 genes in UBT-5, UBE6T-7, UBE6E7T-12, and UE7T-13 cells with neuroectodermal induction at early and late PDs
| Cells/infected genes | No. of PDs | NIa | Expression level of genesb
|
||||
|---|---|---|---|---|---|---|---|
| bmi-1 | TERT | E6 | E7 | Nurr1 | |||
| UBT-5/Bmi1 and TERT | 40 | − | 2.44 | 0.56 | 0.01 | ||
| 75 | − | 2.13 | 0.48 | 0.02 | |||
| 40 | + | 0.88 | 1.93 | 0.90 | |||
| 75 | + | 0.73 | 6.47 | 1.17 | |||
| UBE6T-7/bmi-1, E6, E7, and TERT | 50 | − | 2.31 | 0.04 | 0.61 | 0.02 | |
| 120 | − | 0.55 | 0.29 | 0.21 | 0.02 | ||
| 50 | + | 0.69 | 0.15 | 0.40 | 1.83 | ||
| 120 | + | 0.15 | 1.18 | 0.87 | 2.69 | ||
| UE6E7T-12/E6, E7, and TERT | 33 | − | 0.35 | 0.62 | 0.28 | 0.02 | |
| 130 | − | 0.55 | 0.67 | 0.40 | 0.01 | ||
| 33 | + | 0.62 | 4.05 | 1.49 | 2.91 | ||
| 130 | + | 0.96 | 2.88 | 1.48 | 4.05 | ||
| UE7T-13/E7 and TERT | 33 | − | 0.63 | 1.22 | 0.01 | ||
| 130 | − | 0.49 | 0.87 | 0.01 | |||
| 33 | + | 1.17 | 1.01 | 0.78 | |||
| 130 | + | 2.06 | 3.00 | 8.03 | |||
NI, neuroectodermal induction; +, with NI; −, without NI.
Cells removed from the flask bottom were placed on a coverslip coated with laminin-polylysin. One day after cell passage, the medium was replaced with B27-supplemented Dulbecco's modified Eagle's medium-F12 containing 20 ng/ml of BDNF, 10 ng/ml of bFGF, and 50 ng/ml of NGF to promote neuroectodermal differentiation. RNA was extracted from the cells for quantitative RT-PCR analysis 7 days after induction. The mRNA levels were normalized using the GAPDH gene as a housekeeping gene.
DISCUSSION
Prolongation of the life span of human marrow stromal cells by bmi-1.
This study was undertaken to investigate whether the life span of human marrow stromal cells, a candidate source of cells for therapy, can be prolonged by a cell-cycle associated molecule(s). Introduction of bmi-1, one of the polycomb group genes, clearly down-regulated p16, which is gradually induced as the number of PDs increases, and thus increases dephosphorylated Rb, resulting in the cell's escape from growth arrest or premature senescence, as we intended. This down-regulation of p16 is thought to be mediated via direct binding of bmi-1 to the cis-regulatory element of p16, and thus the binding should be affected by the methylation status or chromatin structure of this cis-regulatory element. At the very least, bmi-1 is functional in terms of decreasing p16 in human stromal cells, and the decrease in p16 leads to escape from premature senescence. This bmi-1 effect on the cell life span has also been reported in human mammary epithelial cells (9), where it occurs via escape from replicative senescence by induction of telomerase activation; however, this effect of bmi-1 on telomerase activation was not detected in our experimental human stromal cell system. In other words, bmi-1 enables human stromal cells to escape from premature senescence but not replicative senescence.
The lack of change in phenotypes of bmi-1-transduced cells, for example, in their differentiation abilities, growth rates, cell surface markers, and gene expression profiles, is rather surprising, because bmi-1 suppresses p16 protein and has been identified as a c-Myc-cooperating gene in murine B- and T-cell lymphomas (15, 37). A critical target of bmi-1 is the Ink4a locus, which encodes p16 and p19 (p14 in humans) (14). Interestingly, bmi-1 is required for maintenance of adult self-renewing hematopoietic stem cells (29) and neural stem cells (25), whereas the simian virus 40 large T antigen oncogene, which prolongs the cell life span, interferes with the differentiation program and transforms cells. The human marrow-derived stromal cells transfected with simian virus 40 large T antigen (12) did not exhibit contact inhibition in vitro and formed tumors within a month when implanted into nude mice (data not shown).
Transdifferentiation of human mesoderm-derived cells into a functional neuronal lineage.
This study was also conducted to determine whether prolongation of the cell life span by hTERT and bmi-1, E6, or E7 would predominate over neurogenic differentiation of marrow stromal cells in vitro. In contrast to our previous study of neurogenic differentiation of immortalized murine marrow-derived stromal cells (22) by demethylating agents, the transdifferentiation of the human stromal cells was limited to neurons, but not astrocytes or oligodendrocytes. This was probably due to the developmentally logical protocol we employed in this study: use of bFGF, NGF, BDNF, and laminin-ornithine coating. The idea for the neuronal differentiation protocol without 5-azacytidine arose from reports of the following: NGF and BDNF support neuron survival and growth (4), and bFGF induces initial differentiation of neural precursor cells and activates transcription factors related to the differentiation of neural precursor cells and embryonic stem cells (19, 31).
The extremely high level of expression of dopaminergic neuron-associated genes, such as nurr1 and wnt5a, in the neuronally differentiated stromal cells, which we accidentally found by the GeneChip analysis and confirmed by RT-PCR, is surprising. wnt5a and nurr1 are involved in the differentiation of midbrain precursors into dopaminergic neurons (20, 39). Although further analysis of functionally differentiated cells is beyond the scope of this study, it is quite interesting that dopaminergic neurons can be generated from marrow-derived stromal cells, since one of the target cells for regenerative medicine is dopaminergic neurons.
Are marrow stromal cells traced back to their default state, i.e., neural lineage, by neurotrophic factors?
The mechanism of the transdifferentiation from marrow stromal cells to neuronal cells remains unresolved. It must be emphasized that the GeneChip analysis showed that the change in gene expression during differentiation is global and drastic: the differentiated cells no longer exhibited the profile of mesenchymal cells or the biphenotypic pattern of neuronal and mesenchymal cells. Contamination of human stromal cell cultures by neural precursor cells is inconceivable, because the cells were subcloned several times after gene transduction and exhibited mesenchymal phenotypes after subcloning. Our previous study of murine stromal cells clearly showed that osteoblasts capable of membranous ossification are likely to differentiate into neuronal lineages but that adipocytes do not (22). The craniofacial membranous bones develop from the neural crest, which is of ectodermal origin. Our finding of in vitro differentiation from mesoderm- to ectoderm-derived cells in this study may be the opposite of the developmental process, i.e., from ectoderm- to mesoderm-derived cells. Development naturally progresses from neural crest cells to terminally differentiated osteoblasts (28). Converting differentiated osteoblasts or marrow stromal cells to neuronal cells, a key future task for any cell-based therapy, would thus oppose the usual direction of cell differentiation. This can now be achieved by exposing stromal cells to neurotrophic factors, at least in vitro.
Are human marrow stromal cells with an extended life span available for cell-based therapy?
Human stromal cells transduced with hTERT and bmi-1, E6, or E7 did not transform according to the classical criteria: they did not generate tumors in immunosuppressed NOD-SCID-interleukin 2 receptor knockout mice, they did not form foci in vitro, and they stopped dividing after confluence (data not shown). However, we cannot rule out the possibility that gene-transduced stromal cells might become tumorigenic in patients several decades after cell therapy. We believe that these gene-modified stromal cells may be used to supply defective enzymes to patients with genetic metabolic diseases, such as neuro-Gaucher disease, Fabry disease, and mucopolysaccharidosis, which have a poor prognosis and are sometimes lethal. The “risk-versus-benefit” balance is essential when applying these gene-modified cells clinically, and the “risk” or “drawback” in this case is transformation of implanted cells.
hTERT is normally expressed in stem cells and in over 90% of human cancers. Ectopic expression of telomerase is sufficient to prevent telomere shortening and can thereby promote indefinite proliferation in human foreskin fibroblasts (5). Several stromal cell strains with extended life spans and maintenance of differentiation capability, such as UBT5, UBE6T7, UE6E7T-12, UE7T-13, and UE6E7-16 cells, were developed based on the above notion. UE6E7-16 cells not transduced with hTERT have a longer life span but enter a “crisis” period in culture at 67 PDs. “Crisis” is the stage in which widespread death occurs in a population of cultured cells or when karyotypic instability develops as a result of fusion of telomere ends, after the cells manage to circumvent senescence or initial blockades. Human cells transduced with bmi-1 or E6 and E7 alone enter a “crisis,” and the cells that spontaneously circumvent premature senescence without gene induction enter replicative senescence. Replicative senescence or crisis may be a tumor suppressor mechanism that avoids the risk of cell transformation after implantation of cells as a source for cell-based therapy.
Even when cells transduced with nononcogenic genes are used for cell-based therapy, the cases of leukemia in SCID patients treated with gene-modified lymphocytes must be taken into account (3). The infected virus genome was integrated into the LMO2 locus in the leukemia cases and resulted in induction of the oncogenic LMO2 gene. Because of these failures, it will take time before gene-modified cells can be used for regenerative medicine. Inhibition of the p16/Rb pathway is sufficient to prolong the life span of cells in cultures of marrow-derived stroma, as shown in this study. The p16/Rb pathway was activated in marrow-derived stromal cells in vitro, the same as in mammary epithelial cells and hepatocytes, but not in foreskin fibroblasts (21). The development of an appropriate culture system without gene transduction to neutralize the p16/Rb braking system, i.e., nonstress medium, will be essential for cell therapy and further experiments.
Supplementary Material
Acknowledgments
We express our sincere thanks to K. Segawa, S. Matsumoto, Y. Okayama, S. Okuyama, and S. Ikeuchi for support throughout the work and to Y. Nakamura, N. Hida, N. Hashimoto, and T. Inomata for providing expert technical assistance. We are grateful to D. A. Galloway (FHCRC, Seattle, Wash.) for pLXSN-16E7 and to Y. Takeuchi (Chester Beatty Laboratories, ICR, United Kingdom) for the FLYA13 cells.
This work was supported in part by a special grant for Advanced Research on Cancer from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan to T.K. and A.U.; a grant from MEXT to A.U.; Health and Labour Sciences Research Grants to A.U.; and a grant from the Organization for Pharmaceutical Safety and Research to A.U.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahn, J. I., K. H. Lee, D. M. Shin, J. W. Shim, J. S. Lee, S. Y. Chang, Y. S. Lee, M. J. Brownstein, and S. H. Lee. 2004. Comprehensive transcriptome analysis of differentiation of embryonic stem cells into midbrain and hindbrain neurons. Dev. Biol. 265:491-501. [DOI] [PubMed] [Google Scholar]
- 2.Allan, E. H., P. W. Ho, A. Umezawa, J. Hata, F. Makishima, M. T. Gillespie, and T. J. Martin. 2003. Differentiation potential of a mouse bone marrow stromal cell line. J. Cell Biochem. 90:158-169. [DOI] [PubMed] [Google Scholar]
- 3.Allsopp, R. C., H. Vaziri, C. Patterson, S. Goldstein, E. V. Younglai, A. B. Futcher, C. W. Greider, and C. B. Harley. 1992. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 89:10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchelor, P. E., G. T. Liberatore, M. J. Porritt, G. A. Donnan, and D. W. Howells. 2000. Inhibition of brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor expression reduces dopaminergic sprouting in the injured striatum. Eur. J. Neurosci. 12:3462-3468. [DOI] [PubMed] [Google Scholar]
- 5.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 6.Brazelton, T. R., F. M. Rossi, G. I. Keshet, and H. M. Blau. 2000. From marrow to brain: expression of neuronal phenotypes in adult mice. Science 290:1775-1779. [DOI] [PubMed] [Google Scholar]
- 7.Brewer, G. J., J. R. Torricelli, E. K. Evege, and P. J. Price. 1993. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci Res. 35:567-576. [DOI] [PubMed] [Google Scholar]
- 8.Castelo-Branco, G., J. Wagner, F. J. Rodriguez, J. Kele, K. Sousa, N. Rawal, H. A. Pasolli, E. Fuchs, J. Kitajewski, and E. Arenas. 2003. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc. Natl. Acad. Sci. USA 100:12747-12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimri, G. P., J. L. Martinez, J. J. Jacobs, P. Keblusek, K. Itahana, M. Van Lohuizen, J. Campisi, D. E. Wazer, and V. Band. 2002. The bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 62:4736-4745. [PubMed] [Google Scholar]
- 10.Faux, C. H., A. M. Turnley, R. Epa, R. Cappai, and P. F. Bartlett. 2001. Interactions between fibroblast growth factors and Notch regulate neuronal differentiation. J. Neurosci. 21:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gojo, S., N. Gojo, Y. Takeda, T. Mori, H. Abe, S. Kyo, J. Hata, and A. Umezawa. 2003. In vivo cardiovasculogenesis by direct injection of isolated adult mesenchymal stem cells. Exp. Cell Res. 288:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 13.Imabayashi, H., T. Mori, S. Gojo, T. Kiyono, T. Sugiyama, R. Irie, T. Isogai, J. Hata, Y. Toyama, and A. Umezawa. 2003. Redifferentiation of dedifferentiated chondrocytes and chondrogenesis of human bone marrow stromal cells via chondrosphere formation with expression profiling by large-scale cDNA analysis. Exp. Cell Res. 288:35-50. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs, J. J., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164-168. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs, J. J., B. Scheijen, J. W. Voncken, K. Kieboom, A. Berns, and M. van Lohuizen. 1999. bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13:2678-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, Y., D. Henderson, M. Blackstad, A. Chen, R. F. Miller, and C. M. Verfaillie. 2003. Neuroectodermal differentiation from mouse multipotent adult progenitor cells. Proc. Natl. Acad. Sci. USA 100(Suppl. 1):1854-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kageyama, R., and T. Ohtsuka. 1999. The Notch-Hes pathway in mammalian neural development. Cell Res. 9:179-188. [DOI] [PubMed] [Google Scholar]
- 18.Kageyama, R., T. Ohtsuka, and K. Tomita. 2000. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cell 10:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Kalyani, A. J., T. Mujtaba, and M. S. Rao. 1999. Expression of EGF receptor and FGF receptor isoforms during neuroepithelial stem cell differentiation. J. Neurobiol. 38:207-224. [PubMed] [Google Scholar]
- 20.Kim, J. H., J. M. Auerbach, J. A. Rodriguez-Gomez, I. Velasco, D. Gavin, N. Lumelsky, S. H. Lee, J. Nguyen, R. Sanchez-Pernaute, K. Bankiewicz, and R. McKay. 2002. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature 418:50-56. [DOI] [PubMed] [Google Scholar]
- 21.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 22.Kohyama, J., H. Abe, T. Shimazaki, A. Koizumi, K. Nakashima, S. Gojo, T. Taga, H. Okano, J. Hata, and A. Umezawa. 2001. Brain from bone: efficient “meta-differentiation” of marrow stroma-derived mature osteoblasts to neurons with Noggin or a demethylating agent. Differentiation 68:235-244. [DOI] [PubMed] [Google Scholar]
- 23.Le, W. D., P. Xu, J. Jankovic, H. Jiang, S. H. Appel, R. G. Smith, and D. K. Vassilatis. 2003. Mutations in NR4A2 associated with familial Parkinson disease. Nat. Genet. 33:85-89. [DOI] [PubMed] [Google Scholar]
- 24.Makino, S., K. Fukuda, S. Miyoshi, F. Konishi, H. Kodama, J. Pan, M. Sano, T. Takahashi, S. Hori, H. Abe, J. Hata, A. Umezawa, and S. Ogawa. 1999. Cardiomyocytes can be generated from marrow stromal cells in vitro. J. Clin. Investig. 103:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molofsky, A. V., R. Pardal, T. Iwashita, I. K. Park, M. F. Clarke, and S. J. Morrison. 2003. bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochi, K., G. Chen, T. Ushida, S. Gojo, K. Segawa, H. Tai, K. Ueno, H. Ohkawa, T. Mori, A. Yamaguchi, Y. Toyama, J. Hata, and A. Umezawa. 2003. Use of isolated mature osteoblasts in abundance acts as desired-shaped bone regeneration in combination with a modified poly-dl-lactic-co-glycolic acid (PLGA)-collagen sponge. J. Cell Physiol. 194:45-53. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto, T., T. Aoyama, T. Nakayama, T. Nakamata, T. Hosaka, K. Nishijo, T. Nakamura, T. Kiyono, and J. Toguchida. 2002. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 295:354-361. [DOI] [PubMed] [Google Scholar]
- 28.Olsen, B. R., A. M. Reginato, and W. Wang. 2000. Bone development. Annu. Rev. Cell Dev. Biol. 16:191-220. [DOI] [PubMed] [Google Scholar]
- 29.Park, I. K., D. Qian, M. Kiel, M. W. Becker, M. Pihalja, I. L. Weissman, S. J. Morrison, and M. F. Clarke. 2003. bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423:302-305. [DOI] [PubMed] [Google Scholar]
- 30.Sano, M., A. Umezawa, H. Abe, A. Akatsuka, S. Nonaka, H. Shimizu, M. Fukuma, and J. Hata. 2001. EAT/mcl-1 expression in the human embryonal carcinoma cells undergoing differentiation or apoptosis. Exp. Cell Res. 266:114-125. [DOI] [PubMed] [Google Scholar]
- 31.Santa-Olalla, J., and L. Covarrubias. 1995. Epidermal growth factor (EGF), transforming growth factor-alpha (TGF-alpha), and basic fibroblast growth factor (bFGF) differentially influence neural precursor cells of mouse embryonic mesencephalon. J. Neurosci. Res. 42:172-183. [DOI] [PubMed] [Google Scholar]
- 32.Satoh, J., and Y. Kuroda. 2002. The constitutive and inducible expression of Nurr1, a key regulator of dopaminergic neuronal differentiation, in human neural and non-neural cell lines. Neuropathology 22:219-232. [DOI] [PubMed] [Google Scholar]
- 33.Scheffner, M., K. Munger, J. M. Huibregtse, and P. M. Howley. 1992. Targeted degradation of the retinoblastoma protein by human papillomavirus E7-E6 fusion proteins. EMBO J. 11:2425-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrock, E., T. Veldman, H. Padilla-Nash, Y. Ning, J. Spurbeck, S. Jalal, L. G. Shaffer, P. Papenhausen, C. Kozma, M. C. Phelan, E. Kjeldsen, S. A. Schonberg, P. O'Brien, L. Biesecker, S. du Manoir, and T. Ried. 1997. Spectral karyotyping refines cytogenetic diagnostics of constitutional chromosomal abnormalities. Hum. Genet. 101:255-262. [DOI] [PubMed] [Google Scholar]
- 35.Sekiguchi, T., and T. Hunter. 1998. Induction of growth arrest and cell death by overexpression of the cyclin-Cdk inhibitor p21 in hamster BHK21 cells. Oncogene 16:369-380. [DOI] [PubMed] [Google Scholar]
- 36.Terai, M., T. Uyama, T. Sugiki, X. K. Li, A. Umezawa, and T. Kiyono. 2005. Immortalization of human fetal cells: the life span of umbilical cord blood-derived cells can be prolonged without manipulating p16INK4a/RB braking pathway. Mol. Biol. Cell 16:1491-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Lohuizen, M., S. Verbeek, B. Scheijen, E. Wientjens, H. van der Gulden, and A. Berns. 1991. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65:737-752. [DOI] [PubMed] [Google Scholar]
- 38.Wright, W. E., and J. W. Shay. 1992. The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol. 27:383-389. [DOI] [PubMed] [Google Scholar]
- 39.Zetterstrom, R. H., L. Solomin, L. Jansson, B. J. Hoffer, L. Olson, and T. Perlmann. 1997. Dopamine neuron agenesis in Nurr1-deficient mice. Science 276:248-250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.