Abstract
The POU transcription factor Oct-3/4 has been shown to be critical for maintaining embryonic stem (ES) cell character. However, the molecular mechanisms underlying its function remain elusive. We have previously shown that among the POU transcription factor family of proteins, Oct-3/4 alone is able to bind to the regulatory region of the UTF1 gene bearing a variant octamer sequence together with Sox-2. Here, we demonstrate using Oct-3/4-Oct-6 chimeras that there is a precise correlation between the ability of proteins to form a complex on the UTF1 enhancer with Sox-2 and the ability to maintain the stem cell state in ES cells. Different chimeric proteins show differential abilities to form a Sox-2-containing complex on the UTF1 regulatory region, with a decrease in efficiency of the complex formation accompanied by a decrease in the level of UTF1 expression and the rate of cell proliferation. Overexpression of UTF1 in these slow-growing cells was able to restore their proliferation rate to wild-type levels. Moreover, UTF1 was also observed to have an effect on teratoma formation. These results suggest a molecular pathway by which Oct-3/4 induces rapid proliferation and tumorigenic properties of ES cells through activation of the UTF1 gene.
Embryonic stem (ES) cells are derived from the inner cell mass of the blastocyst-stage embryo and are capable of growing indefinitely as an established cell line (11, 20). ES cells are pluripotent, meaning that they have the remarkable capability to differentiate into all cell types (7). Unlike other types of stem cells, ES cells are highly proliferative (40), a property that is thought to be involved in maintaining homogeneity. Somatic stem cells, such as hematopoietic stem cells, grow slowly compared to ES cells and do not expand without significant accompanying differentiation (1). Because of these properties, ES cells are regarded as a major potential source of material for future stem cell therapy. However, the molecular mechanisms governing these characteristics are largely unknown. A better understanding at the molecular level of what induces and maintains the stem cell state of ES cells is essential for moving towards the implementation of ES cell-based therapies.
In murine ES cells, cytokine leukemia inhibitory factor is required for the maintenance of stem cell state (38, 48) and functions by activating the transcription factor STAT-3 (21, 28). In addition to STAT-3, a number of other transcription factors, such as Oct-3/4 (25, 29), Sox-2 (3), Nanog (10, 23), and FoxD3 (12), are also essential for maintaining the stem cell state of ES cells. Although the mechanisms by which these factors function are largely unknown, Oct-3/4 is the best characterized among them. Oct-3/4 is essential for maintaining pluripotency of inner cell mass cells of mouse blastocysts (25) and must be expressed at a critical level to preserve the stem cell state (29). When Oct-3/4 levels drop below 50% of the normal level, ES cells dedifferentiate into trophectoderm cells, whereas elevated levels of the factor cause differentiation into primitive endoderm cells. Oct-3/4 contains a DNA binding POU domain as well as two transcriptional activation domains at the amino- and carboxy-terminal ends of the protein (8, 31) that have potent transcription-stimulating activities in ES cells. However, Oct-3/4 mutants lacking both transcriptional activation domains are still able to rescue ES cells in a stem cell state when the protein is fused to the heterologous transactivation domain of Oct-2, although wild-type Oct-2 does not exert any activity (30). These results indicate that the POU-DNA binding domain confers specificity of biological activity, whereas the two transcriptional activation domains simply provide generic transcription-stimulating activities in ES cells.
Thus, to elucidate the molecular basis of Oct-3/4 function in preservation of the stem cell state in ES cells, we focused on the molecular properties of the DNA binding domain. We previously demonstrated through analysis of a regulatory region of the UTF1 gene encoding a transcriptional cofactor specific to pluripotent cells that Oct-3/4 is able to participate in complex formation with Sox-2 on the regulatory region bearing a nonconsensus, variant octamer sequence, 5′-ACTAGCAT-3′ (26, 32). Here, we show that this specific DNA binding activity of Oct-3/4 is indeed important for maintaining the stem cell state in ES cells. Moreover, our data reveal that the Oct-3/4 target gene UTF1 functions to support rapid proliferation of ES cells.
MATERIALS AND METHODS
Culturing ZHBTc4 ES cells.
ZHBTc4 cells in which Oct-3/4 expression can be controlled with the Tet-off system were cultured as described by Niwa et al. (29). These ES cells, which carry zeocin and blasticidin S drug-resistant genes knocked into the Oct-3/4 locus, were usually cultured in the presence of zeocin and blasticidin S to eliminate cells that spontaneously lost stem cell properties. However, the cells shown in Fig. 5C were cultured in the absence of these antibiotics.
FIG. 5.
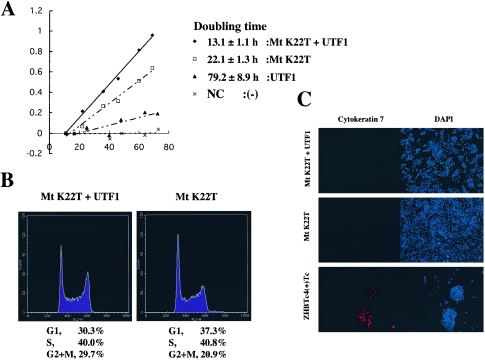
UTF1 supports rapid cell growth of ES cells. (A) The rapid proliferation phenotype characteristic of ES cells was restored to the slow-growing rescued cells by overexpression of UTF1. The UTF1 expression vector (CAG/UTF1/IRES/HIS) or empty vector (CAG/IRES/HIS) was stably integrated into ES cells whose stem cell state was maintained by MtK22T with the aid of histidinol, as described in Materials and Methods. UTF1 and (−) represent differentiated ZHBTc4 cells in which UTF1 and empty vector, respectively, had been stably integrated. After stable selection, these cells were subjected to Tc treatment for 96 h before their cell growth rate was characterized. The cell proliferation rate of these ES cells was measured as with Fig. 4A. NC, not calculable. (B) Examination of the effect of UTF1 on the cell cycle control with FACS analysis. The UTF1-overexpressing and control MtK22T-rescued cells in A were used for FACS analysis. Data shown at the bottom of panels represent averages from five independent experiments. (C) The UTF1-overexpressing and control MtK22T-rescued cells were cultured in the absence of zeocin and blasticidin S, and the numbers of cytokeratin 7-positive cells were compared by immunocytochemistry. The differentiation-induced ZHBTc4 ES cells were also used as a positive control.
Rescue experiments.
Genes encoding chimera proteins and puromycin N-acetyltransferase were placed under the control of the CAG promoter and expressed bicistronically using an internal ribosomal entry site (IRES). The rescue experiments with these expression vectors were performed as described by Niwa et al. (30).
UTF1 overexpression.
For UTF1 overexpression in rescued cells, the mouse UTF1 and histidinol dehydrogenase genes (43) were bicistronically expressed under a CAG promoter using IRES. This construct (pCAG/mUTF1/IRES/HIS) or empty vector (pCAG/IRES/HIS) was electroporated into MtK22T-rescued cells and cultured in selection medium containing 5 mM histidinol (Sigma). The resulting stable transfectants were pooled and expanded for further analysis. The parental ZHBTc4 ES cells were also used for stable integration of the UTF1-expressing or empty vector and then directly injected into nude mice (see Fig. 6A). However, these cells were subjected to tetracycline (Tc) treatment for 96 h before the cell growth rate analysis was performed (see Fig. 5A).
FIG. 6.
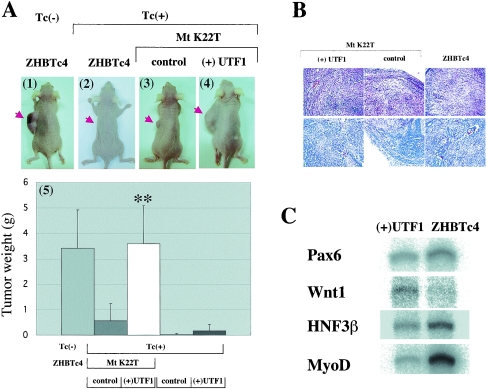
(A) The UTF1 shows a prominent effect on teratoma formation. Panels 1 and 2: injection of ZHBTc4 ES cells into nude mice which had been supplied with Tc-free (panel 1) or Tc-containing (panel 2) drinking water. Panels 3 and 4: tumors in Tc-treated mice derived from injection of MtK22T-rescued ES cells in which empty (panel 3) and UTF1 expression (panel 4) vectors were stably integrated. Panel 5: summary of teratoma formation by the indicated cells. Data shown in the panel were obtained from eight independent injections. The (+)UTF1 and control represent stable integration of UTF1 and empty vectors into the indicated cells, respectively. Double asterisk, P < 0.01 when compared to MtK22T-rescued cells in which the empty vector had been integrated. (B) Histological analysis of teratomas. Teratomas resulting from ES cells containing MtK22T alone, Mt K22T plus UTF1, and the parental ZHBTc4 ES cells were subjected to histological analysis with hematoxylin-eosin staining procedures. (C) Analysis of differentiation marker gene expression in teratomas. RNA was prepared from teratomas derived from UTF1-overexpressing MtK22T-rescued cells and parental ZHBTc4 ES cells, and expression levels of the indicated differentiation marker genes were analyzed by RNase protection.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed as described by Wells and Farnham (45) using polyclonal anti-Oct-3/4 and anti-Oct-6 antibodies or preimmune sera, which had been generated as described previously (32). In brief, chromatin DNA cross-linked with 4% formaldehyde was sonicated to an average length of 600 bp and then incubated with anti-Oct-3/4 or anti-Oct-6 antibodies or preimmune sera at 4°C for 16 h. Antibody complexes were recovered with protein A Sepharose (Amersham Bioscience) and washed twice with buffer A (50 mM Tris-HCl [pH 8.0], 2 mM EDTA, 0.2% Sarkosyl) and four times with buffer B (0.1 M Tris-HCl [pH 9.0], 0.5 M LiCl, 1% NP-40, 1% deoxycholic acid). The antibody complexes were then eluted with 50 mM NaHCO3 and 1% sodium dodecyl sulfate, and the eluted DNA was de-cross-linked at 67°C for 16 h. Subsequently, DNA was treated with proteinase K, extracted with phenol chloroform, and ethanol precipitated. The purified DNA samples were used as templates for PCRs with specific primers. Primers used are as follows: UTF1 enhancer, 5′-AATACTCTGGGCCGTGTGAA-3′ and 5′-TCCACACCACAAGTCAGTAC-3′; FGF-4 enhancer, 5′-TTAGCTCGCTTCAGGGAATG-3′ and 5′-CCAACACTCTTGGAGCCTA-3′; β-actin promoter, 5′-AGTGACTCTCTGTCCATTCA-3′ and 5′-TAGGCCCAGATGTACAGGAA-3′. For the experiments with anti-Oct-6 antibody, we stably introduced an Oct-6 expression vector into ES cells to bring its expression to a level comparative to that of Oct-3/4, since ES cells express very low levels of endogenous Oct-6.
EMSA.
cDNAs encoding chimeric proteins and Sox-2 were subcloned into the pCEP4 expression vector (Clontech) and introduced into HeLa cells by transient transfection. The whole-cell extract preparation and subsequent electrophoretic mobility shift assay (EMSA) analysis were done as described previously (26). The probe sequences used for EMSA were 5′-ggcacttcACTAGCATAACAATGagggctc-3′ (WT) and 5′-ggcacttc ATTAGCATAACAA TGagggctc-3′ (consensus) (uppercase indicates key nucleotides for interaction with DNA binding proteins, and flanking sequences are in lowercase). The sequence used for the analyses shown in Fig. 4C, panel 3, was 5′-cttcACTAGCATag-3′, which bears the variant octamer sequence but lacks the Sox binding site.
FIG. 4.
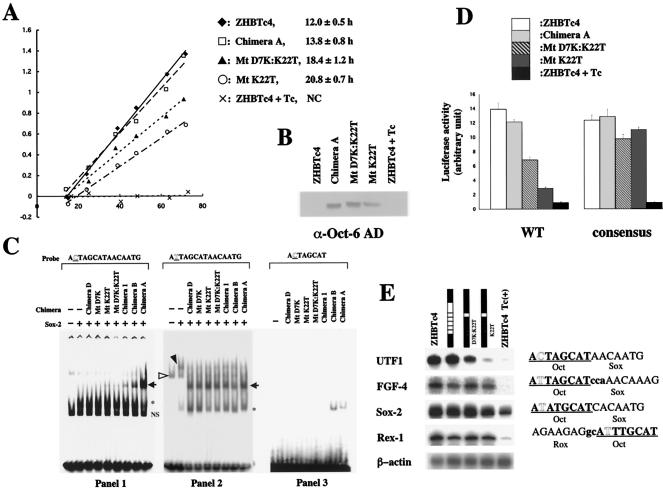
MtK22T fails to support rapid proliferation due to its diminished ability to sustain elevated UTF1 expression in ES cells. (A) ES cells rescued with MtK22T showed slower proliferation than the parental ES cells. Cells (1 × 104) of the parental ES cell line (ZHBTc4) or those bearing either chimera A, MtD7K:K22T, or MtK22T were transferred to tissue culture dishes. Subsequently, cells were trypsinized at the indicated times, and the total number of cells was counted. ZHBTc4 ES cells induced to differentiate with Tc for 96 h were also used for the analysis. NC, not calculable. (B) Western blot analysis of chimeric proteins expressed in the rescued ES cells. Whole-cell extracts were prepared from the indicated ES cells and used for Western blot analysis using an anti-Oct-6 antibody. (C) The biologically active chimeric proteins display different levels of complex formation on the UTF1 regulatory region bearing a variant octamer sequence. The expression vectors for the indicated chimeric proteins, Sox-2, or empty vector were introduced into HeLa cells, and whole-cell extracts were prepared from these cells. For the experiments shown in panels 1 and 2, these extracts were mixed and EMSA was performed as described in Materials and Methods, while extract containing Sox-2 was not used in panel 3. Arrow indicates the Oct-3/4-Sox-2-UTF1 regulatory element ternary complex. NS indicates nonspecific band, while an asterisk indicates the bands representing binding of either Sox-2 or chimeric protein to the probes. (All of the chimeric proteins can bind to probe bearing the variant octamer through the AT-rich Sox-2 binding site. For details, see reference 26). The open and solid triangles indicate Oct-1 and Oct-1/Sox-2 complex bound to the probe, respectively. (D) MtK22T exhibits much less potent activity than wild-type Oct-3/4 in activating the UTF1 regulatory enhancer. The two types of tk-Luc reporter plasmids (tk-Luc with wild-type UTF1 regulatory region or with the UTF1 enhancer bearing the consensus octamer sequence) were independently introduced into three different types of ES cells bearing either chimera A, Mt D7K:K22T, or Mt K22T. These reporters were also introduced in parental ZHBTc4 ES cells cultured in the absence or presence of Tc. After 48 h posttransfection, whole-cell extracts were prepared and luciferase activities were measured. The activity of these ES cells in which tk-Luc reporter plasmid with no regulatory enhancer had been introduced was arbitrarily set to 1. The data shown were obtained from four independent experiments with comparable results. (E) The UTF1 gene shows differential expression levels depending on the type of chimeric protein expressed. The expression levels of UTF1 as well as other Oct-3/4 target genes in parental ZHBTc4 ES cells cultured in the absence or presence of Tc or carrying one of the depicted chimeric proteins was analyzed by RNase protection assay. Enhancer core sequences of the genes are shown on the right, and putative octamer binding sites are underlined.
RNase protection assay.
Preparation of RNA from ES cells and RNase protection assays were performed as described previously (26). For detection of the various mRNA species, the following portions of the cDNA sequences were used: Nanog, bp 54 to 254; FoxD3, bp 1112 to 1336; BMP4, bp 1003 to 1221; BMPRII, bp 2915 to 3132; BMPRIa, bp −141 to 44; Id1, bp 4 to 181; Id2, bp 3 to 169; Id3, bp −6 to 203; UTF1, bp 226 to 544; Sox-2, bp 480 to 905; Rex-1, bp 75 to 351; ERas, bp −46 to 179; GATA4, bp 1187 to 1560; HNF3β, bp 376 to 604; Brachyury (T), bp 1763 to 1921; Pax6, bp 1045 to 1311; Wnt1, bp 862 to 1113; p21, bp 1 to 303; p27, bp 466 to 672; FGF-4, bp 2588 to 2784; and β-actin probe, bp 903 to 1023. The numbers represent the nucleotide position when the adenine nucleotide of the initiating methionine codon is set to +1.
Immunostaining.
Immunostaining was performed using an anti-Sox-2 (Chemicon) or anti-cytokeratin 7 (Chemicon) antibody together with appropriate Alexa-Fluor dye-conjugated secondary antibodies (Molecular Probes) as described by Miyagi et al. (24). The cells shown in Fig. 3D are counterstained with 4′,6′-diamidino-2-phenylindole.
FIG. 3.
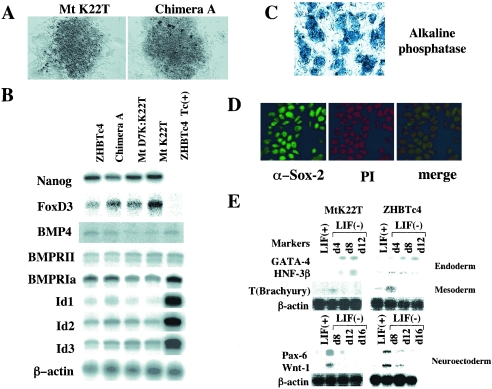
The ES cell state of cells rescued with the chimeric protein MtK22T. (A) Morphology of ZHBTc4 ES cells in which Mtk22T or chimera A was expressed in the place of wild-type Oct-3/4. Rescue experiments were performed as for Fig. 2 with the indicated chimeric protein expression vectors. The obtained colonies were observed under a microscope. (B) Examination in rescued cells of the expression levels of marker genes known to play an important role in maintaining pluripotency. RNA was prepared from the parental ES cells (ZHBTc4) cultured in the absence or presence of Tc. RNA was also prepared from ZHBTc4 ES cells whose stem cell state was maintained due to the expression of either chimera A, MtD7K:K22T, or MtK22T. Subsequently, expression levels of genes shown in the panel were analyzed by RNase protection, as described in Materials and Methods. (C) The confirmation of ES-cell status of the rescued cells with alkaline phosphatase staining. ES cells expressing MtK22T were cultured on a feeder layer and subjected to alkaline phosphatase staining procedures, as described in Materials and Methods. In these experiments, feeder layer cells served as a negative control. (D) The ES cells rescued with MtK22T possessed Sox-2 in their nuclei. ES cells bearing MtK22T were analyzed by immunostaining with anti-Sox-2 antibody and counterstained with 4′,6′-diamidino-2-phenylindole to confirm the nuclear localization of the Sox-2 protein. (E) Potential of rescued cells to differentiate into all three germ layers. Rescued cells bearing MtK22T or parental ZHBTc4 ES cells were differentiated in vitro by inducing embryoid body formation, as described in Materials and Methods. The expression of differentiation marker genes was analyzed by RNase protection, as with panel B.
Detection of alkaline phosphatase-positive cells.
Cells were cultured on feeder cells, which serve as a negative control. These cells were fixed with 4% formaldehyde and incubated with a mixture of Nitro Blue Tetrazolium and 5-bromo-4-chloro-3-indolylphosphate in an alkaline solution (0.1 M NaHCO3) containing 1 mM MgCl2.
In vitro differentiation.
The parental ZHBTc4 ES cells and MtK22T-rescued cells were differentiated in vitro according to the method of Robertson (36). In brief, the embryoid bodies were formed on bacterial-grade plates in the absence of leukemia inhibitory factor for 4 days, and the embryoid bodies were then replated on tissue culture grade plates. For examination of ectoderm-specific gene expression, the embryoid bodies were further cultured on bacterial-grade plates in the presence of 0.5 μM all-trans retinoic acid for an additional 4 days. Subsequently, these embryoid bodies were replated on tissue culture plates in the absence of retinoic acid.
Transfection and luciferase assay.
Cells were transfected in 24-well dishes with ptk-luciferase reporter plasmids (0.18 μg) containing the UTF1 regulatory region from +991 to +1798 (WT) or the fragment whose variant octamer sequence was converted to the canonical (consensus) sequence using Fugene (Roche Diagnostics). The pRL/CMV vector (Promega) (0.02 μg) was included as an internal control. After 48 h of transfection, transcription levels were determined by the dual-luciferase system according to the manufacturer's instructions (Promega).
Analysis of cell cycle kinetics.
For flow-cytometry analysis of DNA, cells were detached from culture dishes by trypsinization, and after centrifugation, cell pellets were resuspended in a solution of 50 mM sucrose, 8 mM sodium citrate (pH 7.6), 16-μg/ml propidium iodide, and 0.4 mM EDTA. These cells were then treated with 0.2-mg/ml RNase for 15 min on ice. Subsequently, fluorescence emitted from the propidium iodide-DNA complex in each nucleus was measured by fluorescence-activated cell sorting with a FACS Caliber device (Becton Dickinson).
Teratoma formation.
The nude mice were pretreated with Tc (10 μg/ml) in their drinking water for 2 weeks and then continuously treated with Tc after injection. The MtK22T-rescued cells (1 × 107), in which UTF1-expressing (pCAG/mUTF1/IRES/HIS) or empty (pCAG/IRES/HIS) vector was stably introduced, were subcutaneously injected into Tc-pretreated mice. The parental ZHBTc4 cells with stably integrated UTF1-expressing or empty vector were also injected into Tc-treated mice. The teratoma shown in panel 1 of Fig. 6A was obtained with parental ZHBTc4 cells injected into a mouse that had not been subjected to Tc treatment.
RESULTS
The UTF1 enhancer specifically recruits Oct-3/4 in vivo.
We have previously shown using in vitro DNA binding assays that octamer factors can interact with a UTF1 regulatory region bearing a variant octamer sequence. However, detailed mutagenesis analysis showed that with the exception of Oct-3/4, these factors fail to bind to the 5′ half of the element due to a sequence mismatch and instead bind to an AT-rich Sox factor-binding site, competing with Sox-2 for binding to the UTF1 enhancer. In contrast, Oct-3/4 can bind to the UTF1 regulatory region together with the Sox-2 protein (26).
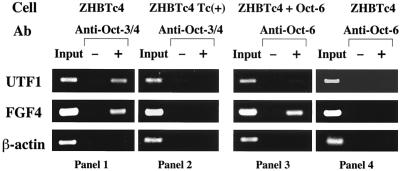
To examine the physiological relevance of these findings, we performed ChIP assays. In addition to the UTF1 regulatory region, we used an enhancer for the fibroblast growth factor 4 (FGF-4) gene, which contains consensus Octamer and Sox binding sequences (50) and regulates expression in ES cells (19), as well as the ubiquitously expressed β-actin promoter. We immunoprecipitated ES cell chromatin using an anti-Oct-3/4 antibody and performed PCR using primers recognizing the UTF1, FGF-4, and β-actin regulatory regions. As expected, the UTF1 and FGF-4 regulatory regions were efficiently amplified, whereas the β-actin promoter region was not (Fig. 1, panel 1). However, when we instead used an antibody recognizing the closely related family octamer factor Oct-6 for the immunoprecipitation of chromatin from cells overexpressing Oct-6, only the FGF-4 enhancer region was amplified (panel 3). These results indicate that the UTF1 regulatory region specifically recruits Oct-3/4 and not Oct-6 in vivo.
FIG. 1.
The UTF1 enhancer specifically recruited Oct-3/4 in vivo. ZHBTc4 ES cells were maintained in the undifferentiated state, except in the experiments represented in the second panel, in which cells were induced to differentiate by Tc treatment. An Oct-6 expression vector was stably integrated in cells to boost Oct-6 expression to a level comparable to that of Oct-3/4 for the experiments shown in the third panel. Cross-linked chromatin from ES cells was immunoprecipitated with anti-Oct-3/4 or anti-Oct-6 antibodies or corresponding preimmune-sera (−). Precipitated DNA was purified and subjected to PCR to amplify the indicated regulatory regions. Genomic DNA was also prepared from the supernatant of a reaction in which no antibody was added and then diluted 300 times with H2O and used as a control for PCRs.
Binding of monomeric Oct-3/4 or Oct-6 to the AT-rich Sox binding sequence of the enhancer, which was observed in our previous in vitro DNA binding study, was not detected by the ChIP assay, suggesting that this binding does not occur in vivo. Control experiments using an anti-Oct-3/4 antibody to immunoprecipitate DNA from Oct-3/4-deficient differentiated ES cells confirmed that none of the three regulatory regions we tested were amplified (Fig. 1, panel 2). Similarly, none of these regulatory regions, including the FGF-4 enhancer, could be amplified following immunoprecipitation using an anti-Oct-6 antibody from chromatin of normal ZHBTc4 ES cells, in which Oct-6 protein was not overexpressed (panel 4).
The ability to form a complex with Sox-2 on the UTF1 enhancer correlates with the ability to sustain ES cells in a stem cell state.
In order to determine the region of Oct-3/4 responsible for maintaining the stem cell state in ES cells, we made a series of constructs encoding chimeric proteins containing portions of both Oct-3/4 and Oct-6 and analyzed their biological functions using the complementation assay developed by Niwa et al. (30). ZHBTc4 ES cells used in the method carry inactivated alleles of Oct-3/4 due to gene targeting but harbor a tTA-dependent Oct-3/4 transgene, allowing these cells to maintain the stem cell state in the absence of Tc but not in the presence of Tc. In the assay, Oct-3/4 or a related protein expression vector is introduced into these ES cells, and the function of these transgenes is examined by their ability to generate ES cell colonies in the presence of Tc.
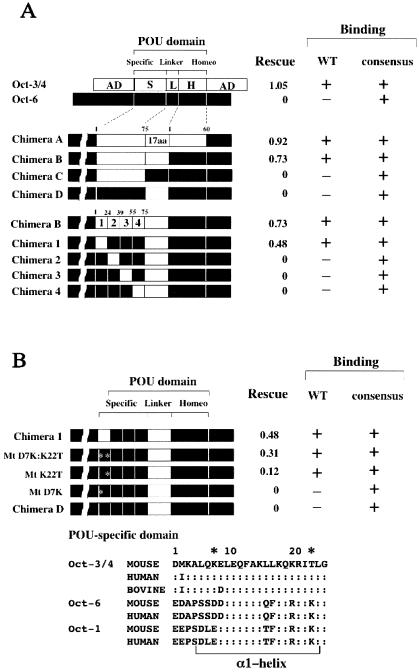
We found that transgenic expression of Oct-3/4 but not Oct-6 was able to support ES cell colony formation (Fig. 2). Moreover, chimeric protein A, in which the POU DNA binding domain of Oct-6 was replaced by that of Oct-3/4, was able to rescue the stem cell state, confirming previous results from Niwa et al. (30 [see introduction for details]). The POU DNA binding domain consists of three structurally separable domains: the POU-specific domain, the POU homeodomain, and the linker region. We have previously shown that the POU-specific domain and the linker region of Oct-3/4 are required for ternary complex formation with Sox-2 on the UTF1 regulatory region containing a variant octamer element, but the POU homeodomain is interchangeable with that of Oct-6 (26). Indeed, chimera B, which bears the POU-specific domain and linker region of Oct-3/4, could form a complex with Sox-2 on the UTF1 regulatory sequence. More importantly, we found that this chimeric protein could rescue the stem cell state of ES cells. Furthermore, replacement of either the POU-specific domain or the linker portion of Oct-3/4 by the corresponding domains of Oct-6 (chimeras C and D) resulted in a complete loss of Oct-3/4 biochemical and biological activities. Since crystallographic analysis has shown that the POU-specific domain is composed of four alpha-helices (16), we generated chimeric proteins swapping each of these helices for the corresponding helix sequence in Oct-6 (chimeras 1 to 4). Along with the linker region, the first alpha-helix of Oct-3/4 was essential for recognition of the variant octamer sequence, but other portions could be replaced by Oct-6 sequences. We also noted that exactly the same domains of Oct-3/4 played a crucial role in sustaining stem cell identity in ES cells.
FIG. 2.
Correlation between the ability of Oct-3/4-Oct-6 chimeric proteins to form a complex with Sox-2 on the UTF1 enhancer sequence carrying a variant octamer motif and to maintain ES cell state. (A) Functional characterization of Oct-3/4-Oct-6 chimeric proteins. The open and solid boxes represent Oct-3/4 and Oct-6 regions, respectively. Amino acids in the POU-specific domain and homeodomain are numbered separately (for details, see reference 13). Amino acid sequences of Oct-3/4 and Oct-6 linker portions are shown in the same reference (13). The POU-specific domain can be divided into four helices, abbreviated as 1, 2, 3, and 4. The potential of the chimeric proteins to maintain the self-renewal of ES cells was analyzed by the rescue experiment, as described in Materials and Methods. Rescue indices were determined by arbitrarily setting the values (number of colonies) obtained with ZHBTc4 ES cells cultured in the absence of Tc as 1, and relative values obtained from expression of chimeric proteins were calculated. The DNA binding ability of chimeric proteins was analyzed with EMSA using as probes the wild-type UTF1 enhancer element (WT) or an enhancer in which the variant octamer sequence was converted to the canonical octamer sequence (consensus). Since the reactions were performed in the presence of Sox-2, formation of ternary complexes (Chimera/Sox-2/DNA) was monitored. (B) Identification of a critical amino acid in the POU-specific domain that is required for Oct-3/4-specific activity. The biochemical and biological properties of the depicted Oct-3/4-Oct-6 chimeric mutant proteins were characterized as in panel A. The amino acid sequences of α-helix 1 of the POU-specific domains of three different octamer factors from various species are shown at the bottom. Double dots represent amino acids in Octamer factors that are identical to those in corresponding positions of mouse Oct-3/4.
Although no similarity in amino acid sequence is evident in the linker portion of the octamer family of proteins, the POU-specific domain is highly conserved (see bottom of Fig. 2B), allowing the identification of the amino acids likely to be critical for Oct-3/4-specific function. We focused on lysine 7 and threonine 22 of the POU-specific domain, since these two amino acids are unique to Oct-3/4, while the corresponding amino acids in Oct-1 and Oct-6 are identical or similar to one another but distinct from those in Oct-3/4. To examine the roles of these two amino acids, we generated expression vectors for mutant proteins shown in Fig. 2B. We found that Mt D7K:K22T and Mt K22T could rescue the stem cell state of ES cells, while Mt D7k and chimera D failed to do so. These results indicate that threonine 22 of Oct-3/4 is, along with the linker portion, critical for maintaining the stem cell state of ES cells. We also characterized the DNA binding activities of these mutant proteins and found a perfect correlation between the ability to form a complex with Sox-2 on the UTF1 regulatory sequence bearing a variant octamer motif and the ability to sustain the stem cell state in ES cells.
ES cells expressing the chimeric protein Mt K22T show hallmark characteristics of ES cells.
Colonies of cells produced by ES cells bearing chimeric Oct proteins consisted of densely packed cells and exhibited typical ES cell colony morphology (Fig. 3A). In order to determine whether these cells indeed possess ES cell-specific properties, we prepared RNA from cell colonies induced by chimera A, Mt D7K:22T, and Mt K22T, as well as parental ZHBTc4 ES cells cultured in the absence or presence of Tc, and examined the expression of genes known to play crucial roles in maintaining the stem cell state of ES cells. As shown in Fig. 3B, Nanog and FoxD3 showed equivalent expression levels in all cell populations except in Tc-treated parental ZHBTc4 ES cells. In addition, since bone morphogenetic protein signals were recently shown to play a role in the maintenance of ES cell pluripotency via induction of inhibitor of differentiation (Id) genes (49), we examined expression of these genes in our cell colonies. Rescued cells and parental ZHBTc4 ES cells cultured in the absence of Tc had expression levels equivalent to those of BMP4, BMP4 receptor, and Id genes. We further confirmed that MtK22T-rescued cells are positive for alkaline phosphatase, which is expressed in undifferentiated ES cells (Fig. 3C). Moreover, immunocytochemistry revealed that the Sox-2 protein, a marker for pluripotent ES cells (3), is localized to the nucleus in cells rescued with MtK22T (Fig. 3D). Finally, we examined whether rescued cells retained their pluripotency by inducing differentiation in the presence or absence of retinoic acid (see Materials and Methods) and examining the expression levels of differentiation marker genes. Rescued cells indeed have the potential to differentiate into endodermal, mesodermal, and ectodermal cells (Fig. 3E), although they tend to differentiate into nonmesodermal cells more readily than the parental cells.
UTF1 expression is reduced in rescued ES cells with slow-growing phenotype.
We observed in our rescue experiments that the proliferation rate of rescued ES cells varied according to the proportion of Oct-3/4 sequence in the chimeric proteins. ES cells expressing the MtK22T chimeric protein showed the lowest growth rate among the four types of ES cells examined (Fig. 4A), although fully differentiated ES cells grew much slower than the rescued cells. Western blot analysis using an anti-Oct-6 antibody showed that the chimeric proteins were produced at comparable levels (Fig. 4B), suggesting that differences in growth rate were not due to variable expression levels. We show that the ability of chimeric proteins to form a complex with Sox-2 on the UTF1 regulatory element with the variant octamer sequence declined as the proportion of Oct-3/4 sequence in the proteins declined (Fig. 4C, panel 1), although they were able to form Oct-Sox complexes on a probe bearing the consensus octamer sequence just as efficiently as wild-type Oct-3/4 (Fig. 4C, panel 2). In addition to the variant octamer sequence (5′-ACTAGCAT-3′), we have recently identified several new octamer-like sequences from random oligonucleotide mixtures that serve as specific binding sites for Oct-3/4 (27). We chose two sequences (5′-ATCAGCAT-3′ and 5′-GTTAGCAT-3′) to test for DNA-protein binding and found that unlike the case of the variant octamer sequence found in the UTF1 regulatory region, there were no differences in the abilities of the different chimeric proteins to bind to these sequences (data not shown). We also examined the intrinsic activity of each chimeric protein to recognize the variant octamer sequence by using a probe which contains the variant octamer but lacks the Sox binding site. From these analyses, we found that only chimera A and chimera B were able to bind to the variant octamer, but none of other chimeric proteins, including MtD7K:K22T and MtK22T, could exert such activity (Fig. 4C, panel 3). Thus, the ability of chimeric proteins to rescue ES cells in the stem cell state was only correlated with the ability to bind to the UTF1 regulatory sequence together with Sox-2 but not in the absence of the Sox-2 protein.
We next examined whether the UTF1 enhancer is able to function in the rescued ES cells as well as in ES cells bearing the wild-type Oct-3/4 protein. For these experiments, we made two tk promoter/luciferase reporter plasmids, one carrying the original UTF1 enhancer sequence and the other bearing an enhancer in which the variant octamer sequence was converted to the consensus sequence. These reporter plasmids were individually introduced into the ES cell lines shown in Fig. 4A. We found that the UTF1 enhancer bearing the consensus octamer sequence was able to support a high level of gene expression in all of the ES cell lines, except the differentiated ES cells (Fig. 4D, ZHBTc4 + Tc). In contrast, there was a perfect correlation between the ability of chimeric proteins to form a complex on the UTF1 enhancer with Sox-2 and the transcriptional activation of the enhancer with the original sequence, indicating that the strength of UTF1 enhancer function depends on the ability of the chimeric protein to bind to the UTF1 enhancer together with Sox-2, although the activity of the enhancer was in all cases higher than was observed with differentiated ES cells.
Furthermore, we examined the level of endogenous UTF1 gene expression in these cells. As shown in Fig. 4E, the attenuated DNA binding activity of chimeric proteins was accompanied by a decrease in the endogenous UTF1 expression level, although these ES cells retained higher levels of the expression than differentiated ES cells. This change in UTF1 expression was specific, since the expression levels of other Oct-3/4 downstream genes, such as FGF-4, Sox-2, and Rex-1, were unaltered, suggesting that only genes regulated by enhancers containing the variant octamer sequence show differential expression in response to activation by different Oct-3/4-Oct-6 chimeric proteins.
UTF1 can induce rapid proliferation of ES cells.
As demonstrated above, the level of UTF1 expression in ES cells rescued with the chimeric protein MtK22T is quite low, and such ES cells grow considerably more slowly than the parental ES cells, suggesting that UTF1 may participate in controlling the proliferation rate of ES cells. To examine this possibility, we overexpressed UTF1 in MtK22T-rescued cells and analyzed the growth rate of these cells. As shown in Fig. 5A, rescued cells had almost normal cell growth rates that were comparable to that of parental ZHBTc4 ES cells. We also noted that UTF1 was able to promote cell cycle progression even in differentiated ZHBTc4 parental ES cells, suggesting that the effect of UTF1 is independent of the differentiation state of ES cells. We examined the cell cycle state of these ES cells by fluorescence-activated cell sorting (FACS) and found that UTF1-overexpressing Mt K22T-rescued cells showed fewer cells in G1 phase than did control Mt K22T-rescued UTF1-deficient cells (Fig. 5B). Although changes in cell cycle were somewhat subtle, consistent results were obtained from five independent experiments. Furthermore, a significant increase in the number of phospho-histone H3-positive cells which represent M-phase cells were observed in the UTF1-overexpressing ES cells compared to UTF1-deficient rescued cells (data not shown). We also found that the numbers of terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling-positive cells were equivalent between the control and UTF1-overexpressing ES cells (data not shown), implying that the UTF1 does not affect cell viability by inhibiting apoptosis. Together, these results suggest that UTF1 promotes proliferation of ES cells.
Next, we examined whether UTF1 prevents ES cells from undergoing spontaneous differentiation or dedifferentiation, since apparent growth-promoting activity could instead be the result of such an activity. We compared the number of cytokeratin 7-positive cells in UTF1-overexpressing and UTF1-deficient rescued cells by immunocytochemistry, since ZHBTc4 ES cells tend to dedifferentiate into trophectodermal cells (29). However, as shown in Fig. 5C, there was no noticeable difference between these two different ES cell populations, eliminating the possibility that UTF1 elevates the number of ES cells by preventing spontaneous loss of the ES cell state.
UTF1 shows a prominent effect on teratoma formation in nude mice.
We next examined the effect of UTF1 expression on teratoma formation of ES cells, since the ability of ES cells to proliferate rapidly appears to be a key feature of this process (15, 40). Since the tet-off system can be manipulated in vivo by administering Tc in the drinking water of mice (22), we examined whether tumor formation by ZHBTc4 ES cells also could be regulated with Tc in nude mice. We found that ES cells were able to produce large tumors when injected subcutaneously into nude mice without Tc treatment (Fig. 6A, panel 1). However, these cells did not generate tumors in Tc-treated mice (Fig. 6A, panel 2), suggesting that Oct-3/4 expression was correctly regulated by Tc in nude mice. We next injected MtK22T-rescued ES cells into Tc-treated mice and found that as for the parental ZHBTc4 ES cells, rescued ES cells in most cases failed to produce visible teratomas in nude mice. Although tumors were observed for these cells in three out of eight animals (Fig. 6A, panel 3), they were considerably smaller than those obtained with ZHBTc4 ES cells in Tc-free mice. In contrast, we found that MtK22T rescued cells in which UTF1 is overexpressed were able to produce large tumors (Fig. 6A, panel 4) that were comparable to those produced by ZHBTc4 ES cells, indicating that UTF1 expression promotes teratoma formation, likely through its ability to promote ES cell proliferation. We also examined the effect of UTF1 expression in the absence of MtK22T on teratoma formation, since UTF1 promoted growth in differentiated ZHBTc4 cells in vitro (Fig. 5A). We found that these cells did not promote significant teratoma formation (Fig. 6A, panel 5). Histological analysis indicated that all of these teratomas contained multiple differentiated cell types, such as epithelial tissue and muscle, with no obvious differences between the different tumors shown in Fig. 6A, including those generated by cells with MtK22T alone (Fig. 6B). Moreover, RNase protection assays showed that like teratomas obtained with the parental ZHBTc4 ES cells, those obtained with UTF1-overexpressing MtK22T-rescued cells contained endodermal, mesodermal, and ectodermal cells, although marker genes show differential expression levels in these two different teratomas (Fig. 6C).
DISCUSSION
Oct-3/4 is expressed in pluripotent ES cells but not in their differentiated derivatives (31, 37). Gene targeting analysis has clearly demonstrated the essential role of this factor in the maintenance of the stem cell state in ES cells (25, 29). However, the molecular basis of Oct-3/4 function in ES cells remains largely unknown. Domain mapping analyses between Oct-3/4 and Oct-6 confirmed the previous results demonstrated by Niwa et al. (30) that Oct-3/4 transactivation domains do not play any specific roles in rescuing ES cells and these domains can be replaced by heterologous transactivation domains, such as that putatively present in Oct-6. This chimeric protein was comparable to wild-type Oct-3/4 in its ability to induce expression of ES cell-specific genes, FGF-4 and UTF1. These results were somewhat unexpected based on our previously published data (26) demonstrating that the Oct-3/4 transactivation domain was required for activating FGF-4 and UTF1 enhancers in COS cells. We presume that these discrepancies are due to differential expression in ES cells and COS cells of transcriptional cofactors required for transcriptional activation by the Oct-6 transactivation domain. Supporting this notion is our observation that these enhancers were more strongly activated in ES cells than in COS cells transiently cotransfected with Oct-Sox expression vectors and a reporter plasmid.
Niwa et al. (30) showed that the Oct-3/4 POU DNA binding domain plays an indispensable role in maintaining the stem cell state in ES cells. In the present study, we show that the ability of Oct-3/4 to form a complex with Sox-2 on the UTF1 regulatory region carrying a variant octamer sequence is crucial for the maintenance of the stem cell state in ES cells. These results suggest that activation of UTF1 expression is an important mechanism by which Oct-3/4 maintains the ES cell state. This is further supported by the fact that attenuation of Oct-3/4 function, as measured by rescue index and cell growth rate, by replacement of certain functional domains with the corresponding regions of Oct-6 was accompanied by a decline in UTF1 expression. Moreover, overexpression of UTF1 in these slow-growing ES cells restored their proliferation rate to normal, although we have no direct evidence that UTF1 is also involved in other aspects of ES cell character, such as self-renewal and pluripotency.
Our detailed domain swapping analysis allowed us to identify a single amino acid (threonine 22) in the POU-specific domain that is crucial for formation of the ternary complex between Oct-3/4, Sox-2, and the UTF1 regulatory element. Although it is not known at present how this amino acid is involved in ternary complex formation, it is noteworthy that it is conserved in the zebrafish octamer factor POU2, the orthologue of mammalian Oct-3/4 (9). Moreover, recent experiments have shown that this protein exhibits weak but noticeable activity in the complementation assay (H.N., unpublished data). The variant octamer sequence in the UTF1 regulatory region differs from the canonical sequence at the position that interacts with the POU homeodomain (16), suggesting that this critical amino acid in the POU-specific domain may not interact directly with the variant sequence. Recently, Remenyi et al. (35) suggested a model for Oct-3/4-Sox-2 interaction based on their analysis of the Oct-1-Sox-2 complex. In this model, Sox-2 utilizes distinct protein surfaces for interaction with Oct-3/4 on FGF-4 and UTF1 regulatory regions. Specifically, they propose that the carboxy-terminal end of the HMG domain of Sox-2 is involved in Oct-3/4 interaction on the FGF-4 enhancer, while a segment of helix 3 participates in the interaction with Oct-3/4 when they are bound to the UTF1 regulatory region. The same surface patch (amino acids 18 to 30) of the POU-specific domain of Oct-3/4 is supposed to mediate interaction with Sox-2 on both of these regulatory enhancers. It is tempting to speculate that threonine 22, which is in the middle of this surface patch, facilitates ternary complex formation through direct interaction with helix 3 of Sox-2. In line with this notion, we found that neither chimera Mt D7k:K22T nor Mt K22T could recognize the variant octamer sequence in the UTF1 regulatory region in the absence of Sox-2, but it was able to interact with the sequence in the presence of Sox-2 (Fig. 4C).
Our data also show that the linker portion of Oct-3/4 plays an important role in biochemical and biological properties specific to Oct-3/4. However, we do not know at present how this linker portion is involved in these activities. The contribution of the linker portion to DNA binding specificity in Oct proteins was previously demonstrated (2, 18, 44), with changes in the length of the Oct-1 linker portion and/or replacements of the linker with a corresponding region from another Oct factor altering DNA binding specificities. Therefore, it is tempting to speculate that the Oct-3/4 linker portion may also possess specificity for the variant octamer sequence.
ES cells possess three distinct but related properties: pluripotency, self-renewal, and rapid cell growth. Oct-3/4 appears to be involved in all three of these aspects of ES cell maintenance (29). So far, a number of genes have been identified as downstream targets of Oct-3/4 in ES cells, including FGF-4 (50), Rex-1 (4), Sox-2 (42), Osteopontin (6), Fbx15 (41), and Nanog (17). In addition, Oct-3/4 was recently shown to participate in its own gene expression (33). Although the functions of these genes in maintenance of the stem cell state are in some cases not clear, functional analyses of these genes allow us to envisage a schema about the Oct-3/4-Sox-2 complex-mediated transcriptional network involved in sustaining the ES cell state. In this schema, the Oct-3/4-Sox-2 complex participates in supporting its own gene expression to secure a large amount of the protein complex in ES cells. The complex then controls Nanog expression for ES-cell self-renewal and pluripotency, while UTF1 is assigned to the execution of rapid proliferative property and teratoma-forming activity of ES cells. Thus, at this stage, it is definitely important to identify genes which are under the control of Nanog and UTF1 to move towards the complete understanding of the stem cell state of ES cells at a molecular level.
Among the genes that are specifically expressed in pluripotent ES cells, ERas has been shown to support their rapid cell growth (40). Since this function appears to be related to that of the UTF1 protein described in this study, it is tempting to speculate that UTF1, which possesses the hallmark characteristics of a transcriptional coactivator (32), functions to promote cell growth by directly elevating the level of ERas gene expression.
How does the rapid proliferation sustained by UTF1 come into play during early embryogenesis? It is known that like ES cells, the pluripotent epiblast cells in early mouse embryos divide very rapidly. Indeed, the cell cycle of these cells at 6.5 days postcoitum is estimated to be as short as 4.4 h, whereas mesodermal cells that differentiate from these cells during gastrulation grow rather slowly in comparison, with a mean cell cycle at 7.0 days postcoitum of about 22.2 h (for details, see reference 14). As a result of this rapid proliferation, epiblast cells are able to expand rapidly to produce a large number of cells, which is undoubtedly necessary for embryos to grow and successfully establish the basic body plan, particularly during gastrulation, in which epiblast cells must respond to a variety of differentiation signals (39). Moreover, numerous types of experimental data have rather directly suggested the involvement of rapid proliferation of pluripotent cells in early embryos for proper development. For example, it has been demonstrated that blastocysts whose total number of cells is substantially lower than that of normal ones do not develop in utero at a normal frequency and exhibit lower rates of full-term development than controls (34, 46, 47). Furthermore, Boiani et al. (5) have demonstrated that blastocysts obtained from oocytes with transplanted somatic cell nuclei had substantially fewer cells than normal blastocysts, which may be a major reason for the lower frequency of normal development of cumulus cell clones. Thus, these data suggest that UTF1 participates in the elevation of the embryonic survival rate by promoting cell cycle progression in early embryos.
Acknowledgments
We are indebted to Yuko Wada for her technical assistance.
This work was supported in part by the Ministry of Education, Science, Sports, and Culture, in particular by a Ministry grant to the Saitama Medical School Research Center for Genomic Medicine. This work was also performed as a part of the Rational Evolutionary Design of Advanced Biomolecules Project, the Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, supported by the Japan Science and Technology Agency. M.N. was also supported by the Kato Memorial Bioscience Foundation and the Sankyo Foundation of Life Science.
REFERENCES
- 1.Antonchuk, J., G. Sauvageau, and R. K. Humphries. 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109:39-45. [DOI] [PubMed] [Google Scholar]
- 2.Aurora, R., and W. Herr. 1992. Segments of the POU domain influence one another's DNA-binding specificity. Mol. Cell. Biol. 12:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avilion, A. A., S. K. Nicolis, L. H. Pevny, L. Perez, N. Vivian, and R. Lovell-Badge. 2003. Multipotent cell lineages in early mouse development depend on Sox-2 function. Genes Dev. 17:126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boiani, M., S. Eckardt, N. A. Leu, H. R. Scholer, and K. J. McLaughlin. 2003. Pluripotency deficit in clones overcome by clone-clone aggregation: epigenetic complementation? EMBO J. 22:5304-5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botquin, V., H. Hess, G. Fuhrmann, C. Anastassiadis, M. K. Gross, G. Vriend, and H. R. Scholer. 1998. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 12:2073-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley, A., M. Evans, M. H. Kaufman, and E. Robertson. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell line. Nature 309:255-256. [DOI] [PubMed] [Google Scholar]
- 8.Brehm, A., K. Ohbo, W. Zwerschke, V. Bontquin, P. Jansen-Durr, and H. R. Scholer. 1999. Synergism with germ line transcription factor Oct-4: viral oncoproteins share the ability to mimic a stem cell-specific activity. Mol. Cell. Biol. 19:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess, S., G. Reim, W. Chen, N. Hopkins, and M. Brand. 2002. The zebrafish spiel-ohne-grenzen (spg) gene encodes the POU domain protein Pou2 related to mammalian Oct-4 and is essential for formation of the midbrain and hindbrain, and for pre-gastrula morphogenesis. Development 129:905-916. [DOI] [PubMed] [Google Scholar]
- 10.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643-655. [DOI] [PubMed] [Google Scholar]
- 11.Evans, M. J., and M. H. Kaufman. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154-156. [DOI] [PubMed] [Google Scholar]
- 12.Hanna, L. A., R. K. Foreman, I. A. Tarasenko, D. S. Keesler, and P. A. Labosky. 2002. Requirement for FoxD3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 16:2650-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herr, W., and M. A. Cleary. 1995. The POU doman: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 9:1679-1693. [DOI] [PubMed] [Google Scholar]
- 14.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hwang, W. S., Y. J. Ryu, J. H. Park, E. G. Lee, J. M. Koo, H. Y. Jeon, B. C. Lee, S. K. Kang, S. J. Kim, C. Ahn, J. H. Hwang, K. Y. Park, J. B. Cibelli, and S. Y. Moon. 2004. Evidence of a pluripotent human embryonic stem cell line derived from a cloned blastocyst. Science 303:1669-1674. [DOI] [PubMed] [Google Scholar]
- 16.Klemm, J. D., M. A. Rould, R. Aurora, W. Herr, and C. O. Pabo. 1994. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77:21-32. [DOI] [PubMed] [Google Scholar]
- 17.Kuroda, T., M. Tada, H. Kubota, H. Kimura, S. Hatano, H. Suemori, N. Nakatsuji, and T. Tada. 2005. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25:2475-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, P., X. He, M. R. Gerrero, M. Mok, A. Aggarwal, and M. G. Rosenfeld. 1993. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 7:2483-2496. [DOI] [PubMed] [Google Scholar]
- 19.Ma, Y. G., E. Rosfjord, C. Huebert, P. Wilder, J. Tiesman, D. Kelly, and A. Rizzino. 1992. Transcriptional regulation of the murine k-FGF gene in embryonic cell lines. Dev. Biol. 154:45-54. [DOI] [PubMed] [Google Scholar]
- 20.Martin, G. R. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78:7634-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda, T., T. Nakamura, K. Nakao, T. Arai, M. Katsuki, T. Heike, and T. Yokota. 1999. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18:4261-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayford, M., M. E. Bach, Y.-Y. Huang, L. Wang, R. D. Hawkins, and E. R. Kandel. 1996. Control of memory formation through regulated expression of a CaMKII transgene. Science 274:1678-1683. [DOI] [PubMed] [Google Scholar]
- 23.Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631-642. [DOI] [PubMed] [Google Scholar]
- 24.Miyagi, S., T. Saito, K. Mizutani, N. Masuyama, Y. Gotoh, A. Iwama, H. Nakauchi, S. Masui, H. Niwa, M. Nishimoto, M. Muramatsu, and A. Okuda. 2004. The Sox-2 regulatory regions display their activities in two distinct types of multipotent stem cells. Mol. Cell. Biol. 24:4207-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. R. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 26.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453-5465. (Erratum, 21:978, 2001.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimoto, M., S. Miyagi, T. Katayanagi, M. Tomioka, M. Muramatsu, and A. Okuda. 2003. The embryonic Octamer factor 3/4 displays distinct DNA binding specificity from those of other Octamer factors. Biochem. Biophys. Res. Commun. 302:581-586. [DOI] [PubMed] [Google Scholar]
- 28.Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12:2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa, H., J.-I. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 30.Niwa, H., S. Masui, I. Chambers, A. G. Smith, and J.-I. Miyazaki. 2002. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell. Biol. 22:1526-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto, K., H. Okazawa, A. Okuda, M. Sakai, M. Muramatsu, and H. Hamada. 1990. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60:461-472. [DOI] [PubMed] [Google Scholar]
- 32.Okuda, A., A. Fukushima, M. Nishimoto, A. Orimo, T. Yamagishi, Y. Nabeshima, M. Kuro-o, Y.-I. Nabeshima, K. Boon, M. Keaveney, H. G. Stunnenberg, and M. Muramatsu. 1998. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 17:2019-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okumura-Nakanishi, S., M. Saito, H. Niwa, and F. Ishikawa. 2005. Oct-3/4 and Sox-2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 280:5307-5317. [DOI] [PubMed] [Google Scholar]
- 34.Ozil, J. P. 1983. Production of identical twins by bisection of blastocysts in the cow. J. Reprod. Fertil. 69:463-468. [DOI] [PubMed] [Google Scholar]
- 35.Remenyi, A., K. Lins, L. J. Nissen, R. Reinbold, H. R. Scholer, and M. Wilmanns. 2003. Crystal structure of POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17:2048-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, E. J. 1987. Embryo-derived stem cell lines, p 71-122. In E. J. Robertson. (ed.), Teratocarcinoma and embryonic stem cell: practical approach. IRL press, Oxford, United Kingdom.
- 37.Scholer, H. R., S. Ruppert, N. Suzuki, K. Chowdhury, and P. Gruss. 1990. New type of POU domain in germ line-specific protein Oct-4. Nature 344:435-439. [DOI] [PubMed] [Google Scholar]
- 38.Smith, A. G., J. K. Heath, D. D. Donaldson, G. G. Wong, J. Moreau, M. Stahl, and D. Rogers. 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336:688-690. [DOI] [PubMed] [Google Scholar]
- 39.Snow, M. H. L. 1977. Gastrulation in the mouse: growth and regionalization of epiblast. J. Embryol. Exp. Morphol. 42:293-303. [Google Scholar]
- 40.Takahashi, K., K. Mitsui, and S. Yamanaka. 2003. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature 423:541-545. [DOI] [PubMed] [Google Scholar]
- 41.Tokuzawa, Y., E. Kaiho, M. Maruyama, K. Takahashi, K. Mitsui, M. Maeda, H. Niwa, and S. Yamanaka. 2003. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomioka, M., M. Nishimoto, S. Miyagi, T. Katayanagi, N. Fukui, H. Niwa, M. Muramatsu, and A. Okuda. 2002. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic. Acids Res. 30:3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tucker, R. M., and D. T. Burke. 1996. Transgenic mice for the establishment of histidinol-resistant embryonic fibroblast feeder layers. FASEB J. 10:1641-1645. [DOI] [PubMed] [Google Scholar]
- 44.van Leeuwen, H. C., M. J. Strating, M. Rensen, W. de Laat, and P. C. van der Vliet. 1997. Linker length and composition influence the flexibility of Oct-1 DNA binding. EMBO J. 16:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells, J., and P. J. Farnham. 2002. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 26:48-56. [DOI] [PubMed] [Google Scholar]
- 46.Willadsen, S. M. 1981. The development capacity of blastomeres from 4- and 8-cell sheep embryos. J. Embryol. Exp. Morphol. 65:165-172. [PubMed] [Google Scholar]
- 47.Willadsen, S. M., and C. Polge. 1981. Attempts to produce monozygotic quadruplets in cattle by blastomere separation. Vet. Rec. 108:211-213. [DOI] [PubMed] [Google Scholar]
- 48.Williams, R. J., D. J. Hilton, S. Pease, T. A. Willson, C. L. Stewart, D. P. Gearing, E. F. Wagner, D. Metcalf, N. A. Nicola, and N. M. Gough. 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336:684-687. [DOI] [PubMed] [Google Scholar]
- 49.Ying, Q.-L., J. Nichols, I. Chambers, and A. Smith. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115:281-292. [DOI] [PubMed] [Google Scholar]
- 50.Yuan, H., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of FGF-4 enhancer requires the synergistic activation of Sox2 and Oct-3. Genes Dev. 9:2635-2645. [DOI] [PubMed] [Google Scholar]