Abstract
Putative TTAGGG repeat-binding factor (TRF) homologues in the genomes of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major were identified. They have significant sequence similarity to higher eukaryotic TRFs in their C-terminal DNA-binding myb domains but only weak similarity in their N-terminal domains. T. brucei TRF (tbTRF) is essential and was shown to bind to duplex TTAGGG repeats. The RNA interference-mediated knockdown of tbTRF arrested bloodstream cells at G2/M and procyclic cells partly at S phase. Functionally, tbTRF resembles mammalian TRF2 more than TRF1, as knockdown diminished telomere single-stranded G-overhang signals. This suggests that tbTRF, like vertebrate TRF2, is essential for telomere end protection, and this also supports the hypothesis that TRF rather than Rap1 is the more ancient DNA-binding component of the telomere protein complex. Identification of the first T. brucei telomere DNA-binding protein and characterization of its function provide a new route to explore the roles of telomeres in pathogenesis of this organism. This work also establishes T. brucei as an attractive model for telomere biology.
Telomeres are specialized protein-DNA complexes at the ends of eukaryotic chromosomes. Telomere DNA generally consists of simple, repetitive, TG-rich sequences that are maintained by telomerase (5) and end with a single-stranded G-rich overhang (78). A specialized telomere structure, the T loop, formed by the invasion of telomere G overhangs into the telomeric double-stranded region, in mammals, hypotrichous ciliates, and Trypanosoma brucei has been identified (26, 51, 53). It is hypothesized that hiding the telomere single-stranded G overhangs in a T-loop structure helps to protect the telomere ends.
Proteins that bind to duplex or single-stranded telomere DNA are integral components of the telomere complex and play critical roles in both telomere length regulation and end protection (36). In mammalian cells, two paralogues, TTAGGG repeat-binding factor 1 (TRF1) and TRF2, bind duplex TTAGGG repeats (4, 9, 12). Both TRF1 and TRF2 have a C-terminal myb motif for DNA binding (3, 9) and an upstream TRF homology (TRFH) domain for homodimerization (24). However, the N termini are quite different: TRF1 is acidic, while TRF2 is basic. Mammalian TRF1 negatively regulates telomere length through a telomerase-dependent pathway (61, 75), whereas TRF2 is involved in both telomere length regulation (38, 61) and telomere end protection (17). Removal of TRF2 from telomeres by overexpression of a dominant-negative mutant of TRF2 resulted in an at least 30% loss of telomere G-overhang signal and nonhomologous end joining (NHEJ)-dependent chromosome end fusions (60, 76). Exactly how TRF2 protects chromosome ends is not clear, but TRF2 has been shown to promote the T-loop formation in vitro (63). In fission yeast, Taz1, a TRF homologue (42), binds the duplex telomere DNA (15). It contains a C-terminal myb motif and homodimerizes (15, 62). It is involved in both negative telomere length regulation and chromosome end protection (15, 16, 25, 49). Similar to the case with TRF2, loss of Taz1 resulted in NHEJ-mediated chromosome end fusions in G1 phase (25), and Taz1 promotes T-loop formation in vitro (68).
The duplex telomere DNA-binding factor in budding yeasts is Rap1 (44). Although Rap1 also negatively regulates telomere length and protects chromosome ends (13, 46), it is not a TRF homologue (42). The only TRF ortholog in Saccharomyces cerevisiae is TBF1, which is not an integral component of the yeast telomere complex, and its role in telomere biology is not clear (8, 39). Both mammalian and fission yeast telomere complexes contain a Rap1 ortholog, but this is tethered to telomeres through interaction with TRF2 or Taz1 (35, 42, 56).
Trypanosoma brucei is a protozoan parasite that causes sleeping sickness in humans and nagana in cattle in Africa. This parasite regularly switches the variant surface glycoprotein gene (VSG) expressed when it is in a mammalian host, thus evading eradication by the host immune system (2). Although there are approximately 1,000 VSG genes and pseudogenes in T. brucei (70), each cell expresses a single VSG from a polycistronic expression site (ES), located at a subtelomeric region and transcribed by RNA polymerase I (27). There are ∼20 ESs (54), but only 1 is active at a time. All identified ESs have similar structures, with the promoter located ∼40 to ∼60 kb upstream of the telomere, followed by several expression-site-associated genes, ending with the VSG gene, which can be within 2 kb of the telomere repeats (1, 33, 43, 57). Elongation of ES transcription is regulated so that only one ES is fully active (72). In addition, in a silent ES, the transcription of a promoter-driven reporter gene decreases closer to the telomere (31). In the midgut of the tsetse, the insect vector of T. brucei, VSG genes are not transcribed and VSG is replaced by members of the procyclin family of surface glycoproteins, whose nontelomeric genes are also transcribed by RNA polymerase I.
The T. brucei genome consists of 11 pairs of essential megabase chromosomes (0.9 to 5.7 Mb), a few intermediate chromosomes (200 to 900 kb), and ∼100 minichromosomes (50 to 100 kb), all carrying two telomeres (23). T. brucei telomeres consist of TTAGGG repeats (6, 71), are maintained by telomerase (10; O. Dreesen et al., unpublished data), end with single-stranded G overhangs, and form T loops (51). Of the more conserved multifunctional complexes that have been implicated in telomere function in other organisms, Mre11 and Ku homologues are present in trypanosomes, and the deletion of either subunit of the Ku complex results in telomere shortening (14, 34, 58, 66).
The fact that active VSG genes are subtelomeric and that telomeric silencing occurs in T. brucei suggests that telomeres may play a role in antigenic variation. In addition, trypanosomes diverged from vertebrates a few hundred million years ago (64), yet T. brucei and vertebrates share many similarities in telomere structures, which makes trypanosomes interesting models for telomere biology. Previous attempts to identify telomere-binding factors yielded one activity with apparent high affinity for the single-stranded G overhang (11) and two that appeared to prefer subtelomeric sequences (21, 22). None of these studies led to the identification of any protein, so the composition and function of the T. brucei telomere complex remain unknown.
Here we report the identification and characterization of T. brucei TRF (tbTRF), the first telomere duplex DNA-binding factor in T. brucei. tbTRF is a homologue of vertebrate TRFs. We demonstrate that tbTRF is an integral component of the telomere complex throughout the cell cycle. tbTRF has a similar telomere end protection function as vertebrate TRF2, as reduced tbTRF binding leads to a great reduction of telomere G-overhang signals. The evolutionary implications of our results will be discussed.
MATERIALS AND METHODS
Cloning of tbTRF, RNA interference (RNAi) expression, and knockout constructs.
The human TRF1 (hTRF1) myb domain was used in BLAST searches of the emerging T. brucei genome database to identify Tb10.389.0680. Two oligonucleotides were designed to anneal to the starting and ending sites of Tb10.389.0680. Using genomic DNA from the Lister 427 strain as the template, the tbTRF open reading frame (ORF) was amplified by PCR. Several independent PCR products were used to clone the tbTRF ORF into pLew111, pET-15b, and pGEX-4T-2 vectors and multiple sequencing from both directions generated consistent results (GenBank accession no. AY910010).
The C-terminal two-thirds of the tbTRF ORF was inserted into the pZJMβ vector (50) at the XbaI site to generate a tbTRF RNAi construct. The hygromycin- or puromycin-resistant fragments flanked by tbTRF 5′ and 3′ untranslated regions were inserted into pBluescript SK to make two tbTRF deletion constructs.
Cell culture, RNAi induction, purification of live cells, isolation of total mRNA, genomic DNA, and preparation of whole cell extract and DNA plugs.
Lister 427 procyclic and bloodstream antigenic type MITat 1.2 clone 221a T. brucei was used. Cells harboring the T7 polymerase and the tetracycline (Tet) repressor, single marker (bloodstream) and 29-13 (procyclic) (80), were used for conditional expression of the tbTRF double-stranded RNA (dsRNA) and epitope-tagged tbTRFs. Bloodstream and procyclic cells were cultured in HMI-9 medium and SDM-79 medium, respectively, with appropriate drug selection.
For tbTRF RNAi induction, doxycycline was added to bloodstream and procyclic cells at 100 ng/ml and 500 ng/ml, respectively. To eliminate dead bloodstream cells, the population was loaded onto a BBSCG {0.050 M Bicine [N,N-bis(2-hydroxyethyl)glycine], 0.077 M glucose, 0.050 M NaCl, 0.005 M KCl, 0.005 M CaCl2, pH 8.0}-equilibrated Whatman DE52-cellulose column at no more than 109 cells per 1-ml column. The live cells in the flowthrough fraction were used for various analyses. RNA was isolated with RNA STAT-60 (TEL-TEST, Inc.). Protein extracts were obtained by lysing cells with 2× sodium dodecyl sulfate (SDS) Laemmli buffer (0.1 M Tris-Cl, pH 6.8, 6% SDS, 20% glycerol, 0.004% bromophenol blue). Genomic DNA was isolated as published previously (19). DNA plugs were prepared as published previously (32).
Antibody generation.
A glutathione S-transferase (GST)-tbTRF recombinant protein was used to immunize two New Zealand rabbits, generating AB 1260 and AB 1261. A His6-tbTRF recombinant protein was partially purified from Escherichia coli, coupled to CNBr-activated Sepharose 4B beads, and used to purify the rabbit sera.
In-gel hybridization and quantification of telomere G-overhang signal.
Undigested chromosomes were separated by use of a rotating agarose gel electrophoresis (RAGE) apparatus (Stratagene), as described previously (32). After imaging, the ethidium bromide-stained gel was dried at room temperature. Hybridization with end-labeled (CCCATT)4 or (TTAGGG)4 and subsequent washes were carried out at 50°C. The probe was kept at −20°C after the first hybridization. The washed gel was exposed to a phosphorimager screen for ∼2 days, with the exact durations noted. After a scanning of the phosphorimager, the gel was denatured, neutralized, hybridized with the same probe, and washed at 55°C. Gel exposure to the phosphorimager was repeated for 2 h. The signals were quantified with ImageQuant. Telomere G-overhang signal levels were calculated by dividing the amounts of signals obtained before denaturation by the amounts of those obtained after denaturation. The change in the telomere G-overhang level was quantified with Microsoft Excel by normalizing G-overhang levels at different time points to that at zero time.
Indirect immunofluorescence (IF)/fluorescence in situ hybridization (FISH), delta-vision imaging, and chromatin immunoprecipitation (ChIP).
IF, combined with FISH, was carried out as described previously (45). Cell cycle stages were determined by cytology (47, 48). Cell images were captured by using a DeltaVision image restoration microscope (Applied Precision/Olympus), deconvoluted by using measured point spread functions, and edited with Photoshop. ChIP was carried out as described previously (45), with AB 1260 and its preimmune serum.
Electrophoretic mobility shift assay (EMSA).
Partially purified baculovirus-expressed hTRF2 was a generous gift from Titia de Lange (76). Partially purified GST-Ty1 was a gift from Jenny Li (Li and Cross, unpublished data). The (TTAGGG)12 probe was prepared as published previously (81). Proteins were mixed with 1 μl 80% glycerol, 0.5 μl 1 μg/μl sheared E. coli DNA, 2 μl 150 mM Tris-HCl (pH 9.0), 0.5 μl 100 ng/μl β-casein, and 1 ng labeled (TTAGGG)12 probe, plus corresponding competitors, incubated at room temperature for 30 min, loaded onto a 0.6% agarose gel, and separated at 200 V in 0.1× Tris-borate-EDTA. The agarose gel was dried at 60°C for 1 h and exposed to a phosphorimager screen. Permutated TTAGGG repeats cloned into the pSXneo vector (81) were generous gifts from Titia de Lange and were used as competitors. Specifically, pSXneo plasmid containing (T2AG2C)25, (T3AG3)55, (T2AG5)140, and (T2AG3I)140 repeats were used at ∼5-, ∼10-, and ∼35-fold molar excesses to probe in competition assays.
Yeast two-hybrid analysis.
All TRF fragments were cloned into pBTM116 and pACT2 vectors at BamHI/EcoRI sites. These constructs were transformed into the yeast two-hybrid reporter strain L40. A liquid assay using ONPG (o-nitrophenyl-β-d-galactopyranoside) as the substrate was used to quantify the β-galactosidase activity. For each yeast strain, at least three independent clones were tested and multiple assays were performed. The average values and standard deviations were calculated from the combined results.
Nucleotide sequence accession number
The sequence of the tbTRF ORF has been assigned GenBank accession number AY910010.
RESULTS
Identification of tbTRF as a homologue of vertebrate TRF.
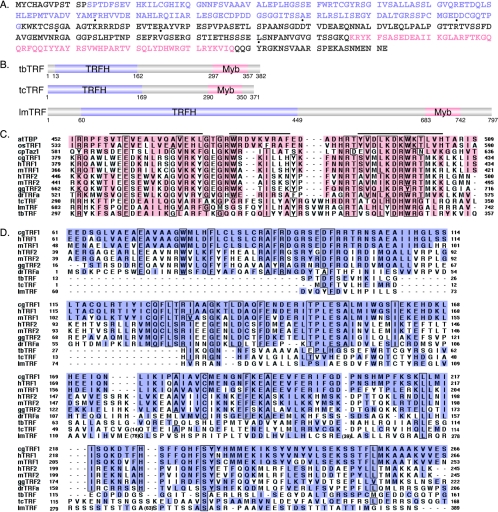
We identified, in the emerging T. brucei genome sequence, a single open reading frame (Tb10.389.0680) encoding a C-terminal myb motif that showed significant similarity to the myb motif of telomeric TTAGGG-binding factors in other organisms (Fig. 1A and C). This myb domain has all three signature helices (Fig. 1C). It is most conserved at the third helix, where the protein is predicted to contact DNA (29, 55). We identified homologues of Tb10.389.0680 in the genome sequences of Trypanosoma cruzi and Leishmania major (Fig. 1B), two parasites related to T. brucei and the causative agents of Chagas' disease and leishmaniasis, respectively. The myb domains in these three proteins are almost identical (Fig. 1C).
FIG. 1.
tbTRF is a true TRF homologue. (A) Protein sequence of tbTRF in strain Lister 427. TRFH and myb domains are in light purple and beige, respectively. The sequence outside these two domains is in dark gray. Dotted amino acids differ from Tb10.389.0680 in the Sanger T. brucei database (strain TREU 927, clone 4). (B) Domain structures of T. brucei TRF, T. cruzi TRF, and L. major TRF. TRFH and myb domains and their positions are marked. (C) ClustalX alignment of the myb domains in various TRFs. The three signature helices are indicated by bars on top of the aligned sequences. The identities/similarities (%) between tbTRF and other TRFs in the myb domain are as follows: for Arabidopsis thaliana telomere binding protein (atTBP), 18.2/42.9; for Oryza sativa TRF1 (osTRF1), 15.6/37.7; for Cryptosporidium parvum Taz1 (cpTaz1), 16.9/35.4; for Cricetulus griseus TRF1 (cgTRF1), 20.8/36.4; for hTRF1, 19.5/36.4; for mouse TRF1 (mTRF1), 22.1/37.7; for hTRF2, 18.2/32.5; for mouse TRF2 (mTRF2), 15.6/33.8; for Gallus gallus TRF2 (ggTRF2), 18.2/37.7; for Danio rerio TRFa (drTRFa), 20.8/37.7; for T. cruzi TRF (tcTRF), 46.8/59.7; and for L. major TRF (lmTRF), 41.6/53.2. The identity/similarity between hTRF1 and hTRF2 myb domains is 45.6/60.3. (D) ClustalX alignment of TRFH domains. Numbers in parentheses indicate amino acid insertions. The identities/similarities (%) between tbTRF and other TRFs in the TRFH domain are as follows: for cgTRF1, 11.5/31.1; for hTRF1, 11.5/31.1; for mTRF1, 10.8/33.1; for hTRF2, 12.2/42.6; for mTRF2, 11.5/40.5; for ggTRF2, 8.8/39.2; for drTRFa, 8.8/36.5; for tcTRF, 27.7/50.7; and for lmTRF, 23.6/43.9. The identity/similarity between hTRF1 and hTRF2 TRFH domains is 26.3/58.0. Homologous sequences are shaded and identical residues are boxed in panels C and D.
In addition, the N termini of Tb10.389.0680 and its T. cruzi and L. major homologues show weak but discernible sequence similarity with the TRFH domain in other TRFs (Fig. 1D). We therefore named these newly identified ORFs as TRF ORFs. tbTRF has a very short neutral N terminus, unlike either mammalian TRF. Although the myb domain of tbTRF seems more closely related to that of vertebrate TRF1 (Fig. 1C), the DNA-binding activity could be detected only in vitro by using hTRF2 DNA-binding conditions (see below). The TRFH domain of tbTRF is more closely related to the vertebrate TRF2 than TRF1 (Fig. 1D). Thus, it is impossible to determine whether tbTRF is a TRF1 or TRF2 homologue based solely on its sequence. By careful examination of T. brucei genome sequence data, we could not identify any additional TRF homologues.
After identifying tbTRF, we cloned the gene from T. brucei Lister 427, the preferred strain for genetic studies. When it was compared to the Sanger Institute Tb10.389.0680 sequence, which was obtained from the official genome strain TREU 927/4, we found 11 nucleotide differences and 6 amino acid differences (Fig. 1A). tbTRF is predicted to be a 41.5-kDa protein. Northern analysis and Western blotting indicated that neither the mRNA nor the protein abundance of tbTRF differs between the bloodstream and procyclic life cycle stages (data not shown).
tbTRF is an integral component of the T. brucei telomere complex.
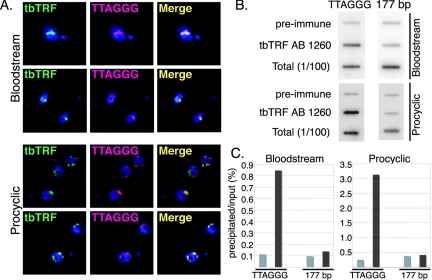
The sequence similarity between tbTRF and other TRFs strongly suggested that tbTRF is a telomere-binding factor. To test this hypothesis, we examined the association of tbTRF and telomere DNA by using several approaches. First, we observed that tbTRF colocalized with telomere DNA at all stages of the cell cycle in both bloodstream and procyclic cells, when endogenous tbTRF was detected by using rabbit antibodies (AB 1260 and AB 1261; see Materials and Methods) in combined IF/FISH (Fig. 2A). We also independently tested the localization of Ty1-epitope-tagged tbTRF expressed from its endogenous locus and ectopic Ty1-tbTRF and green fluorescent protein (GFP)-tbTRF expressed from an rRNA gene locus. In all cases, tagged tbTRF proteins colocalized with the TTAGGG DNA throughout the cell cycle in both life cycle stages (data not shown).
FIG. 2.
tbTRF is an integral component of T. brucei telomeres. (A) IF combined with FISH indicates that tbTRF colocalizes with TTAGGG repeats. Endogenous tbTRF (green) is detected by AB 1261 (bloodstream) and AB 1260 (procyclic). TTAGGG repeats (pink) are detected by hybridization with a CCCUAA probe. DNA is stained with DAPI (4′,6′-diamidino-2-phenylindole) (blue). Two sets of images are shown for each life cycle stage. (B) The TTAGGG repeat DNA coimmunoprecipitates with tbTRF. DNA precipitated by ChIP with AB 1260 or preimmune serum, together with the input DNA sample (1%), was hybridized with either a TTAGGG-repeat probe or a 177-bp repeat probe as indicated. (C) Quantification of the ChIP results shown in panel B. Light gray bars represent precipitations with preimmune serum; dark gray bars represent precipitations with AB 1260. Probes used in different hybridizations were indicated at the bottom.
Nearly one-third of the T. brucei genome consists of minichromosomes, which consist primarily of 177-bp repeats, subtelomeric VSG genes, and telomere repeats (79). The small size of minichromosomes makes it impossible to distinguish telomere signals from 177-bp DNA signals by FISH. Thus, although tbTRF colocalized with the TTAGGG signal, it was possible that tbTRF actually bound DNA elements on minichromosomes other than the TTAGGG repeats. To exclude this possibility, we carried out ChIP analysis. A TTAGGG repeat probe was used to detect the precipitated telomere DNA and a 177-bp repeat probe was used as a control. As shown in Fig. 2B and C, the TTAGGG repeat DNA was enriched 10-fold by use of anti-tbTRF AB 1260 compared to preimmune serum, whereas little 177-bp repeat DNA was precipitated by either serum.
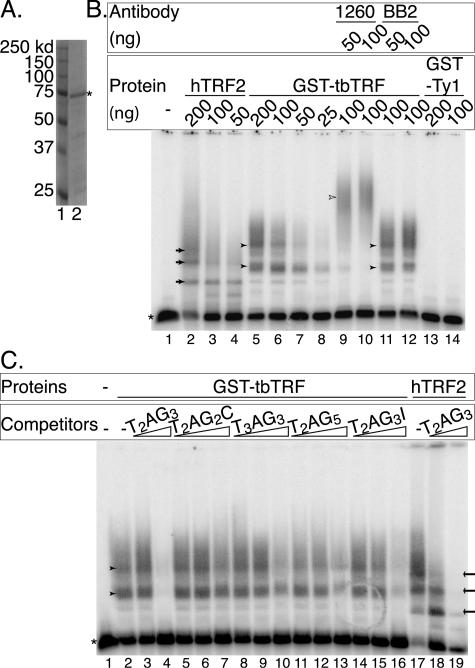
IF/FISH and ChIP data strongly suggest that tbTRF binds telomere DNA in vivo. Subsequently, we tested whether tbTRF binds to TTAGGG repeats in vitro by using EMSA. GST-tbTRF was expressed and partially purified from E. coli (Fig. 3A). It bound to a double-stranded (TTAGGG)12 probe (Fig. 3B, lanes 5 through 12) but not to single-stranded (TTAGGG)4 or (CCCTAA)4 oligonucleotides (data not shown). This binding was not due to the GST protein, as a partially purified GST-Ty1 recombinant protein did not have any DNA-binding activity when tested under the same conditions (Fig. 3B, lanes 13 and 14). In addition, the GST-tbTRF-DNA complex was supershifted by anti-tbTRF AB 1260 (Fig. 3B, lanes 9 and 10) and AB 1261 (data not shown) but not by anti-Ty1 monoclonal antibody BB2 (Fig. 3B, lanes 11 and 12). The multiple binding activities exhibited by GST-tbTRF are probably due to multimerization of GST-tbTRF. tbTRF bound to duplex TTAGGG repeats under the same conditions used for vertebrate TRF2 (T. de Lange, personal communication) but did not bind under the conditions used for TRF1 (81) (data not shown), suggesting that the tbTRF myb domain contacts telomere DNA in a similar way to vertebrate TRF2 rather than TRF1. By competition analysis, it was also clear that tbTRF preferred TTAGGG repeats rather than DNA repeats containing permutations of TTAGGG (Fig. 3C and data not shown).
FIG. 3.
tbTRF binds to double-stranded TTAGGG repeats. (A) Coomassie-stained SDS-polyacrylamide gel electrophoresis of GST-tbTRF. Lane 1, molecular mass marker; lane 2, partially purified fraction from cells expressing GST-tbTRF. The asterisk indicates the GST-tbTRF recombinant protein. Molecular masses are indicated to the left. (B) The partially purified GST-tbTRF protein binds to double-stranded (TTAGGG)12 DNA. Partially purified baculovirus-expressed hTRF2 (lanes 2 through 4) and partially purified GST-Ty1 (lanes 13 and 14) are used as controls. The amounts of proteins used are indicated at the top of the gel. AB 1260 (anti-tbTRF) or BB2 (anti-Ty1) and the amounts used are indicated. Open arrowhead indicates the band supershifted by AB 1260. (C) tbTRF has a higher affinity for TTAGGG repeats. EMSA was carried out with the labeled (TTAGGG)12 probe using either 100 ng of GST-tbTRF (lanes 2 through 16) or 100 ng of hTRF2 (lanes 17 through 19). Competitor sequences are indicated. The amounts of nonradioactive TTAGGG repeats used were 0.5 and 2 μg; the amounts of other nonradioactive competitors used were 0.25 μg, 0.5 μg, and 2 μg (Materials and Methods). The asterisk indicates the free probe, and arrows indicate the bands shifted by hTRF2. Arrowheads indicate the band shifted by GST-tbTRF.
tbTRF interacts with itself.
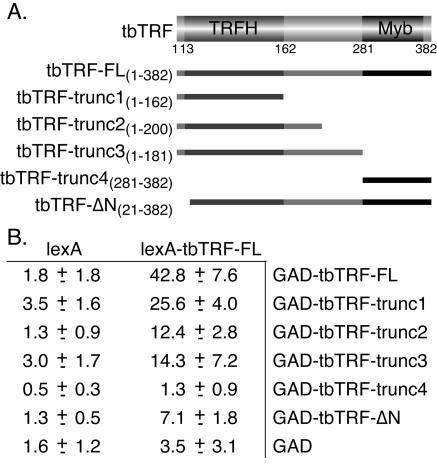
At least two myb motifs are normally required for DNA binding (55). TRF1, TRF2, and Taz1 all form homodimers (3, 9, 62). We therefore investigated whether the same applied to tbTRF by using yeast two-hybrid interaction assays. Full-length wild-type tbTRF was fused to either a LexA binding domain (lexA-tbTRF-FL) or a Gal4 activation domain, and both fusion proteins were expressed in a yeast two-hybrid reporter strain, L40 (77), which had the LexA binding sites placed upstream of a lacZ reporter gene. Both a colony-lift filter assay and a liquid assay using ONPG as the substrate (Materials and Methods) showed that these cells have high β-galactosidase activity (Fig. 4B and data not shown). On the other hand, yeast cells harboring only one fusion construct did not display high β-galactosidase activity, although lexA-tbTRF-FL had a very weak transcription activation activity by itself (Fig. 4B). These data suggest that tbTRF can interact with itself. To determine which domain is necessary for self-interaction, the Gal4 activation domain was fused to parts of tbTRF and tested with lexA-tbTRF-FL (Fig. 4A). The tbTRFH domain was sufficient for this self-interaction (Fig. 4B). The N-terminal 20 amino acids were critical for self-interaction, as the deletion of this small region greatly reduced the β-galactosidase activity. Western blots showed that all fusion proteins except one were expressed at similar levels in yeast cells. tbTRF-trunc4 was expressed at a higher level (data not shown).
FIG. 4.
tbTRF interacts with itself. (A) Deletion mutants of tbTRF used in two-hybrid analysis. Amino acid numbers (inclusive) are given. (B) Yeast two-hybrid analysis results showing that tbTRF interacts with itself. β-Galactosidase activity units measured by liquid assay with ONPG as the substrate are given as averages ± standard deviations calculated from multiple experiments (Materials and Methods). GAD, Gal4 activation domain.
tbTRF is essential.
To analyze the functions of tbTRF, we attempted to generate tbTRF null mutants. Two deletion constructs were generated. Both could efficiently establish single-allele deletions when transfected into wild-type cells. tbTRF+/− cells grew normally (data not shown). However, despite repeated attempts, we failed to establish double-knockout cell lines for tbTRF. In contrast, after introducing an ectopic Ty1-tbTRF into the tbTRF+/− cells, we were able to delete the second tbTRF allele. Similar results were obtained for both bloodstream and procyclic cells. These data strongly suggested that tbTRF is essential.
As attempts to obtain tbTRF−/− cells were fruitless, we turned to RNAi techniques in single marker (bloodstream) and 29-13 (procyclic) cells harboring T7 RNA polymerase and Tet repressor (80). A tbTRF RNAi construct was generated in the pZJMβ vector (50) and targeted into an rRNA gene locus in tbTRF+/− cells. Expression of tbTRF double-stranded mRNA was controlled by opposing Tet-inducible T7 promoters flanking the tbTRF fragment in pZJMβ. The single allele of endogenous tbTRF in these cells was either untagged (six bloodstream clones and one procyclic cell pool) or tagged with GFP (three bloodstream clones) or Ty1 (three bloodstream clones) at its N terminus. In total, 12 independent bloodstream clones and 1 procyclic pool were analyzed, and the results were consistent.
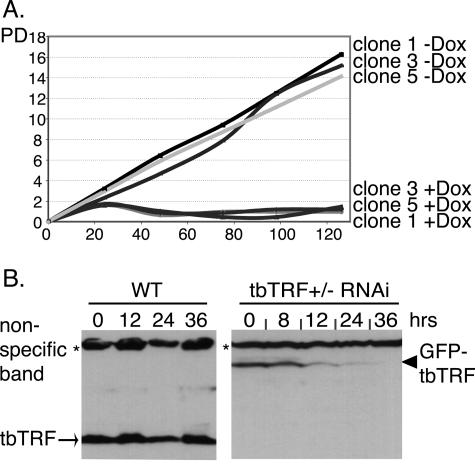
Bloodstream cells stopped proliferating within 24 h of tbTRF dsRNA induction (Fig. 5A), consistent with tbTRF being essential. Procyclic cells, which have a longer doubling time, stopped proliferating within 2 days (data not shown). In bloodstream cells, tbTRF mRNA level dropped by ∼50% within 12 h of RNAi induction and then decreased moderately during the next 24 h (data not shown). In addition, we observed a continuous decrease in tbTRF protein levels between 12 and 36 h after RNAi induction (Fig. 5B and data not shown), further suggesting that the growth arrest was due to dilution or loss of tbTRF. Similar phenotypes in procyclic cells were observed (data not shown).
FIG. 5.
tbTRF is essential. (A) Growth curves for three independent bloodstream tbTRF+/− tbTRF-RNAi clones (1, 3, and 5) under both induced (+Dox) and uninduced (−Dox) conditions. The population doublings (PD) and hours after induction (y axis) are indicated. (B) tbTRF protein level decreases after induction of tbTRF RNAi. Whole-cell protein extracts were prepared from bloodstream wild-type (WT) or tbTRF+/− tbTRF-RNAi cells that express GFP-tbTRF from tbTRF endogenous locus. Western blotting was carried out with tbTRF antibody 1261.
tbTRF RNAi resulted in a strong and acute growth phenotype. Thus, the following analyses were carried out at only the earliest time points: within 36 h after the addition of doxycycline in bloodstream trypanosomes and 6 days in procyclics. In addition, to exclude the possibility of analyzing dead cell phenotypes, we removed any dead bloodstream cells before analysis. Because bloodstream cells are covered with VSG molecules, live cells pass unimpeded through a DE52 cellulose anion-exchange column, while negatively charged dead cells, having shed their VSG coats, are trapped. The following results were obtained with purified live bloodstream cells unless indicated. Similar results were obtained with procyclic cells.
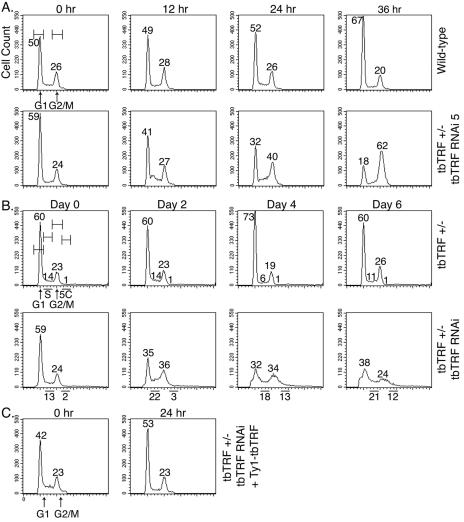
To determine whether cells were arrested at a particular stage of the cell cycle, cells were fixed, stained with propidium iodide and analyzed by fluorescence-activated cell sorting (FACS) at different times after RNAi induction. The population of bloodstream G1 cells dropped from ∼60% to ∼35%, while the population of G2/M cells increased from ∼22% to ∼60% (Fig. 6A), indicating a G2/M-phase arrest. Wild-type cells, in contrast, did not vary significantly. In procyclic cells, however, we found more S-phase and 5C cells but fewer G1-phase cells, whereas the number of G2/M phase cells increased only transiently (Fig. 6B), indicating a partial S-phase arrest.
FIG. 6.
tbTRF RNAi leads to cell cycle arrest. Cells were fixed at the indicated times after tbTRF RNAi induction and stained with propidium iodide for FACS analysis. (A) Bloodstream wild-type cells or clone 5 of tbTRF+/− tbTRF-RNAi cells were analyzed. G1 and G2/M peaks and gates are marked in the first panel. The percentage of cells in these two phases among the population is indicated next to each peak. (B) Procyclic tbTRF+/− or tbTRF+/− tbTRF-RNAi cells were analyzed. G1 and G2/M peaks plus S phase and cells with 5C content were marked in the first panel. “5C” is used to represent cells with DNA contents equivalent to five copies of haploid DNA. The percentages of cells that are in each phase among the population are indicated. (C) Ectopic Ty1-tbTRF expression complemented the tbTRF RNAi growth arrest phenotype. FACS analysis of bloodstream tbTRF+/− tbTRF-RNAi cells that also carry an ectopic Tet-inducible Ty1-tbTRF is shown.
tbTRF RNAi reduced telomeric G-overhang signals.
To understand what triggers growth arrest due to tbTRF RNAi, we examined telomere structures in cells that have lower levels of tbTRF proteins. Telomere Southern blots showed that telomere lengths did not change during the short period after tbTRF RNAi was induced (data not shown). We then tested whether tbTRF RNAi affected the telomere G-overhang structure. Intact chromosomes were separated by RAGE, and telomere G overhangs were detected by hybridization of the native gel with a (CCCTAA)4 probe. As a control, the same gel was subsequently denatured and hybridized with the same probe. As there are ∼100 minichromosomes in T. brucei and, under the conditions used, they all migrate at the same position on a RAGE gel, the G-overhang signals are most prominent on minichromosomes. The telomere G-overhang level was therefore calculated only for minichromosomes, by dividing the amount of native hybridization signals by the amount of postdenaturation hybridization signals.
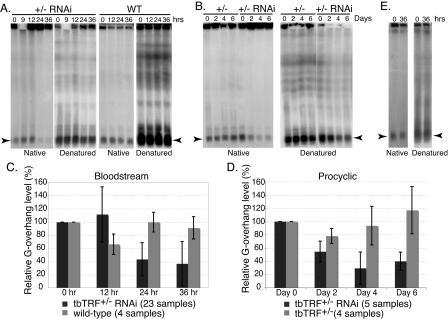
In tbTRF RNAi cells, the G-overhang signal diminished dramatically (Fig. 7), indicating that tbTRF is essential for this telomere end structure. In all bloodstream cells, the G-overhang level decreased after 24 h (Fig. 7A). Thirty-six hours after the induction, the G-overhang level was reduced by at least 50%. In many cases, only 20% of the original signal remained. Similar results were obtained from procyclic cells (Fig. 7B).
FIG. 7.
Telomere G-overhang signal is reduced by tbTRF RNAi. DNA plugs were prepared from cells at indicated hours (A and E) or days (B) after tbTRF RNAi induction. Intact chromosomal DNA was separated by RAGE, and the dried gel was hybridized with the end-labeled (CCCTAA)4 either before or after denaturation as indicated. (A) DNA plugs were prepared from bloodstream wild-type (WT) or tbTRF+/− tbTRF-RNAi cells. (B) DNA plugs were prepared from procyclic tbTRF+/− or tbTRF+/− tbTRF-RNAi cells. (C and D) Quantified telomere G-overhang level changes in bloodstream (C) and procyclic (D) tbTRF+/− tbTRF-RNAi cells. The telomere G-overhang levels at different time points after tbTRF RNAi induction were normalized to those at the zero time point. The averages of relative telomere G-overhang level (the G-overhang signal at any time/the G-overhang signal at zero time) ± standard deviations are shown for wild-type cells (light grey bars in panel C), tbTRF+/− cells (light grey bars in panel D), or tbTRF+/− tbTRF-RNAi cells (dark grey bars in panels C and D). The numbers of samples included in each calculation are indicated at the bottom of each panel. (E) DNA plugs were prepared from bloodstream tbTRF+/− tbTRF-RNAi cells with an ectopic Ty1-tbTRF. In panels A, B, and E, arrowheads indicate the signals from minichromosomes.
The total telomere DNA signal also decreased with tbTRF RNAi, though to a lesser degree. Since the G-overhang level is normalized to the total TTAGGG hybridization signals, this loss of total telomere DNA has already been taken into account in calculating the G-overhang signal. The seemingly higher level of telomere DNA loss observed in procyclic cells (Fig. 7B) may result from dead cells that are impossible to remove from the population. To quantify the loss of G-overhang signals in tbTRF RNAi cells more accurately, we ran numerous RAGE experiments with independent DNA plugs from multiple tbTRF RNAi cell lines, using the exactly same conditions. The data are thus statistically significant (Fig. 7C and D).
As additional controls, the same DNA plugs were separated by RAGE and hybridized with an end-labeled (TTAGGG)4 probe under native conditions, and no signal was observed on any of the chromosomes (data not shown). In addition, DNA plugs were treated with ExoI, a 3′ single-stranded DNA exonuclease, before in-gel hybridization with the (CCCTAA)4 probe. ExoI treatment removed all the signals we observed previously (data not shown), further confirming that they were attributable to telomere G-strand overhangs.
Mammalian TRF2 is also critical for telomere G-overhang maintenance, and removal of TRF2 from telomeres resulted in ligase IV-mediated chromosome end fusions (60, 76). Thus, we tested whether tbTRF RNAi caused chromosome end fusions. As T. brucei chromosomes do not condense in mitosis, chromosome end fusions cannot be visualized by cytology. However, telomere DNA is susceptible to Bal31 degradation unless chromosome ends are fused and the telomere repeats become internal (18). T. brucei chromosome ends are frequently marked by subtelomeric VSG genes, making it convenient to follow individual telomere ends with specific VSG probes. In our cells, VSG BR2 has two subtelomeric copies and one nontelomeric copy, and VSG 224 has a single subtelomeric copy. Genomic DNA was isolated at different times after tbTRF RNAi induction, treated with Bal31 for different times, and then digested with either EcoRI or BglI, to release VSG BR2- or VSG 224-containing terminal fragments, respectively. However, after multiple attempts, even with long exposures, we could not detect any Bal31-refractory DNA fragment appearing after tbTRF RNAi induction (data not shown). We also did not observe any new telomeric bands in Southern analysis of DNA from tbTRF RNAi cells (data not shown). These observations suggested that chromosome end fusions were not a frequent event in T. brucei, even after the drastic reduction of telomere G overhangs.
The tbTRF RNAi phenotypes were complemented by ectopic Ty1-tbTRF.
To further verify that the phenotypes we observed were caused by a reduction of tbTRF protein, we introduced a Tet-inducible ectopic Ty1-tagged tbTRF into tbTRF RNAi cells. Adding doxycycline to these cells induces expression of tbTRF dsRNA and ectopic expression of Ty1-tbTRF from a T7 promoter. These cells grew normally after induction, suggesting that the production of excess tbTRF could overcome the effects of RNAi. The cell cycle profile was the same as those for wild-type and uninduced cells (Fig. 6C). In addition, the telomere G-overhang signal after 36 h of induction was similar to that in cells without induction (Fig. 7F). Western analysis showed that the ectopic Ty1-tbTRF expression compensated for the decrease in endogenous tbTRF protein level (data not shown).
DISCUSSION
Identification of tbTRF as a homologue of TTAGGG repeat-binding factor.
T. brucei diverged from the vertebrate branch of the evolutionary tree hundreds of million years ago (64). The sequence conservation between T. brucei and vertebrate proteins is low, even for normally highly conserved proteins, such as tubulin and histones. Thus, it is much more difficult to identify telomere binding factors by data mining in T. brucei than in higher eukaryotes. Nevertheless, proteins binding to TTAGGG repeats are likely to have similar DNA-binding motifs. Our identification of tbTRF using only the hTRF1 myb domain as query and the fact that the putative DNA-contact helix is the most conserved region support this hypothesis. TRFH domains are much less conserved among various TRFs. There is also little sequence similarity among TRFs outside TRFH and myb domains. Members of the Rap1 telomere protein family also show sequence similarities within just a few well-defined domains (35, 42, 56).
We demonstrated that tbTRF is a true TRF homologue based on its functions. First, tbTRF is a duplex telomere DNA-binding factor. It is interesting that tbTRF has higher affinity for double-stranded TTAGGG repeats than for any permutations of TTAGGG, suggesting that tbTRF binding is very selective. We noticed that ChIP with anti-TRF antibody precipitated less TTAGGG DNA in bloodstream than in procyclic cells. Approximately 13% of thymidine in telomeres is modified to β-d-glucosyl-hydroxymethyluracil (J) in the bloodstream, but not in the procyclic stage of T. brucei (73, 74), and two J-binding proteins have been identified (20, 59). Thus, tbTRF may bind the J-containing repeats with lower affinity, which also appears to be the case for hTRF1 expressed in T. brucei (52). However, the details of tbTRF DNA-binding specificity remain to be determined. Second, like mammalian TRFs and fission yeast Taz1, tbTRF interacts with itself, probably as a homodimer. This self-interaction is expected to be essential for its DNA-binding activity, as, in general, at least two myb motifs are required for DNA binding and tbTRF only has one. In fact, a myb deletion mutant of tbTRF, when overexpressed in bloodstream cells, acted as a dominant-negative mutant and led to acute cell growth arrest (unpublished observations). Third, tbTRF is also essential for telomere G-overhang structure, as is vertebrate TRF2.
Several lines of evidence suggest that tbTRF is more closely related to TRF2 than TRF1. First, although the alignment of the myb domains indicated that tbTRF is more similar to TRF1 than TRF2, the optimum in vitro DNA-binding conditions for tbTRF are identical to those for hTRF2. Second, the alignment of TRFH domains indicated that tbTRF resembles TRF2 more than TRF1. Third, tbTRF protects the telomere G-overhang structure, as does TRF2. Finally, T. brucei telomerase elongates telomeres at a steady rate, and no significant change in the telomere elongation rate of cells overexpressing tbTRF was observed (data not shown), suggesting that tbTRF does not contribute to telomere length regulation to a significant degree in the presence of telomerase, in contrast to TRF1. However, whether tbTRF may accelerate the telomere-shortening rate in telomerase negative cells, similar to hTRF2, is not clear. This can be examined when the telomerase knockout cells are available in the near future (Dreesen et al., unpublished data).
tbTRF is essential for telomere end structure.
We showed that tbTRF is essential for stability of the telomere G overhang. The reduction of the telomere G-overhang level in tbTRF RNAi cells was quite dramatic. In many clones, there was a 60 to 80% decrease in G-overhang signals. We also observed variations in the telomere G-overhang levels in different tbTRF RNAi cells. This was not completely unexpected. The RNAi vector is targeted to an rRNA gene spacer region, and T. brucei has multiple dispersed copies of rRNA genes, so an RNAi construct will integrate into different sites in different clones, which may exhibit different expression levels. Based on this consideration, we analyzed 12 independent bloodstream clones and a procyclic pool. For each clone, multiple DNA samples were examined to compensate for fluctuations and reveal mild yet significant effects.
Knocking down tbTRF resulted in growth arrest. Since unprotected telomeres in mammalian cells trigger DNA damage responses and result in ATM/p53-dependent apoptosis or senescence-like cell cycle arrest (37, 65, 76), this result suggests that T. brucei also has a DNA repair machinery that responds to telomere damage. tbTRF RNAi arrested bloodstream and procyclic cells at different stages of the cell cycle, suggesting that the checkpoint mechanisms differ in these two life cycle stages. Consistent with this possibility, different cyclins are used in bloodstream and procyclic stages to control cell-cycle progress (28, 69). Interestingly, the overexpression of hTRF1 in procyclic T. brucei also resulted in S-phase arrest, though the phenotypes are not exactly the same as with tbTRF RNAi (52). It was proposed that hTRF1 partially displaces endogenous TTAGGG-binding factor(s) in procyclic cells. The similar growth arrest phenotypes in these two situations are consistent with this hypothesis.
Chromosome end fusions are a hallmark of unprotected telomere ends in yeasts, plants, and mammals. In most cases, these fusions are mediated through the NHEJ pathway and are ligase IV dependent (25, 30, 60). However, extensive Southern analysis did not detect any chromosome end fusions in tbTRF RNAi cells but would not be sensitive enough to detect infrequent fusions. PCR amplification of telomere end fusion products has been successfully applied in telomerase-deficient Arabidopsis thaliana cells (30). However, the average telomere length in the Lister 427 strain of T. brucei is 15 kb (51), and more than 200 telomere ends are present in each nucleus. Therefore, PCR amplification of telomere fusion products by using primers annealing to the few single-copy subtelomeric VSG genes is not a practical option. Another possibility would be that the NHEJ machinery is very inefficient compared to homologous recombination in T. brucei (7, 67). T. brucei is a diploid, and it is likely that low-frequency NHEJ events are masked by the high efficiency of homologous recombination events (41). In fission yeast, chromosome end fusions occur only in haploid Taz1 deletion cells when the cells are arrested in G1, when the NHEJ pathway is used more frequently than in G2 (25).
A conserved telomere complex.
Two types of telomere complexes have been described. One is represented by vertebrates and fission yeast, in which TRF homologues bind duplex telomere DNA and Rap1 orthologs are tethered to telomeres through interactions with TRF homologues (42). Another is represented by budding yeast, in which Rap1 binds to the duplex telomere DNA directly and a TRF homologue is absent from the telomere complex. It has been proposed that the common ancestor is probably a TRF DNA-binding telomere complex (42). The characterization of telomere complexes in T. brucei, which represents a very ancient and distinct evolutionary branch (64), would shed more light on how telomere complexes evolved. Confirmation that tbTRF is the only duplex telomere DNA-binding factor would support the hypothesis that a TRF-like telomere complex represents the ancestral scenario. It would be even more revealing if a Rap1 ortholog in T. brucei can be identified and shown to be tethered to telomeres through interaction with tbTRF. The fact that there was only one tbTRF in T. brucei that was identified further raises the possibility that the ancestral telomere complex contains only one TRF protein rather than two and that vertebrate TRF1 evolved much later than TRF2. It is worth noting that both TRF1 and TRF2 exist in chickens and Xenopus laevis (40; C. Price personal communication; F. Ishikawa, personal communication), while only Taz1 has been found in fission yeast. The evolution of TRF proteins will become clearer when TRF complexes are identified in more organisms.
The identification of tbTRF will facilitate studies of the role of telomeres in antigenic variation in T. brucei. In addition, any telomere component that is conserved from T. brucei to mammals is likely to be of great interest, and the conserved features of the telomere complex in T. brucei and vertebrates make T. brucei an attractive model for telomere study.
Acknowledgments
We greatly appreciate Titia de Lange for her insightful discussions. We also thank her for generously providing baculovirus-expressed hTRF2 protein and competitors for EMSA. The members of the Cross lab are thanked for their discussions, suggestions, technical support, and comments on the manuscript. Microscopy was carried out at the RU Bio-Imaging Resource Center with guidance from Alison North.
This work was supported by National Institutes of Health grant AI50614.
REFERENCES
- 1.Alexandre, S., M. Guyaux, N. B. Murphy, H. Coquelet, A. Pays, M. Steinert, and E. Pays. 1988. Putative genes of a variant-specific antigen gene transcription unit in Trypanosoma brucei. Mol. Cell. Biol. 8:2367-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, J. D., and R. McCulloch. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1-70. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi, A., S. Smith, L. Chong, P. Elias, and T. de Lange. 1997. TRF1 is a dimer and bends telomeric DNA. EMBO J. 16:1785-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilaud, T., C. Brun, K. Ancelin, C. E. Koering, T. Laroche, and E. Gilson. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17:236-239. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn, E. H. 1991. Telomeres. Trends Biochem. Sci. 16:378-381. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, E. H., and P. B. Challoner. 1984. Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell 36:447-457. [DOI] [PubMed] [Google Scholar]
- 7.Blundell, P. A., G. Rudenko, and P. Borst. 1996. Targeting of exogenous DNA into Trypanosoma brucei requires a high degree of homology between donor and target DNA. Mol. Biochem. Parasitol. 76:215-229. [DOI] [PubMed] [Google Scholar]
- 8.Brigati, C., S. Kurtz, D. Balderes, G. Vidali, and D. Shore. 1993. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol. Cell. Biol. 13:1306-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17:231-235. [DOI] [PubMed] [Google Scholar]
- 10.Cano, M. I., J. M. Dungan, N. Agabian, and E. H. Blackburn. 1999. Telomerase in kinetoplastid parasitic protozoa. Proc. Natl. Acad. Sci. USA 96:3616-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cano, M. I. N., J. J. Blake, E. H. Blackburn, and N. Agabian. 2002. A Trypanosoma brucei protein complex that binds G-overhangs and co-purifies with telomerase activity. J. Biol. Chem. 277:896-906. [DOI] [PubMed] [Google Scholar]
- 12.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663-1667. [DOI] [PubMed] [Google Scholar]
- 13.Conrad, M. N., J. H. Wright, A. J. Wolf, and V. A. Zakian. 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63:739-750. [DOI] [PubMed] [Google Scholar]
- 14.Conway, C., R. McCulloch, M. L. Ginger, N. P. Robinson, A. Browitt, and J. D. Barry. 2002. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J. Biol. Chem. 277:21269-21277. [DOI] [PubMed] [Google Scholar]
- 15.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385:744-747. [DOI] [PubMed] [Google Scholar]
- 16.Cooper, J. P., Y. Watanabe, and P. Nurse. 1998. Fission yeast taz1 protein is required for meiotic telomere clustering and recombination. Nature 392:828-831. [DOI] [PubMed] [Google Scholar]
- 17.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 18.de Lange, T., and P. Borst. 1982. Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature 299:451-453. [DOI] [PubMed] [Google Scholar]
- 19.de Lange, T., L. Shiue, R. M. Myers, D. R. Cox, S. L. Naylor, A. M. Killery, and H. E. Varmus. 1990. Structure and variability of human chromosome ends. Mol. Cell. Biol. 10:518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiPaolo, C., R. Kieft, M. Cross, and R. Sabatini. 2005. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol. Cell 17:441-451. [DOI] [PubMed] [Google Scholar]
- 21.Eid, J. E., and B. Sollner-Webb. 1995. ST-1, a 39-kilodalton protein in Trypanosoma brucei, exhibits a dual affinity for the duplex form of the 29-base-pair subtelomeric repeat and its C-rich strand. Mol. Cell. Biol. 15:389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eid, J. E., and B. Sollner-Webb. 1997. ST-2, a telomere and subtelomere duplex and G-strand binding protein activity in Trypanosoma brucei. J. Biol. Chem. 272:14927-14936. [DOI] [PubMed] [Google Scholar]
- 23.Ersfeld, K., S. E. Melville, and K. Gull. 1999. Nuclear and genome organization in Trypanosoma brucei. Parasitol. Today 15:58-63. [DOI] [PubMed] [Google Scholar]
- 24.Fairall, L., L. Chapman, H. Moss, T. de Lange, and D. Rhodes. 2001. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8:351-361. [DOI] [PubMed] [Google Scholar]
- 25.Ferreira, M. G., and J. P. Cooper. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7:55-63. [DOI] [PubMed] [Google Scholar]
- 26.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 27.Gunzl, A., T. Bruderer, G. Laufer, B. Schimanski, L. C. Tu, H. M. Chung, P. T. Lee, and M. G. Lee. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2:542-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammarton, T. C., M. Engstler, and J. C. Mottram. 2004. The Trypanosoma brucei cyclin, CYC2, is required for cell cycle progression through G1 phase and for maintenance of procyclic form cell morphology. J. Biol. Chem. 279:24757-24764. [DOI] [PubMed] [Google Scholar]
- 29.Hanaoka, S., A. Nagadoi, and Y. Nishimura. 2005. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 14:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heacock, M., E. Spangler, K. Riha, J. Puizina, and D. E. Shippen. 2004. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23:2304-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horn, D., and G. A. M. Cross. 1997. Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J. 16:7422-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horn, D., and G. A. M. Cross. 1997. Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol. Biochem. Parasitol. 84:189-201. [DOI] [PubMed] [Google Scholar]
- 33.Horn, D., C. Spence, and A. K. Ingram. 2000. Telomere maintenance and length regulation in Trypanosoma brucei. EMBO J. 19:2332-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janzen, C. J., F. Lander, O. Dreesen, and G. A. M. Cross. 2004. Telomere length regulation and transcriptional silencing in KU80-deficient Trypanosoma brucei. Nucleic Acids Res. 32:6575-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanoh, J., and F. Ishikawa. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11:1624-1630. [DOI] [PubMed] [Google Scholar]
- 36.Kanoh, J., and F. Ishikawa. 2003. Composition and conservation of the telomeric complex. Cell. Mol. Life Sci. 60:2295-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlseder, J., D. Broccoli, Y. M. Dai, S. Hardy, and T. de Lange. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283:1321-1325. [DOI] [PubMed] [Google Scholar]
- 38.Karlseder, J., A. Smogorzewska, and T. de Lange. 2002. Senescence induced by altered telomere state, not telomere loss. Science 295:2446-2449. [DOI] [PubMed] [Google Scholar]
- 39.Koering, C. E., G. Fourel, E. Binet-Brasselet, T. Laroche, F. Klein, and E. Gilson. 2000. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 28:2519-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konrad, J. P., W. Mills, D. J. Easty, and C. J. Farr. 1999. Cloning and characterisation of the chicken gene encoding the telomeric protein TRF2. Gene 239:81-90. [DOI] [PubMed] [Google Scholar]
- 41.Leal, S., A. Acosta-Serrano, J. Morris, and G. A. M. Cross. 2004. Transposon mutagenesis of Trypanosoma brucei identifies glycosylation mutants resistant to concanavalin A. J. Biol. Chem. 279:28979-28988. [DOI] [PubMed] [Google Scholar]
- 42.Li, B., S. Oestreich, and T. de Lange. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101:471-483. [DOI] [PubMed] [Google Scholar]
- 43.Lips, S., P. Revelard, and E. Pays. 1993. Identification of a new expression site-associated gene in the complete 30.5 kb sequence from the AnTat 1.3A variant surface protein gene expression site of Trypanosoma brucei. Mol. Biochem. Parasitol. 62:135-138. [DOI] [PubMed] [Google Scholar]
- 44.Longtine, M. S., N. M. Wilson, M. E. Petracek, and J. Berman. 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr. Genet. 16:225-239. [DOI] [PubMed] [Google Scholar]
- 45.Lowell, J. E., and G. A. M. Cross. 2004. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J. Cell Sci. 117:5937-5947. [DOI] [PubMed] [Google Scholar]
- 46.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 47.Matthews, K. R., and K. Gull. 1994. Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J. Cell Biol. 125:1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews, K. R., and K. Gull. 1994. Cycles within cycles: the interplay between differentiation and cell division in Trypanosoma brucei. Parasitol. Today 10:473-476. [DOI] [PubMed] [Google Scholar]
- 49.Miller, K. M., and J. P. Cooper. 2003. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell 11:303-313. [DOI] [PubMed] [Google Scholar]
- 50.Morris, J. C., Z. Wang, M. E. Drew, K. S. Paul, and P. T. Englund. 2001. Inhibition of bloodstream form Trypanosoma brucei gene expression by RNA interference using the pZJM dual T7 vector. Mol. Biochem. Parasitol. 117:111-113. [DOI] [PubMed] [Google Scholar]
- 51.Munoz-Jordan, J., G. A. M. Cross, T. de Lange, and J. D. Griffith. 2001. T-loops at trypanosome telomeres. EMBO J. 20:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munoz-Jordan, J. L., and G. A. M. Cross. 2001. Telomere shortening and cell cycle arrest in Trypanosoma brucei expressing human telomeric repeat factor TRF1. Mol. Biochem. Parasitol. 114:169-181. [DOI] [PubMed] [Google Scholar]
- 53.Murti, K. G., and D. M. Prescott. 1999. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl. Acad. Sci. USA 96:14436-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro, M., and G. A. M. Cross. 1996. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol. Cell. Biol. 16:3615-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogata, K., S. Morikawa, H. Nakamura, A. Sekikawa, T. Inoue, H. Kanai, A. Sarai, S. Ishii, and Y. Nishimura. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639-648. [DOI] [PubMed] [Google Scholar]
- 56.Park, M. J., Y. K. Jang, E. S. Choi, H. S. Kim, and S. D. Park. 2002. Fission yeast Rap1 homolog is a telomere-specific silencing factor and interacts with Taz1p. Mol. Cells 13:327-333. [PubMed] [Google Scholar]
- 57.Pays, E., P. Tebabi, A. Pays, H. Coquelet, P. Revelard, D. Salmon, and M. Steinert. 1989. The genes and transcripts of an antigen expression site from T. brucei. Cell 57:835-845. [DOI] [PubMed] [Google Scholar]
- 58.Robinson, N. P., R. McCulloch, C. Conway, A. Browitt, and J. D. Barry. 2002. Inactivation of Mre11 does not affect VSG gene duplication mediated by homologous recombination in Trypanosoma brucei. J. Biol. Chem. 277:26185-26193. [DOI] [PubMed] [Google Scholar]
- 59.Sabatini, R., N. Meeuwenoord, J. H. van Boom, and P. Borst. 2002. Recognition of base J. in duplex DNA by J.-binding protein. J. Biol. Chem. 277:958-966. [DOI] [PubMed] [Google Scholar]
- 60.Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch, and T. de Lange. 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12:1635. [DOI] [PubMed] [Google Scholar]
- 61.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spink, K. G., R. J. Evans, and A. Chambers. 2000. Sequence-specific binding of Taz1p dimers to fission yeast telomeric DNA. Nucleic Acids Res. 28:527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stansel, R. M., T. de Lange, and J. D. Griffith. 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20:5532-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevens, J. R., and W. Gibson. 1999. The molecular evolution of trypanosomes. Parasitol. Today 15:432-437. [DOI] [PubMed] [Google Scholar]
- 65.Takai, H., A. Smogorzewska, and T. de Lange. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13:1549-1556. [DOI] [PubMed] [Google Scholar]
- 66.Tan, K. S., S. T. Leal, and G. A. M. Cross. 2002. Trypanosoma brucei MRE11 is non-essential but influences growth, homologous recombination and DNA double-strand break repair. Mol. Biochem. Parasitol. 125:11-21. [DOI] [PubMed] [Google Scholar]
- 67.Ten Asbroek, A. L. M. A., C. Mol, R. Kieft, and P. Borst. 1993. Stable transformation of Trypanosoma brucei. Mol. Biochem. Parasitol. 59:133-142. [DOI] [PubMed] [Google Scholar]
- 68.Tomaska, L., S. Willcox, J. Slezakova, J. Nosek, and J. D. Griffith. 2004. Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J. Biol. Chem. 279:50764-50772. [DOI] [PubMed] [Google Scholar]
- 69.Tu, X., and C. C. Wang. 2004. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 279:20519-20528. [DOI] [PubMed] [Google Scholar]
- 70.Van der Ploeg, L. H., D. Valerio, T. de Lange, A. Bernards, P. Borst, and F. G. Grosveld. 1982. An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res. 10:5905-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van der Ploeg, L. H. T., A. Y. C. Liu, and P. Borst. 1984. Structure of the growing telomeres of trypanosomes. Cell 36:459-468. [DOI] [PubMed] [Google Scholar]
- 72.Vanhamme, L., P. Poelvoorde, A. Pays, P. Tebabi, H. Van Xong, and E. Pays. 2000. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Microbiol. 36:328-340. [DOI] [PubMed] [Google Scholar]
- 73.van Leeuwen, F., A. Dirks-Mulder, R. W. Dirks, P. Borst, and W. Gibson. 1998. The modified DNA base β-d-glucosyl-hydroxymethyluracil is not found in the tsetse fly stages of Trypanosoma brucei. Mol. Biochem. Parasitol. 94:127-130. [DOI] [PubMed] [Google Scholar]
- 74.van Leeuwen, F., E. R. Wijsman, E. Kuyl-Yeheskiely, G. A. van der Marel, J. H. van Boom, and P. Borst. 1996. The telomeric GGGTAA repeats of Trypanosoma brucei contain the hypermodified base J. in both strands. Nucleic Acids Res. 24:2476-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 76.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 77.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 78.Wellinger, R. J., and D. Sen. 1997. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 33:735-749. [DOI] [PubMed] [Google Scholar]
- 79.Wickstead, B., K. Ersfeld, and K. Gull. 2004. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 14:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wirtz, E., S. Leal, C. Ochatt, and G. A. M. Cross. 1999. A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 81.Zhong, Z., L. Shiue, S. Kaplan, and T. de Lange. 1992. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 12:4834-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]