Abstract
MRGX is one of the members of MORF4/MRG family of transcriptional regulators, which are involved in cell growth regulation and cellular senescence. We have shown that MRGX and MRG15 associate with Rb in nucleoprotein complexes and regulate B-myb promoter activity. To elucidate the functions of MRGX and to explore its potential role in modulating cell growth in vivo, we have generated MrgX-deficient mice. Characterization of the expression pattern of mouse MrgX demonstrated it was ubiquitously expressed in all tissues of adult mice and also during embryogenesis and overlapped with its homolog Mrg15. MRGX and MRG15 proteins localize predominantly to the chromatin fraction in the nucleus, although a small amount of both proteins localized to the nuclear matrix. Whereas disruption of Mrg15 results in embryonic lethality, absence of MrgX did not impair mouse development and MrgX null mice are healthy and fertile. MrgX-deficient and wild-type mouse embryonic fibroblasts (MEFs) also had similar growth rates and showed no differences in cell cycle-related gene expression in response to serum stimulation. Mrg15 expression in MrgX-deficient tissues and MEFs was not upregulated compared with wild-type tissues and MEFs. MRG15 is highly conserved with orthologs present from humans to yeast and is essential for survival of mice. In contrast, MRGX, which evolved later, is expressed only in vertebrates, suggesting that the lack of phenotype of MrgX-deficient mice is secondary to a compensatory effect by the evolutionarily conserved MRG15 protein but not vice versa.
Normal human fibroblasts do not divide indefinitely in vitro but finally enter a terminally nondividing state after undergoing several rounds of division (11). This process is referred to as cellular or replicative senescence and is believed to be an antitumor mechanism (32, 35). In studies to identify cellular senescence-related genes, we isolated a novel gene family, of which three members, MORF4 (mortality factor on human chromosome 4), MRG15 (MORF4-related gene on human chromosome 15), and MRGX (MORF4-related gene on human X chromosome), are expressed (2). MORF4 induces senescence in a subset of human tumor cell lines, whereas MRGX and MRG15 do not induce senescence. These three proteins share common motifs which are seen in many transcriptional regulators such as helix-loop-helix (HLH) and leucine zipper (LZ) regions. MRG15 has an N-terminal chromodomain region (2, 3) that has been identified in other chromatin-related proteins and is involved in recognition of methylated histone H3 and RNA binding (1, 4, 7, 16-18). MRGX has a unique N-terminal sequence which has no homology to other proteins. MORF4 lacks the N-terminal regions of both MRG15 and MRGX. We have speculated that the differences in the N-terminal sequences of these three proteins may confer functional differences. Both MRGX and MRG15 interact with Rb and a novel protein, PAM14, and activate or repress the B-myb (Mybl2) promoter in a cell type-dependent manner (34). The HLH and LZ regions are important for both the activation and repression activities and therefore require the common C-terminal motif in both proteins. However, in the case of MRG15, the N-terminal chromodomain-containing region of the protein is also required for activation of the B-myb promoter and it activates versus represses, as does MRGX, this promoter in EJ cells (20, 29). MRG15 has been shown to be one of the components of the human NuA4 histone acetyltransferase multiprotein complex that includes TIP60, which is the catalytic subunit in the complex (5, 6, 8, 9, 15). Although Cai et al. have identified MRGX as a component of the human NuA4 complex (5, 6), other groups have not and additional biochemical analysis is needed to verify this.
Since MRGX is present only in vertebrates, whereas MRG15 is a highly conserved protein with orthologs in yeast to humans (3), MRGX may be involved in higher-order functions in mammalian cells whereas MRG15 is required for more fundamental processes. In fact, the Mrg15-deficient phenotype is embryonic lethal in mice (33), with null embryos exhibiting a runted phenotype and Mrg15 null mouse embryonic fibroblasts (MEFs) displaying a clear growth deficit (33).
In this study, we have generated MrgX-deficient mice to study the function of MrgX and to explore its possible role in modulating cell growth in vivo. We demonstrate that MrgX is expressed ubiquitously in adult mouse tissues and during embryogenesis and its expression pattern is similar to that of Mrg15. We have also found that MrgX null mice (MrgX−/− female and MrgX− male) are viable and healthy and appear to have normal fertility. Analysis of MEFs has demonstrated that MrgX-deficient cells have the same growth rate and express genes related to cell cycle initiation and progression in a manner equivalent to control littermates. The results suggest that MRGX is not essential for either development or cell proliferation and that its function is likely compensated for by MRG15.
MATERIALS AND METHODS
Northern blot analysis.
Mouse MrgX cDNA was PCR amplified from a brain cDNA library of adult C57BL/6J mice using the primers MMRGX-5′ (5′-GGCTTTCTATGGCGGTTGGAGGAG-3′) and MMRGX-3′ (5′-AGACAATAGTGAGCGGTCAGTAGA-3′). The amplified fragment was subcloned into the EcoRV site of pBluescript II and the sequence confirmed. A mouse RNA Master Blot (no. 7771-1; Clontech, Palo Alto, CA) was hybridized using as probe a fragment of the mouse MrgX-specific sequence amplified by PCR using the plasmid mentioned above as a template. The primers were MMRGX-5′ and MMRGX-9 (5′-CTTAAAGTTGTCTTCTTCAGCAGA-3′), and the probe DNA contained a 188-bp 5′ untranslated region and a 66-bp coding sequence.
RNA was isolated from tissues or MEFs using the Trizol reagent (Invitrogen). Frozen tissue samples were homogenized with a 1.5-ml pellet pestle, 1 ml of Trizol was added to the tissue sample, and the suspension was passed thorough a 21-gauge needle to shear the DNA. RNA was extracted according to the manufacturer's instructions. Total RNA (20 μg) was resolved on a 1% agarose gel and then transferred to Hybond-N+ nylon membrane. Blots were hybridized in NorthernMax hybridization buffer (Ambion, Austin, TX) at 42°C overnight with an MrgX- or Mrg15 (as control)-specific probe (36). The plasmids which contain mouse cyclin E1 (Ccne1), cyclin D1 (Ccnd1), Mybl2, and Myc fragment were kindly provided by Nicholas J. Dyson. The blot was washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 65°C twice for 10 min and then washed with 0.2× SSC-0.1% SDS at 65°C twice for 15 min.
Nuclear protein fractionation.
HeLa cells (9.4 × 106) were harvested by trypsin treatment and washed with phosphate-buffered saline (PBS). Protein fractionation was performed by a previously published method (31). In brief, pelleted cells were suspended in 800 μl of RSB buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 1× Protease Inhibitor Cocktail Set I [no. 539131; Calbiochem]), and the cytoplasmic membrane of the cells was disrupted by being passed through a 25-gauge needle 20 times. We confirmed microscopically that over 95% of the cells were effectively disrupted by this treatment. The nuclei were collected by centrifugation at 6,800 × g for 3 min and washed twice with RSB buffer. The pelleted nuclei were suspended in DNase I buffer (10 mM Tris-HCl [pH 7.6], 2.5 mM MgCl2, 0.5 mM CaCl2, 0.5% Triton X-100, 1× Protease Inhibitor Cocktail Set I) supplemented with 4 mM vanadyl ribonucleoside complex (RNase inhibitor; Fluka no. 94742) and 100 U of DNase I (New England BioLabs no. M0303S) and incubated at 30°C for 50 min. After incubation, 100 μl of 1 M ammonium sulfate (final concentration, 0.25 M) was added, and the lysate was centrifuged at 6,800 × g for 3 min and collected (nuclear fraction 1). The pellet was suspended in DNase I buffer supplemented with 2 M NaCl, incubated on ice for 10 min and centrifuged at 6,800 × g for 3 min, and the lysate was collected (nuclear fraction 2). The pellet was suspended in DNase I buffer, RNase A (100 μg/ml) and RNase T1 (40 U/ml) were added, and the mixture was incubated at room temperature for 1 h. The lysate was centrifuged at 6,800 × g for 3 min and collected (nuclear fraction 3). The pellet was dissolved in 1× SDS sample buffer (nuclear fraction 4). We adjusted loading amounts by cell number (corresponding to 5 × 105 cells). Nuclear proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane (Bio-Rad).
Construction of the targeting vector.
The intronic fragment of the mouse MrgX gene was amplified by PCR using 129S6/SvEv tail DNA as a template. The primers for PCR were MMRGX-1 (5′-TGGAAGGGAAAGAAGGAACATTGT-3′) and MMRGX-2 (5′-TCAGCCCGTGCCCTTTTCTTCCG-3′). The amplified fragment (1.1 kb) was subcloned into the EcoRV site of pBluescript II (Stratagene), confirmed by sequencing, and used as a probe for screening of genomic clones. Three independent MrgX genomic clones were isolated from a 129S6/SvEv mouse embryonic stem (ES) cell genomic library (Stratagene, La Jolla, CA).
The targeting vector to inactivate MrgX contained a 3.4-kb EcoRI-ClaI fragment of the MrgX gene for the 5′ homology arm, a loxP site, a 2.2-kb ClaI-HindIII fragment of the MrgX gene, a PGK-neo selectable marker cassette floxed by loxP sites, a 3.0-kb HindIII-BamHI fragment of the MrgX gene for the 3′ homology arm, and an MC1tk expression cassette for negative selection. Twenty-five micrograms of KpnI-linearized targeting vector was electroporated into 107 AB2.2 ES cells. ES clones were selected in culture medium containing G418 and 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5′-iodouracil. DNA from the clones was analyzed by Southern blotting, targeted ES cell clones were expanded and injected into C57BL/6J blastocysts, and chimeric males were mated with C57BL/6J females as previously described (24). Germ line transmission was obtained from chimeras derived from two independent ES cell clones (MrgX-126-B7 and MrgX-126-D5).
To produce a MrgX-deficient mouse line, mice which had a recombinant floxed allele of the MrgX gene were mated with EIIa-cre transgenic mice which express Cre recombinase under the control of the adenovirus EIIa promoter (19).
The generation of Mrg15+/− mice, which were used for double-knockout (DKO) analysis, has been previously reported by our group (33).
Genotyping.
Either tail DNA, DNA from the yolk sac, or DNA from a part of the embryo digested with EcoRV or EcoRI (for MrgX) or BamHI (for Mrg15) at 37°C overnight was submitted to Southern analysis to determine the genotype of the embryos or mice. The digested DNAs were separated on 0.7% agarose gel and transferred to Hybond-N+ (Amersham Pharmacia) using an alkaline transfer method. The membrane was probed with the 3′ external or 5′ internal probe (for MrgX) using Rapid Hybridization solution (Amersham Pharmacia). The Mrg15 probe for genotyping has been described previously (33). Genotyping of the EIIa-cre transgene was performed by PCR using 5′-CCGGGCTGCCACGACCAA-3′ and 5′-GGCGCGGCAACACCATTTTT-3′ primers.
Western blot analysis.
Tissues from adult mice were lysed with TNESV buffer (20 mM Tris-HCl [pH 7.5], 1% NP-40, 1 mM EDTA, 150 mM NaCl, 10% glycerol, 1× Protease Inhibitor Cocktail Set I [Calbiochem]). The lysates were kept on ice for 30 min and centrifuged at 20,000 × g for 15 min. The protein concentration of the supernatants was determined by the Bradford method (Bio-Rad). The total proteins (100 μg) were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. MRG15 protein was detected using rabbit anti-MRG15 C-terminal antibody raised by our laboratory. Mouse anti-β-actin antibody (Abcam AC-15) was used as a loading control.
Antibodies used to detect MRG15, MRGX, and lamin A/C in nuclear protein fractionated preparations were rabbit anti-MRG15 C-terminal antibody (1:1,000; antigen, SASDYEVAPPEYHRKAV), rabbit anti-MRGX antibody (1:1,000; antigen, KQGSQPRGQQSA), and goat anti-lamin A/C antibody (1:500; Santa Cruz, sc-6215). The anti-MRG15 and anti-MRGX antibodies were affinity purified using an antigen column, and their specificity was confirmed by Western blotting using lysates of cells expressing hemagglutinin-tagged MRG15 or MRGX. Anti-MRG15 or anti-MRGX and anti-hemagglutinin antibodies detected the same protein band.
Generation of MEFs and analysis of growth properties.
We bred MrgX− males with MrgX+/− females to isolate13.5-day-old (E13.5) embryos. After removal of the head and abdominal organs, each embryo was washed with PBS and minced, and the tissue was placed in a 15-ml conical tube. After centrifugation, 1 ml of trypsin solution (0.25% trypsin-0.005% EDTA in PBS) was added to the pelleted tissue, which was digested on ice overnight. Trypsin was inactivated by the addition of Eagle's minimum essential medium (Invitrogen) containing 10% fetal bovine serum (FBS), 2 mM glutamine, 0.1 mM nonessential amino acids, and 50 μg of gentamicin per ml. After pipetting several times, the cells were plated into one T75 tissue culture flask and incubated at 37°C until confluent. We designated this culture PD 0.
For the growth study, equal numbers of cells (1 × 105) from PD 0 MEFs were plated into 60-mm-diameter tissue culture dishes and maintained in 5% CO2 at 37°C and counted every 24 h (in triplicate) using a Coulter Counter. We tested at least two independent MEF lines for each genotype for the growth study.
For gene expression analysis, PD 0 MEFs (1 × 106) were plated into 100-mm-diameter tissue culture dishes and maintained at 37°C for 2 days. The cells were washed twice with PBS, 0.1% FBS-containing medium was added, and the cells were maintained at 37°C for 3 days. The cells were then stimulated by the addition of 10% FBS-containing medium, and total RNA was isolated at various points thereafter.
RESULTS AND DISCUSSION
MrgX mRNA expression during mouse development and in adult mice.
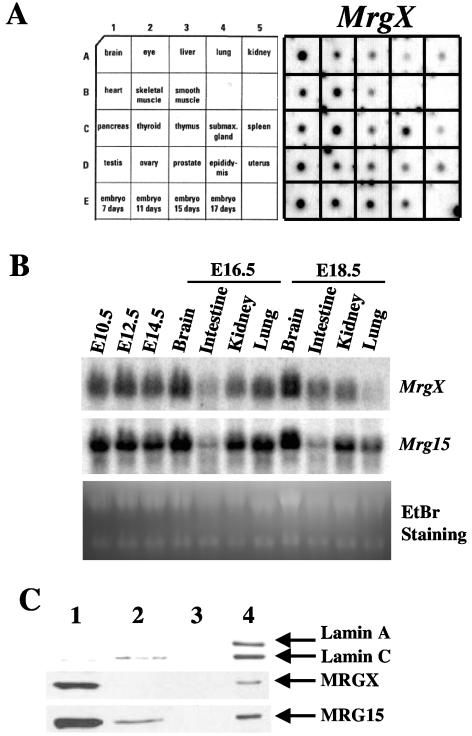
To determine expression of mouse MrgX, we performed dot blot and Northern blot analyses of embryonic and adult tissues. MrgX mRNA was ubiquitously expressed in embryos beginning at 7 days and in adult tissues (Fig. 1A), and the expression pattern was very similar to that of Mrg15 mRNA (36). Mrg15 mRNA appears to be more highly expressed in adult testis based on the hybridization signal.
FIG. 1.
Expression of MrgX mRNA and protein. (A) MrgX mRNA expression in adult tissues and embryos. A mouse RNA Master Blot was hybridized using an MrgX-specific probe. (B) MrgX and Mrg15 mRNA expression during mouse development. MrgX and Mrg15 mRNA expression in embryos and embryonic tissues was determined by Northern blotting. EtBr staining of RNAs (28S and 18S) indicated an equal amount of loading of the various samples. (C) Subnuclear fractionation analysis of MRGX and MRG15 proteins in HeLa cells. Nuclear proteins from HeLa cells were fractionated as reported previously (31). The amount of sample loaded in each lane was based on cell number (protein from 5 × 105 cells per lane). MRGX and MRG15 proteins were detected by Western blotting. The presence of lamins A and C served as a marker for the nuclear matrix fraction. Lanes: 1, 0.25 M (NH4)2SO4 extracted fraction after DNase I treatment; 2, 2 M NaCl extracted fraction; 3, extracted fraction after RNase A plus RNase T1 treatment; 4, nuclear matrix (pelleted) fraction.
Next, we analyzed MrgX versus Mrg15 mRNA expression in embryos and embryonic tissues at later stages of embryogenesis (Fig. 1B) and determined that MrgX, as well as Mrg15, is highly expressed in early stages of embryos similar to that observed from dot blot analyses. MrgX and Mrg15 mRNA expression in embryonic brain was high, and this did not change at later stages (compare E16.5 and E18.5). Expression of MrgX and Mrg15 in intestine and kidney was moderate, and there was no major change in the expression pattern between E16.5 and E18.5. Both MrgX and Mrg15 were moderately expressed in lung at E16.5, but expression decreased slightly at E18.5. These results suggest that the differential expression of MrgX and Mrg15 in different tissues at various stages of embryogenesis may reflect gene expression regulation by these proteins and impact on development. The most interesting fact is that the expression pattern of MrgX in embryonic tissues is generally similar to that of Mrg15.
Nuclear protein fractionation analysis of MRGX and MRG15.
To determine the localization of MRGX in cells, we transfected a plasmid construct which encoded MRGX fused with enhanced green fluorescent protein into HeLa cells. Enhanced green fluorescent protein-MRGX localized to the nucleus, similar to MRG15, as described before (data not shown) (36). Next, we determined MRGX and MRG15 protein localization in the nucleus following biochemical protein fractionation. Most of the MRGX and MRG15 protein was eluted by 0.25 M (NH4)2SO4 solution following DNase I treatment (Fig. 1C, fraction 1), which causes removal of chromatin from nuclei together with many other nuclear proteins (over 80% of nuclear proteins) (12). Thus, most of the MRGX and MRG15 protein appears to be present in the nucleoplasm and/or weakly attached to chromatin. Interestingly, a small amount of MRGX and MRG15 localized to the nuclear matrix fraction, in which lamins A and C were localized (Fig. 1C, fraction 4). Recently, it has become evident that the nuclear lamina, which is a filamentous nuclear structure involved in many nuclear activities including transcription, RNA splicing, DNA replication, chromatin organization, cell cycle regulation, cell differentiation, and apoptosis (23, 28, 38), is dysregulated or mutated in genes in a wide range of human diseases such as cancer (26, 27) and Hutchinson-Gilford progeria syndrome (10, 30). The latter premature-aging syndrome is also observed in mutant mice with genetically modified lamin A (25). We do not know the physiological implications of the nuclear matrix localization of MRGX and MRG15 proteins at this time. We have reported that MRGX and MRG15 are both associated with Rb (20, 29, 34) and some fraction of Rb is known to be associated with the nuclear matrix (21). The hypophosphorylated form of Rb binds to lamin A/C and is anchored by binding to lamina-associated polypeptide 2α-lamin A/C complexes (22). It has been speculated that the lamin/lamina-associated polypeptide 2α complex may tether Rb molecules to a promoter and form an effective silencing complex. Thus, one possible explanation for the nuclear matrix localization of MRGX and MRG15 is that they are tethered with Rb in the nuclear matrix and are involved in Rb-related functions.
Disruption of mouse MrgX by gene targeting.
The genomic structure of the mouse MrgX gene is very similar to that of the human MrgX that is deposited in GenBank (accession no. AL049610). The 5′ untranslated region is divided by at least three exons, and the coding sequence of both the mouse and human MrgX is encoded by only one exon. We decided to use a conditional-knockout system for the generation of MrgX-deficient ES lines because the ES cell line (AB2.2) that we use is of male origin and has just one X chromosome. If MrgX was essential for cell growth, and we used a conventional knockout method, we would obtain no positive ES clones from screening. The strategy we used is depicted in Fig. 2A. The coding exon of the mouse MrgX gene was flanked by two loxP sites so that Cre recombinase-mediated excision of this entire region would result in loss of production of MRGX. The linearized targeting construct was electroporated into ES cells, followed by drug selection. We screened 93 drug-resistant clones by Southern blotting using external 5′ and internal 3′ probes, and 35 clones (37.6%) displayed the correct targeting pattern on both sides. ES cells from two independent clones were injected into blastocysts to generate mice bearing the MrgX recombinant floxed allele. This allele was successfully germ line transmitted (Fig. 2B, MX9-1). The resulting MrgXFlox/Flox females and MrgXFlox males were healthy and fertile, indicating that the recombinant allele had no overt effects on the mice, and MrgX mRNA was expressed at the same level as in the wild type in MrgXFlox male mice (Fig. 2C).
FIG. 2.
Disruption of the mouse MrgX locus by gene targeting. (A) Genomic structure of the mouse MrgX gene and the targeting vector used to generate the conditionally targeted allele of MrgX. The targeting vector was constructed by flanking a 2.2-kb region containing the MrgX coding exon by two loxP sites with a PGK-neo cassette. The regions of homology consist of a 3.4-kb region for the 5′ arm and a 3-kb region for the 3′ arm, respectively. The MC1tk cassette was located next to the 3′ homology arm. The probe fragments for genotyping are indicated as shaded boxes at the bottom. Abbreviations: E5, EcoRV; E1, EcoRI; H3, HindIII; C1, ClaI; B1, BamHI. (B) Southern blot analysis of tail DNA from pups at weaning from breeding pairs containing the recombinant (floxed) allele (MX9-1) and the MrgX-deleted [Δ (−)] allele (MX13-1). Genomic DNAs were digested with EcoRV or EcoRI and hybridized with 5′ or 3′ probes, respectively. WT, wild type. (C) Northern blot analysis of MrgX expression in wild-type (+), MrgX− (−), and MrgXFlox (floxed allele, F) males. EtBr staining of RNAs (28S and 18S) was used to demonstrate equal loading of samples.
To generate MrgX-deficient mice, we crossed MrgXFlox/+ females with adenovirus EIIa promoter-driven deleter (EIIa-cre) males (19). Cre recombinase effectively deleted the coding exon of the mouse MrgX gene from the MrgX recombinant allele and produced an MrgX deletion (Δ) allele (Fig. 2B, MX13-1). We obtained MrgX-deficient mice (MrgX−/− females and MrgX− males) at the expected ratio from a combination of mating pairs, indicating no impairment in mouse development.
We confirmed that there was no expression of MrgX in tissues from adult MrgX− males (Fig. 2C) and MrgX−/− females (data not shown) by Northern blotting. The MrgX-deficient mice were healthy and fertile, and we did not observe any obvious abnormalities after 1 year. This suggests that MrgX is not required during embryogenesis or in adult mice under normal conditions.
Cell growth and expression of cell cycle-related genes in MrgX-deficient MEF in response to serum stimulation.
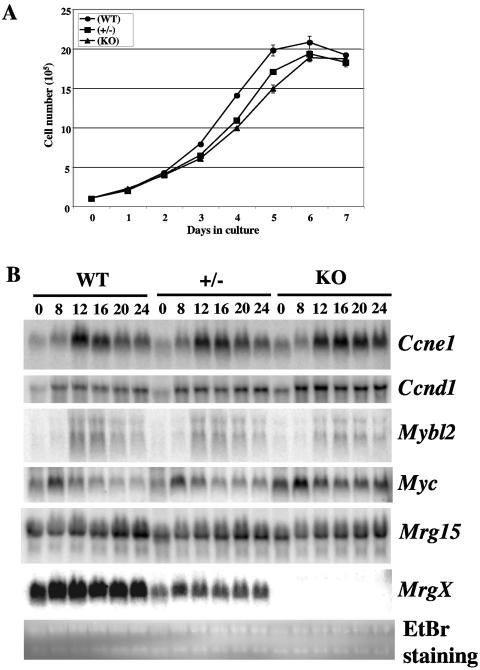
We have found that Mrg15-deficient MEFs have a growth defect compared with control cells (33) and that p21Sdi1/Cip1/Waf1 is upregulated in these MEFs at earlier passages. To determine whether MRGX had a similar effect on cell growth, we bred MrgX+/− females with MrgX− males and isolated MEFs from E13.5 embryos. Daily cell counts revealed no significant differences in the growth rates of wild-type (WT), MrgX+/− (+/−), and MrgX-deficient (knockout [KO]) MEFs (Fig. 3A).
FIG. 3.
MEF analyses. (A) Growth kinetics of MEFs. MEFs (1 × 105) were plated in 60-mm-diameter dishes and counted every 24 h. This result is representative of two independent experiments. (B) Expression patterns of cell cycle-regulated genes by Northern blot analysis. Quiescent MEFs were stimulated by adding 10% FBS-containing medium, and total RNA was isolated at the indicated time points (hours after stimulation). The expression levels of cyclin E1 (Ccne1), cyclin D1 (Ccnd1), Mybl2, Myc, Mrg15, and MrgX mRNAs were detected by Northern blotting. EtBr staining of RNAs (28S and 18S) served as a control for equal amounts of loading of samples. WT, wild type.
Since both MRGX and MRG15 interact with Rb, we then determined whether MrgX-deficient MEFs responded to serum stimulation with expected changes in expression of the cell cycle-related genes encoding cyclin E1 (Ccne1), cyclin D1 (Ccnd1), B-myb (Mybl2), and c-myc (Myc), which are known to be expressed at G1/S. The expression of all four genes in wild-type (WT), MrgX+/− (+/−), and MrgX-deficient (KO) MEFs was low in the quiescent state (0 h) and increased in a time-dependent manner after serum stimulation (Fig. 3B). Again, no difference was observed in the expression patterns of the three genotypes. Cyclin E1 expression is upregulated at G0 in Rb-deficient MEFs because Rb repression of cyclin E1 expression is abrogated (13, 14). However, cyclin E1 expression remained repressed in MrgX-deficient MEFs at G0 (0 h). The expression levels of Mrg15 remained constant during this time and, importantly, there was no difference in Mrg15 expression levels among the three genotypes. We confirmed that half the amount of MrgX transcript and no MrgX transcript was expressed in MrgX+/− and MrgX-deficient MEFs, respectively, using the same membrane. Ethidium bromide (EtBr) staining of 28S and 18S rRNAs was used as a loading control (Fig. 3B).
Collectively, these data indicate that MRGX is not essential for cell growth control, and this is most likely due to a compensatory effect of MRG15.
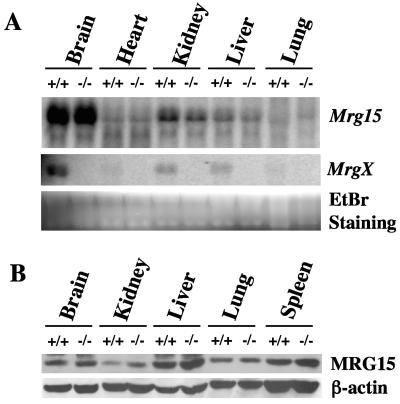
Mrg15 expression in MrgX-deficient mice.
The lack of phenotype of MrgX deficiency indicates that MRG15 or some other protein(s) must compensate for the function(s) of MrgX in deficient mice. We therefore determined Mrg15 expression in tissues from adult MrgX-deficient mice. Mrg15 mRNA expression in brain, heart, kidney, liver, and lung tissues from MrgX−/− mice was the same as that of the wild type (Fig. 4A) despite the confirmation that MrgX mRNA was not detected in tissues from MrgX null mice. MRG15 protein levels in brain, kidney, liver, lung, and spleen tissues were also not different in wild-type versus MrgX null mice (Fig. 4B). β-Actin demonstrated equal loading in the various samples. These results indicate that Mrg15 mRNA, as well as protein, was not upregulated in the absence of MrgX.
FIG. 4.
Mrg15 expression in MrgX-deficient mice. (A) Mrg15 and MrgX mRNA expression in various tissues from wild-type (+/+) and MrgX−/− (−/−) mice. Expression of mRNA was determined by Northern blotting. No MrgX transcript was present in MrgX−/− tissues. EtBr staining of RNAs (28S and 18S) was the control for equal amounts of loading of samples. (B) MRG15 protein in various tissues from wild-type (+/+) and MrgX−/− (−/−) mice was determined by Western blot analysis. Equal amounts of loading of wild-type and MrgX−/− samples was confirmed using an anti-β-actin antibody. These results are representative of analyses of multiple male or female mice.
Overlapping functions of MrgX and Mrg15 in early embryogenesis.
To investigate the possibility of functional redundancy of MrgX and Mrg15 during embryogenesis, we crossed Mrg15+/− MrgX− males and Mrg15+/− MrgX−/− females, collected embryos at the indicated embryonic stages, and determined the genotype of the embryos (Table 1). We did not obtain Mrg15−/− MrgX− or MrgX−/− (DKO) embryos at E14.5, the stage at which we were able to identify Mrg15 single-KO embryos occurring at the expected Mendelian frequency (33). DKO embryos were present at E10.5 and were smaller than their littermates. The results indicate that Mrg15 and MrgX have overlapping functions during early mouse embryogenesis.
TABLE 1.
Genotype analysis of Mrg15+/− MrgX− male × Mrg15+/− MrgX−/− female intercrosses
| Age | No. with genotype of:
|
No. of litters | ||
|---|---|---|---|---|
| Mrg15+/+MrgX− or MrgX−/− | Mrg15+/−MrgX− or MrgX−/− | Mrg15−/−MrgX− or MrgX−/− | ||
| E10.5 | 2 | 2 | 2a | 1 |
| E11.5 | 5 | 10 | 0 | 2 |
| E14.5 | 4 | 13 | 0 | 3 |
Note that these embryos were smaller than other littermates.
MRGX protein shares common motifs in the C-terminal area with MRG15 such as the HLH and LZ (2, 3), although both proteins have specific and different sequences in the N-terminal region. The common motifs between MRGX and MRG15 are important for protein-protein interactions, such as Rb and HDAC1 (20, 34, 37), and transcriptional regulation of the B-myb promoter. Thus, it is very likely that the MrgX deficiency is compensated for by MRG15, although Mrg15 expression (both mRNA and protein levels) in MrgX-deficient mice and MEFs is not upregulated.
Mrg15 null embryos are lethal at E14.5, and MEFs exhibit growth defects. In contrast, MrgX deficiency has no obvious phenotype in development, adult mice, and MEF cultures. MRG15 is highly conserved evolutionarily from yeast to humans, although it is not known whether it has a fundamental function(s) in cellular processes in all of these organisms. MRGX is present only in mammals, and therefore the possibility exists that it evolved to perform some very specific and subtle function(s) in mammals that we have not yet discovered. For example, because MRGX is highly expressed in the brain, it may be important for more sophisticated brain functions such as learning and memory. Thus, the MrgX-deficient mouse model has the potential to determine additional MORF4/MRG family gene functions in vivo in the future.
Acknowledgments
We thank Simona Varani for ES cell technology, Bisong Haupt for ES cell injection, Johanna Echigo and Julio Agno for maintaining mice, Meihua Song for purification of antibodies, Emiko Tominaga for MEF and HeLa cell cultures, and James R. Smith for discussion. We also thank all members of the Smith group at UTHSCSA for many valuable discussions and comments.
This work was supported by NIH grants P01AG2752 (O.M.P.S.) and CA60651 (to M.M.M.), the Ellison Medical Foundation (O.M.P.S.), and the American Federation for Aging Research (K.T.).
REFERENCES
- 1.Akhtar, A., D. Zink, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407:405-409. [DOI] [PubMed] [Google Scholar]
- 2.Bertram, M. J., N. G. Berube, X. Hang-Swanson, Q. Ran, J. K. Leung, S. Bryce, K. Spurgers, R. J. Bick, A. Baldini, Y. Ning, L. J. Clark, E. K. Parkinson, J. C. Barrett, J. R. Smith, and O. M. Pereira-Smith. 1999. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 19:1479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram, M. J., and O. M. Pereira-Smith. 2001. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene 266:111-121. [DOI] [PubMed] [Google Scholar]
- 4.Brehm, A., K. R. Tufteland, R. Aasland, and P. B. Becker. 2004. The many colours of chromodomain. Bioessays 26:133-140. [DOI] [PubMed] [Google Scholar]
- 5.Cai, Y., J. Jin, L. Florens, S. K. Swanson, T. Kusch, B. Li, J. L. Workman, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280:13665-13670. [DOI] [PubMed] [Google Scholar]
- 6.Cai, Y., J. Jin, C. Tomomori-Sato, S. Sato, I. Sorokina, T. J. Parmely, R. C. Conaway, and J. W. Conaway. 2003. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278:42733-42736. [DOI] [PubMed] [Google Scholar]
- 7.Cavalli, G., and R. Paro. 1998. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol. 10:354-360. [DOI] [PubMed] [Google Scholar]
- 8.Doyon, Y., and J. Cote. 2004. The highly conserved and multifunctional NuA4 complex. Curr. Opin. Genet. Dev. 14:147-154. [DOI] [PubMed] [Google Scholar]
- 9.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Cote. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriksson, M., W. T. Brown, L. B. Gordon, M. W. Glynn, J. Singer, L. Scott, M. R. Erdos, C. M. Robbins, T. Y. Moses, P. Berglund, A. Dutra, E. Pak, S. Durkin, A. B. Csoka, M. Boehnke, T. W. Glover, and F. S. Collins. 2003. Recurrent de nono point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423:293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 12.He, D., J. A. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera, R. E., V. P. Sah, B. O. Williams, T. P. Makela, R. A. Weinberg, and T. Jacks. 1996. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell. Biol. 16:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurford, R. K. J., D. Cobrinik, M.-H. Lee, and N. Dyson. 1997. pRb and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 15.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 16.Jones, D. O., I. G. Cowell, and P. B. Singh. 2000. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22:124-137. [DOI] [PubMed] [Google Scholar]
- 17.Koonin, E. V., S. Zhou, and J. C. Lucchesi. 1995. The chromo superfamily: new members, duplication of the chromodomain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 23:4229-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 19.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung, J. K., N. G. Berube, A. Venable, S. Ahmed, N. Timchenko, and O. M. Pereira-Smith. 2001. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 276:39171-39178. [DOI] [PubMed] [Google Scholar]
- 21.Mancini, M. A., B. Shan, J. A. Nickerson, S. Penman, and W.-H. Lee. 1994. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc. Natl. Acad. Sci. USA 91:418-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markiewicz, E., T. Dechat, R. Foisner, R. A. Quinlan, and C. J. Hutchison. 2002. Lamin A/C binding protein LAP2α is required for nuclear anchorage of retinoblastoma protein. Mol. Biol. Cell 13:4401-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattout-Drubezki, A., and Y. Gruenbaum. 2003. Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell. Mol. Life Sci. 60:2053-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matzuk, M. M., M. J. Finegold, J. J. Su, A. J. W. Hsueh, and A. Bradley. 1992. Alpha-inhibin is a tumor suppressor gene with gonadal specificity in mice. Nature 360:313-319. [DOI] [PubMed] [Google Scholar]
- 25.Mounkes, L. C., S. Kozlov, L. Hernandez, T. Sullivan, and C. L. Stewart. 2003. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature 423:298-301. [DOI] [PubMed] [Google Scholar]
- 26.Mounkes, L. C., and C. L. Stewart. 2004. Aging and nuclear organization: lamins and progeria. Curr. Opin. Cell Biol. 16:322-327. [DOI] [PubMed] [Google Scholar]
- 27.Muchir, A., and H. J. Worman. 2004. The nuclear envelope and human disease. Physiology 19:309-314. [DOI] [PubMed] [Google Scholar]
- 28.Nickerson, J. A. 2001. Experimental observations of a nuclear matrix. J. Cell Sci. 114:460-474. [DOI] [PubMed] [Google Scholar]
- 29.Pardo, P. S., J. K. Leung, J. C. Lucchesi, and O. M. Pereira-Smith. 2002. MRG15 a novel chromodomain protein is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem. 277:50860-50866. [DOI] [PubMed] [Google Scholar]
- 30.Pollex, R. L., and R. A. Hegele. 2004. Hutchinson-Gilford progeria syndrome. Clin. Genet. 66:375-381. [DOI] [PubMed] [Google Scholar]
- 31.Ran, Q., R. Wadhwa, O. Bischof, S. Venable, J. R. Smith, and O. M. Pereira-Smith. 2001. Characterization of a novel zinc finger gene with increased expression in nondividing normal human cells. Exp. Cell Res. 263:156-162. [DOI] [PubMed] [Google Scholar]
- 32.Smith, J. R., and O. M. Pereira-Smith. 1996. Replicative senescence: implications for in vivo aging and tumor suppression. Science 273:63-67. [DOI] [PubMed] [Google Scholar]
- 33.Tominaga, K., B. Kirtane, J. G. Jackson, Y. Ikeno, T. Ikeda, C. Hawks, J. R. Smith, M. M. Matzuk, and O. M. Pereira-Smith. 2005. MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 25:2924-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tominaga, K., J. K. Leung, P. Rookard, J. Echigo, J. R. Smith, and O. M. Pereira-Smith. 2003. MRGX: a novel transcriptional regulator that exhibits activation or repression of B-myb promoter in a cell type dependent manner. J. Biol. Chem. 278:49618-49624. [DOI] [PubMed] [Google Scholar]
- 35.Tominaga, K., A. Olgun, J. R. Smith, and O. M. Pereira-Smith. 2002. Genetics of cellular senescence. Mech. Ageing Dev. 123:927-936. [DOI] [PubMed] [Google Scholar]
- 36.Tominaga, K., and O. M. Pereira-Smith. 2002. The genomic organization, promoter position and expression profile of the mouse MRG15 gene. Gene 294:215-224. [DOI] [PubMed] [Google Scholar]
- 37.Yochum, G. S., and D. E. Ayer. 2002. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol. Cell. Biol. 22:7868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zink, D., A. H. Fischer, and J. A. Nickerson. 2004. Nuclear structure in cancer cells. Nature Rev. Cancer 4:677-687. [DOI] [PubMed] [Google Scholar]