Abstract
Epstein-Barr virus (EBV) genomes persist indefinitely in latently infected human cells, in part due to their ability to stably segregate during cell division. This process is mediated by the viral EBNA1 protein, which tethers the viral episomes to the cellular mitotic chromosomes. We have previously identified a mitotic chromosomal protein, human EBNA1 binding protein 2 (hEBP2), which binds to EBNA1 and enables EBNA1 to partition EBV-based plasmids in Saccharomyces cerevisiae. Using an RNA silencing approach, we show that hEBP2 is essential for the proliferation of human cells and that repression of hEBP2 severely decreases the ability of EBNA1 and EBV-based plasmids to bind mitotic chromosomes. When expressed in yeast, hEBP2 undergoes the same cell cycle-regulated association with the mitotic chromatin as in human cells, and using yeast temperature-sensitive mutant strains, we found that the attachment of hEBP2 to mitotic chromosomes was dependent on the Ipl1 kinase. Both RNA silencing of the Ipl1 orthologue in human cells (Aurora B) and specific inhibition of the Aurora B kinase activity with a small molecule confirmed a role for this kinase in enabling hEBP2 binding to human mitotic chromosomes, suggesting that this kinase can regulate EBV segregation.
Epstein-Barr virus (EBV) infects most people worldwide and stably persists for the life of the host through latent infection of B lymphocytes and epithelial cells (40). Due to the immortalization effects of the viral latency proteins, latent infection predisposes the host to develop a variety of malignancies. Only one viral protein, EBNA1, is required to maintain the viral episomes in proliferating latently infected cells, as it plays important roles in both the replication and the segregation of the EBV episomes during cell division (51). EBNA1 contributes to DNA replication by binding to specific sequences in the dyad symmetry element of the latent origin of replication, oriP, while the segregation of the EBV episomes requires EBNA1 binding to the 20 tandem recognition sites in the family of repeats (FR) element of oriP (30, 39). The interaction of EBNA1 with the dyad symmetry and FR sequences occurs through the DNA binding and dimerization domain of EBNA1, which is located between amino acids 459 and 607 (2, 45). In addition to this domain, the replication and segregation functions of EBNA1 require N-terminal sequences of EBNA1; replication appears to involve multiple redundant regions of the EBNA1 N terminus, while segregation function is absolutely dependent on the Gly-Arg-rich sequence between amino acids 325 and 376 and moderately affected by amino acids 8 to 67 (33, 42, 49).
The concept that EBNA1 mediated the segregation of EBV episomes by tethering them to the cellular mitotic chromosomes stemmed from observations that EBNA1, EBV episomes, and oriP-containing constructs were all tightly associated with mitotic chromosomes and that the association of oriP plasmids with mitotic chromosomes was EBNA1 dependent (17, 20, 27, 38, 43). Mutational analyses of EBNA1 also supported the chromosomal tethering model of segregation, as mutations in EBNA1 that negatively affected chromosome attachment had similar effects on the segregation function of EBNA1 (48, 49). In addition, Hung et al. (25) showed that the EBNA1 N-terminal region responsible for chromosome attachment and oriP plasmid maintenance could be functionally replaced by the chromosome binding sequences of high-mobility group 1 and histone H1.
While EBNA1 is thought to attach to mitotic chromosomes by binding one or more cellular protein components, the nature of these cellular proteins and the mechanism by which their association with EBNA1 is regulated has not been clear. A few years ago, we identified a previously uncharacterized cellular protein that we called EBNA1 binding protein 2 (EBP2) from a two-hybrid screening for human proteins that specifically bind EBNA1 (42). This protein is conserved in eukaryotes, and the Saccharomyces cerevisiae homologue, which is strictly localized to the nucleolus, was subsequently shown to play an essential role in rRNA processing (24, 47). Mapping of the human EBP2 (hEBP2) binding region on EBNA1 showed that efficient hEBP2 binding required amino acids 325 to 376 and was moderately affected by amino acids 8 to 67 (42, 49). The ability of EBNA1 to bind hEBP2 correlated with its ability to associate with mitotic chromosomes and segregate oriP plasmids, suggesting that hEBP2 might play a role in this process. The cellular localization of hEBP2 was also consistent with this hypothesis, since hEBP2, which is nucleolar in interphase (7), is found all over the condensed chromosomes in mitosis, much like EBNA1 (48).
Direct evidence that hEBP2 could function with EBNA1 to segregate oriP plasmids came from reconstitution experiments with S. cerevisiae. In this system, yeast-replicating plasmids containing the EBV FR element stably segregate only in the presence of both EBNA1 and hEBP2, and mutations in EBNA1 that affect oriP plasmid segregation in human cells have similar effects on the segregation of the FR-containing plasmids in yeast (29, 49). The ability of hEBP2 to function in EBNA1-mediated plasmid partitioning was shown to require two hEBP2 domains, a central coiled-coil domain that binds to mitotic chromosomes in both yeast and human cells and a C-terminal domain that binds to EBNA1 (28). When expressed on its own in yeast, hEBP2 was found to associate with the entire mass of mitotic chromatin, whereas EBNA1 associated with the mitotic chromatin only in the presence of hEBP2 (28). These results indicate that the role of hEBP2 in this segregation system is in tethering EBNA1 to the mitotic chromosomes and that EBNA1-mediated plasmid segregation in yeast occurs through chromosome attachment, as it does in humans.
While the data show that EBNA1 can partition plasmids by binding to hEBP2 on mitotic chromosomes, several important questions remain unanswered. First, since hEBP2 is only one of many protein components of human mitotic chromosomes, the relative importance of this protein as an EBNA1 attachment point in human cells is unclear, as there could be other chromosomal proteins that are bound by EBNA1. Secondly, the mitotic chromosomal component to which hEBP2 itself binds is unknown, as is the mechanism by which hEBP2 redistributes from its specific localization in the nucleolus in interphase to extensive association with the condensed chromosomes in mitosis. In this paper, we show that the silencing of hEBP2 expression in human cells has pronounced effects on the ability of EBNA1 and oriP plasmids to associate with mitotic chromosomes. We also show that the attachment of hEBP2 to mitotic chromosomes is dependent on the Ipl1/Aurora kinase in the yeast-reconstituted segregation system and in human cells.
MATERIALS AND METHODS
Immunofluorescence microscopy of human cells.
Log-phase cells were adhered to poly-l-lysine-coated coverslips and prepared for immunofluorescence microscopy as described previously (49). hEBP2 was detected with rabbit antibody raised against highly purified hEBP2 (48), followed by fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (Invitrogen). EBNA1 was detected with the mouse monoclonal antibody OT1x (kindly provided by Jaap Middeldorp) followed by Texas Red-conjugated goat anti-mouse antibody (Molecular Probes). In all cases, DNA was visualized by DAPI (4′,6′-diamidino-2-phenylindole; 25 ng/μl) staining. Microscopy was performed at ×400 magnification using a Leica DMR microscope and OpenLab software.
HeLa, 293T, and C33A cells expressing EBNA1 were generated by transfecting the cells with an oriP plasmid expressing EBNA1 (pc3oriPE) (42) and growing the cells under G418 selection (300 μg/ml) for this plasmid. For mitotic chromosome spreads, cells were blocked in mitosis with Colcemid (0.1 μg/ml) for 16 h and chromosomes were prepared using the methanol:acetic acid method described by Kapoor and Frappier (28). The chromosomes were then stained for hEBP2 and EBNA1 (when present) and counterstained with DAPI. Some spreads were stained for B23 instead of EBNA1, using goat anti-B23 (Santa Cruz) and donkey anti-goat Cy3 (Chemicon) antibodies. All chromosome spreads were analyzed at ×630 magnification.
Western blots on fractionated human cell lysates.
Cells transfected with siRNA constructs (or the control construct) were blocked in mitosis with Colcemid (0.1 μg/ml medium) for 16 (hEBP2 silencing experiments) or 6 (Aurora kinase silencing experiments) hours. Equal cell numbers from each sample were then harvested by mitotic shake-off and lysed in 100 μl of Buffer A (20 mM Tris HCl [pH 7.5], 75 mM KCl, 30 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol, 0.5 mM EDTA,1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine). Fifty microliters of the cell lysate was removed (sample W), and the remaining 50 μl of lysate was spun at 10,000 rpm for 10 min at 4°C in a microcentrifuge. The supernatant (sample S) was removed, and the pellet was resuspended in 50 μl of Buffer A (sample P). Twenty-five microliters of each W, S, and P sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotted using rabbit antibody against hEBP2 or OT1x monoclonal antibody against EBNA1. In control experiments, samples were probed with mouse anti-histone H1 (Santa Cruz), goat anti-B23 (Santa Cruz), or mouse anti-Nap1 (26) (kindly provided by Yukio Ishimi). Membranes were developed with an ECL Plus system (Amersham Biosciences) and, for quantification purposes, chemifluorescence was measured using a Typhoon 9400 variable mode imager (Amersham Biosciences) set at Laser Blue 457. The resulting bands were quantified with Image-Quant 5.0 software.
RNA silencing.
HeLa, 293T, and C33A cells that do or do not express EBNA1 were plated at 5 × 105 cells per 6-cm dish 24 h prior to transfection with the ESCORT reagent according to the manufacturer's instructions (Sigma). Cells were separately transfected with 2 μg of pSilencer 2.1 U6 plasmid (Ambion) expressing hairpin RNA designed to silence either hEBP2 ([AA]GCTAAGCAACTGCGAGCAC), Aurora A ([AA]ATGCCCTGTCTTACTGTCA), Aurora B ([AA]CGCGGCACTTCACAATTGA), Aurora C ([AA]GTCGCAGATAGAGAAGGAA) or green fluorescent protein (GFP; no silencing control). After transfection, the cells were grown under hygromycin (150 μg/ml) selection for pSilencer until the target protein was silenced (3 days for hEBP2, 4 days for Aurora B, 5 days for Aurora A, and 6 days for Aurora C). G418 was also added to EBNA1-expressing cells to maintain the EBNA1 expression plasmid. To determine silencing efficiency, equal amounts of cell lysates prepared before and after transfection were analyzed by Western blotting. Blots were probed with anti-USP7 rabbit serum as a loading control (22) and with either rabbit anti-hEBP2 (48), goat anti-Aurora A (Santa Cruz), anti-Aurora B (Abcam), or anti-Aurora C antibodies (Abgent) and then with goat anti-rabbit (Bio-Rad Laboratories) or donkey anti-goat (Santa Cruz) antibodies. Blots from EBNA1-expressing HeLa cells were also probed with EBNA1 monoclonal antibody OT1x. Fractionation of cell lysates was performed (as described above) as soon as silencing of the target protein was observed (3 to 6 days posttransfection, as indicated above).
C666-1 cells.
The EBV-transformed nasopharyngeal carcinoma cell line, C666-1 (9), was kindly supplied by Lo Kwok Wai and Fei-fei Lui. C666-1 cells were grown in RPMI and transfected with pSilencer plasmids expressing hairpin RNA from hEBP2 or GFP as described above except that Lipofectamine 2000 (Invitrogen) was used. Four days posttransfection, an aliquot of the cells was analyzed for hEBP2 expression. When hEBP2 silencing was confirmed, the rest of the cells were blocked in mitosis with Colcemid and then fractionated and analyzed for EBNA1 expression as described above.
Southern blots on fractionated human cell lysates.
HeLa cells were transfected with 5 μg of pc3oriPE and 2 μg pSilencer expressing small interfering RNA (siRNA) against GFP or hEBP2 as described above. After confirming efficient silencing, cells were blocked for 16 h in Colcemid (0.1 μg/ml), harvested by mitotic shake-off, lysed, and separated into soluble and chromosomal pellet fractions as described above for Western blots. pc3oriPE plasmids from the whole-cell, supernatant, and pellet fractions prepared from 2.5 × 106 cells were isolated by Hirts extraction and ethanol precipitation. The plasmids were then linearized, Southern blotted, and quantified by PhosphorImager analysis using ImageQuant software.
ZM447439 treatment.
Log-phase HeLa cells or Raji EBV-transformed Burkitt's lymphoma cells were treated with 2 μM ZM447439 (a gift from AstraZeneca) as previously described (11). After 2 h, Colcemid was added, and cells were incubated for an additional 16 h. Mitotic cells were collected by mitotic shake-off, and lysates were fractionated and Western blotted for EBNA1 as described above.
Yeast strains and mitotic chromosome spreads.
S. cerevisiae strains are listed in Table 1. For temperature-sensitive strains, permissive and restrictive temperatures were 23°C and 37°C (39°C for smc1-1), respectively. To obtain yeast expressing hEBP2, yeast strains were transformed with pR425/PGK.hEBP2 (28) or pRS424GPD.hEBP2 (YBL31-9-3c). pRS424GPD.hEBP2 contains the hEBP2 gene in the XmaI site of the pRS424GPD plasmid (37). Yeast mitotic cells were obtained as follows. Condensin mutants were first grown until log phase (optical density at 600 nm [OD600], 0.2 to 0.4) in selective medium to minimize plasmid loss and then transferred to yeast extract-peptone-dextrose (YPD) medium containing 20 μg of nocodazole (Sigma)/ml of YPD for 3 h at 23°C. The cultures were then either shifted to 37°C for 30 min to inactivate condensin or kept at 23°C (control). For the cohesin and ipl1 mutants, log-phase cultures (OD600, 0.2 to 0.4) under selective conditions were synchronized in G1 phase by transferring them to YPD-3 μM α-factor (Sigma) for 3 h at 23°C (or 37°C for the inactivation of Ipl1 in the G1 experiment). The cells were then washed in YPD and rearrested in mitosis by the addition of 20 μg/ml nocodazole 30 min later. The cells were then kept at 23°C (control) or shifted to 37°C (39°C for 5dAS98) for a further 3 h. JHY93 was blocked in mitosis with nocodazole for 3 h at 30°C. In all cases, the extent of the mitotic arrest was monitored by microscopy; typically, >70% of the cells were large budded with a single undivided nucleus. Chromatin spreads were prepared as described previously (28, 36) and were stained with rabbit anti-hEBP2 and fluorescein isothiocyanate-conjugated goat anti-rabbit antibodies. G1 spreads for the ipl1 temperature-sensitive (ts) strain was also stained with mouse anti-NOP1 (kindly provided by John Aris) and Texas Red-conjugated goat anti-mouse antibodies. All spreads were counterstained with DAPI (1 μg/ml) and visualized at ×1,000 magnification using a Leica DMR microscope and OpenLab software.
TABLE 1.
Yeast strains in this study
| Strain | Mating type | Genotype | Background | Reference |
|---|---|---|---|---|
| BLY07 | MATa | ycg1-2:KAN ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2-801 ade2-101 | S288c | Lavoie et al. (31) |
| 1aAS330 | MATa | smc2-8 ura3 leu2 lys2 his3 ade2 | S288c | Freeman et al. (13) |
| YBL31-9-3c | MATa | mcd1-1 trp1 his3Δ200 | S288c | This study |
| 5dAS98 | MATa | smc1-1 ade2 his3 leu2 trp1 ura3 | S288c | Strunnikov et al. (44) |
| K5824 | MATa | smc3-42 ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 GAL [psi+] (ura negative) | W303 | Michaelis et al. (36) |
| YBL69-4-6d | MATa | ipl1-321 trp1Δ63 ade2 leu2 his3Δ200 ura3-52 ade2-101 his3Δ1 leu2-3,112 trp1-289 lys2-801 | Other | This study |
| JHY93 | MATa | Δ(hht1-hhf1) Δ(hht2-hhf2)/CEN TRP1 hht2-5(H3-S10,28A) HHF2 | S288c | Lavoie et al. (31) |
RESULTS
Localization of hEBP2 in human cells.
hEBP2 is localized to the nucleolus in interphase human cells, but in mitosis, when the nucleolus does not exist, we have found hEBP2 to be tightly associated with the condensed cellular chromosomes in both epithelial and B-lymphocyte-derived cell lines (28, 48). When log-phase HeLa (cervical carcinoma), Raji (EBV-positive Burkitt's lymphoma), and BL41 (EBV-negative B-lymphocyte) cells were stained with polyclonal antibodies specific for hEBP2, hEBP2 exhibited nucleolar staining in all interphase cells and localized to the condensed chromosomes in all mitotic cells (Fig. 1). These results are consistent with the previously described cell-cycle-dependent localization of GFP-tagged hEBP2 in HeLa cells (28). As we analyzed hEBP2 localization with an increasing number of different cell lines, we made the surprising observation that, while hEBP2 was readily detected by immunofluorescence in every human cell line we tested during interphase, hEBP2 could be detected only in a subset of lines once the cells entered mitosis. When log-phase 293T (embryonic kidney cells transformed with SV40 and adenovirus oncogenes) and C33A (cervical carcinoma) cells were stained with the same anti-hEBP2 antibody, hEBP2 was readily visualized in the nucleolus in all interphase cells but was undetectable in mitotic cells (Fig. 1, left and middle panels). Consistent with the results for whole cells, when chromosome spreads were prepared from cells blocked in mitosis with Colcemid, intense hEBP2 staining was detected on the mitotic chromosomes from HeLa, BL41, and Raji cells, but no staining was observed for the chromosome spreads from 293T or C33A (Fig. 1, right panel). These observations are unlikely to derive from a staining artifact, as GFP-tagged hEBP2 in HeLa and C33A cells showed similar staining patterns in interphase, but in mitosis, GFP-hEBP2 could be detected only in HeLa cells (reference 28 and data not shown). In addition, the B23 and histone H1 proteins were readily visualized by immunofluorescence on the mitotic chromosomes from both 293T and C33A cells (data not shown). B23 is a nucleolar ribosome assembly protein that, like hEBP2, associates with the cellular chromosomes in mitosis (12).
FIG. 1.
Immunofluorescence images of hEBP2 in human cell lines. Log-phase cells from the indicated cell lines were fixed and stained with antibody against hEBP2 and counterstained with DAPI. Interphase and mitotic cells are as indicated (magnification, ×400). Mitotic chromosome spreads prepared from cells blocked in mitosis were also stained with hEBP2 antibody and counterstained with DAPI. These images were captured using ×630 magnification. All images were photographed using the same exposure time.
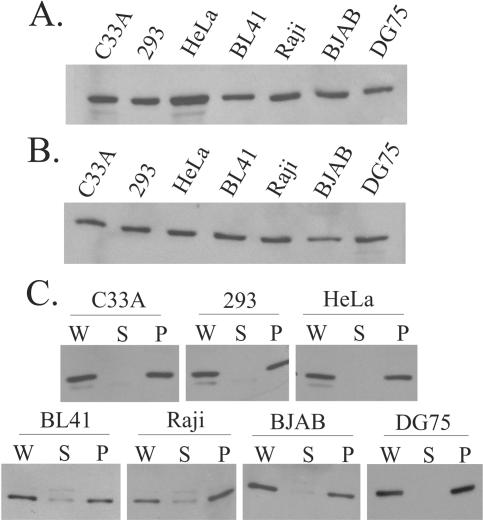
Differences in detection of hEBP2 in different mitotic cells by microscopy could reflect overall lower hEBP2 expression levels in the cells that do not exhibit mitotic chromosome staining relative to those that do or could be due to degradation of hEBP2 at the onset of mitosis in cells where hEBP2 is not detected in mitosis. To investigate the first possibility, Western blot analysis was performed, using the same anti-hEBP2 antibody that was used for microscopy, on equal amounts of whole-cell lysates prepared from the log-phase cells described above and from two additional EBV-negative B-cell lines (DG75 and BJAB). As shown in Fig. 2A, no significant differences in the levels of hEBP2 in any of the cell lines were observed. To address the second possibility, similar Western blot analyses were performed with whole-cell lysates from cells blocked in mitosis and collected by mitotic shake-off, but no significant differences in the levels of hEBP2 were observed (Fig. 2B). These results indicate that the failure to visualize hEBP2 by microscopy in some mitotic cells is not due to low levels of hEBP2 in mitosis.
FIG. 2.
hEBP2 expression levels and localization in fractionated cell lysates. Equal amounts of cell lysates from log-phase cells (A) or from cells blocked in mitosis with Colcemid (B) were analyzed by Western blotting using hEBP2 antibody. (C) Lysates were prepared from mitotically blocked cells, and these whole-cell lysates (W) were separated into chromosomal pellet (P) and soluble (S) fractions by centrifugation. Aliquots of each sample from equal cell numbers were analyzed by Western blotting using anti-hEBP2 antibody.
Since we could clearly detect hEBP2 in mitosis in all cell lines by Western blotting, we used this detection method in combination with biochemical fractionation of the cells to investigate the localization of hEBP2 in mitotic HeLa, C33A, 293T, BL41, Raji, BJAB, and DG75 cells. To this end, the cells were blocked in mitosis, and whole-cell lysates were prepared under conditions in which the chromatin remains intact (19). These lysates were then separated by low-speed centrifugation into a pellet fraction, which contains the chromatin and cell debris, and a soluble protein fraction. Western blot analyses of the cellular fractions for hEBP2 showed that virtually all of the hEBP2 in all seven cell lines localized to the chromatin pellet fraction (Fig. 2C), as did the known chromosomal proteins, B23 and histone H1 (see Fig. 5B). As expected, the nucleosome assembly protein 1 (NAP1), which has not been shown to associate with mitotic chromatin, remained in the soluble fraction (see Fig. 5B). Together, these results suggest that, while hEBP2 is refractory to detection by microscopic approaches on mitotic chromosomes in a subset of cell lines tested (e.g., C33A and 293T), it is nonetheless present and bound to chromatin.
FIG. 5.
Effect of hEBP2 silencing on the biochemical fractionation of EBNA1 and oriP plasmids. (A) Whole-cell lysates (W) were prepared from equal cell numbers of mitotic HeLa, 293T, and C33A cells expressing EBNA1 before (+) and after (−) transfection with the hEBP2 silencing plasmid. Lysates were separated into chromosomal pellet (P) and soluble protein (S) fractions and then were analyzed by Western blotting using EBNA1 antibody. (B) Lysates prepared from mitotic HeLa cells, before (+) and after (−) hEBP2 silencing, were fractionated as for panel A and then analyzed by Western blot using antibodies against Nap1, histone H1, and B23. (C) The localization of oriP plasmids was determined for each HeLa cell fraction shown in panel A by Southern blotting. The percentage of the plasmid that localized to each fraction is indicated below each lane. (D) Lysates were prepared from mitotic C666-1 cells before (+) and after (−) silencing of hEBP2 and fractionated as for panel A. Western blots probed with EBNA1 antibodies are shown.
Effects of hEBP2 silencing on cell viability and mitotic chromosome attachment of EBNA1.
We have previously shown that hEBP2 can enable EBNA1 to segregate EBV-based plasmids in yeast by tethering EBNA1 to the chromosomes in mitosis, suggesting that EBNA1 might also attach to human mitotic chromosomes by binding hEBP2 (28, 29). To assess this possibility, we designed a hairpin RNA to silence hEBP2 expression and expressed it from a plasmid in HeLa cells that had been engineered to express EBNA1. After transfection, the cells were grown under selection for the silencing plasmid and analyzed for hEBP2 expression. Optimal silencing of hEBP2 was observed 3 days posttransfection (as determined by Western blotting) and continued until approximately 7 days posttransfection (data not shown). We examined the viability of HeLa cells with silenced hEBP2 by plating cells transfected with the hEBP2 silencing plasmid on slides and monitoring the doubling of the cells by microscopy. Cells that had taken up the silencing vector proliferated for the first 6 to 7 days posttransfection and then stopped dividing around the seventh day. Since the silencing of hEBP2 took 2 to 3 days after transfection to occur, the cells lacking hEBP2 were able to undergo approximately four doublings before they were no longer viable. This growth profile is very similar to the slow-stop phenotype seen for S. cerevisiae with a yeast EBP2 (yEBP2) temperature-sensitive mutant, which reflects the essential role of yEBP2 in generating ribosomes (24). Examination of the hEBP2-silenced cells both before and after growth arrest did not reveal any obvious alterations to the cell morphology, nor were gross changes in chromosome structure detected in mitotic chromosome spreads (data not shown).
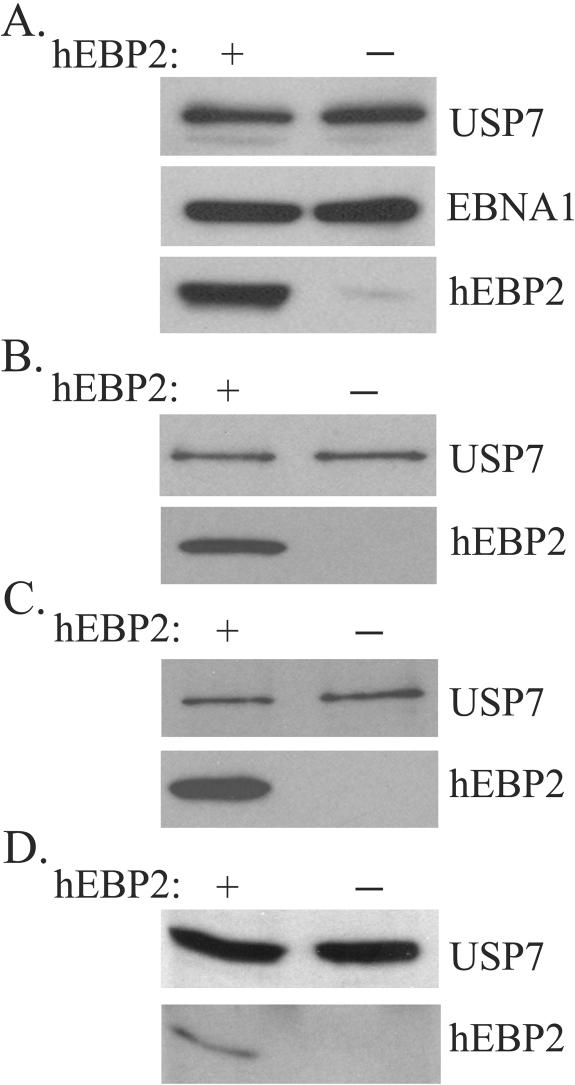
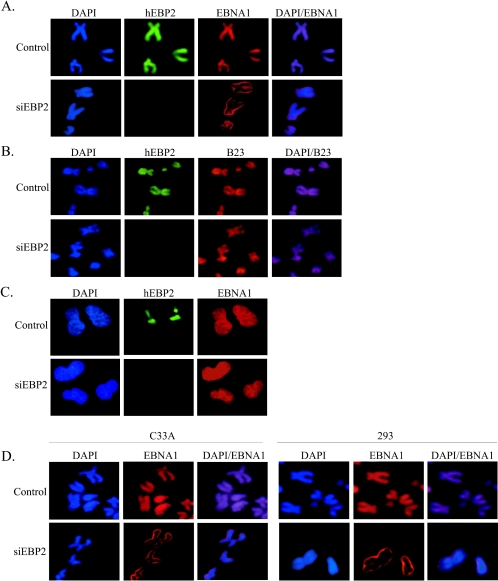
To assess an in vivo role for hEPB2 in EBNA1-mediated segregation, we examined the ability of EBNA1 to associate with mitotic chromosomes in the absence of hEBP2. These analyses were performed at 3 days posttransfection, when silencing and cell viability were optimal. Western blot analyses showed that, while hEBP2 was effectively silenced by this construct, the levels of EBNA1 or the host nuclear ubiquitin-specific protease USP7 (used as a loading control) were not affected (Fig. 3A). Immunofluorescence staining of the cells after hEBP2 silencing showed that hEBP2 was not detectable in 75% of the cells (data not shown). We then used this population of cells to compare EBNA1 localization in cells in which hEBP2 expression was and was not silenced. To examine the effects of hEBP2 expression on EBNA1 binding to mitotic chromosomes, we stained mitotic chromosome spreads with antibodies specific for EBNA1 and hEBP2 (Fig. 4A). In the control cells in which hEBP2 was still present, both hEBP2 and EBNA1 were observed all over the chromosomes. In the cells in which hEBP2 expression had been silenced, hEBP2 could not be detected on the chromosomes and EBNA1 staining was dramatically decreased, such that residual staining was observed predominantly at the outer edges of the chromosomes. The overlay of the DAPI and EBNA1 staining showed that this residual staining in the absence of hEBP2 is actually outside the DAPI-stained DNA, whereas EBNA1 staining is coincident with the DAPI staining on chromosomes containing hEBP2. Identical results were obtained with mitotic cells collected from log-phase cultures in the absence of Colcemid treatment (data not shown). This effect was not observed with the cellular chromosomal protein B23, which gave strong staining coincident with the DAPI staining with and without hEBP2 expression (Fig. 4B), indicating that hEBP2 silencing specifically affected EBNA1. While the images shown in Fig. 4 are from HeLa cells that express EBNA1 at levels approximately eightfold higher than in the EBV-transformed Raji cell line, the same effect of hEBP2 silencing on EBNA1 localization was observed using HeLa cells stably expressing EBNA1 at levels approximately fourfold lower than in Raji cells (data not shown). Hence, the results obtained are not an artifact of EBNA1 overexpression. The possible effect of hEBP2 silencing on EBNA1 localization in interphase was also examined by staining log-phase cells for hEBP2 and EBNA1, but, as expected, no obvious effect was observed. As shown in Fig. 4C, EBNA1 gave a pan-nuclear staining pattern both in the presence and absence of hEBP2. The results indicate that hEBP2 is specifically required for the efficient interaction of EBNA1 with the condensed chromosomes in mitosis.
FIG. 3.
hEBP2 repression by RNA silencing. Whole-cell lysates prepared from log-phase HeLa (A), 293T (B), or C33A (C) cells expressing EBNA1 or from EBV-positive C666-1 cells (D) were analyzed by Western blotting for USP7 (loading control), EBNA1 (A only), and hEBP2 after transfection with pSilencer expressing hEBP2 (−) or GFP (+) hairpin RNA.
FIG. 4.
hEBP2 silencing specifically affects EBNA1 attachment to mitotic chromosomes. (A and B) HeLa cells expressing EBNA1 were transfected with the hEBP2 silencing plasmid and then blocked in mitosis with Colcemid for 16 h. Mitotic chromosome spreads were stained with anti-hEBP2 rabbit antibody and either anti-EBNA1 mouse antibody (A) or anti-B23 mouse antibody (B) and counterstained with DAPI. EBNA1 localization is shown for cells expressing (control) or lacking (siEBP2) hEBP2. Overlays of the EBNA1 and DAPI staining (A) or B23 and DAPI staining (B) are also shown. (C) Interphase HeLa cells from log-phase cultures. (D) Effect of hEBP2 silencing on EBNA1 in C33A and 293T cells. Cells were prepared as described for panel A. Since hEBP2 is not visible in these cells in mitosis, the EBNA1 staining in untransfected cells (control) is compared to the pattern seen in the majority of the cells after transfection with the hEBP2 silencing plasmid (siEBP2). All images were captured using the same exposure time.
Given the apparent differences in the accessibility of hEBP2 on mitotic chromosomes in C33A and 293T cells compared to HeLa cells, we also examined the effect of hEBP2 silencing on EBNA1 binding to mitotic chromosomes from C33A and 293T cells expressing EBNA1, as described above for HeLa cells. These experiments have the complication that we cannot visualize hEBP2 in mitosis in these cells by microscopy in order to know which cells are expressing or not expressing hEBP2. However, by examining log-phase cells, we found that hEBP2 was silenced with similar efficiency as in HeLa cells, as approximately 75% of the cells transfected with the hEBP2 silencing construct lacked the nucleolar staining typical of hEBP2 (data not shown). Efficient silencing of hEBP2 in C33A and 293T cells was also seen by Western blotting (Fig. 3B and 3C). When we compared EBNA1 staining on mitotic chromosome spreads before and after silencing hEBP2, an obvious decrease was seen in EBNA1 staining on the mitotic chromosomes prepared from approximately 75% of the C33A or 293T cells after the cells had been transfected with the hEBP2 silencing plasmid (Fig. 4D). As we observed for HeLa cells, the residual EBNA1 staining seen on mitotic C33A and 293T chromosomes from hEBP2-silenced cells formed a ring around the chromosomes which was outside of the DAPI staining. These results indicate that hEBP2 is important for EBNA1 attachment to mitotic chromosomes in C33A and 293T cells, as it is in HeLa cells.
To further investigate and quantify the effects of hEBP2 silencing on EBNA1 binding to mitotic chromosomes, we fractionated mitotically blocked cells, before and after silencing hEBP2, into chromosomal pellet and soluble fractions and compared the localization of EBNA1 by Western blotting (Fig. 5A). As expected, virtually all of the EBNA1 in the HeLa, C33A, and 293T cells fractionated with the chromosomal pellet when hEBP2 was expressed. Upon silencing of hEBP2, however, a significant proportion of the EBNA1 shifted to the soluble protein fraction, indicating that it no longer bound to the chromosomes. The silencing of hEBP2 had no effect on the localization of control proteins, however, as the fractionation pattern of the chromatin-associated B23 and histone H1 proteins, as well as the soluble NAP1 protein, were unchanged (Fig. 5B). The amount of EBNA1 in soluble and pellet fractions with and without hEBP2 silencing was quantified for multiple experiments in each cell line, and the average numbers are shown in Table 2. After transfection with the hEBP2 silencing plasmid, 56% of the EBNA1 in HeLa cells, 42% in 293T cells, and 38% in C33A cells was soluble. When the background of approximately 25% of the cells that still express hEBP2 is taken into account, these numbers indicate that, in the cells in which hEBP2 is silenced, 75%, 56%, and 51% of the EBNA1 is not chromosome bound in HeLa, 293T, and C33A cells, respectively. These numbers are also in keeping with the degree of EBNA1 staining observed by microscopy. Therefore EBP2 plays a major role in EBNA1 binding to mitotic chromosomes in all three cell lines.
TABLE 2.
Effect of hEBP2 silencing on EBNA1 localization and Aurora B silencing on hEBP2 localization
| Cell type | EBNA1 localizationa
|
hEBP2 localizationb
|
||||||
|---|---|---|---|---|---|---|---|---|
| hEBP2 +ve
|
hEBP2 −ve
|
AurB +ve
|
AurB −ve
|
|||||
| Supernatant | Pellet | Supernatant | Pellet | Supernatant | Pellet | Supernatant | Pellet | |
| HeLa | 14 | 86 | 56 (75)c | 44 | 4 | 96 | 24 (32)d | 76 |
| 293 | 17 | 83 | 42 (56) | 58 | 3 | 97 | 13 (17) | 87 |
| C33A | 15 | 85 | 38 (51) | 62 | 17 | 83 | 42 (56) | 58 |
Percentage of EBNA1 detected in the soluble and chromosomal pellet fractions of mitotic cell lysates by Western blotting before (hEBP2 +ve) and after (hEBP2 −ve) transfection with the hEBP2 silencing plasmid. Values are averages from three separate experiments.
Percentage of hEBP2 detected in the soluble and chromosomal pellet fractions of mitotic cell lysates by Western blotting before (AurB +ve) and after (AurB −ve) transfection with the Aurora B silencing plasmid. Values are averages from two to five separate experiments
The numbers in parentheses in this column are the percentages of EBNA1 in the soluble fractions when only cells in which hEBP2 are silenced are considered (75% of the cell population).
The numbers in parentheses in this column are the percentages of hEBP2 in the soluble fractions when only cells in which Aurora B are silenced are considered (75% of the cell population).
Effect of hEBP2 silencing on mitotic chromosome attachment of oriP plasmids.
Since cells lacking hEBP2 will double only four times, the ability of EBNA1 to segregate oriP plasmids in these cells cannot be definitively addressed, as segregation effects of EBNA1 begin to become detectable only after 3 doublings and are typically measured by assessing oriP plasmid maintenance after 14 or more doublings (42, 49, 50). However, the effect of hEBP2 silencing on the attachment of oriP plasmids to the cellular mitotic chromosomes could be determined, since this could be measured immediately upon silencing of hEBP2. To this end, whole-cell lysates were prepared from mitotic HeLa cells cotransfected with the oriP plasmid expressing EBNA1 and the silencing plasmid for hEBP2 or GFP. Lysates were separated into chromosomal pellet and soluble fractions, and oriP plasmids in each fraction were quantified by Southern blotting (Fig. 5C). As expected, most of the plasmids were found in the chromosomal pellet in cells expressing hEBP2. However, silencing of hEBP2 caused a notable shift of the plasmids into the soluble fraction that was similar in magnitude to that observed for EBNA1, indicating that hEBP2 is important for the association of oriP plasmids with mitotic chromosomes.
Effect of hEBP2 silencing in EBV-transformed cells.
To extend our results showing release of EBNA1 from mitotic chromosomes upon hEBP2 silencing to EBV-transformed cells, we silenced hEBP2 in the C666-1 EBV-transformed nasopharyngeal carcinoma cell line (9) (Fig. 3D). This line was chosen since EBV-transformed B cells are not well suited for siRNA experiments due to low transfection efficiency. As shown in Fig. 5D, EBNA1 pelleted with the mitotic chromosomes in the presence of hEBP2 but approximately 50% of the EBNA1 was released from the mitotic chromosomes to the soluble fraction upon silencing of hEBP2. Therefore, hEBP2 plays a similar role in tethering EBNA1 to mitotic chromosomes in EBV-transformed as it does in EBNA1-expressing cell lines.
hEBP2 binding to mitotic chromosomes is dependent on Ipl1/Aurora kinases.
Having established that the presence of hEBP2 on mitotic chromosomes is important for the attachment of EBNA1 and oriP plasmids to these chromosomes, we next addressed how hEBP2 itself binds to chromosomes. We have previously shown that hEBP2 binds to mitotic chromosomes through its central coiled-coil domain, but the component of the chromosomes recognized by this domain is not known (28). We have also shown that when hEBP2 is expressed in budding yeast, it associates with the cellular chromatin only through the nucleolar region in interphase but binds over the entire chromatin mass in mitosis (28). A similar cell cycle-dependent redistribution of hEBP2 occurs in human cells, indicating that the mitotic chromosome attachment of hEBP2 is subject to the same regulation in human and yeast cells. Therefore, we took advantage of a series of yeast ts mutants to gain insight into the requirements for the redistribution of hEBP2 to the mitotic chromosomes. Experiments were performed using yeast ts mutants compromised in mitotic chromosome structure and function, notably in several subunits of the essential cohesin (mcd1-1, smc1-1, and smc3-42) and condensin (smc2-8, ycg1-2, and ycs4-2) complexes, which disrupt sister chromatid cohesion and/or mitotic chromosome condensation (13, 18, 31, 36, 44, 46). Each yeast strain was transformed with a hEBP2-expressing plasmid, blocked in mitosis, and then incubated at either the permissive or restrictive temperature. Chromatin spreads were then prepared and stained for hEBP2. Neither the loss of chromosome condensation nor sister chromatid cohesion had any detectable effect on hEBP2 binding to mitotic chromosomes (Fig. 6A). Furthermore, as the chromatin association of the entire cohesin or condensin complexes is dependent on the activity of both their SMC and non-SMC components (31, 36, 46), it is unlikely that any of these proteins serve as the attachment points for hEBP2.
FIG. 6.
hEBP2 association with yeast mitotic chromosomes. (A) Yeast ts mutants (as indicated) containing a hEBP2 expression plasmid were arrested in mitosis and then incubated at either the restrictive (R) or permissive (P) temperature (except H3 S10,28A, which is at 30°C). Chromosome spreads were stained for hEBP2 and with DAPI. (B) G1-arrested ipl1 cells expressing hEBP2 were incubated at either the restrictive or permissive temperature. Chromosome spreads were stained for hEBP2 with Nop1 as a nucleolar marker and counterstained with DAPI. All images were captured using ×1000 magnification and the same exposure time.
In addition to condensin and cohesin mutants, we also examined the possible requirement for the Ipl1 kinase in mitotic chromosome binding by hEBP2, using a yeast strain with a ts mutation that inactivates this enzyme (ipl1-321) (3). Ipl1 is known to play several key roles in mitosis, through phosphorylation of kinetochore components, histone H3, and theYcg1 condensin subunit (3, 8, 23, 32). The ipl1 strain was synchronized in G1 and then released into a mitotic block at either the permissive or restrictive temperature prior to the spreading of the mitotic chromatin on slides and the staining of it for hEBP2. While hEBP2 was observed bound to the chromatin mass prepared at the permissive temperature, it was not detected on the chromatin prepared at the restrictive temperature (Fig. 6A). Since the expression level of hEBP2 does not change at the restrictive temperature, the failure to detect hEBP2 on the chromosomes indicates that Ipl1 function is required for hEBP2 to bind to mitotic chromosomes. In contrast, hEBP2 association with the nucleolar chromatin was unaffected by the loss of any residual Ipl1 activity in G1 cells (Fig. 6B). Therefore, Ipl1 is not required for hEBP2 localization to the nucleolus but is required for the redistribution of hEBP2 over the mitotic chromosomes.
Since histone H3 is known to be phosphorylated by Ipl1 in mitosis and is widely distributed over the chromosome, as is hEBP2, we investigated the possibility that hEBP2 may be interacting with phosphorylated H3. To this end, we monitored hEBP2 chromatin association in a yeast strain containing a nonphosphorylatable histone H3 mutant, which lacks the conserved serine 10 and 28 residues (H3 S10,28A) (23, 31). However, disruption of these phosphorylation sites on histone H3 had no obvious effect on the ability of hEBP2 to attach to the yeast mitotic chromatin (Fig. 6A).
In humans, there are three homologues of Ipl1, Aurora kinases A, B, and C (6, 14). Since Aurora B has the most functional similarity to Ipl1, we first investigated its involvement in hEBP2 attachment to human mitotic chromosomes. We transfected HeLa, 293T, and C33A cells with a plasmid-expressing hairpin RNA designed to silence Aurora B and confirmed the decreased expression of Aurora B by Western blot analysis of cell lysates (Fig. 7A). We then compared the localization of hEBP2 in mitosis, with and without Aurora B silencing, by Western blot analysis of fractionated cell lysates (Fig. 7B). We consistently found that silencing of Aurora B caused a proportion of the hEBP2 to shift from the chromosomal pellet into the soluble fraction, indicating a role for this kinase in the mitotic chromosome attachment of hEBP2. This effect was seen with all three cell lines tested to various degrees, with the weakest effect seen with 293T cells (Table 2). The effect of Aurora B silencing on hEBP2 solubilization was specific to mitosis, as no change in hEBP2 fractionation was detected in log-phase cells upon Aurora B silencing (data not shown). The specificity of the Aurora B effect on hEBP2 was also reflected in the finding that down-regulation of Aurora A and Aurora C by siRNA had no detectable effect on the ability of hEBP2 to attach to mitotic chromosomes (Fig. 7C and D), nor did treating cells with staurosporine, an inhibitor of several kinases, including protein kinase C, CDK2, CDK4, and calcium-calmodulin-dependent kinase (data not shown) (34).
FIG. 7.
Effect of Aurora B silencing and inhibition on hEBP2 localization in human cells. (A) Western blot showing Aurora B expression in comparison to USP7 (loading control), in HeLa, 293T, and C33A cells before (+) and after (−) silencing of Aurora B. (B) Lysates (W) were prepared from mitotically blocked cells before (+) and after (−) silencing of Aurora B and separated into chromosomal pellet (P) and soluble (S) protein fractions. Samples were analyzed by Western blotting using anti-hEBP2 antibody. (C) Western blot showing Aurora A or Aurora C expression, as indicated, in HeLa cells before (+) and after (−) Aurora A or Aurora C silencing, respectively. (D) Lysates were prepared from mitotic HeLa cells before (+) or after (−) silencing of Aurora A, B, or C, as indicated, and then separated into soluble (S) and pellet (P) fractions. Western blots were performed using hEBP2 antibody. (E) HeLa or Raji whole-cell lysates (W) were prepared before (−) or after (+) the addition of ZM447439 and separated into soluble and pellet fractions. Western blots were performed using hEBP2 antibody.
The contribution of Aurora B to the mitotic chromosome association of hEBP2 was further investigated using a small molecule (ZM447439) previously shown to specifically inhibit the kinase activity of Aurora A and B, giving cellular effects consistent with a specific inhibition of Aurora B function (11). We used this drug to treat HeLa cells and determined its effect on hEBP2 localization to mitotic chromosomes by biochemical fractionation. As shown in Fig. 7E, ZM447439 treatment gave the same effect as Aurora B silencing, causing release of a proportion of hEBP2 from the mitotic chromosomes. This verifies that the effect of the Aurora B siRNA treatment was indeed due to loss of Aurora B and, further, shows that it is the kinase activity of Aurora B that is important for hEBP2 relocalization to the mitotic chromosomes. We also used this drug to determine if Aurora B plays a role in hEBP2 binding to mitotic chromosomes in EBV-transformed B cells. To this end, we treated the EBV-positive Raji Burkitt's lymphoma cells with ZM447439 and monitored hEBP2 in mitotic cell fractions. As in HeLa cells, ZM447439 treatment resulted in incomplete attachment of hEBP2 to the mitotic chromosomes as observed by the appearance of hEBP2 in the soluble cell fraction, a result that was consistent in multiple experiments.
DISCUSSION
The attachment of EBNA1 to the host chromosomes in mitosis is a crucial step in the segregation and stable persistence of EBV episomes. We have previously found that the ability of EBNA1 to bind hEBP2 correlates with its ability to attach to mitotic chromosomes and to segregate oriP-containing plasmids (48, 49). We have also shown that hEBP2 enables EBNA1-mediated plasmid segregation in yeast when coexpressed with EBNA1 and that it functions to tether EBNA1 to the mitotic chromosomes in that system (28, 29). We now show that hEBP2 plays a major role in the attachment of EBNA1 to mitotic chromosomes in human cells and, as expected, the loss of EBNA1 from the chromosomes leads to the dissociation of oriP plasmids from the chromosomes. In cells in which hEBP2 expression had been silenced, 50 to 75% of the EBNA1 in three different cell lines no longer associated with the mitotic chromosomes. The remaining EBNA1 staining seen on the mitotic chromosomes after EBP2 silencing was localized predominantly to just outside the DAPI staining and therefore is qualitatively different than the DAPI-coincident EBNA1 staining seen in the presence of hEBP2. The EBNA1 signal observed outside the DAPI staining may simply reflect nonspecific sticking of EBNA1 to the chromosomes or may indicate an alternative mechanism of attachment through RNA tails or DNA loops at the chromosome periphery. There also appears to be some residual EBNA1 staining in the body of the mitotic chromosomes in the absence of hEBP2, which may indicate that a proportion of EBNA1 can attach to mitotic chromosomes through other chromosomal proteins. Unlike the effects in mitosis, hEBP2 silencing had no obvious effect on EBNA1 localization in interphase, suggesting that the two proteins do not interact until the onset of mitosis. This is consistent with the limited overlap of hEBP2 and EBNA1 staining in interphase, where hEBP2 is nucleolar and EBNA1 is found throughout the nucleus, and with our previous mutational analyses of EBNA1 that indicated that hEBP2 binding was not responsible for the localization of a proportion of the EBNA1 to the nucleolus (49).
During the course of our studies, we made the unexpected observation that, in mitosis, hEBP2 was detectable by microscopy in a cell line-specific manner. This was not an artifact of our antibody, since GFP-tagged hEBP2 behaved similarly. Since hEBP2 is a highly conserved essential protein in eukaryotes, it would not be expected to behave fundamentally differently in different cell lines. Indeed, Western blot analyses showed that hEBP2 is present in similar amounts in mitosis in all cell lines examined and that its localization in fractionated cells is consistent with mitotic chromosome binding and not with localization in the soluble portion of the cell. In addition, silencing of hEBP2 was found to severely decrease EBNA1 attachment to mitotic chromosomes in all cell lines, providing additional evidence for the association of hEBP2 with the chromosomes. The reason that hEBP2 is clearly seen on metaphase chromosomes by microscopy in some cells lines and detected on the chromosomes in other cell lines only by Western blotting is not known but most likely reflects the masking of hEBP2 by another protein. While this suggests that hEBP2 is not very accessible in metaphase in some cell types, the effects of hEBP2 silencing on EBNA1 localization shows that hEBP2 is accessible to EBNA1 in all cell lines examined and, in keeping with this observation, EBNA1 can mediate the segregation of oriP plasmids in HeLa, 293, and C33A cells to similar degrees (K. Shire, H. Wu, and L. Frappier, unpublished data). The ability of EBNA1 to access hEBP2 in all cell lines examined is likely due to the time at which the initial EBNA1-hEBP2 interaction occurs. Nucleoli in mammalian cells disintegrate during early prophase, and the nucleolar proteins redistribute, many becoming associated with the chromosome periphery (12, 15, 21). It is expected that EBNA1 and hEBP2 would associate during this redistribution in early prophase. The timing of the association and dissociation of EBNA1 and hEBP2 with each other and with cellular chromosomes will be important to address in future studies.
We have also investigated the mechanism by which hEBP2 itself attaches to chromosomes in mitosis. Two lines of evidence suggest that this interaction is likely to occur through hEBP2 binding to an as-yet-undefined chromosomal protein as opposed to binding directly to the DNA. First, the central domain of hEBP2 that is responsible for the mitotic chromosome interaction is predicted to be a coiled-coil domain (28), and coiled-coil domains in other proteins typically mediate protein interactions (1). Secondly, the interaction of hEBP2 with chromatin is more extensive in mitosis than in interphase, whereas the DNA itself would be more accessible in interphase. We have also tested the possibility that hEBP2 could be tethered to the mitotic chromosomes through RNA tails, as reported for some proteins (21), but find no detectable release of hEBP2 from the chromosomes upon RNase treatment (H. Wu and L. Frappier, unpublished data).
A paper by Sears et al. (41) recently reported that hEBP2 was not important for EBNA1-mediated segregation in human cells, largely based on the observations that (i) multimerization of the EBNA1 N-terminal region in the absence of the main hEBP2 binding region (325 to 376) could result in some ability of EBNA1 to maintain plasmids, (ii) the fusion of hEBP2 to the EBNA1 DNA binding domain was insufficient to stably maintain oriP plasmids in human cells, and (iii) that hEBP2 was not visualized on mitotic chromosomes using a monoclonal antibody. However, there are several problems with the interpretation of these experiments. While the first set of experiments may indicate that EBNA1 can be engineered to segregate plasmids in different ways, it does not address the mechanism of attachment of the wild-type EBNA1 protein, which does not have a multimerized N terminus. In the context of the native EBNA1 protein, deletion of the 325-to-376 hEBP2 binding region abrogates the segregation activity of EBNA1 without affecting its DNA replication function (42). In the second set of experiments, while the hEBP2-EBNA1 fusion protein was not able to stably maintain oriP plasmids, it also was unable to replicate them. Since the stable maintenance of oriP plasmids depends on their ability to replicate, no conclusions can be made about the segregation activity of this fusion protein. In the third set of experiments, failure to detect hEBP2 on mitotic chromosomes from 293 cells could be due to the inability to stain for hEBP2 as reported here; however, we also note that there is no published evidence that the monoclonal antibody used is specific for hEBP2. This antibody was raised against a crude nucleolar extract (7), and there has been no published study to show that it recognizes hEBP2 as opposed to other nucleolar proteins of a similar size. The fact that this antibody detects a protein that is not associated with the chromosomes in mitosis (something we have never seen by staining with hEBP2-specific antibodies or expressing GFP-tagged EBP2 in the many cell lines tested) suggests that it is recognizing a nucleolar protein that is distinct from hEBP2.
Based on sequence similarity of EBNA1 residues 41 to 54 and 329 to 350 with the AT hooks of HMG1, Sears et al. (41) propose that these regions of EBNA1 bind directly to DNA to mediate the attachment of EBNA1 to mitotic chromosomes. However, these EBNA1 sequences have not been shown to directly bind DNA in the context of the folded EBNA1 protein. Furthermore, the dissociation of EBNA1 from the chromosomes upon EBP2 silencing, as shown here, strongly argues against this model as the predominant route of EBNA1 attachment to mitotic chromosomes. The AT hook model of chromosome attachment is also not consistent with reports that the majority of EBNA1 does not bind cellular chromosomes in interphase (10), when the DNA would be most accessible, and that deletion of the first putative AT hook (residues 34 to 52) has no effect on the ability of EBNA1 to attach to mitotic chromosomes or segregate EBV-based plasmids (49). However, it is possible that the residual EBNA1 staining seen on mitotic chromosomes in the absence of hEBP2 may be due to an AT-hook interaction with exposed DNA loops in the chromosome periphery.
In order to examine some of the possible protein requirements for hEBP2 binding to mitotic chromosomes, we took advantage of the fact that hEBP2 expressed in yeast undergoes the same cell cycle-regulated association with the host chromatin as it does in human cells (28). Using a series of ts yeast mutants, we found that hEBP2 attachment to the mitotic chromosomes was dependent neither on the presence of condensin or cohesin complexes on the chromosomes nor on proper chromosome condensation or cohesion. In this respect, the segregation of EBV plasmids in yeast is different than that of yeast 2μm plasmids, which directly interact with the yeast cohesion complex through the Mcd1 subunit (35). The interaction of hEBP2 with the chromosomes in mitosis was dependent, however, on the Ipl1 kinase. Ipl1 is known to phosphorylate several proteins, including kinetochore components, the Ycg1 component of condensin and histone H3, and a consensus phosphorylation site has been identified as [RK]X[TS][ILV] (3, 8, 23, 32). Our results suggest that in mitosis, Ipl1 must phosphorylate either hEBP2 itself or the chromosomal protein to which hEBP2 binds in order to trigger the interaction of hEBP2 with the mitotic chromosomes. To test the first possibility, we searched hEBP2 for Ipl1 consensus sites but found no matches to this sequence. Therefore, we currently favor the second possibility, that the Ipl1 requirement is due to phosphorylation of another chromosomal protein. This target protein would have to be widely distributed over the chromosomes in order to result in the observed hEBP2 staining pattern. While the known Ipl1/Aurora B target, histone H3, fits this distribution, yeast strains containing histone H3 N-terminal tail mutants lacking the known Ipl1/Aurora B target sites (serines 10 and 28) (16, 23) were not defective in hEBP2 binding to the mitotic chromosomes (Fig. 6 and data not shown). While these results may indicate that H3 is not the attachment site for hEBP2, we cannot rule out the possibility that it might play a redundant role in mediating Ipl1-dependent hEBP2 interactions. On that note, it is interesting that the cellular protein (Brd4) recently shown to mediate the mitotic chromosome attachment of the bovine papillomavirus E2 protein, and hence the segregation of bovine papillomavirus genomes, attaches to chromosomes through modified (acetylated) histone tails (5, 52).
Ipl1 belongs to the Aurora family of kinases, of which there are three in humans, Aurora A, B, and C (6, 14). Considerable evidence indicates that Aurora B is functionally equivalent to Ipl1. Like Ipl1, Aurora B in several organisms has been shown to phosphorylate histone H3 and kinetochore components and to play essential roles in chromosome condensation, segregation, and cytokinesis (6). Since Ipl1 is essential for the attachment of hEBP2 to mitotic chromosomes in yeast, we examined the involvement of Aurora B in hEBP2 binding to human mitotic chromosomes. We consistently observed that down-regulation of Aurora B by RNA silencing or inhibition of Aurora B kinase activity with ZM447439 led to decreased hEBP2 binding to the mitotic chromosomes and increased hEBP2 in the soluble fraction. This effect was seen for all four cell lines tested (including EBV-transformed B cells) to various degrees, and the degree of the effect did not correlate with the ability to visualize hEBP2 in mitosis by microscopy. The effect of Aurora B silencing on hEBP2 localization in human cells was not as dramatic as the effect of Ipl1 inactivation on hEBP2 localization in yeast. This may be due to different degrees of down-regulation of these proteins due to the different techniques used. While the Ipl1 ts mutation would cause the inactivation of all Ipl1 in the cell in a single cell cycle, the RNA silencing approach used for Aurora B is likely to result in a low residual level of catalytically active Aurora B, which may be sufficient to promote hEBP2 attachment to the chromosomes. Similarly, treatment with ZM447439 may not completely inactivate all of the Aurora B kinase in the cell. Alternatively, the results may indicate that there is some functional redundancy between Aurora B and other kinases, although these do not appear to be Aurora A or C. The role of Aurora B in hEBP2 relocalization fits with the fact that Aurora B levels and activity peak in G2/M and that Aurora B localizes to the chromosomes during early prophase (4), where it could phosphorylate a chromosomal component that is then bound by hEBP2. Identifying this chromosomal component will be an important part of future studies.
Acknowledgments
We thank Mark Grunstein for yeast strains expressing histone H3 tail mutants, Lo Kwak Wai and Fei-fei Lui for C666-1 cells, and AstraZeneca UK Limited for ZM447439.
This work was supported by grants from the Canadian Institutes of Health Research to L.F and B.D.L. P.K. is a research student of the National Cancer Institute of Canada, supported with funds provided by the Terry Fox run. L.F. is a Canada Research Chair in Molecular Virology, and B.D.L is a CIHR New Investigator.
REFERENCES
- 1.Adamson, J. G., N. E. Zhou, and R. S. Hodges. 1993. Structure, function and application of the coiled-coil protein folding motif. Curr. Opin. Biotechnol. 4:428-437. [DOI] [PubMed] [Google Scholar]
- 2.Ambinder, R. F., M. Mullen, Y. Chang, G. S. Hayward, and S. D. Hayward. 1991. Functional domains of Epstein-Barr nuclear antigen EBNA-1. J. Virol. 65:1466-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman, and A. W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, J. R., L. Anderson, Y. Zhu, K. Mossie, K., L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, C. S. M. Chan, M. Novotny, D. J. Slamon, and G. D. Plowman. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brannon, A. R., J. A. Maresca, J. D. Boeke, M. A. Basrai, and A. A. McBride. 2005. Reconstitution of papillomavirus E2-mediated plasmid maintenance in Saccharomyces cerevisiae by the Brd4 bromodomain protein. Proc. Natl. Acad. Sci. USA 102:2998-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmena, M., and W. C. Earnshaw. 2003. The cellular geography of Aurora kinases. Nat. Rev. 4:842-854. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, A., J. W. Freeman, and H. Busch. 1987. Identification and partial characterization of a Mr 40,000 nucleolar antigen associated with cell proliferation. Cancer Res. 47:1123-1129. [PubMed] [Google Scholar]
- 8.Cheeseman, I. M., S. Anderson, M. Jwa, E. M. Green, J. Kang, J. R. Yates, C. S. M. Chan, D. G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111:163-172. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, S. T., D. P. Huang, A. P. Hui, K. W. Lo, C. W. Ko, Y. S. Tsang, N. Wong, B. M. Whitney, and J. C. Lee. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int J. Cancer 83:121-126. [DOI] [PubMed] [Google Scholar]
- 10.Daikoku, T., A. Kudoh, M. Fujita, Y. Sugaya, H. Isomura, and T. Tsurumi. 2004. In vivo dynamics of EBNA1-oriP interaction during latent and lytic replication of Epstein-Barr virus. J. Biol. Chem. 279:54817-54825. [DOI] [PubMed] [Google Scholar]
- 11.Ditchfield, C., V. L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S. S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2 and Cenp-E to kinetochores. J. Cell Biol. 161:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundr, M., U. T. Meier, N. Lewis, D. Rekosh, M.-L. Hammarskjöld, and M. Olson. 1997. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma 105:407-417. [DOI] [PubMed] [Google Scholar]
- 13.Freeman, L., L. Aragon-Alcaide, and A. Strunnikov. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149:811-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giet, R., and C. Prigent. 1999. Aurora/Ipl1p-related kinases, a new oncogenic family of mitotic serine-threonine kinases. J. Cell Sci. 112:3591-3601. [DOI] [PubMed] [Google Scholar]
- 15.Goessens, G. 1984. Nucleolar structure. Int. Rev. Cytol. 87:107-158. [DOI] [PubMed] [Google Scholar]
- 16.Goto, H., Y. Yasui, E. A. Nigg, and M. Inagaki. 2002. Aurora-B phosphorylates histone H3 at serine 28 with regard to the mitotic chromosome condensation. Genes Cells 7:11-17. [DOI] [PubMed] [Google Scholar]
- 17.Grogan, E. A., W. P. Summers, S. Dowling, D. Shedd, L. Gradoville, and G. Miller. 1983. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology and chromosome binding. Proc. Natl. Acad. Sci. USA 80:7650-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock, R. 1974. Interphase chromosomal deoxyribonucleoprotein isolated as a discrete structure from cultured cells. J. Mol. Biol. 86:649-663. [DOI] [PubMed] [Google Scholar]
- 20.Harris, A., B. D. Young, and B. E. Griffin. 1985. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J. Virol. 56:328-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez-Verdun, D., and T. Gautier. 1994. The chromosome periphery during mitosis. Bioessays 16:179-185. [DOI] [PubMed] [Google Scholar]
- 22.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen 1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, J.-Y., Z.-W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 24.Huber, M., J. Dworet, K. Shire, L. Frappier, and M. McAlear. 2000. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem. 275:28764-28773. [DOI] [PubMed] [Google Scholar]
- 25.Hung, S. C., M.-S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishimi, Y., and A. Kikuchi. 1991. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J. Biol. Chem. 266:7025-7029. [PubMed] [Google Scholar]
- 27.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588.11313483 [Google Scholar]
- 28.Kapoor, P., and L. Frappier. 2003. EBNA1 partitions Epstein-Barr virus plasmids in yeast by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J. Virol. 77:6946-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavoie, B., E. Hogan, and D. Koshland. 2002. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 156:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavoie, B. D., E. Hogan, and D. Koshland. 2004. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 18:76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meggio, F., A. Donella Deana, M. Ruzzene, A. M. Brunati, L. Cesaro, B. Guerra, T. Meyer, H. Mett, D. Fabbro, P. Furet, et al. 1995. Different susceptibility of protein kinases to staurosporine inhibition. Kinetic studies and molecular bases for the resistance of protein kinase CK2. Eur. J. Biochem. 234:317-322. [DOI] [PubMed] [Google Scholar]
- 35.Mehta, S. V., X. M. Yang, C. S. Chan, M. J. Dobson, M. Jayram, and S. Velmurugan. 2002. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation. J. Cell Biol. 158:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaelis, C., R. Ciosk, and K. Naysmith. 1997. Cohesions: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 37.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176:563-574. [DOI] [PubMed] [Google Scholar]
- 39.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 40.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 41.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78:11487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shire, K., D. F. J. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson, K., A. McGuigan, and C. Huxley. 1996. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol. Cell. Biol. 16:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strunnikov, A. V., V. L. Lrionov, and D. Koshland. 1993. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 123:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summers, H., J. A. Barwell, R. A. Pfuetzner, A. M. Edwards, and L. Frappier. 1996. Cooperative assembly of EBNA1 on the Epstein-Barr virus latent origin of replication. J. Virol. 70:1228-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tóth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsujii, R., K. Miyoshi, A. Tsuno, Y. Matsui, A. Toh-e, T. Miyakawa, and K. Mizuta. 2000. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-RNA processing and ribosomal subunit assembly. Genes Cells 5:543-553. [DOI] [PubMed] [Google Scholar]
- 48.Wu, H., D. F. J. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of the Epstein-Barr virus EBNA1 protein. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yates, J. L., and S. M. Camiolo. 1988. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells 6:197-205. [Google Scholar]
- 51.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 52.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]