Abstract
The engineered TadA variants used in cytosine base editors (CBEs) present distinctive advantages, including a smaller size and fewer off-target effects compared to cytosine base editors that rely on natural deaminases. However, the current TadA variants demonstrate a preference for base editing in DNA with specific motif sequences and possess dual deaminase activity, acting on both cytosine and adenosine in adjacent positions, limiting their application scope. To address these issues, we employ TadA orthologs screening and multi sequence alignment (MSA)-guided protein engineering techniques to create a highly effective cytosine base editor (aTdCBE) without motif and adenosine deaminase activity limitations. Notably, the delivery of aTdCBE to a humanized mouse model of Duchenne muscular dystrophy (DMD) mice achieves robust exon 55 skipping and restoration of dystrophin expression. Our advancement in engineering TadA ortholog for cytosine editing enriches the base editing toolkits for gene-editing therapy and other potential applications.
Subject terms: Protein design, CRISPR-Cas9 genome editing, Targeted gene repair
Engineered TadA variants in cytosine base editors (CBEs) offer advantages like smaller size and reduced off-target effects. Here, authors develop an advanced CBE (aTdCBE) through ortholog screening and protein engineering, successfully enabling exon skipping in Duchenne muscular dystrophy model.
Introduction
DNA base editors composed of Cas9 nickase and deaminases can perform specific single base conversion on target sequences without requiring DNA double-stranded breaks1. The main effector domains of the cytosine base editor (CBE) and adenine base editor (ABE) are cytidine and adenine deaminase, catalyzing the base conversion from C·G to T·A and A·T to G·C, respectively. Cytidine deaminases derived from proteins such as APOBEC, AID, and CDA naturally possess cytosine deamination activity1–3. The adenosine deaminases TadA of ABEs were engineered from tRNA-specific adenosine deaminase for adenosine base editing of DNA4,5. After removing uracil glycosylase inhibitor (UGI) from CBEs, C-to-G base editors (CGBEs) were developed to catalyze the conversion of bases from C.G to T.A6,7.
By introducing stop codons (CAA/CAG/CGA to TAA/TAG/TGA), CBEs have shown great potential in the treatment of common diseases such as T-cell acute lymphoblastic leukemia8, Hepatitis B9,10, and acquired immunodeficiency syndrome11. Although high on-target DNA editing efficiency can be achieved with traditional CBEs12, they can also exhibit different types of DNA and RNA off-target effects13–16. Compared to natural cytidine deaminases with high intrinsic single-stranded DNA (ssDNA) affinity, TadA of ABE exhibited undetectable guide-independent off-target effects at DNA and RNA levels17. Previous studies reported that the TadA8e variant derived from E. coli TadA exhibited partial cytidine deaminase activity for TC sequences within the editing window18. Recently, various CBEs, including Td-CBEmax and TadCBEd, have been developed by engineering TadA8e into a cytosine deaminase, demonstrating the engineering plasticity of TadA for versatile base editors development19–21. In addition, TadA homologues from different species were introduced with two amino acid mutations, exhibiting efficient cytidine deaminase activity22. However, these TadA-based CBEs still have defects such as motif bias, unwanted adenosine deaminase activity or insufficient cytosine editing activity. Moreover, unwanted A-to-G base editing can cause the termination codon to become TGG, limiting the disruption effect of the target gene. Therefore, screening more TadA variants or orthologs are expected to further improve the functionality of CBEs.
Here, we utilize both TadA orthologs screening and protein mutagenesis strategies to obtain TadA variants from Acinetobacter junii with the most potent activity for cytosine base editing. Furthermore, by optimizing its activity through protein amino acid substitution, we obtain aTdCBE with better performance. We then establish a genetically humanized DMD mouse model and demonstrate the potential of exon skipping and dystrophin restoration using AAV delivery, which may inform future pre-clinical research. This study provides a practical strategy for the development of base editors with distinctive features, which is valuable for expanding the functional diversity of gene editing tools and their potential applications for gene editing therapy and research use.
Results
Screening of TadA orthologs using fluorescent reporter system
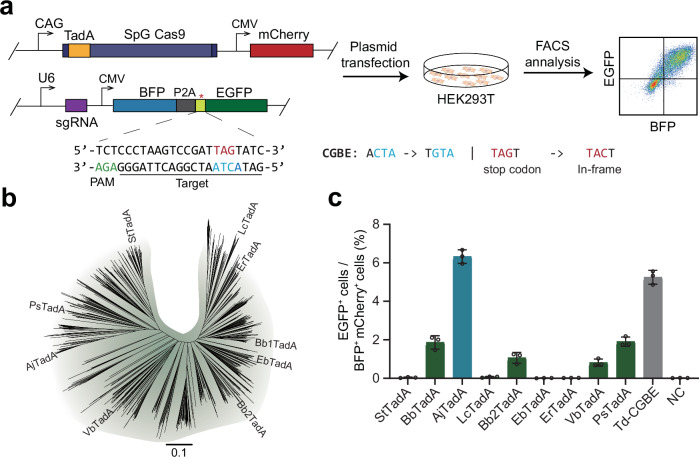
To sensitively detect DNA base editing activity in mammalian cells, we designed a fluorescent reporter system, termed as BFP-*EGFP which contains a BFP and an inactive EGFP carrying stop mutation that can be corrected through C-to-G base editing (Fig. 1a). Due to the weakest base editing preference for ACT motif by cytosine base editor based on the ecTadA variant21, we introduced ACT in the 5th to 7th positions of the target sequence to screen for variants that can efficiently edit AC motif sequences. When the target sequence is edited by cytosine deaminase of CGBEs, the TAG termination codon within mutant *EGFP will be corrected into TAC to achieve normal translation of EGFP protein.
Fig. 1. Screening for TadA orthologs with cytosine deaminase activity in mammalian cells.
a The experimental workflow for detecting base editing activity with fluorescence reporter system in HEK293T cells. b The Phylogenetic Tree of TadA orthologs. c Screening for TadA orthologs using fluorescence reporter system. Data are presented as means ± S.D. Values and error bars represent mean and S.D., n = 3 independent biological replicates. All of the above base editors do not contain UGI. PAM stands for protospacer adjacent motif. NC represents negative control. Source data are provided as a Source Data file.
Given that ABE2.1 constructed by ecTadA carrying four amino acid mutations of D108N/A106V/D147Y/E155V can produce effective editing at multiple genomic sites, we introduced these four mutations into different TadA orthologs to construct base editors1. Based on different sequence homology, we synthesized nine representative TadA orthologs with four amino acid substitutions, and constructed base editors together with SpG Cas9 (Fig. 1b). We also used Td-CGBE as control to edit the BFP-*EGFP reporter by co-transfection of CGBE-mCherry and reporter plasmids targeted by the *EGFP-specific sgRNA in HEK293T cells. Two days after transfection, we observed that about 5% of successfully transfected cells had EGFP activated by Td-CGBE (Fig. 1c). Notably, we found that a TadA ortholog from Acinetobacter junii showed the highest editing activity among the nine TadA orthologs, thereby named AjTadA.v1(Fig. 1c). We further used AjTadA.v1 to target PIK3CA loci and only found that AjTadA.v1 has the cytosine deamination activity (Supplementary Fig. 1).
Engineering of AjTadA based on multiple sequence alignment
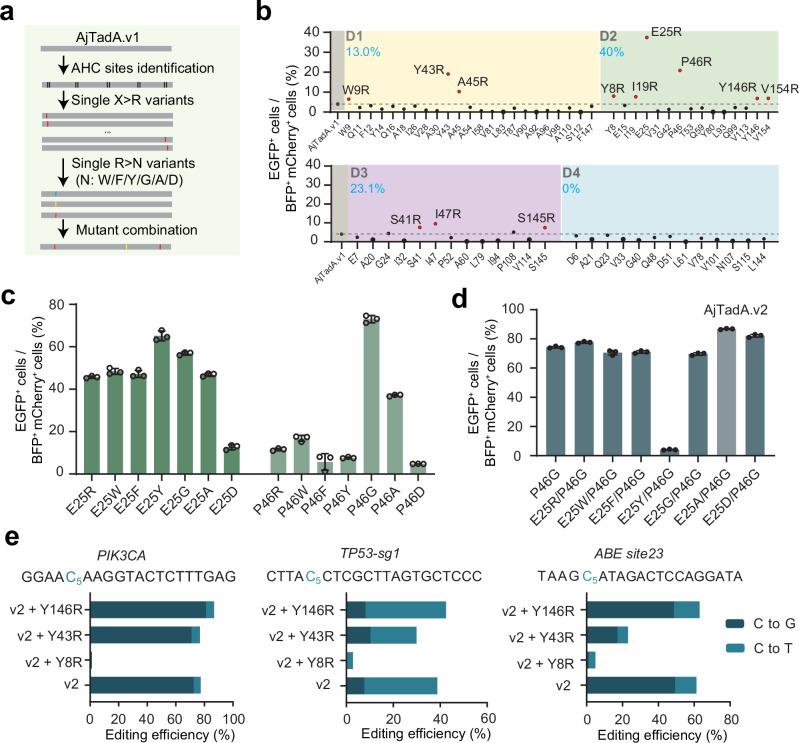
Multiple sequence alignment (MSAs) is commonly used for phylogenetic inference23, protein structure prediction24,25, functional effects of mutations prediction26,27. Through multiple sequence alignment, we identified 20 highly conserved amino acid sites with over 99% conservation in the TadA family (Supplementary Fig. 2). Highly conserved residues are under greater selection pressure throughout the evolution process to be indispensable for normal deaminase activity of TadA and survival advantage of the organism. We assume that adjacent amino acids of highly conserved sites (AHC) can assist highly conserved amino acids in performing their functions, and replacing these AHC amino acids may enhance or decrease their activity. Substitutions of amino acid residues with arginine have been shown to improve editing activity of RNA-guided nucleases by improving the binding ability of proteins and nucleic acids, including Cas12, IscB and Fanzor28–31. We wonder whether this strategy can be applied to improve the editing efficiency of deaminases. Then we performed scanning mutagenesis and substituted all non-positively charged AHC residues with arginine (Fig. 2a, Supplementary Fig. 2). We divided AHC amino acids with distances of 1, 2, 3, and 4 from highly conserved amino acids into four groups: D1, D2, D3, and D4. In total, 65 arginine single substitutions of AjTadA.v1 were screened using BFP-*EGFP reporter assay. Based on the threshold of over 1.5-fold for the EGFP signal increased by AjTadA variants relative to the AjTadA.v1 controls, we identified 12 variants with enhanced base editing activity (Fig. 2b). Remarkably, 40% (6/15) of D2 variants showed enhanced activity. In addition, E25R and P46R variants showed over 4-fold improvement relative to AjTadA.v1 (Fig. 2b).
Fig. 2. Engineering of AjTadA to improve base editing efficiency in mammalian cells.
a The experimental workflow for engineering of AjTadA. b Substitutions of non-positively charged amino acids of AjTadA at the AHC sites. Each dot represents activity for a single variant. c Substitutions of 25E and 46 P of AjTadA.v1 with seven amino acid residues. Values represent mean, n = 2 independent biological replicates. d Combination of mutations in 25E and 46 P of AjTadA.v1. Values and error bars represent means ± S.D., n = 3 independent biological replicates. e Combination of single amino acid mutation with AjTadA.v2 at three endogenous loci. Values and error bars represent means ± S.D., n = 3 independent biological replicates. All of the above base editors do not contain UGI. Source data are provided as a Source Data file.
To test whether replacing these two sites with other amino acids can further enhance activity, we selected six different type of amino acids and found that the P46G variant performed best, producing over 60% of EGFP positive cells (Fig. 2c). We then combined P46G with E25R, E25W, E25F, E25Y, E25G, E25A, or E25D in AjTadA.v1 for reporter assay and found that the variant carrying P46G/E25A exhibited the highest editing activity, named AjTadA.v2 (Fig. 2d). Considering the upper detection limit of the fluorescent reporter system, we next selected three different endogenous gene loci to further evaluate the editing activity of the variants. On the basis of AjTadA.v2, we further introduced amino acid substitutions in different regions (Y8R, Y43R, Y146R) that performed well in the first round of screening. We found that the variant carrying P46G/E25A/Y146R showed relatively high editing activity at all three endogenous sites. We further fused AjTadA.v2-Y146R with SpCas9 and UGI to obtain a cytosine editor named as aTdCBE (Fig. 2e).
ATdCBE enables robust genomic editing in mammalian cells with broad target scope and high specificity
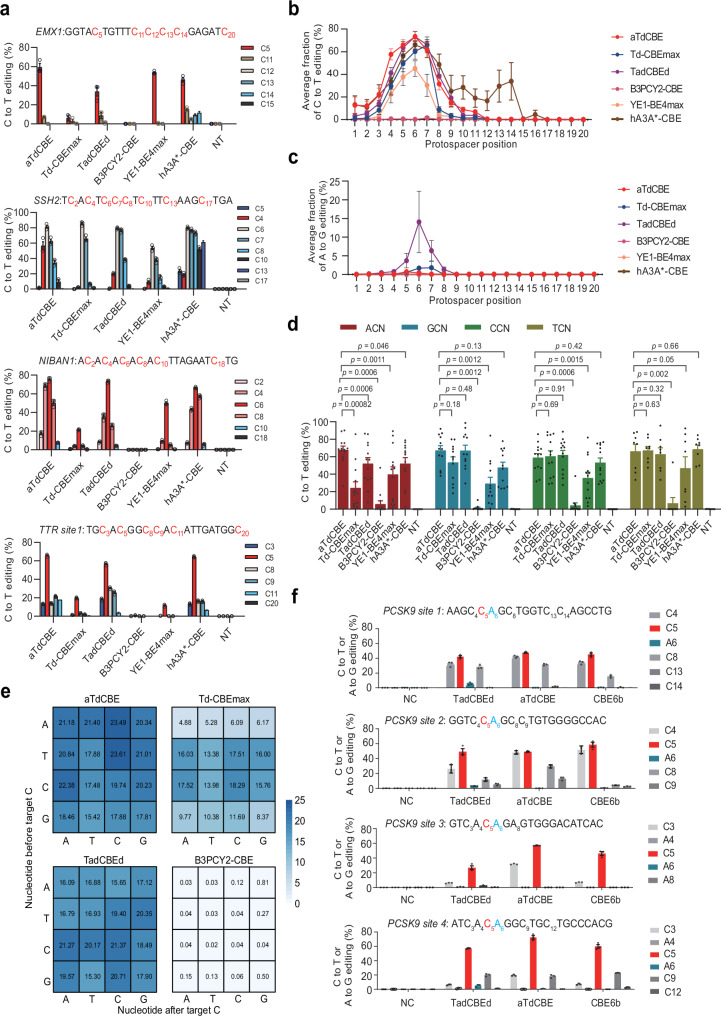
In order to compare the editing efficiency of our aTdCBE with other CBEs, Td-CBEmax19, TadCBEd21, B3PCY2-CBE22, hA3AW104A(hA3A*)-CBE32 and YE1-BE4max33 were selected due to their diverse properties, including differences in editing efficiency, off-target effects, substrate preferences, and applicability across various cell types and model organisms. By including these specific CBEs in our study, we aimed to provide a comprehensive analysis of the current state-of-the-art cytidine base editing tools and facilitate a comparative assessment of their performance under standardized experimental conditions. We selected 25 different genomic loci for activity assessment and found that aTdCBE and TadCBEd showed comparable cytosine editing efficiency (Fig. 3a, Supplementary Fig. 3). It is noteworthy that aTdCBE exhibited no significant adenosine deaminase activity. However, TadCBEd had detectable adenosine deaminase activity at multiple sites (Supplementary Fig. 4). At almost all loci, aTdCBE and TadCBEd performed better than Td-CBEmax, B3PCY2-CBE, hA3A*-CBE and YE1-BE4max. Moreover, the non-evolved TadA deaminase B3PCY2 only produces effective editing for very few targets. Except for hA3A*-CBE, the aTdCBE and TadCBEd exhibit a broad cytosine editing window than other cytosine base editors, approximately between target position 4 and 7 (Fig. 3b). In addition, TadCBEd exhibits weak editing activity towards adenosine in the fifth to seventh positions of the target sequence (Fig. 3c).
Fig. 3. ATdCBE enables robust genomic base editing in mammalian cells.
a Comparison of C-to-T conversion efficiency for endogenous loci by base editors derived from aTdCBE, Td-CBEmax, TadCBEd, B3PCY2-CBE, hA3A*-CBE and YE1-BE4max in HEK293T cells. b Base editing activity window plots showing mean C-to-T editing at all tested target positions. c Base editing activity window plots showing mean A-to-G editing at all tested target positions. d Comparison of motif preference for 25 endogenous loci by base editors derived from aTdCBE, Td-CBEmax, TadCBEd, B3PCY2-CBE, hA3A*-CBE and YE1-BE4max in HEK293T cells. The editing window of CBEs was primarily concentrated between bases 4–7. Consequently, the analysis focused on preference motifs exhibiting higher editing efficiency within this range. e High-throughput library experiments to evaluate the motif preference of aTdCBE, Td-CBEmax, TadCBEd and B3PCY2-CBE. f Introducing premature termination codons into the PCSK9 coding region using CBEs. The numbers in the grid represent the average editing efficiency of cytosines with corresponding motifs at positions 4–7 for each sgRNA. Data are presented as means ± S.D. Values represent n = 3 independent biological replicates. P-values determined by one-sided Mann-Whitney U-test and adjusted by Benjamini-Hochberg procedure. All of the above base editors contain UGI. NC represents negative control. Source data are provided as a Source Data file.
Previous studies have shown that CBE editors YE1-BE4max and TadCBEd based on Apopec and ecTadA have weaker editing abilities for GC and AC motif in library experiments, respectively. Consistent with previous results, the average editing efficiency of Apobec and ecTadA in GC and AC context of endogenous loci is indeed weaker than in other motifs, respectively (Fig. 3d). Another ecTadA-derived cytosine base editor Td-CBEmax exhibits weak editing efficiency in both AC and GC. To verify the motif preference of the TadA deaminases, we performed high-throughput library experiments using a library containing 11,868 paired sgRNA34. Consistent with previous results, the aTdCBE has better editing efficiency for AC sites (Fig. 3e).
We then tested the effects of aTdCBE, TadCBEd, and recently reported CBE6b35 on introducing the PCSK9 termination codon, using four sites containing the CAG codon (Fig. 3f). We found that TadCBEd produces A-to-G editing at all four sites, while aTdCBE and CBE6b hardly cause A-to-G editing. The aTdCBE performs better at the AC motif site than aTdCBE and CBE6b. The aTdCBE, TadCBEd and CBE6b can generate C-to-T editing efficiencies of up to approximately 72%, 57% and 60% for these CAG codon sites, respectively.
We further evaluated the specificity of aTdCBE, Td-CBEmax, TadCBEd, B3PCY2-CBE, and YE1-BE4max in HEK293T cells at two target sites respectively. After deep-sequencing analysis, we found that all the base editors displayed low level of off-target effects at the predicted off-target sites by Cas-OFFinder (Supplementary Fig. 5a). To detect nuclease-independent off-target effect of aTdCBE and TadCBEd, we performed the orthogonal R-loop assay33. Using five previous SaCas9 target sites, we found that aTdCBE exhibited comparably low nuclease-independent off-target events with TadCBEd (Supplementary Fig. 5b).
TadA8e was previously reported to have significant RNA off-target editing, and the introduction of V106W mutations resulted in significant reduction of RNA editing with slightly decreased on-target editing activity36. To investigate whether TadA variants with cytosine deaminase activity can cause transcriptome-wide off-target effects, we conducted RNA-seq to evaluate RNA off-target effect for aTdCBE and TadCBEd. Compared to the HEK293T control without the base editor transfection, both aTdCBE and TadCBEd showed a slight off-target effect on cytosine in RNA, while TadCBEd showed lower off-target effects (Supplementary Fig. 5c).
Due to the engineered AjTadA was obtained through C-to-G fluorescence reporter screening. To investigate whether the edited product of engineered AjTadA exhibits C-to-G preference compared to other deaminase enzymes, we removed the UGIs of aTdCBE and TadCBEd performed better than Td-CBEmax, B3PCY2-CBE, hA3A*-CBE and CBE6, and generated 6 CGBEs. Compared to the effect of sites and sequences on editing product preference, the effect of deaminase variants on C-to-G preference is relatively low (Supplementary Fig. 6).
Local muscle administration of cytosine base editor aTdCBE restores dystrophin expression
DMD is a fatal X-linked recessive disease affecting 1 out of 3500-5000 newborn males resulting from thousands of pathogenic mutations in the human X chromosome-linked DMD gene37. While there are thousands of documented clinical mutations, most DMD-causing mutations occur in a “hotspot” region encompassing exons 45 to 55 of the DMD gene, and skipping of exon 55 can provide therapeutic benefits to approximately 2% of DMD patients38. Engineering TadA with broad targeting scope and stringent cytosine deaminase activity may be a potential strategy for gene editing correction of DMD.
To evaluate the in vivo activity of cytosine base editors, we firstly compared the cytosine base editors in HEK293T cells, and found that only aTdCBE and TadCBEd exhibited high activity at exon 55 splice acceptor site (SAS) (Supplementary Fig. 7). We further generate a genetically humanized DMD mouse model (DMD∆E54 mdx mice) (Fig. 4a, Supplementary Fig. 8a). Sequencing of the RT-PCR product confirmed proper splicing between human DMD exon53 and exon55 in the DMDΔE54 mdx mice (Supplementary Fig. 8b). Immunostaining and western blot revealed a complete loss of dystrophin expression in DMDΔE54 mdx mice (Supplementary Fig. 8c, d). Additionally, muscular histology, creatine kinase (CK) activity and motor function also suggested that DMDΔE54 mdx mice presented severe DMD symptoms (Supplementary Fig. 8e, f, g).
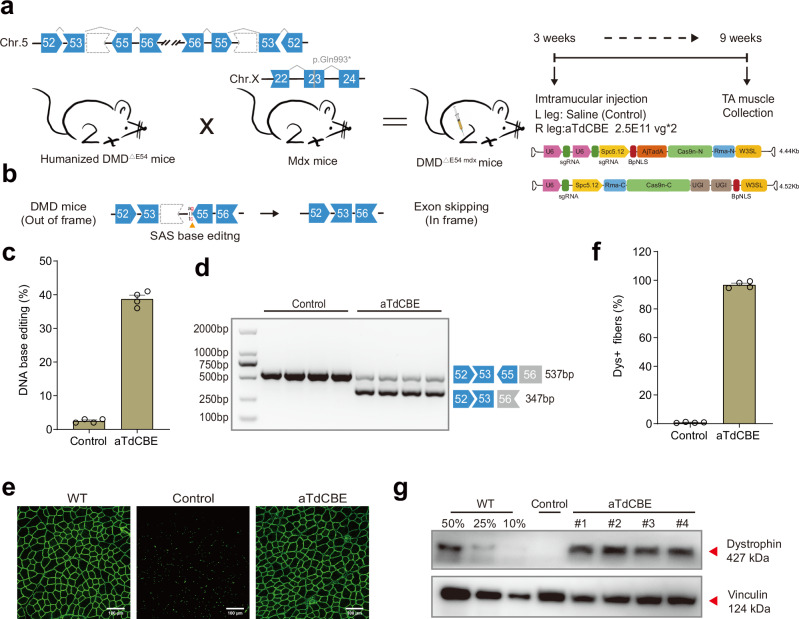
Fig. 4. ATdCBE treatment robustly rescued dystrophin expression in TA six weeks after AAV injection.
a Overview for the in vivo intramuscular (IM) injection of the AAV9-aTdCBE construct into the tibialis anterior (TA) of the right leg of 3-week-old DMDΔ54 mdx mice. The DMDΔE54 mdx mice were derived from the mating of the humanized DMD∆E54 mice with mdx mice. Left leg was injected with saline as a control. Black arrows indicate time points for tissue collection after injection. AjTadA.v2-Y146R, UGIs, Cas9n split by Rma intein and gRNA were packaged into 2 separated AAV particles. A muscle-specific promoter Spc5.12 was used to drive AjTadA-Cas9n-N or Cas9n-C. b Schematic illustrating exon skipping strategies to restore the correct open reading frame (ORF) of the DMD transcript. The shape and color of the boxes representing DMD exons indicate the reading frame. Specifically, the deletion of exon 54 in the DMD gene results in a premature stop codon in exon 55 (depicted in red). The in-frame ORF can be achieved by editing exon 55 splice acceptor sites (SAS) (ag base pair). c The DNA editing efficiency was analyzed after 6-week treatment. d RT-PCR products from muscle of DMDΔE54 mdx mice were analyzed by gel electrophoresis. e Dystrophin (Abcam, ab15277) is shown in green. Scale bar, 100 μm. f Quantification of Dys+ fibers in cross sections of TA muscles. g Western blot analysis of dystrophin (Sigma, D8168) and vinculin (CST, 13901S) expression in TA muscles 6 weeks after injection with AAV-aTdCBE or saline. For comparative analyses, wild-type (WT) mice were derived from crosses between STOCK Tg (DMD) 72Thoen/J mice (#018900) and mdx mice, which carry a c.2977 C > T, p.Gln993* mutation in exon 23 on Chr.X. Dilutions of protein extract from WT mice were used to standardize dystrophin expression (25%, 50% and 75%). Each band (#1-4) represents an individual mouse sample. Vinculin was used as the loading control. Data are represented as mean ± S.E.M (n = 4 independent biological replicates). Each dot represents an individual mouse. Source data are provided as a Source Data file.
Given the vector size beyond the genome packaging capacity of AAV, we have developed a strategy where each fragment of the base editor is expressed individually by two separate AAV vectors. The Rhodothermus marinus (Rma) intein facilitated the precise autocatalytic splicing of the two fragments, thereby reconstructing the full-length, active form of aTdCBE within the target cells. Then, we performed intramuscular (IM) injection of dual-AAV9 particles in tibialis anterior (TA) muscle of 3-week-old DMDΔE54 mdx mice. Six weeks after IM injection, TA muscle samples were collected for subsequent analysis (Fig. 4a). Deletion of exon 54 in the DMD gene resulted in the introduction of a downstream premature stop codon in exon 55, resulting in the production of a nonfunctional truncated dystrophin protein. The DMD open reading frame (ORF) can be restored by editing exon 55 SAS, allowing for the splicing of exons 53 to 56 in the case of exon 55 skipping (Fig. 4b). Our findings revealed that aTdCBE exhibited over 40% DNA base editing efficiency (Fig. 4c). RT-PCR results indicated successful splicing alteration to skip human DMD exon 55 following aTdCBE-induced base conversion, as confirmed by gel analysis (Fig. 4d). Furthermore, immunostaining results demonstrated a remarkable rescue of dystrophin expression following local injection of aTdCBE (Fig. 4e, Supplementary Fig. 9). The percentage of dystrophin-positive fibers after aTdCBE treatment reached up to 99% of the wildtype level (Fig. 4f). To quantify the level of dystrophin restoration, we conducted western blot analysis, revealing that aTdCBE system restored 60% of dystrophin expression (Fig. 4g). Together, these results indicate that aTdCBE-derived cytosine base editor, as a highly effective base editing tool with broad targeting scope, provides a promising approach for basic research and therapeutic applications.
Discussion
In this study, through TadA orthologs screening and MSA-based protein engineering, we obtained the cytosine base editor aTdCBE with robust cytosine editing activity in mammalian cells. We found that by replacing adjacent amino acids of conserved residues with arginine, over 18% variants achieved enhanced activity with more than 1.5 fold. Compared to other strategies, protein engineering methods used in our study are much more convenient and have the potential to be applied to the engineering of other proteins. Compared with the previous cytosine base editors, aTdCBE has significant advantages in editing target sequences with AC motif. In addition, the aTdCBE carrying no significant adenosine deaminase activity avoids generating unwanted editing products in applications. Although these TadA deaminases exhibited slight RNA off-target of C-to-U compared to the control, their specificity can be further improved by mutant modification or insertion of deaminases into the Cas9 protein in the future36,39. Considering its high efficiency with DNA base editing and high specificity at DNA levels, aTdCBE may have more advantages in application compared to other cytosine base editors. The aTdCBE has been employed in in vivo gene editing therapy studies for DMD in mice, and has demonstrated that aTdCBE is an efficient strategy to modulate exon skipping and restores dystrophin expression. Overall, aTdCBE offers a platform with highly efficient DNA base editing in mammalian cells, broadening applications in fundamental research, and has the potential to be applied in the field of gene editing therapy.
Methods
Study approval
Exclusively male mice were utilized for all experiments, including grip strength tests, creatine kinase (CK) analysis, and AAV injections. All animal experiments were performed and approved by the Institutional Animal Care and Use Committee (IACUC) of HuidaGene Therapeutics Inc., Shanghai, China and Lingang Laboratory, Shanghai, China.
Computational analysis of TadA orthologs
Firstly, we downloaded 15,167 TadA protein sequences from NCBI database. We further used BLASTP (v2.2.21) to remove redundant proteins with identity over than 90%40. Then, we performed multiple sequence alignment using MAFFT (v7.429)41. MEGA11 were used to construct phylogenetic tree42. “Calculate_AHC.pl” were used to identify highly conservative residues and AHC residues. MCL (v14-137)43 was used to cluster the redundant proteins with identity over than 70%, and nine TadA orthologs were randomly selected from different clusters for experimental screening.
Plasmid constructions
Human Codon-optimized orthologous TadAs were synthesized commercially (GenScript Co., Ltd) and cloned to generate pT7_NLS-TadA-Cas9-NLS_pA_pCBH_mCherry_pA plasmid by NEBuilder (New England Biolabs). All sequences are listed in Supplementary Data 1. The dual-AAV delivery system was designed to express two separate fragments of a base editor, which are subsequently spliced into the full-length protein via the Rhodothermus marinus (Rma) intein, as described in a previous study44,45. The intein sequences, specifically the N- and C-terminal segments, were synthesized by Genewiz (Suzhou, China). These segments were then integrated into the 573 and 574 amino acid residues of the aTdCBE backbones using Gibson cloning of PCR-amplified inserts.
Mammalian cell culture, transfection and flow cytometry analysis
The HEK293T cells (ATCC, CRL-3216) were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco, 11965-092) supplemented with 10% fetal bovine serum (Gibco, 10099-141 C), and 1% Pen-Strep-Glutamine (100×) (Gibco, 10378-016) at 37 °C with 5% CO2 in a cell incubator. For TadA variants screening, HEK293T cells cultured in 24-well plates were co-transfected with 1.0 μg of tagBFP-*EGFP reporter plasmid and TadA-mCherry plasmid in a molar ratio of 1:1 with Polyetherimide (PEI). After 48 h, mCherry, BFP and EGFP fluorescence were analyzed by Beckman CytoFlex flow-cytometer. To evaluate genome editing in endogenous sites, cells were harvested at 48 h after transfection and sorted by BD FACS Aria III flow cytometer. FACS data were analyzed with FlowJo X (v10.0.7).
Detection of gene editing frequency
20 μL of lysis buffer with proteinase K (Vazyme Biotech) were used to lysis about ten thousand sorted cells following the manufacturer’s manual. Targeted amplifications were produced by Phanta Max Super-Fidelity DNA Polymerase (Vazyme Biotech). For targeted amplicon sequencing, PCR reactions were performed using primers with different barcodes (Supplementary Data 2). The DNA products were purified with Gel extraction kit (Omega) and analyzed by 150 bp paired-end reads Illumina NovaSeq 6000 platform (Genewiz Co. Ltd.). The deep sequencing data were first de-multiplexed by Cutadapt (v.2.8) based on sample barcodes. The de-multiplexed reads were then processed by CRISPResso2 for the quantification of editing efficiency, including indels, A-to-G or C-to-T conversions at each target site46.
High-throughput library experiments
The HEK293T cells previously constructed using 11,868 pairs of sgRNA lentivirus plasmid library was used to detect motif preference in the base editor34. For each 10 cm dish, 35 μg plasmids that encode CBEs and mCherry were transfected using PEI. After 48 h, transfected cells were harvested using FACS followed by genomic DNA extraction. The PCR products were sequenced using a 150 bp paired-end Illumina NovaSeq 6000 platform (Genewiz Co. Ltd.). High-throughput sequencing datasets were processed using CRISPResso2 to calculate editing efficiency of each target. The target sites were excluded with a coverage depth of less than 100 in each sample. Cytosines in positions 4–7 of the target sequences were used to statistically analyze motif preferences.
Off-target analysis with in-silico prediction
To evaluate the specificity of TadA base editors, the Cas-OFFinder was employed to predict the potential off-target sites as described previously47. Search queries covered both Cas9 spacer sequence and PAM of the on-target site. The PAM of research was set to “NGG” and the mismatches were set to less than 5. All other parameters were left as default. The potential off-target sites were amplified and deep sequenced for analysis (Supplementary Data 3).
Orthogonal R-loop assay
Orthogonal R-loop assay was performed to detect the nuclease-independent off-target editing as described previously33. 1.5 μg plasmids that encode aTdCBE/TadCBEd and an on-target sgRNA for aTdCBE/TadCBEd, along with plasmids expressing dSaCas9 and a SaCas9 sgRNA that targets the genome locus previously reported were co-transfected using PEI. After 48 h, transfected cells were harvested using FACS followed by genomic DNA extraction with 20 μL of freshly prepared lysis buffer (Vazyme) with proteinase K added. The targeted loci by dSaCas9 were amplified and deep sequenced.
Generation of humanized DMD∆E54 mdx mice
Mice were housed in a barrier facility with a 12 h light/dark cycle and maintained in compliance with the guidelines outlined in the Instructive Notions with Respect to Caring for Laboratory Animals issued by the Ministry of Science and Technology of China. To generate the humanized DMD∆E54 mice, we employed the CRISPR/Cas9 system on the embryos obtained from mating STOCK Tg(DMD)72Thoen/J male and female mice (#018900). Specifically, we designed two sgRNAs targeting the flanking introns of human DMD exon 54 on Chr.5. The sequences of these sgRNAs are gRNA1: gTTTCTGCAAGTGCAGAGAGG and gRNA2: GGTGTGTGGAGTGAGATACT. Each sgRNA template was appended with the T7 promoter sequence (TAATACGACTCACTATAg) for efficient transcription. The PCR product was then purified directly using the Omega gel extraction kit (Omega, D2500-02), and the templates were used for in vitro transcription with the MEGAshortscript T7 Kit (Invitrogen, AM1354). The sgRNAs were purified using a MEGAclear Kit (Invitrogen, AM1908) and eluted with nuclease-free water. The concentration of target sgRNA was measured using a NanoDrop instrument. For cytoplasmic injection, spCas9 mRNA (100 ng/μl), sgRNA-L (50 ng/μl) and sgRNA-R (50 ng/μl) were mixed and then injected into fertilized eggs using a FemtoJet microinjector (Eppendorf) with constant flow settings. The injected zygotes were cultured in KSOM medium for 12 h and surgically transferred to the oviduct of recipient mice 24 h after estrus was observed. Genomic DNA from the tail tissue of founder (F0) mice was isolated according to manufacturer’s instructions for the OMEGA Kit (Omega, D3396-02) for PCR, followed by gel electrophoresis. All sequences are listed in Supplementary Data 4.
AAV9 production and delivery to DMDΔE54 mdx mice
AAVs used in this study were produced by HuidaGene Therapeutics Co., Ltd. The transfection process involved achieving a confluency of 70–90%, after which the media was replaced with fresh pre-warmed growth media prior to transfection. For each 15 cm dish, a mixture of 20 μg of pHelper, 10 μg of pRepCap and 10 μg of GOI plasmid was transferred dropwise to the cell media. Following a three-day incubation, the AAVs were purified using iodixanol density gradient centrifugation. The DMDΔE54 mdx mice were derived by mating the humanized DMD∆E54 mice with mdx mice carrying stop mutation in mouse exon 23 on Chr.X. In the case of intramuscular injection, 3-week-old DMD∆E54 mdx mice were anesthetized, and their tibialis anterior (TA) muscle was injected with 50 μL of AAV9 (2.5 × 1011 vg per virus) preparations or with an equivalent volume of saline solution. Tissues were divided into distinct segments for targeted assessment. Specifically, the distal region was allocated for evaluating DNA editing and exon skipping efficiency, the middle portion was dedicated to Western blot analysis of dystrophin expression, and the proximal segment was reserved for immunofluorescent analysis of dystrophin levels at six weeks after treatment.
Western blot analysis
The samples were homogenized using RIPA buffer supplemented with protease inhibitor cocktail. The lysate supernatants were quantified using a Pierce BCA protein assay kit (Thermo Fisher Scientific, 23225) and adjusted to an identical concentration using H2O. Equal amounts of the sample were mixed with NuPAGE LDS sample buffer (Invitrogen, NP0007) and 10% β-mercaptoethanol, then boiled at 70 °C for 10 min. Ten µg of total protein per lane was loaded into 3% to 8% tris-acetate gels (Invitrogen, EA03752BOX) and electrophoresed for 1 h at 200 V. Protein was transferred onto a PVDF membrane under wet conditions at 350 mA for 3.5 h. Subsequently, the membrane was blocked in 5% non-fat milk in TBST buffer and then incubated with primary antibody to label the specific protein. After washing three times with TBST, the membrane was incubated with an HRP-conjugated secondary antibody specific to the IgG of the species of primary antibody against dystrophin (Sigma, D8168) or vinculin (CST, 13901S). Finally, the target proteins were visualized using Chemiluminescent substrates (Invitrogen, WP20005).
Histology and Immunofluorescence
Tissue samples were collected and immersed into preconditioned 4% paraformaldehyde. The fixed tissues underwent dehydration through a series of alcohol concentrations, followed by treatment with xylene and embedding in melted paraffin wax. Subsequently, the paraffin-embedded tissues were deparaffinized using xylene, followed by a series of alcohol washes ranging from high to low concentrations, and finally placed in distilled water. For hematoxylin and eosin (H&E) staining, the slides were stained with hematoxylin for 3–8 min, followed by color separation using acid water and ammonia water. After dehydration using 70% and 90% alcohol for 10 min each, the tissues were stained in eosin staining solution for 1–3 min, and dehydrated in ascending alcohol solutions (50%, 70%, 80%, 95%, 100%). Coverslips were then mounted onto the glass slides with neutral resin.
For Sirius red staining, the slides were stained with picrosirius red for one hour, washed in two changes of acidified water. Physical removal of most of the water from the slides was accomplished by vigorous shaking. Then, slides were dehydrated in three changes of 100% ethanol, cleared in xylene, and finally mounted in neutral resin.
For immunofluorescence, the tissues were embedded in optimal cutting temperature (OCT) compound and snap-frozen in liquid nitrogen. Serial frozen cryosections (10 µm) were fixed for two hours at 37 °C followed by permeabilization with PBS + 0.4%Triton-X for 30 min. After washing with PBS, the samples were blocked with 10% goat serum for 1 h at room temperature. Next, the slides were incubated overnight at 4 °C with primary antibodies against dystrophin (Abcam, ab15277) and spectrin (Millipore, MAB1622). The next day, samples were extensively washed with PBS and incubated with compatible secondary antibodies (Alexa Fluor® 488 AffiniPure donkey anti-rabbit IgG (Jackson ImmunoResearch labs, 711-545-152) or Alexa Fluor 647 AffiniPure donkey anti-mouse IgG (Jackson ImmunoResearch labs, 715-605-151)) and DAPI for 3 h at room temperature. Following a 15-minute wash with PBS, the slides were sealed with fluoromount-G mounting medium. All images were visualized using Nikon C2. The number of Dys+ muscle fibers is represented as a percentage of total spectrin-positive muscle fibers.
RNA-seq for off-target analysis
To quantify the transcriptome deaminases off-target edits, HEK293T cells were cultured in 10 cm dishes with 80% confluence and transfected with 35 μg plasmids containing base editors and gRNA. After 48 h, about 600,000 transfected cells were sorted by FACS, and RNA was extracted using Trizol (Ambion) for RNA-seq library preparation. An RNA-seq library was generated with a TruSeq Stranded Total RNA library preparation kit according to the standard protocol. The transcriptome libraries were sequenced using a 150 bp paired-end Illumina NovaSeq 6000 platform (Genewiz Co. Ltd.).
The calculation analysis referred to previously published methods36. Trimmomatic (v.0.39-2) were using to filter the RNAseq raw data48. The clean reads were aligned to the hg38 reference genome with Hisat2 (v.2.2.1)49. RNA editing sites were calculated using REDItools (v1.2.1) with “-e -d -p -u -m 60 -T 5-5 -W -n 0.0” parameters50. The edited adenosines divided by total adenosines and the edited cytosines divided by total cytosines were calculated separately.
Statistics & reproducibility
All cell experimental results are presented as mean ± s.d, while all animal experimental results are presented as mean ± s.e.m. The one-sided Mann-Whitney U test or unpaired two-tailed Student’s t-test were utilized for comparisons. The number of independent biological replicates are shown in figure legends. No data were excluded from the analyses. We randomly selected cells for test group and control group. DMD mice used for gene editing therapy were allocated to control or AAV9 treated group randomly.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by HUIDAGENE Therapeutics Inc, Project of National Natural Science Foundation of China (82271048 to J.H., 82301215 to G.Z.), Science and Technology Commission of Shanghai Municipality (22S11900200 and 23XD1420500 to J.H.), Seed Industry Vitalization Program of Jiangsu Province (JBGS[2021]025 to E.Z.), Jiangsu Provincial Key Research and Development Program (BE2021372 to E.Z.) and the Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-SCAB-202301 to T.Y.).
Author contributions
Y.Z. conceived the project. G.L., X.D., J.L., J.H. and G.Z. designed experiments. X.D., J.L. and G.L. performed protein engineering and endogenous sites base editing assay. T.L. and G.Z. performed the off-target assay. Y.Z. performed bioinformatics analysis. E.Z. and T.Y. performed the high-throughput library experiments. G.L., Y.Yu., and Y.W. performed in vivo virus injection, tissue dissection, histological immunostaining and muscle function experiments. H.Z., J.Z., Z.Z., H.W., S.C., and Y.Yu. assisted with experiments. Y.Z., J.H., and C.X. supervised the whole project. Y.Z., C.X., G.L. and Y.Yuan. wrote the manuscript with data contributed by all authors who participated in the project.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The deep-seq data and RNA-seq data generated in this study have been deposited in the NCBI database under accession code PRJNA1061823. Source data are provided with this paper.
Code availability
The scripts and codes used in this manuscript are available at zenodo.org (accession code: 13337534).
Competing interests
X.D, G.L. and Y.Z. have submitted a patent application to the China National Intellectual Property Administration pertaining to the deaminase engineering aspect of this work (application number PCT/CN2024/078613). The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guoling Li, Xue Dong, Jiamin Luo, Tanglong Yuan, Tong Li, Guoli Zhao.
Change history
10/18/2024
A Correction to this paper has been published: 10.1038/s41467-024-53171-y
Contributor Information
Erwei Zuo, Email: zuoerwei@caas.cn.
Chunlong Xu, Email: xucl@lglab.ac.cn.
Jinhai Huang, Email: jinhaihuang@fudan.edu.cn.
Yingsi Zhou, Email: yingsizhou@huidagene.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52485-1.
References
- 1.Komor, A. C. et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science353, aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Ma, Y. et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods13, 1029–1035 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porto, E. M. et al. Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov.19, 839–859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao, D. et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol.39, 35–40 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Kurt, I. C. et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol.39, 41–46 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiesa, R. et al. Base-edited CAR7 T cells for relapsed T-Cell acute lymphoblastic leukemia. N. Engl. J. Med.389, 899–910 (2023). [DOI] [PubMed] [Google Scholar]
- 9.Smekalova, E. M. et al. Cytosine base editing inhibits hepatitis B virus replication and reduces HBsAg expression in vitro and in vivo. Mol. Ther. Nucleic Acids35, 102112 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou, H. et al. Efficient silencing of hepatitis B virus S gene through CRISPR-mediated base editing. Hepatol. Commun.6, 1652–1663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knipping, F. et al. Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Mol. Ther.30, 130–144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anzalone, A. V. et al. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol.38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science364, 289–292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science364, 292–295 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Zhou, C. et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature571, 275–278 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Grünewald, J. et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature569, 433–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan, N. et al. Cytosine base editors induce off-target mutations and adverse phenotypic effects in transgenic mice. Nat. Commun.14, 1784 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H. S. et al. Adenine base editors catalyze cytosine conversions in human cells. Nat. Biotechnol.37, 1145–1148 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Chen, L. et al. Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing. Nat. Biotechnol.41, 663–672 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Lam, D. K. et al. Improved cytosine base editors generated from TadA variants. Nat. Biotechnol.41, 1686–1697 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neugebauer, M. E. et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat. Biotechnol.41, 673–685 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, S. et al. TadA orthologs enable both cytosine and adenine editing of base editors. Nat. Commun.14, 414 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogden, T. H. et al. Multiple sequence alignment accuracy and phylogenetic inference. Syst. Biol.55, 314–328 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopf, T. A. et al. Mutation effects predicted from sequence co-variation. Nat. Biotechnol.35, 128–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frazer, J. et al. Disease variant prediction with deep generative models of evolutionary data. Nature599, 91–95 (2021). [DOI] [PubMed] [Google Scholar]
- 28.McGaw, C. et al. Engineered Cas12i2 is a versatile high-efficiency platform for therapeutic genome editing. Nat. Commun.13, 2833 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, H. et al. An engineered xCas12i with high activity, high specificity, and broad PAM range. Protein Cell.14, 538–543 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito, M. et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature620, 660–668 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han, D. et al. Development of miniature base editors using engineered IscB nickase. Nat. Methods20, 1029–1036 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Wang, X. et al. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol.36, 946–949 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Doman, J. L. et al. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol.38, 620–628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan, T. et al. Deep learning models incorporating endogenous factors beyond DNA sequences improve the prediction accuracy of base editing outcomes. Cell Discov.10, 20 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, E. et al. Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference. Nat. Commun.15, 1697 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol.38, 883–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, T. C. et al. Therapeutic approaches for Duchenne muscular dystrophy. Nat. Rev. Drug. Discov.22, 917–934 (2023). [DOI] [PubMed] [Google Scholar]
- 38.Bladen, C. L. et al. The TREAT-NMD DMD global database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat.36, 395–402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, Y. et al. A Cas-embedding strategy for minimizing off-target effects of DNA base editors. Nat. Commun.11, 6073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul, S. F. et al. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 41.Nakamura, T. et al. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics34, 2490–2492 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura, K. et al. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol.38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enright, A. J. et al. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res.30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy, J. M. et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng.4, 97–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin, M. et al. Correction of human nonsense mutation via adenine base editing for Duchenne muscular dystrophy treatment in mouse. Mol. Ther. Nucleic Acids35, 102165 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol.37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bae, S. et al. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolger, A. M. et al. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim, D. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flati, T. et al. HPC-REDItools: a novel HPC-aware tool for improved large scale RNA-editing analysis. Bmc. Bioinforma.21, 353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The deep-seq data and RNA-seq data generated in this study have been deposited in the NCBI database under accession code PRJNA1061823. Source data are provided with this paper.
The scripts and codes used in this manuscript are available at zenodo.org (accession code: 13337534).