Abstract
There is a growing interest in the creation of engineered condensates formed via liquid–liquid phase separation (LLPS) to exert precise cellular control in prokaryotes. However, de novo design of cellular condensates to control metabolic flux or protein translation remains a challenge. Here, we present a synthetic condensate platform, generated through the incorporation of artificial, disordered proteins to realize specific functions in Bacillus subtilis. To achieve this, the “stacking blocks” strategy is developed to rationally design a series of LLPS-promoting proteins for programming condensates. Through the targeted recruitment of biomolecules, our investigation demonstrates that cellular condensates effectively sequester biosynthetic pathways. We successfully harness this capability to enhance the biosynthesis of 2’-fucosyllactose by 123.3%. Furthermore, we find that condensates can enhance the translation specificity of tailored enzyme fourfold, and can increase N-acetylmannosamine titer by 75.0%. Collectively, these results lay the foundation for the design of engineered condensates endowed with multifunctional capacities.
Subject terms: Industrial microbiology, Synthetic biology, Metabolic engineering, Translation
Biomolecular condensates can serve as synthetic organelle-like compartments within prokaryotes. Here, the authors report a synthetic condensate platform, generated through the incorporation of artificial, disordered proteins, and demonstrate control of metabolic flux and protein translation in Bacillus subtilis.
Introduction
Microbial cell factories can be constructed and remodeled using synthetic biology toolboxes for the enhanced biosynthesis of valuable products, such as fine chemicals, functional nutraceuticals, or pharmaceuticals1–4. Engineering organelles for the compartmentalization of biosynthetic pathways is a commonly reported strategy in synthetic biology5,6. In eukaryotes, organelles surrounded by lipid bilayer membranes can create an isolated environment that can be bioengineered by targeting desired components7,8. Extensive research has demonstrated increased reaction kinetics for natural product synthesis by packaging enzymatic reactions within endogenous organelles9,10. In contrast, the lack of universal organelle structures in prokaryotes limits the implementation of compartmentalization strategies. To this end, protein-shelled microcompartments have been rationally designed and engineered in prokaryotes as artificial organelles11,12. However, most of these compartmentalization strategies are limited by the diffusion of biomolecules of interest13,14.
Biomolecular condensates play a critical role in regulating cellular behaviors15–17. Generally, the condensates can be formed through a liquid-liquid phase separation (LLPS) process of intrinsically disordered proteins (IDPs) or other biomolecules, and are based on multivalent interactions18,19. Compared with the membrane-bound organelles or protein-shelled microcompartments, the liquid-like condensates can exchange biomolecules freely with the exterior20,21. For instance, spider silk-driven condensates and RNA-based condensates have been formed to achieve the acceleration of reaction kinetics for the efficient biosynthesis of target compounds in Escherichia coli22,23. Meanwhile, the development of de novo protein design has provided a potential route to use designer protein for controlling biological processes24,25. However, de novo design and functionalization of genetically encoded proteins that form cellular condensates for engineering cellular metabolism or protein translation remains an ongoing challenge.
In this study, we present an engineerable approach to multi-functional biomolecular condensates formed by the de novo designer disordered proteins in Bacillus subtilis. First, we develop the “stacking blocks” strategy for bottom-up de novo design of an octapeptide FW1 and create a series of synthetic IDPs (SIDPs) that comprised repeats of FW1 motif and variants with rational amino acid mutations to modulate the formation and physical properties of condensates. A subset of this series is then used to assemble cellular micron-size condensates with multiple functionalities by manipulating the localization of targeted molecules to synthetic condensates, including a metabolism control system and a spatially separated orthogonal translation system. As a result, the metabolism control system leads to a 2.23-fold increase in the titer of 2′-fucosyllactose (2′-FL) for five-enzyme co-localization. Furthermore, cells expressing spatially separated orthogonal translation system produce more N-acetylmannosamine (ManNAc), with an improvement of 75.0% by enhancing the translation specificity of tailored enzyme with noncanonical amino acid (ncAA). Our de novo SIDPs system developed here can serve as a robust toolkit for a broad range of applications in synthetic biology and opens up a promising avenue for transcending the boundaries that separate prokaryotic and eukaryotic systems.
Results
De novo design of synthetic biomolecular condensates
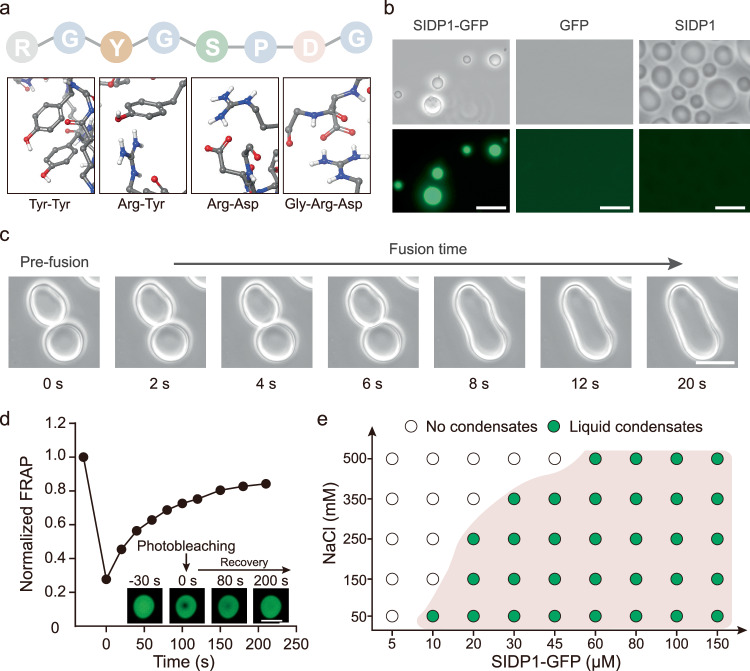
The “stacking blocks” strategy was developed to generate the SIDP that promotes molecular condensation. Specifically, each amino acid was treated as a building block, and they were assembled into a peptide as minimal repeat. Then, the SIDP with several repeats of peptide was obtained, containing various categories of amino acids that have been shown to promote LLPS behavior through intrachain interactions, such as pi-pi interactions, pi-cation, cation-anion, and hydrogen bonding26. To do this, glycine (G) and proline (P) were selected as fixed anchor points to separate other amino acids due to their unique structures and significance. Subsequently, we gave the sequence an overall net-zero charge by incorporating arginine (R) and aspartic acid (D) into the peptide sequence, as they can also drive LLPS via intrachain interactions. Considering that the R side chain can interact with the side chains of aromatic amino acids, we incorporated tyrosine (Y) into the designed sequence. Finally, we incorporated serine (S), which has the highest occurrence probability in natural IDPs, as it can form hydrogen bonds with water (Supplementary Fig. 1). Building on these design principles, the 8-mer peptide RGYGSPDG termed FW1 was obtained (Fig. 1a). By using molecular dynamics simulations, the octapeptide FW1 presented values of collapse degree (2.1) and clustering degree (0.95), suggesting that FW1 has high aggregation capabilities. Inspired by the observed molecular weights of native IDPs such as RGG, SIDP1 comprising 20 repeats of the FW1 unit was then expressed and purified by using recombinant methods for their experimental characterization in vitro (Supplementary Fig. 2 and Supplementary data 1). Both SIDP1 and SIDP1-GFP (N-terminal fusion with GFP) formed micron-size spherical droplets. At the same time, GFP alone produced dispersed fluorescence and no liquid droplets were observed (Fig. 1b). In addition, the SIDP1-GFP droplets tended to fuse into larger ones (Fig. 1c). In a fluorescence recovery after photobleaching (FRAP) study, the fluorescent signal of the bleached droplet could be recovered to about 83% with a characteristic recovery time of ~210 s (Fig. 1d). These results indicated that SIDP1-driven droplets have typical liquid-like features with high fluidity and high dynamics.
Fig. 1. Construction and characterization of synthetic condensate formed by SIDP1-GFP in vitro.
a Expected interactions promoting LLPS of 8-mer peptide FW1 into synthetic condensate, and the representative interactions are presented on the line below. b The SIDP1-GFP and SIDP1 protein at protein concentration of 50 μM formed condensates in vitro (150 mM NaCl, pH = 7.4, 25 oC, without PEG2000). GFP was used as a negative control. c Light microscopy images showing two individual condensates tend to fuse together. d Fluorescence recovery of a partially photobleached droplet (n = 3). The inset shows representative images of photobleaching on the droplet. e A regime diagram illustrates the formation of SIDP1-GFP condensates at different protein and salt concentrations. Scale bars = 10 μm. Data are presented as mean ± s.d. of three biologically independent replicates. Source data provided as a Source Data file.
Subsequently, the physicochemical conditions that have been previously shown to affect in vitro LLPS behavior were examined for further characterization of the SIDP1-GFP phase boundary. As shown in Fig. 1e, SIDP1-GFP formed liquid-like droplets at protein concentrations <10.0 µM (50 mM NaCl, pH = 7.4, 25 °C). Higher SIDP1-GFP concentrations promoted phase behavior, whereas higher NaCl concentrations favored miscibility. Unexpectedly, the protein solution exhibited lower critical solution temperature (LCST) phase behavior. They were transparent at lower temperature, but upon heating they phase separated and become visibly turbid (Supplementary Fig. 3 and Supplementary Movie 1). In addition, the different concentrations of the macromolecular crowder polyethylene glycol (PEG2000) were used to mimic the different intracellular physiological crowding contexts. As expected, more molecular crowding agent could further reduce the fluidity of SIDP1-GFP droplets (Supplementary Fig. 4). In summary, these observations indicated that the SIDP1-GFP droplets are tunable and could be manipulated in vitro.
To achieve programable physical features of the synthetic condensates, we sought to create a series of FW1 sequence variants by adjusting the composition and location of amino acids. We first investigated the impact of the arrangement of amino acids on condensate properties with the amino acid composition remaining unchanged. The variants SIDP2-GFP and SIDP3-GFP that had the sequences (RGGDSPYG)20 and (RGSPYGDG)20 can phase separate at a lower concentration of ~8 and 6 μM (150 mM NaCl, pH = 7.4, 25 °C), respectively. Meanwhile, they also showed a lower mobility than SIDP1-GFP. By contrast, the variants SIDP4-GFP and SIDP5-GFP that had the sequences (GYPSDGRG)20 and (DGRGSPYG)20 performed similar recovery over the time periods interrogated by FRAP when compared with SIDP1-GFP (Supplementary Fig. 5). When the aromatic amino acid Y was replaced with alanine (A), the variant (SIDP6) exhibited only subtle improvements in fluorescence recovery. However, the SIDP7-GFP (replacing Y with threonine) showed arrested recovery over the time periods studied by FRAP (Supplementary Fig. 6a). Besides, amorphous and irreversible structures were observed when an additional aromatic amino acid was positioned at the end of the FW1 sequence (Supplementary Fig. 6b). Strikingly, omitting the R at the N-terminal position or replacing with other types of amino acids such as lysine (K), E, A, and T completely prevents phase separation, as no condensates were observed, even at a protein concentration of 150 µM (Supplementary Fig. 6c). These results indicated that cation-anion interaction between the side-chains of R and D plays a crucial role in driving condensate formation. Besides, although both K and R can participate in cation-anion interaction, only R side chain can form pi–pi interaction with the aromatic amino acid side chains for facilitating the formation of condensates more easily due to its guanidium group. Overall, these results suggested that not only the composition of amino acids but also the arrangement of amino acids, influences the formation and properties of condensates.
Formation and functionalization of synthetic condensates inside bacterial cells
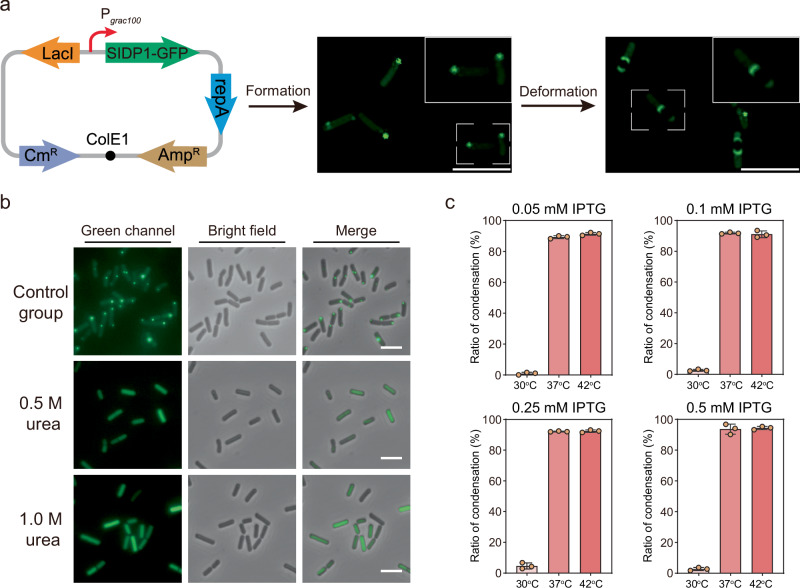
To examine whether these condensates enable the formation of condensates in prokaryotes and how they can be used to achieve desired function, the selected SIDP1 genetically fused with GFP was overexpressed in Bacillus subtilis, a model microorganism widely used in the biosynthesis of nutraceuticals and fine chemicals27. Fluorescence imaging revealed that the expression of the SIDP1-GFP fusion as condensates mainly localized near the middle or pole regions of cells, while GFP alone produced fluorescence dispersed through the cytosol (Fig. 2a and Supplementary Fig. 7). Interestingly, the fluorescent condensates underwent deformation due to the spatial compression caused by the spore, providing preliminary evidence that the cellular SIDP1-GFP condensates are liquid-like (Fig. 2a). Besides, the cellular condensates could be disrupted by treating the cells with 0.5 or 1.0 M urea, because low concentration urea can dissolve liquid-like condensates but not solid-like condensates, supporting that the condensates formed in B. subtilis are dynamic (Fig. 2b and Supplementary Fig. 8)22,28. Furthermore, we observed a fluorescence recovery and excellent solubility of SIDP1-GFP condensates in B. subtilis, providing further evidences that the condensates are liquid-like rather than inclusion bodies (Supplementary Fig. 9 and Fig. 10).
Fig. 2. Liquid-like properties of cellular synthetic condensate.
a The fluorescence microscopy images of B. subtilis overexpression SIDP1-GFP fusion protein. All cells were cultivated at 37 °C and induced with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) for gene expression. b Characterization of the synthetic cellular condensate by urea perturbation after induction for 2 h. c Measurement of the ratio of synthetic condensates in B. subtilis under different SIDP1-GFP expression level and temperature (n = 3). Scale bars = 5 μm. Data are presented as mean ± s.d. of three biologically independent replicates. Source data provided as a Source Data file.
Interestingly, live imaging growing cells confirmed that the temperature-responsive phase behavior is also observed in vivo. Specifically, when the concentration of the inducer isopropyl-β-d-thiogalactoside (IPTG) was 0.05 mM, there was almost no observable LLPS within B. subtilis at 30 °C, while at higher temperature (37 or 42 °C), the ratio of condensation was significantly improved to 90.5% and 92.4%, respectively. This trend was consistent in different expression levels of SIDP1-GFP (Fig. 2c and Supplementary Fig. 11). Notably, when compared with the control, the cell growth (OD600) was decreased by 19.4% after SIDP1-GFP overexpression for 2 h, whereas the cell morphology was not substantially altered (Supplementary Fig. 12).
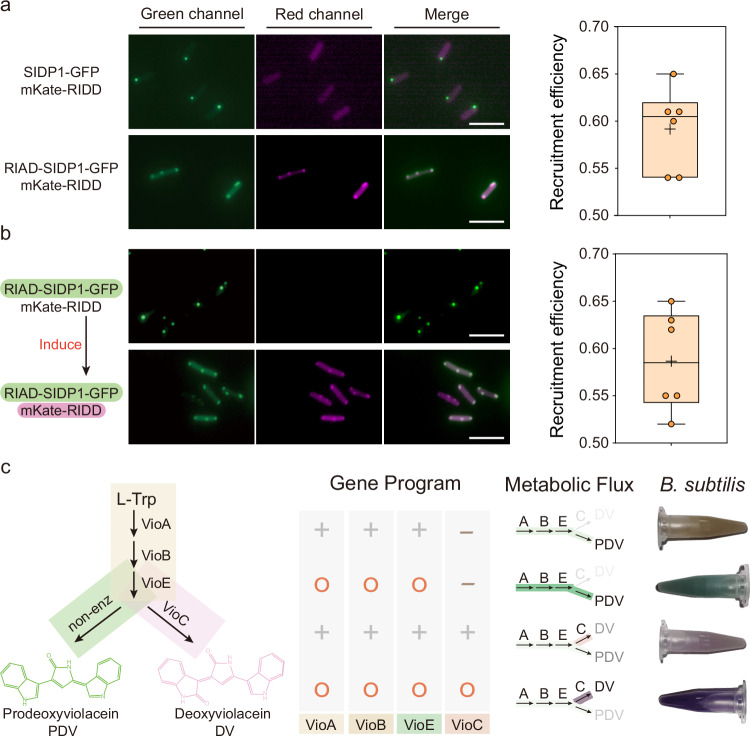
Having established cellular condensate in B. subtilis, we next sought to explore the potential of using synthetic condensates to achieve intended functions. First, the cognate interaction motif RIAD from the cAMP-dependent protein kinase and RIDD from the A kinase-anchoring proteins were chosen to test whether larger molecules can interact with condensates due to their small size and strong binding affinity29. Specifically, the RIAD and RIDD were encoded on the N-terminus of the SIDP1-GFP protein and the C-terminus of mKate, respectively. As shown in Fig. 3a and Supplementary Fig. 13, SIDP1-GFP fused with RIAD formed condensates and ~59.2% of mKate protein was recruited to the condensates. In contrast, the mKate protein was uniformly distributed in the cytoplasm due to a lack of RIAD peptide in SIDP1-GFP. However, these results could not prove that client protein can be recruited after a SIDP1 condensate has formed, as the client protein in the previous experiment are co-expressed and could bind in the cytoplasm before the phase separation takes place. To obtain a better understanding of the system, we transformed another plasmid regulated by the xylose operon that encodes the mKate-RIDD. We first allowed the expression of SIDP1-GFP at 37 °C for a sufficient duration to enable the formation of condensates, and then we removed the IPTG induction medium and replaced it by a xylose-containing medium. We observed that after IPTG induction, ~58.7% of mKate protein remained recruited to the cellular condensates (Fig. 3b). Besides, as shown in Supplementary Fig. 14, when the order of IPTG and xylose induction was exchanged, the condensates still maintained a similar recruitment efficiency (56.0%). These results suggested that the cellular condensates fused with specific interaction motifs can serve as synthetic crucibles for co-localizing with client proteins cooperated by corresponding peptide tags.
Fig. 3. Engineered intracellular condensates with intended function.
a Measuring recruitment efficiency of target fluorescent protein within condensates upon co-expression of RIAD-SIDP1-GFP and mKate-RIDD (n = 6 biologically independent cells). SIDP1-GFP protein without RIAD peptide was used as a negative control. b Measuring recruitment efficiency of mKate-RIDD (driven by xylose operon) within RIAD-SIDP1-GFP formed condensates (driven by IPTG). The first three columns indicate the dual-channel imaging and merging images of RIAD-SIDP1-GFP and mKate-RIDD. The fourth column represents the ratio of mKate fluorescence intensity of condensates to that of the whole cell (n = 6 biologically independent cells). The box plots in (a) and (b) show the mean value as a black cross, the center as a black line, box extending between the 25th and 75th percentiles, and whiskers indicating minimum and maximum data points. c Validation of the potential function of cellular condensates for controlling metabolic flux. The leftmost column represents the metabolic pathways of deoxyviolacein (DV) and prodeoxyviolacein (PDV). Cellular condensate programs flexibly redirect the metabolic flux of the DV/PDV biosynthesis pathway, resulting in a significant change in cell color. 1.5 mL culture were harvested by centrifugation. The “+” represents the gene being expressed in the strain, while the “-” indicates non-expression. Additionally, the “O” signifies that the protein is colocalized within the cellular condensate. Scale bars = 5 μm. Source data provided as a Source Data file.
Recently, p-Azido-L-phenylalanine (pAzF)-based bio-orthogonal copper-free click chemistry reaction has been demonstrated to be an applicable approach for examining the permeability of small molecules within cellular condensates30. Therefore, to further examine whether small molecules can diffuse into the synthetic condensates here, the pAzF incorporation system for B. subtilis was constructed and tested by expressing an aaRS/tRNA mutant pair that can incorporate pAzF into amber stop codons31 (Supplementary Fig. 15a). We then designed and expressed SIDP1TAG that carries a unique reactive group (that is, an azide). After incubating live recombinant B. subtilis with 1 g/L Cyanine 5 DBCO for 30 min, we observed fluorescent condensates in vivo after removing excess dye (Supplementary Fig. 15b). These results clearly showed that small molecules can freely permeate through the synthetic SIDP1 condensate.
The ease of both product visualization and quantification makes the deoxyviolacein (DV)/prodeoxyviolacein (PDV) pathway an ideal platform. In previous study, Avalos and Toettcher laboratory has found that light-switchable clustering can be extended to the assembly of functional organelles and control the metabolic flux through the DV pathway in yeast32. Inspired by this, we reconstructed the DV and PDV-producing strains by the heterologous expression of the Vio operon from Chromobacterium violaceum fused with the RIDD peptide to examine the potential in cellular condensates for controlling metabolic flux. As shown in Fig. 3c, the cell color presented significant change as the condensate forms. Without clustering, the color of the strain presented as light brown (VioABE) and pink (VioABEC), respectively. However, in the presence of the SIDP1 condensates, clustering VioABE or VioABEC, the cell color changed from light brown to green and from pink to purple, respectively. Meanwhile, we performed the quantitative measure, and the production of DV and PDV were increased by 59.1% and 135.1%, respectively, when compared with the corresponding control group (Supplementary Fig. 16). These results clearly showed that the synthetic cellular condensate can pave the way to spatially control metabolic flux in B. subtilis.
Synthetic condensates enable orthogonal translation by spatial separation
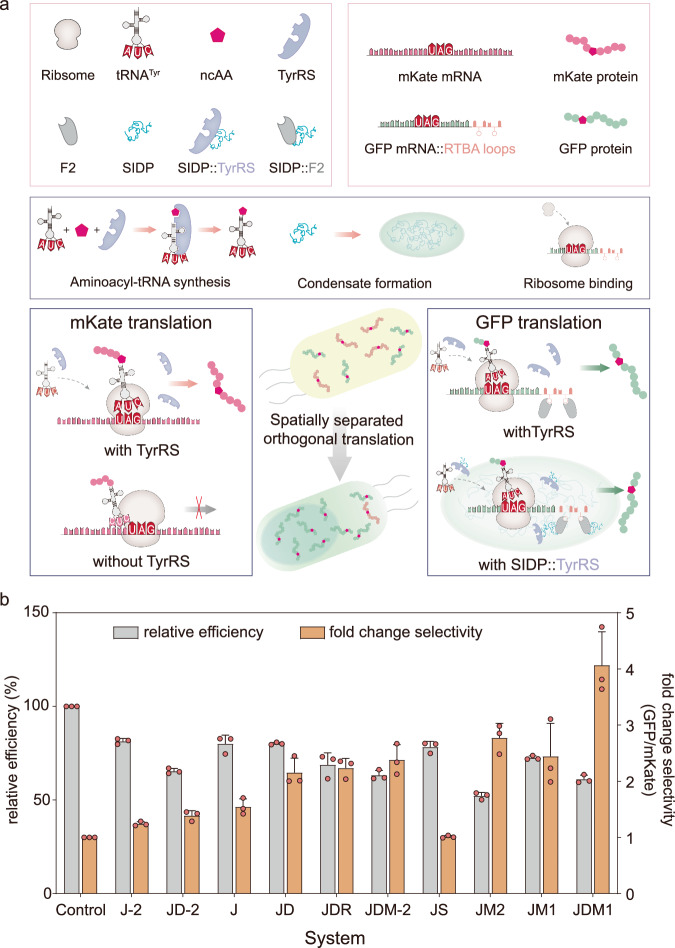
The ability to engineer the translation of ncAAs site-specifically into proteins in living cells can greatly enhance catalytic activities or enable the introduction of additional functionalities into proteins33. However, preventing unwanted modification of nontargeted host proteins and minimizing the impact on the host cell machinery remain challenging. In eukaryotes, the cellular condensates were created to enable codon reassignment of selected mRNAs34,35. Inspired by this, we hypothesized that it is possible to create a spatially orthogonal translation system in B. subtilis by spatially enriching specific components of the genetic code expansion (GCE) machinery within synthetic condensates. To this end, three components were engineered and combined for the construction of the desired condensate, including an mRNA-targeting system (RTBA-thrombin protein), a tRNA/RS suppressor pair that incorporates O-methyl-L-tyrosine (OMeY) into amber stop codons, and the synthetic condensates (Fig. 4a). Specifically, the mRNA-targeting system was designed by fusing six RTBA stem loops to the 3′ untranslated region of selected mRNA, and creating an mRNA::RTBA fusion coding for the target protein. The previous research has demonstrated that the RTBA loops can bind to the thrombin (F2) specifically, and result in a stable and specific mRNA::RTBA–F2 complex in B. subtilis36. In addition, an aaRS/tRNA mutant pair derived from Methanococcus jannaschii that can incorporate OMeY into amber stop codons was chosen and tested due to its simplicity37 (Supplementary Fig. 17). To evaluate the effect of the synthetic condensate in facilitating the orthogonal translation of an RTBA-tagged mRNA, we designed a dual-reporter construct, in which GFP and mKate were driven by the P43 and Pveg promoters, respectively, ensuring that the mRNA ratio between them is constant across all experiments. Stop codons were introduced at permissive sites into the third position of both GFP and mKate. We then evaluated the performance of each designed system in relation to their selectivity and relative efficiency.
Fig. 4. Functionalization of cellular condensates for spatially separated orthogonal translation.
a Spatial separation of the necessary components to enable orthogonal translation to control language of protein translation. Without the synthetic condensate in vivo, the selected gene GFP and other gene such as mKate containing TAG both can be transcribed and translated into a complete fluorescent protein normally. When the genetic code expansion (GCE) machinery was applied within a synthetic condensate, the selected gene GFP containing TAG that targeted to the condensate can still express normally. In contrast, the corresponding stop codon in gene mKate that are not targeted to the condensate should not get translated. TyrRS, aminoacyl-tRNA synthetase; ncAA, noncanonical amino acid; SIDP, synthetic intrinsically disordered protein. b For all experiments, the indicated constructs and dual reporter GFP3TAG::6RTBA and mKate3TAG were co-expressed. The orange bars (normalized to cytoplasmic TyrRS) represent the fold change in the ratios of the mean fluorescence intensities of GFP versus mKate for all the systems tested. The gray bars represent the relative efficiency as defined by the mean fluorescence intensity of GFP for each condition divided by cytoplasmic TyrRS control. The J-2, JD-2, and JDM-2 systems were constructed by using one RTBA based on J, JD, and JDM1 systems, respectively. Data are presented as mean ± s.d. of three biologically independent replicates. Source data provided as a Source Data file.
As shown in Fig. 4b, the simplest assembly system ( J), constructed by fusing F2 to the C-terminus of TyrRS, only showed a minor selectivity gain of 1.54-fold, which is concomitant with a 20.0% decrease in efficiency. Subsequently, we attempted to assemble the TyrRS-F2 in synthetic condensates for gathering the target mRNA translation process spatially, resulting in two advanced system based on J system, named JM1 and JM2, respectively. Meanwhile, to assess the condensate formation in cells, the RIAD and RIDD were fused with JM1 system (constructed by fusing SIDP1 to TyrRS-F2) and GFP, respectively. In contrast to J system fused with RIAD, the JM1 system formed GFP condensates, which indicated the occurrence of synthetic condensates formation in the presence of SIDP1 (Supplementary Fig. 19). As expected, the JM1 system showed an approximate 2.44-fold selectivity gain accompanied by an almost 27.5% decrease in efficiency. The JM2 system (constructed by fusing SIDP3 to TyrRS-F2) showed a selectivity gain of 2.76-fold and a 47.8% decrease in efficiency. Meanwhile, we used fluorescence in situ hybridization (FISH) against tRNATyr, and a single-color GFP3TAG::6RTBA reporter was expressed in B. subtilis. The results showed that tRNATyr can nicely partition into the condensates (Supplementary Fig. 20). Besides, we found that ribosomes distribution throughout the cell but also an enrichment at the synthetic condensates, which suggested that orthogonal translation may happen inside or within proximity of the synthetic condensates (Supplementary Fig. 21).
Meanwhile, we tested another optimized assembly strategy to aggregate the target translation process at the spatial level. By fusing the N-terminal 60 amino acids of the gene divIVA (curvature sensitive membrane binding protein) and divIVAR18C (the mutant of divIVA binding to the chromosome) to the fusion enzyme TyrRS-F2, respectively, two systems named JD and JDR were generated (Supplementary Fig. 22). We observed that both, JD and JDR systems, selectivity increased 2.15-fold and 2.24-fold, respectively. Compared with J system, the efficiency of JD remained unchanged, whereas the efficiency of JDR decreased to 68.6%. Based on the above results, we tested all possible combinations. For all combinations, the best performing system was the fusion of SIDP1 with JD, exhibiting a selectivity fourfold higher and 61.1% efficiency (Fig. 4b and Supplementary Fig. 23). These results demonstrate that the combination of the two assembler strategies (that is, phase separation paired with spatial targeting) yields the highest selectivity, which is consistent with previously reported study34.
To verify that the observed selectivity gain is specific to the RTBA-F2 interaction, we next characterized the system with the F2 component by adjusting the number of RTBA repeats. As shown in Fig. 4b and Supplementary Fig. 24, there was a noticeable decrease in the selectivity gain when the one RTBA was used. We speculated that the reason for this phenomenon is that the target gene fused with one RTBA is more disadvantaged than the one with six RTBA loops in the translational competition of dual-reporter. Besides, no selective orthogonal translation was observed when the F2 was removed in JS system (Fig. 4b). These results suggest that these systems act selectively on the RTBA-tagged mRNA. Additionally, we tested the effect of changing the reporter by replacing the original GFP with CFP, which performed similarly in terms of selectivity gain (Supplementary Fig. 25). These results indicate that it is possible to construct synthetic condensate to control the protein translation in prokaryotes.
Utilizing functional condensates for metabolic control: 2′-fucosyllactose biosynthesis
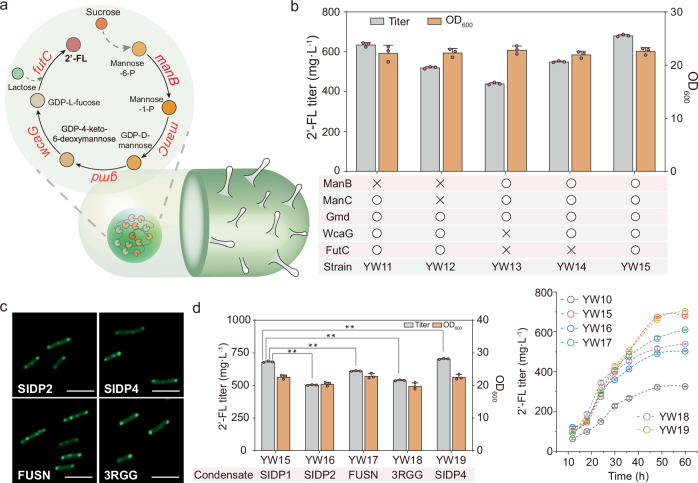
To further verify the universality of functional condensates for controlling metabolic flux, the de novo 2′-FL biosynthesis pathway was used as a model system. 2′-FL is a promising infant formula ingredient and can be synthesized from sucrose17,38 (Fig. 5a). Specifically, the peptide RIDD was fused to the N- or C-terminus of the enzymes of the pathway: phosphomannomutase (encoded by manB), mannose-1-phosphate guanylyltransferase (manC), GDP-mannose 4,6-dehydratase ( gmd), GDP-Lfucose synthase (wcaG) and α-1,2-fucosyltransferase (futC). The strain MT1 constructed in previous works was selected as a starting point, and the genes yesZ and ganA were knocked out, generating the strain MT1-1 as the reference for comparison39.
Fig. 5. Controlling 2′-fucosyllactose biosynthesis by co-localization of targeted cargo enzymes.
a Illustration of the synthetic condensate platform of de novo biosynthesis of 2′-fucosyllactose (2′-FL) in B. subtilis. futC, α1,2-fucosyltransferase; manB, phosphomannomutase; manC, α-D-mannose 1-phosphate guanylyltransferase; gmd, GDP-mannose 6-dehydrogenase; wcaG, GDP-L-fucose synthase. b High-performance liquid chromatography (HPLC) quantification of 2′-FL produced by different recombinant strain with SIDP1 condensate co-localizing diverse pathway enzymes. The “O” signifies that the enzyme is colocalized within the cellular condensate, while “X” represents enzymes that are not recruited. c Fluorescence microscopy images of SIDP2-GFP, SIDP4-GFP, FUSN-GFP, and 3RGG-GFP driven condensates within B. subtilis. d Effects of condensates with virous physical properties on 2′-FL production. Additionally, trends of 2′-FL concentration of the all recombinant strains are shown. Data are presented as mean ± s.d. of three biologically independent replicates. Significance (P value) was evaluated by two-sided t-test. *: p < 0.05, **: p < 0.01. Scale bars = 5 μm. Source data provided as a Source Data file.
First, we tested the impact of the fusion enzymes on the 2′-FL biosynthesis, which moderately affected 2′-FL production (Supplementary Fig. 26). The 2′-FL titer of strain YW6 (with RIDDGmd, RIDDWcaG, and FutCRIDD) and YW9 (with ManBRIDD, ManCRIDD, RIDDGmd, and RIDDWcaG) was decreased by 13.4% and 11.5%, respectively, when compared with MT1-1 (361.1 mg/L). The titer of strain YW10 (with five fusion enzymes) was 328.0 mg/L, which decreased by 9.2% when compared with MT1-1. Then, we induced the sequential co-localization of partial or complete pathway enzymes to regulate 2′-FL biosynthesis based on the SIDP1 condensate. As shown in Fig. 5b, the OD600 of strain YW11 (YW5 with cellular condensates) was 22.2, and the product formation improved 88.3% to 636.1 mg/L when compared with the strain YW5 (337.9 mg/L). Besides, there was a noticeable increase in the production of strain YW12 (YW6 with cellular condensates) and YW14 (YW9 with cellular condensates), with improvements of 66.4% and 72.2%, respectively, when compared with parental strain without forming condensate. The OD600 of strain YW15 (YW10 with cellular condensates) was 22.6, and this strain could produce the highest 2′-FL titer (681.6 mg/L), which is 2.1-fold that of the parental strain YW10.
Then, we sought to use several condensates with different fluidity to regulate 2′-FL biosynthesis. In addition to the above-designed condensates SIDP2 and SIDP4, other widely utilized natural IDPs were also selected and reconstructed for comparison. For example, FUSN is the N-terminal low-complexity domain of the RNA-binding protein FUS, which serves as the crucial domain that contributes to the formation of condensate40. Besides, RGG domain is necessary to promote RNA helicases LAF-1 phase separation and can serve as a protein scaffold to control the 2′-FL biosynthesis in E. coli17,41. Therefore, a corresponding triplet RGG protein (3RGG) and FUSN were also constructed to form the cellular condensates. These above-mentioned proteins were fused with GFP for overexpression within B. subtilis. As expected, the SIDP2, SIDP4, FUSN, and 3RGG-driven condensates were observable and could interact with the target protein in B. subtilis (Fig. 5c and Supplementary Figs. 27–29). The recombinant strains with different synthetic condensates were constructed. As shown in Fig. 5d, the strain YW16 expressing RIAD-SIDP2 exhibited the lowest improvement, increasing the titer of 2′-FL by 53.5% compared with YW10. The OD600 of the strain YW18 expressing RIAD-3RGG decreased by 9.1% compared with YW10, while the 2′-FL titer increased by 64.5% to 539.4 mg/L. The titer of the strain YW17, using FUSN condensate, reached 611.5 mg/L, which was 1.9-fold that of YW10. In addition, the strain YW19 (derived from YW10 and modified with SIDP4 condensates) produced 703.6 mg/L of 2′-FL, which was similar to that obtained by strain YW15. Subsequently, a test experiment was conducted to screen the effect of different inducing time points: 6, 8, 10, 12, and 14 h, to identify the optimal time for 2′-FL biosynthesis. The results show that the best titer was obtained when inducing at 8 h (Supplementary Fig. 30). Under this condition, the 2′-FL titer of the strain YW19 reached 806.5 mg/L, 14.6% increase related to previous condition, which is 2.2-fold that of the original strain MT1-1.
In summary, these results demonstrate the capacity to regulate metabolic flux through spatial control of metabolic pathways at the subcellular scale based on the use of synthetic condensate.
Enhancing the translation specificity of tailored enzyme for increased biosynthesis of N-acetylmannosamine
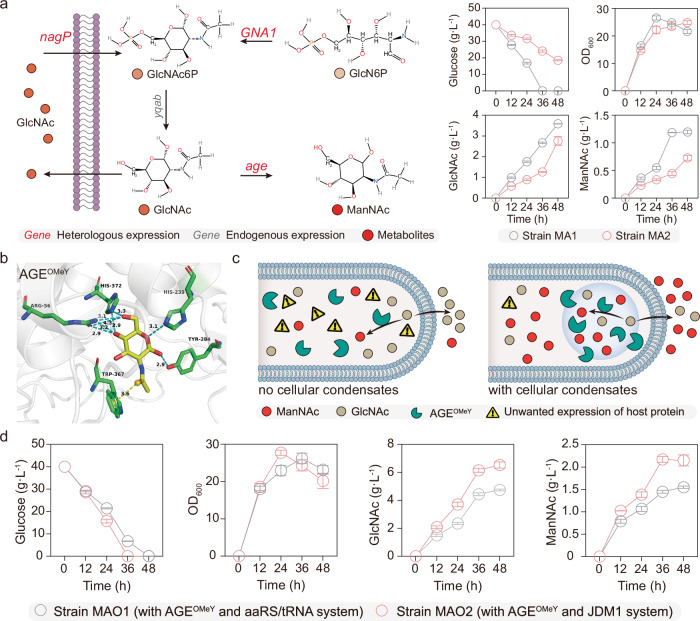
The significant improvement of 2′-FL production showed the potential application of synthetic condensates for controlled biosynthesis in B. subtilis. We then decided to study the biosynthesis of ManNAc in B. subtilis as a proof of concept, which is an important precursor of high value-added pharmaceuticals42. First, the glucosamine-6-phosphate (GlcN6P) N-acetyltransferase (encoded by the gene GNA1) and N-acylglucosamine 2-epimerase (encoded by age) were integrated into the genome of the GlcN6P-producing chassis S5 under the control of the constitutive promoter P43, generating the strain MA1, which could produce 1.2 g/L ManNAc (Fig. 6a). Meanwhile, we hypothesized that it is possible to improve the efficiency the AGE enzyme by combing computational redesign and GCE strategies. To this end, an ncAA OMeY was incorporated into Rosetta for computational design, and the final combined mutant AGEOMeY (M15L, N41D, E57Q, G169N, Q234L, T236N, T269W, N291K, S312A, R323E, Q343P, V373OMeY) was obtained.
Fig. 6. Improving translation specificity of AGEOMeY involving N-acetylmannosamine biosynthesis.
a Shake flask fermentation of ManNAc by strains MA1 and MA2. The gene GNA1 and age were integrated into the genome of the engineered strain S5, generating strain MA1. Besides, the original aaRS/tRNAOMeY system was expressed in MA1, yielding the strain MA2. GlcN6P glucosamine-6-phosphate, GlcNAc N-acetylglucosamine, GlcNAc6P GlcNAc-6-phosphate, ManNAc N-acetylmannosamine, GNA1 GlcN6P N-acetyltransferase, AGE N-acylglucosamine 2-epimerase, yqab, hydrolase-like phosphatase, nagP phosphotransferase system GlcNAc-specific transporter subunit EIICB, OMeY O-methyl-L-tyrosine. b The Rosetta-predicted combined mutant AGEOMeY with GlcNAc bound is shown. The blue dotted line means hydrogen-bonding, and the yellow dotted line means pi interactions. c Illustration of synthetic functional condensates to increase translation specificity of enzyme AGEOMeY for improving the ManNAc biosynthesis. d Trends of cell growth, glucose, GlcNAc, and ManNAc concentration of the strains MAO1 and MAO2. Data are presented as mean ± s.d. of three biologically independent replicates. Source data provided as a Source Data file.
To evaluate the catalytic profiles of AGE wild type (WT) and its mutant AGEOMeY, kinetics were assessed (Table 1 and Supplementary Fig. 31). The results showed that the kcat value of AGEOMeY was 124.7 s−1, 54.2-fold that of WT (2.3 s−1). In addition, the analysis of kinetic parameters indicated that catalytic activity of AGEOMeY increased by 86.0% to 86,078.9 s–1·M–1 compared with the WT enzyme (46,266.9 s–1 M–1). This suggests that the amino acids R56 and H372 of AGEOMeY present more functional interactions with the ligand than that of WT (Fig. 6b and Supplementary Fig. 32).
Table 1.
In vitro kinetic characterization of AGE and mutant activities
| Enzymes | Km (μM) | kcat (s−1) | Vmax (μM min−1) | kcat/Km (s–1 M–1) |
|---|---|---|---|---|
| AGE | 49.2 | 2.3 | 0.4 | 46,266.9 |
| AGEOMeY | 1448.9 | 124.7 | 24.4 | 86,078.9 |
Encouraged by the successful design of AGEOMeY, we subsequently heterologously overexpress it to enhance the ManNAc biosynthesis. The protein expression levels in cells expressing AGEOMeY were both maintained using the ribosome binding site optimization strategy (Supplementary Fig. 33). The titer of strain MAO1 (overexpression of AGEOMeY with aaRS/tRNAOMeY system) increased by 25.0% to 1.5 g/L when compared with MA1. The observed titer improvement was below our expectations. According to our statistical analysis, there are ~596 genes in B. subtilis that utilize amber codon (TAG) as the stop codon (Supplementary data 2). We used metabolic model for simulations and speculated that the aaRS/tRNAOMeY system could interfere with normal cellular metabolism (Supplementary data 3)43. Besides, compared with MA1, the ManNAc titer of strain MA2 (strain MA1 containing aaRS/tRNAOMeY system) decreased by 41.7% to 0.7 g/L, and it was accompanied by a sharp decrease in glucose consumption rate (46.3%), which further validated the above-mentioned hypotheses. As an alternative, the synthetic condensate assembled both aaRS/tRNAOMeY and mRNA-targeting systems (that is, JDM1 system) was chosen for minimization of background incorporation of the ncAAs, and six RTBA loops were also fused to the 3’untranslated region of AGEOMeY (Fig. 6c). As shown in Fig. 6d, the strain MAO2 could produce 2.1 g/L, which was 75.0% higher than that obtained by the strain MA1. These results suggest that we could conclude that synthetic condensates are a valuable tool to regulate both protein translation and metabolite production in prokaryotes.
Discussion
Our work demonstrates that SIDPs comprising repeats of a minimal peptide motif de novo designed via the “stacking blocks” strategy exhibit phase separation in vitro and in living cells. As an alternative to using natural sequences, the de novo designed SIDP system offers several advantages as a synthetic platform to form biomolecular condensates. For instance, when appended as an N-terminal tag to a GFP, the fusion protein SIDP1-GFP produces insoluble fluorescent condensates alone without requiring other interacting species such as nucleic acid. Besides, the synthetic condensates are highly dynamic and behave as liquid-like droplets through a series of in vitro and in-cell experiments. Interestingly, SIDP1 exhibits reversible LCST phase separation, a useful feature for potential thermo-switchable applications that allows induction and dissolution of condensates in microbial cell factories, a feature not available in some native IDPs41. We speculate that hydrophilic to hydrophobic amino acids ratio may influence the LCST phase separation, and it will be interesting to explore the effects of hydrophilic: hydrophobic amino acid ratio on LCST phase boundaries by modulating hydrophilic versus hydrophobic residues contents. In addition, the simplicity of minimal peptide provides the potential to create unique condensates with varying biophysical properties for different applications. By using two simple design parameters (amino acid composition and arrangement) that are genetically encoded at the sequence level, a series of LLPS-promoting artificial proteins with different properties is constructed and characterized. These findings also support the growing evidence of interactions that promote LLPS and present a potential approach to engineer and manipulate native IDPs with customizable properties30. Future work may also explore the relation between the linear arrangement of amino acids and condensate properties at the individual amino acid level by combining machine-learning and synthetic biology technologies to expand the understanding of biomolecular condensate.
The idea of engineering synthetic functional condensates to control intracellular bio-reactions in living cells has gained renewed interest in recent years44,45. As expected, we demonstrate that de novo designed proteins can be used to engineer functional intracellular condensates in B. subtilis. Specifically, the cellular synthetic condensates provide a distinct subcellular compartment that is permeable to small molecules, and can be engineered to selectively recruit proteins of interest. We use these features to construct a system to regulate the metabolic flux of 2′-FL biosynthesis. The significant increase of 2′-FL titer by the compartmentalization of target pathway enzymes confirms the potential of synthetic condensate to control cellular biosynthesis. Furthermore, using different condensates to alter 2′-FL biosynthesis emphasizes how condensate physical properties affect reactions46,47. In the case of using the same pair of peptides (RIAD-RIDD), the higher fluidity of synthetic condensates leads to a greater enhancement in 2′-FL production. We speculate that synthetic condensates with low fluidity may more readily transit to solid aggregates irreversibly, resulting in a decrease in the efficiency of compartmentalized metabolic regulation.
In addition to the metabolism control system, we show that the cellular condensates can function as an orthogonal membraneless organelle to tune translation process in prokaryotes, which goes beyond the previously available studies. Compared with previous studies, such as replacing all selected codons in the host genome, the system developed here offers plug-and-play convenience48. Furthermore, use of orthogonal ribosome to achieve orthogonal translation has also been limited due to low activity and poor orthogonality to wild-type ribosomes, and the rational design of the structure and function for high-performance orthogonal ribosomes remains difficult49,50. Meanwhile, the modular engineering of this system allows for the replacement of the aaRS/tRNA pair or mRNA-targeting system with any other schemes without requiring time-consuming engineering51. Here, the fold change selectivity of fluorescence intensity has demonstrated that the ncAA-modified protein can be expressed selectively in synthetic condensates with minimal disruption to the endogenous metabolism of the cell. This finding is also consistent with previous performances in eukaryotic systems34,35. However, the selectivity observed in our experiments is lower than that obtained in eukaryotic systems, and we speculate that this discrepancy may have occurred due to differences in translational mechanisms between prokaryotes and eukaryotes52. Besides, the cell size could be another crucial factor affecting translation selectivity, as larger cell size in eukaryotes allows for greater spatial separation, which is essential for translation selectivity53. Finally, we reveal that the fluorescent protein can be substituted by enzyme of interest, resulting in a 75.0% enhancement in ManNAc titer, which suggests that our approach is generalizable and will enable the control of other proteins with multiple ncAAs incorporation.
Taken together, we offer a programmable approach for controlled biosynthesis in B. subtilis by targeting of cellular machinery to synthetic condensates. Furthermore, we anticipate that our bottom-up designed SIDPs will bring deeper insights into the biomolecular condensation, and provide a valuable tool in other microbial species to reprogram cellular bio-reactions with desired applications.
Methods
Plasmid construction and genome manipulation
The software SnapGene was carried out to present and design genetic sequences. Primers designed and used for genetic modification in this study are listed in Supplementary Table S1. Oligonucleotides and genes were ordered from GENEWIZ. Gibson Assembly toolkit (NEB, USA) was used to construct all plasmids with specific backbones in E. coli JM109 or BL21 (DE3). The plasmids and sequences of genetic parts used in this study are listed in Supplementary Tables S2 and S3, respectively. In addition, the CRISPR/Cpf1 system was used to perform gene knockout or overexpression in B. subtilis genome.
Strains and cultivation conditions
All recombinant strains constructed and used in this study are provided in Supplementary Table S4. For cell culture, Luria–Bertani (LB) medium (per liter: 10.0 g tryptone, 5.0 g yeast extract, and 5.0 g NaCl) was used unless noted otherwise. For 2′-FL producing strain culture, OSF medium (per liter: 12.5 g K2HPO4·3H2O, 12.0 g yeast extract, 6.0 g tryptone, 2.5 g KH2PO4, 10 mL trace metal solution, 20.0 g sucrose, and 10.0 g lactose) was applied. In addition, sucrose and lactose were replaced with glucose (40.0 g L–1) and glycerol (10.0 g L–1) for ManNAc producing strain culture. Besides, the ncAA-dependent cells were cultured in the LB or OSF medium with 0.25 mM OMeY or 1 mM pAzF. Ampicillin (100 μg mL–1), chloromycetin (30 μg mL–1) and kanamycin (50 μg mL–1) were used for selections in E. coli, and chloromycetin (5 μg mL–1), kanamycin (50 μg mL–1), and zeocin (30 μg mL–1) were used for selections in B. subtilis, unless noted otherwise. All chemicals used in this study were purchased from Sangon Biotech unless otherwise specified.
Deoxyviolacein (DV) or prodeoxyviolacein (PDV)-producing strain was precultured in a shake flask with 10 mL LB medium for 12 h at 37 °C. Then it was inoculated into 50 mL LB with 5% proportion and cultured with shaking at 220 rpm for 24 h. If necessary, 0.5 mM IPTG was added to promote the formation of cellular condensates at 8 h.
To synthesize 2′-FL or ManNAc, single colonies were picked into 10 mL of LB medium in a shake flask and cultured for 10 h at 37 °C. Subsequently, the seed cultures were further inoculated into 30 mL of fermentation medium with a 5% proportion in a 250-mL shake flask and cultured with shaking at 220 rpm (37 °C). Three replicates were performed for each recombinant strain. If necessary, 0.5 mM IPTG was added to promote the formation of cellular condensates at 12 h.
Recombinant protein expression and purification
For artificial disordered protein expression, recombinant cells of E. coli BL21(DE3) harboring relevant plasmids were cultivated at 37 °C in a 500-mL flask containing 50 mL Terrific Broth medium (per liter: 4 mL glycerin, 24 g yeast extract, 12 g peptone, 2.31 g KH2PO4, and 16.42 g K2HPO4·3H2O) at 220 rpm. In addition, the enzyme N-acylglucosamine 2-epimerase (AGE) and its mutant were overexpressed within B. subtilis expressing aaRS/tRNAOMeY system.
When the OD600 reached 0.6–0.8, the recombinant strains were induced with 0.5 mM IPTG and then transferred to 25 °C for 24 h. The cells were harvested at 5000 × g and washed twice with 20 mM Tris (pH 7.4). Then, the resuspended cell pellets were lysed by high-pressure homogenization in the lysis buffer containing 500 mM NaCl, and 20 mM Tris (pH 7.4). After centrifuging at 12,000 × g for 30 min, the supernatant was performed with an ÄKTA pure system (GE Healthcare) with a HisTrap HP column (5 ml, GE Healthcare). The wash buffer contained 500 mM NaCl, 20 mM Tris (pH 7.4), 20 mM imidazole, and the elution buffer contained 500 mM NaCl, 20 mM Tris (pH 7.4), and 500 mM imidazole. Proteins were assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and their concentrations were measured with the BCA protein assay kit (Beyotime, Shanghai, China). Meanwhile, the target proteins with correct molecular weights were cut from the SDS-PAGE. After being treated with trypsin, they were performed on ultrafleXtreme™ MALDI-TOF/TOF (Bruker, Germany) and analyzed by Mascot.
For solubility analysis, recombinant strains harboring relevant plasmids were cultivated at 37 °C in a 500-mL flask containing 50 mL TB medium at 220 rpm for overnight. Pellets were harvested at 5000 × g by centrifugation and washed twice with 20 mM Tris (pH 7.4). Then, the resuspended cell pellets were lysed by high-pressure homogenizer in the lysis buffer containing 500 mM NaCl, and 20 mM Tris (pH 7.4). For separation of cellular soluble and insoluble fraction, suspensions were centrifuged at 14,000 × g for 30 min. The supernatant (soluble fraction) was removed, and the pellets (insoluble fraction) were resuspended in an equal volume of the lysis buffer. For SDS-PAGE, samples were mixed with appropriate volumes of reducing SDS loading dye and heated to 99 °C for 10 min. 20 µL of samples were loaded alongside 5 μL of color pre-stained protein standard (Yeasen, Shanghai) onto 10% or 12% NuPAGE Bis-Tris gels (Thermofisher) and ran until the loading dye reached the bottom of the gel.
Phase separation assay
Phase separation was induced by mixing artificial disordered protein (stored in 500 mM NaCl and 20 mM Tris, pH 7.4) with no-salt buffer (0 mM NaCl and 20 mM Tris, pH 7.4) to obtain a solution containing describe concentration of NaCl (normally at 150 mM) and 20 mM Tris, pH 7.4, unless otherwise stated. Protein concentrations were adjusted as necessary by diluting further with same buffer conditions. Besides, the purified artificial disordered protein was reconstituted into a buffer supplemented with different percentage of PEG 2000 for providing a crowding environment to mimic intracellular environments. The resulting turbid mixture was then loaded onto glass slides and visualized by light and fluorescence microscopy. The same amount of purified GFP (green fluorescent protein) protein was used as the negative control. In addition, the fusion of individual droplet was monitored under a Nikon C-HGF microscope. The turbidity of SIDP1 protein was measured in triplicates at λ = 600 nm using a spectrophotometer (Cary 100 Bio, Varian) at 25 °C, unless stated otherwise. Turbidity was recorded after shaking the sample for 5 s.
Fluorescence recovery after photobleaching analysis
The fluorescence recovery after photobleaching (FRAP) experiments were conducted under the Leica TCS SP8 microscope (Leica Microsystems). The protein droplets were loaded onto glass slides under the microscope and activated at 488 nm with 2% laser power to show appropriate green intensity. Then, the droplet center was irradiated with 100% laser power for ~30 s. After photobleaching, time-lapse images were captured for ~210 s at ~25 °C. The fluorescence intensity within the region of interest (ROI) was monitored and normalized by LAX S.
In vivo FRAP was performed using a Nikon-AX exciting at 488 nm at ~25°C. Bleaching was performed using 8 ms laser burst at 50% laser power, followed by imaging every 30 s for 3 min to record fluorescence recovery. Besides, data analysis was carried out in Nikon-AX. For each bleaching measurement, recovery was normalized relative to the mean fluorescence intensity before bleaching (normalized to 1) and the minimum fluorescence intensity measured immediately after bleaching (normalized to 0).
Protein redesign and characterization
The software AlphaFold2 was utilized to predict the three-dimensional structure of the protein AGE in this study, and the computational tool AutoDock Vina was subsequently used for performing molecular docking with N-acetylglucosamine (GlcNAc) to select the lowest-energy complex for further investigation. Additionally, the params file and rotamer library of ncAA OMeY were created and incorporated into Rosetta for design, and this method was presented at Rosetta commons ((Design with non-canonical amino acids (NCAA) rosettacommons.org)). Meanwhile, the deep learning approach (DLKcat) was employed to obtain a set of 100 homologous sequences with comparable higher value of kcat54. Then, AGE and its homologous sequences were applied for multiple sequence alignment by using software COBALT, and 12 putative residues were identified. Subsequently, the above-mentioned 12 residues mutated to other 20 amino acid residues (including OMeY) virtually, and the penalty constraint scores and total energy were calculated. As a result, a total of 12 mutants (M15L, N41D, E57Q, G169N, Q234L, T236N, T269W, N291K, S312A, R323E, Q343P, V373OMeY) with comparable lower penalty constraint scores and total energy were selected, and we then combined all the mutants to generate the final combined mutant AGEOMeY for further studies.
To measure the kinetic parameters of AGE and its mutant AGEOMeY, a final volume of 1.0 mL containing 100 mM Tris-HCl, 500 mM NaCl, 10 mM MgCl2, 5 mM ATP, 0.15 g L–1 purified protein, and different concentrations of GlcNAc from 2.0 to 200 g L–1, and the pH was adjusted to 7.4. The reactions were carried out at 37 °C for 5 min and stopped by boiling for 10 min. The specific activity was determined by measuring the concentration of the ManNAc formed after 5 min reaction time. The parameter kcat was then evaluated according to the formula: kcat = Vmax/E. E was the enzyme concentration.
Analytical methods
The cell density was measured by ultraviolet spectrophotometer at 600 nm. In addition, to analyze glucose, GlcNAc, ManNAc and 2′-FL, the samples were centrifuged at 12,000 × g for 15 min, and the supernatants were filtered through 0.22 μm membranes. The samples were detected via high-performance liquid chromatography (HPLC) with a Rezex ROA Organic Acid H+ column (Phenomenex, Torrance, CA, USA), a refractive index detector (30 °C) and a diode array detector (210 nm) (Agilent 1260). The mobile phase, flow rate and column temperature were aqueous H2SO4 (5 mM), 0.6 mL min–1 and 55 °C, respectively. The GlcNAc, 2′-FL and ManNAc standards were bought from Sigma-Aldrich.
For the quantitative measure of DV and PDV, 1 mL cells were centrifuged at 5000 × g for 10 min. The cell precipitate was resuspended with 1 mL ethanol and vortexed with quartz sand. The DV production was measured by HPLC equipped with a Symmetry C18 column (4.6 × 250 mm, 5 μm, Waters) and a diode array detector (570 nm) (Agilent 1260). The column was eluted with 70% (v/v) methanol at a flow rate of 1 mL/min at 30 °C. Meanwhile, the PDV production was detected by microplate multimode reader. Specifically, pellets of centrifuged cells were extracted by 200 mL DMSO, vortexed, and then centrifuged again. 100 mL supernatants were transferred into 96-well black plates, and measured with microplate multimode reader at absorbances 588 nm to represent the amounts of PDV production23.
Assay of fluorescence intensity
To examine the performance of cellular condensates for orthogonal translation, the recombinant B. subtilis strains were precultured in LB medium for 8 h and further inoculated into 200 μL of LB medium containing 0.5 mM IPTG with 1% proportion in 96-well plates (Corning, 3603). The 96-well plates were subsequently cultured at 37 °C for 10 h with shaking at 750 rpm. GFP fluorescence (excitation, 488 nm; emission, 523 nm; gain value, 70), mKate fluorescence (excitation, 589 nm; emission, 634 nm; gain value, 100) and OD600 were measured using a microplate multimode reader (Tecan, infinite 200Pro) directly. The relative fluorescence intensity is defined as the ratio of fluorescence intensity without background fluorescence to the OD without background.
The selectivity was defined as the ratio of the mean fluorescence intensities of GFP divided by the mean fluorescence intensities of mKate for all tested systems. Final values were expressed as fold change selectivity relative to that of original TyrRS/tRNA system. Besides, the relative efficiency was defined as the mean fluorescence intensities of GFP of each system divided by the mean fluorescence intensity of the TyrRS/tRNA system (defined as 100% here).
Fluorescence imaging and analysis
For urea perturbation experiments, 1 mL cell culture were harvested at 5000 × g and washed twice with 50 mM PBS buffer (pH 7.4). Then, the cell pellets were resuspended in 50 mM PBS, 50 mM PBS with 0.5 M urea, and 50 mM PBS with 1 M urea, respectively. After incubating at ~25 °C for 20 min, the cells were further washed twice with 50 mM PBS, and were prepared for microscopy imaging.
To test the subcellular localization of proteins of interest, the corresponding plasmid was transformed into B. subtilis and induced by 0.5 mM IPTG or 30 g L–1 xylose. Fluorescence imaging was performed under a Nikon C-HGF microscope fitted with a 100× oil immersion objective. Cells with GFP were observed using a FITC fluorescent filter, and cells with mKate (red fluorescent protein) were observed using a TRITC fluorescent filter. The fluorescence images were then imported into ImageJ for analysis.
The Pearson correlation coefficient (PCC) was determined using the Colocalization Finder via ImageJ. Specifically, the captured images were first transformed into 8-bit type by splitting channels in ImageJ 1.53t to get the images of the corresponding fluorescence channel. A single cell was selected as a ROI. PCCs of the ROIs were then obtained by using the Colocalization Finder plug-in in ImageJ 1.53t to analyze the degree of fluorescence co-localization of the ROIs. For calculating the mean fluorescence intensity of confocal images, the images were transformed into 8-bit type by splitting channels in ImageJ 1.53t and all cells in the visual field are selected by adjusting the threshold. Then, the area and mean gray value of every cell can be calculated by the Analyze Particles plug-in in ImageJ 1.53t. The average fluorescence intensity of the certain channel was the total fluorescence intensity of all cells divided by the total area of the cells.
To calculate the percentage of proteins recruited to the condensates in each cell, images were first automatically thresholded (Otsu’s method) in ImageJ. Then, the total red fluorescence within the condensate fraction (area × mean background-subtracted fluorescence) was divided by the total fluorescence (area × mean background-subtracted fluorescence) of the whole cell to get the ratio of proportion of recruited proteins.
In addition, the super-resolution microscopy with structured illumination was conducted using the Nikon-AX system. The imaging was performed with a CFI SR HP Apochromat TIRF 100× C oil objective. GFP was excited at 488 nm at 40% laser power. Data were acquired at 1 s frame rates, and 15 images were collected. These images were reconstructed in Nis-Elements AR (Version 5.41.01) with the following parameters: illumination modulation contrast 0.5, high resolution noise suppression 1, and out of focus blur suppression 0.15.
Fluorescence in situ hybridization (FISH)
For FISH experiments, cells with orthogonal systems were grown in 2 mL LB medium with 0.25 mM OMeY for 12 h at 37 °C, 220 rpm. 37% formaldehyde was added to the cell culture to a final concentration of 4%, and incubated for 15 min at 37 °C with continued shaking at a low speed. The cells were then harvested by centrifugation for 1 min at 3000 × g and resuspended in 1 mL fixation buffer (4% formaldehyde, 0.1 M potassium phosphate buffer, pH 6.4) at room temperature for 4 h on a rotating platform. The pellets were washed with 0.1 M potassium phosphate buffer (pH 6.4) for three times and further incubated in 1 mL washing buffer (0.1 M potassium phosphate, 1.2 M sorbitol, pH 6.4) with 500 µg lysozyme to convert the cells to spheroplasts for 1 h at 37 °C with continued shaking at a low speed. The spheroplasts were washed twice with 1 mL washing buffer, collected by centrifugation at 3000 × g for 3 min, and resuspend in 50 µL wash buffer. 20 µL of the cell suspension was applied to the glass slide (prepared with 0.01% poly-L-lysine solution) and allowed the cells to adhere at room temperature for 20 min. The excess liquid was aspirated to remove non-adhering cells. The glass slide was washed with 2 × SSC (0.3 M sodium chloride, 0.03 M sodium citrate, pH 7.4). 12 µL hybridization buffer (4 × SSC, 50% formamide, 10% dextran sulfate, 500 mg/mL salmon sperm DNA, 1×Denhardt’s solution) were applied to covered the samples and incubated at 37 °C for 1 h in a humid chamber. The hybridization probe (5′-(Cy5) CCAGCGCCATGCGGATTTAGAGTCCGCCGTTCTGCCC-3′) was added to the hybridization buffer to a final concentration of 0.25 µM. After covering with a cover slip, the glass slide was incubated at 37 °C in the humid chamber overnight. Finally, the glass slide was washed successively on a shaking platform with 50 mL of 2 × SSC, 1 × SSC, and 0.5 × SSC (each for 5 min at room temperature) and was prepared for confocal imaging. The samples were excited at 640 nm, with an emission wavelength range of 662.5–737.5 nm.
Western blot (WB) assay
The purified proteins were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and then electro-transferred to PVDF membranes. The membranes were subsequently incubated with the mouse anti-His antibody and assayed for anti-mouse IgG secondary antibody conjugated to horseradish peroxidase. The images were analyzed by ImageJ.
Statistics and reproducibility
All experiments were independently repeated at least three times unless otherwise specified in this study and the data were expressed as mean ± SD. Statistical data analysis was performed with two-sided t-test by using GraphPad Prism 9 or Excel (Microsoft 365) and statistical significance is indicated as *P < 0.05 and **P < 0.01. No data were excluded from the analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Prof. Dr. Xinliang Feng (Technische Universität Dresden) for critically reading the manuscript and helpful discussion. This work was financially supported by the Key Research and Development Program of China [2022YFC2104903 (L.L.)], and the National Natural Science Foundation of China [31930085 (G.D.)] and [32021005 (L.L.)].
Author contributions
W.Y. and L.L. conceived and designed the project. W.Y. performed most of the experiments. K.J. and D.W. conducted the visual measurements. N.W. and Y.Li contributed to the protein redesign. W.Y., K.J., N.W., J.L., G.D. and J.C. analyzed the results. W.Y., Y.Liu, X.L., R.L.A, and L.L. co-wrote the paper with contributions from all co-authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data generated in this study are provided in the Supplementary Information/Source Data file. Unprocessed microscopy images are available under restricted access due to the difficulty of hosting this data format, access can be obtained by contacting the corresponding author, who will respond within two business week local time. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52411-5.
References
- 1.Huang, J. et al. Complete integration of carbene-transfer chemistry into biosynthesis. Nature617, 403–408 (2023). 10.1038/s41586-023-06027-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumschabl, M. et al. Conversion of CO2 into organic acids by engineered autotrophic yeast. Pric. Natl Acad. Sci. USA119, e2211827119 (2022). 10.1073/pnas.2211827119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang, Y. Q. et al. Manipulation of sterol homeostasis for the production of 24-epi-ergosterol in industrial yeast. Nat. Commun.14, 437 (2023). 10.1038/s41467-023-36007-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scown, C. D. & Keasling, J. D. Sustainable manufacturing with synthetic biology. Nat. Biotechnol.40, 304–306 (2022). 10.1038/s41587-022-01248-8 [DOI] [PubMed] [Google Scholar]
- 5.Jin, K. et al. Compartmentalization and transporter engineering strategies for terpenoid synthesis. Micro. Cell Fact.21, 92 (2022). 10.1186/s12934-022-01819-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dusséaux, S., Wajn, W. T., Liu, Y. X., Ignea, C. & Kampranis, S. C. Transforming yeast peroxisomes into microfactories for the efficient production of high-value isoprenoids. Proc. Natl Acad. Sci. USA117, 31789–31799 (2020). 10.1073/pnas.2013968117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omidvar, M., Zdarta, J., Sigurdardóttir, S. B. & Pinelo, M. Mimicking natural strategies to create multi-environment enzymatic reactors: From natural cell compartments to artificial polyelectrolyte reactors. Biotechnol. Adv.54, 107798 (2022). 10.1016/j.biotechadv.2021.107798 [DOI] [PubMed] [Google Scholar]
- 8.Moon, S. Y., Son, S. H., Oh, S. S. & Lee, J. Y. Harnessing cellular organelles to bring new functionalities into yeast. Biotechnol. Bioproc. E28, 936–948 (2023). 10.1007/s12257-022-0195-5 [DOI] [Google Scholar]
- 9.Ma, Y. S., Li, J. B., Huang, S. W. & Stephanopoulos, G. Targeting pathway expression to subcellular organelles improves astaxanthin synthesis in Yarrowia lipolytica. Metab. Eng.68, 152–161 (2021). 10.1016/j.ymben.2021.10.004 [DOI] [PubMed] [Google Scholar]
- 10.Grewal, P. S., Samson, J. A., Baker, J. J., Choi, B. & Dueber, J. E. Peroxisome compartmentalization of a toxic enzyme improves alkaloid production. Nat. Chem. Biol.17, 96–103 (2021). 10.1038/s41589-020-00668-4 [DOI] [PubMed] [Google Scholar]
- 11.Jiang, S., da Silva, L. C., Ivanov, T., Mottola, M. & Landfester, K. Synthetic silica nano-organelles for regulation of cascade reactions in multi-compartmentalized systems. Angew. Chem. Int. Ed.61, e202113784 (2022). 10.1002/anie.202113784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang, W. et al. Organizing enzymes on self-assembled protein cages for cascade reactions. Angew. Chem. Int. Ed.61, e202214001 (2022). 10.1002/anie.202214001 [DOI] [PubMed] [Google Scholar]
- 13.Bobik, T. A. & Stewart, A. M. Selective molecular transport across the protein shells of bacterial microcompartments. Curr. Opin. Microbiol.62, 76–83 (2021). 10.1016/j.mib.2021.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerfeld, C. A., Aussignargues, C., Zarzycki, J., Cai, F. & Sutter, M. Bacterial microcompartments. Nat. Rev. Microbiol.16, 277–290 (2018). 10.1038/nrmicro.2018.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian, Z. G., Huang, S. C. & Xia, X. X. Synthetic protein condensates for cellular and metabolic engineering. Nat. Chem. Biol.18, 1330–1340 (2022). 10.1038/s41589-022-01203-3 [DOI] [PubMed] [Google Scholar]
- 16.Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol.18, 285–298 (2017). 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan, L., Zhu, Y. Y., Zhang, W. L. & Mu, W. M. Phase-separated synthetic organelles based on intrinsically disordered protein domain for metabolic pathway assembly in Escherichia coli. ACS Nano17, 10806–10816 (2023). 10.1021/acsnano.3c02333 [DOI] [PubMed] [Google Scholar]
- 18.Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell176, 419–434 (2019). 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, H. et al. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci. China Life Sci.63, 953–985 (2020). 10.1007/s11427-020-1702-x [DOI] [PubMed] [Google Scholar]
- 20.Dai, Y., You, L. & Chilkoti, A. Engineering synthetic biomolecular condensates. Nat. Rev. Bioeng.1, 466–480 (2023). 10.1038/s44222-023-00052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Y. et al. Phase-separated multienzyme compartmentalization for terpene biosynthesis in a prokaryote. Angew. Chem. Int. Ed.61, e202203909 (2022). 10.1002/anie.202203909 [DOI] [PubMed] [Google Scholar]
- 22.Wei, S. P. et al. Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol.16, 1143–1148 (2020). 10.1038/s41589-020-0579-9 [DOI] [PubMed] [Google Scholar]
- 23.Guo, H. T. et al. Spatial engineering of E. coli with addressable phase-separated RNAs. Cell185, 3823–3837 (2022). 10.1016/j.cell.2022.09.016 [DOI] [PubMed] [Google Scholar]
- 24.Dai, Y. F. et al. Programmable synthetic biomolecular condensates for cellular control. Nat. Chem. Biol.19, 518–528 (2023). 10.1038/s41589-022-01252-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilditch, A. T. et al. Assembling membraneless organelles from de novo designed proteins. Nat. Chem.16, 89–97 (2024). 10.1038/s41557-023-01321-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose, T., Ninomiya, K., Nakagawa, S. & Yamazaki, T. A guide to membraneless organelles and their various roles in gene regulation. Nat. Rev. Mol. Cell Biol.24, 288–304 (2023). 10.1038/s41580-022-00558-8 [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y. F., Liu, L., Li, J. H., Du, G. C. & Chen, J. Synthetic biology toolbox and chassis development in Bacillus subtilis. Trends Biotechnol.37, 548–562 (2019). 10.1016/j.tibtech.2018.10.005 [DOI] [PubMed] [Google Scholar]
- 28.Qi, X. M., Sun, Y. F. & Xiong, S. D. A single freeze-thawing cycle for highly efficient solubilization of inclusion body proteins and its refolding into bioactive form. Micro. Cell Fact.14, 24 (2015). 10.1186/s12934-015-0208-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang, W. et al. Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux. Nat. Commun.10, 4248 (2019). 10.1038/s41467-019-12247-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dzuricky, M., Rogers, B. A., Shahid, A., Cremer, P. S. & Chilkoti, A. De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem.12, 814–825 (2020). 10.1038/s41557-020-0511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amiram, M. et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol.33, 1272–1279 (2015). 10.1038/nbt.3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao, E. M. et al. Light-based control of metabolic flux through assembly of synthetic organelles. Nat. Chem. Biol.15, 589–597 (2019). 10.1038/s41589-019-0284-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adhikari, A. et al. Reprogramming natural proteins using unnatural amino acids. RSC Adv.11, 38126–38145 (2021). 10.1039/D1RA07028B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinkemeier, C. D., Girona, G. E. & Lemke, E. A. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science363, eaaw2644 (2019). 10.1126/science.aaw2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinkemeier, C. D. & Lemke, E. A. Dual film-like organelles enable spatial separation of orthogonal eukaryotic translation. Cell184, 4886–4903 (2021). 10.1016/j.cell.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng, J. Y. et al. Creating an in vivo bifunctional gene expression circuit through an aptamer-based regulatory mechanism for dynamic metabolic engineering in Bacillus subtilis. Metab. Eng.55, 179–190 (2019). 10.1016/j.ymben.2019.07.008 [DOI] [PubMed] [Google Scholar]
- 37.Tian, R. Z. et al. Titrating bacterial growth and chemical biosynthesis for efficient N-acetylglucosamine and N-acetylneuraminic acid bioproduction. Nat. Commun.11, 5078 (2020). 10.1038/s41467-020-18960-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, Y. Y., Cao, H. Z., Wang, H. & Mu, W. M. Biosynthesis of human milk oligosaccharides via metabolic engineering approaches: current advances and challenges. Curr. Opin. Biotechnol.78, 102841 (2022). 10.1016/j.copbio.2022.102841 [DOI] [PubMed] [Google Scholar]
- 39.Yu, W. W. et al. A pathway independent multi-modular ordered control system based on thermosensors and CRISPRi improves bioproduction in Bacillus subtilis. Nucleic Acids Res.50, 6587–6600 (2022). 10.1093/nar/gkac476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optodroplets. Cell168, 159–171 (2017). 10.1016/j.cell.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster, B. S. et al. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun.9, 2985 (2018). 10.1038/s41467-018-05403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrillo, N. et al. Safety and efficacy of N-acetylmannosamine (ManNAc) in patients with GNE myopathy: an open-label phase 2 study. Genet. Med.23, 2067–2075 (2021). 10.1038/s41436-021-01259-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bi, X. Y. et al. etiBsu1209: a comprehensive multiscale metabolic model for Bacillus subtilis. Biotechnol. Bioeng.120, 1623–1639 (2023). 10.1002/bit.28355 [DOI] [PubMed] [Google Scholar]
- 44.Mayr, C. et al. Frontiers in biomolecular condensate research. Nat. Cell Biol.25, 512–514 (2023). 10.1038/s41556-023-01102-2 [DOI] [PubMed] [Google Scholar]
- 45.Garabedian, M. V. et al. Designer membraneless organelles sequester native factors for control of cell behavior. Nat. Chem. Biol.17, 998–1007 (2021). 10.1038/s41589-021-00840-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galvanetto, N. et al. Extreme dynamics in a biomolecular condensate. Nature619, 876–883 (2023). 10.1038/s41586-023-06329-5 [DOI] [PubMed] [Google Scholar]
- 47.Lee, D. S. W. et al. Size distributions of intracellular condensates reflect competition between coalescence and nucleation. Nat. Phys.19, 586–596 (2023). 10.1038/s41567-022-01917-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, K. H. et al. Defining synonymous codon compression schemes by genome recoding. Nature539, 59–64 (2016). 10.1038/nature20124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson, E. D. et al. Engineered ribosomes with tethered subunits for expanding biological function. Nat. Commun.10, 3920 (2019). 10.1038/s41467-019-11427-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmied, W. H. et al. Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature564, 444–448 (2018). 10.1038/s41586-018-0773-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunkelmann, D. L., Oehm, S. B., Beattie, A. T. & Chin, J. W. A 68-codon genetic code to incorporate four distinct non-canonical amino acids enabled by automated orthogonal mRNA design. Nat. Chem.13, 1110–1117 (2021). 10.1038/s41557-021-00764-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson, G. E., Lalanne, J. B., Peters, M. L. & Li, G. W. Functionally uncoupled transcription-translation in Bacillus subtilis. Nature585, 124–128 (2020). 10.1038/s41586-020-2638-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray, W. T. et al. Nucleoid size scaling and intracellular organization of translation across bacteria. Cell177, 1632–1648 (2019). 10.1016/j.cell.2019.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, F. R. et al. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nat. Catal.5, 662–672 (2022). 10.1038/s41929-022-00798-z [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Source data generated in this study are provided in the Supplementary Information/Source Data file. Unprocessed microscopy images are available under restricted access due to the difficulty of hosting this data format, access can be obtained by contacting the corresponding author, who will respond within two business week local time. Source data are provided with this paper.