Central Illustration
Key Words: aggregation, cardiomyopathy, disease mechanism, p.Arg14del, phospholamban, sarcoplasmic reticulum, SERCA
Highlights
-
•
The p.Arg14del variant of PLN causes cardiomyopathy, leading to severe heart failure. Initially, the primary disease pathology was attributed to SERCA2a superinhibition. However, recent studies showed partial or complete loss of SERCA2a inhibition in PLN-R14del and revealed a more prominent role for abnormal PLN protein distribution as underlying disease mechanism.
-
•
This review will reassess the disease mechanisms of PLN-R14del cardiomyopathy and explore its implications for novel therapeutic approaches. Therapies solely addressing calcium handling may be less effective than anticipated, whereas targeting the PLN-R14del–mediated S/ER cluster formation may offer a more promising therapeutic strategy.
-
•
Additional studies are required to further dissect the underlying molecular mechanisms to uncover novel targets for innovative treatments for PLN-R14del cardiomyopathy.
Summary
The phospholamban (PLN) pathogenic gene variant, p.Arg14del (PLN-R14del), can lead to dilated and arrhythmogenic cardiomyopathy, resulting in heart failure. PLN-R14del cardiomyopathy has been conceptualized as a disease caused by sarco/endoplasmic reticulum calcium adenosine triphosphatase 2a (SERCA2a) superinhibition. However, recent studies raised controversy regarding the effect of PLN-R14del on SERCA activity and revealed a prominent role for abnormal PLN protein distribution and sarco/endoplasmic reticulum disorganization as underlying disease mechanism. Strategies targeting sarco/endoplasmic reticulum malformation may, therefore, prove more effective than SERCA activity modulation. This review reassesses the disease mechanisms of PLN-R14del cardiomyopathy and emphasizes the importance of dissecting the underlying molecular mechanisms to uncover targets for innovative treatments.
The p.Arg14del mutation in the gene encoding phospholamban (PLN) represents the most prevalent pathogenic variant (termed PLN-R14del) identified within PLN and is under extensive investigation. The mutation can cause both dilated cardiomyopathy (DCM) and arrhythmogenic cardiomyopathy (ACM), often resulting in severe heart failure.1, 2, 3 Hence, the clinical cardiac phenotype comprises a combination of cardiac dilatation with reduced ejection fraction, often with malignant ventricular arrhythmias leading to sudden cardiac death.4 At the histological level, severe cardiac fibrosis or fibrofatty-replacement is observed in tissues from patients with end-stage heart failure due to PLN-R14del cardiomyopathy. On a structural level, a distinctive feature is the presence of perinuclear PLN aggregates in cardiomyocytes. These PLN aggregates have not been observed in cardiac tissues of other DCM patients and are therefore regarded as a hallmark of this disease. Importantly, a recent in-depth investigation revealed that these aggregates consist of malformed sarco/endoplasmic reticulum (S/ER) clusters.5 Therefore it is rather the local clustering of S/ER membranes that provides the strong PLN staining and not PLN aggregation per se.
So far, 2 founder PLN-R14del mutations have been identified, both leading to the development of analogous disease phenotypes in carriers. One founder mutation has its origin in Greece, whereas the other emanates from the Netherlands,1,2 exerting an exceptionally pronounced disease impact in the latter country. In the Netherlands, the prevalence of the PLN-R14del mutation is noteworthy, accounting for 15% of DCM and 12% of ACM patients.3,6 The high prevalence can also be attributed to the pathogenic autosomal dominant inheritance pattern of the PLN-R14del allele. Since its discovery, carriers have been identified in the United States,7 Canada,8 China,9 Japan,10 the United Kingdom,11 Norway,11 Spain,12 Germany,13 and Belgium11 as well, and further expansion is anticipated. Because of the high disease prevalence and life-threatening disease severity, a successful treatment is warranted. Unfortunately, there are no disease-specific treatments yet. Instead, patients receive standard heart failure medication, an implantable cardioverter-defibrillator when at risk of sudden cardiac death, or a left ventricular assist device when cardiac function is severely impaired with heart transplantation, often as a last resort.
The Effect of PLN-R14del On Serca Function
PLN localizes to the S/ER membrane and has a regulatory function in cardiomyocyte calcium handling. The S/ER is the major intracellular calcium store and in cardiomyocytes it releases calcium into the cytosol on action potential stimulation via calcium-induced calcium release,14 which involves the opening of S/ER membrane localized ryanodine receptor calcium channels.15 Cytosolic calcium binds to the myofilament proteins to cause sarcomere shortening and cardiomyocyte contraction. To establish subsequent relaxation, cytosolic calcium is transported back into the S/ER by the sarco/endoplasmic reticulum calcium adenosine triphosphatase 2a (SERCA2a) pump.15 PLN negatively regulates SERCA2a activity by diminishing its affinity for calcium. Therefore, PLN is often regarded as a SERCA2a inhibitor. The inhibitory action of PLN can be reduced via beta-adrenergic stimulation, thereby allowing faster relaxation of the heart muscle. This process is mediated via protein kinase A (PKA) induced phosphorylation of PLN at cytosolic residue serine-16, which results in a conformational change in PLN and alteration of the PLN/SERCA2a interaction leading to increased SERCA2a activity. The amino acid arginine at position 14 (R14) of PLN is an essential residue of the PKA consensus phosphorylation motif (R-R-X-S/T),16 and deletion of R14 abolishes PKA phosphorylation of serine-16 in PLN.17 Based on this mechanism, it was hypothesized that the non-phosphorylatable PLN-R14del protein would result in constitutive SERCA2a inhibition, so called SERCA2a superinhibition.
The effect of the p.Arg14del mutation has been evaluated in several mouse models. A transgenic mouse model overexpressing the mutant PLN protein provided evidence for PLN-R14del-induced SERCA superinhibition and thus reduced calcium reuptake into the S/ER.2 However, when PLN-R14del was overexpressed in a PLN-knockout background (in absence of wild-type [WT] PLN, WT-PLN) the SERCA superinhibitory effect was absent.18 In a humanized PLN-R14del mouse model, SERCA inhibition by PLN-R14del was observed in right ventricular (RV) cardiomyocytes only, whereas calcium handling in left ventricular (LV) cardiomyocytes was unaffected by the mutation.19,20 This is surprising because SERCA, PLN, and other known associated proteins were shown to be similarly expressed in the RV and LV.19 Differential gene expression of some ion channels and ion-regulated proteins were observed,19,21 but it will require further investigations to explore whether these differences can explain the PLN-R14del–mediated calcium uptake dissimilarity in LV and RV cardiomyocytes from these mice. The observed abnormal calcium handling in RV cardiomyocytes may cause the increased propensity to arrhythmias in these mice, but it cannot be excluded that other remodeling effects that are also present in these mice may be involved. The use of the human PLN sequence in the above-described mouse model can be advantageous for certain studies, such as the exploration of gene repair. However, a potential drawback is that the interspecies interaction between human PLN and mouse SERCA or other proteins may not precisely mirror the interaction between these proteins in the normal context when species origin is identical. The substitution of the highly conserved N27 amino acid residue in PLN into K27 in primates is in this respect intriguing and human PLN has been shown to cause stronger SERCA2a inhibition in mice.22 Moreover, this residue may be important for interactions with other proteins in a species-specific manner as well.
Opposing findings to the humanized mouse model have been observed in a knock-in mouse with a compatible PLN-R14del mutation in the murine locus, potentially due to these species-specific differences. The ability of mouse PLN to inhibit SERCA2a was lost in this PLN-R14del mouse model, in both heterozygous and homozygous mice.5,23 Moreover, a dose-dependent effect of the mutation was shown, with the strongest diminishment of SERCA2a inhibition in cardiomyocytes derived from homozygous mice and an intermediate effect in cardiomyocytes derived from heterozygous mice.5 Most interestingly, overexpression of the SERCA activator DWORF (dwarf open reading frame, a small S/ER protein) was able to stimulate S/ER calcium uptake in WT mice, but not in homozygous PLN-R14del mice. This observation confirmed the presence of already enhanced SERCA activity in homozygous PLN-R14del mice, instead of SERCA superinhibition. Nevertheless, DWORF strongly delayed heart failure development and extended life span in PLN-R14del mice.5 This study revealed that DWORF overexpression did not exert its cardioprotective effects via SERCA stimulation for this specific disease, instead it diminished PLN-R14del–induced abnormal S/ER formation. It was implied that abnormal clustering of PLN occurred via sequential stages, in which the cluster size increases and starts to disorganize the entire distribution of the S/ER. DWORF overexpression restrained PLN cluster size, thereby diminishing S/ER malformation. By intervening in the process of S/ER malformation, DWORF reduced cardiomyocyte cell death and improved cardiac function in PLN-R14del mice. This indicates that altered calcium handling is not the main driver of disease development. Similarly, selective stimulation of SERCA2a via the small molecule PST3093, was not able to further stimulate S/ER calcium reuptake in mouse heterozygous PLN-R14del cardiomyocytes and in human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) from human patients carrying the PLN Arg.14del variant.23,24 These data suggest that the PLN-R14del protein results in loss of SERCA2a inhibition, rather than generating a SERCA superinhibitor.
A zebrafish model has also been generated with deletion of the R14 codon of PLN.25 However, the situation in zebrafish is more complex due to gene duplication. Two different Pln genes (Plna and Plnb) encoding 2 different protein isoforms are present in zebrafish, which are less conserved to mouse and human PLN.26 In the published study, the R14-deletion was introduced only in Plna. In these zebrafish, an increase in diastolic calcium level, prolongation of the calcium transient, and an increase in τ, the time constant for calcium decay, were observed.25 The nonselective SERCA2a activator, istaroxime, which also stimulates the cardiac Na+/K+ adenosine triphosphatase, was able to improve τ in these zebrafish cardiomyocytes.25 This appears to contrast observations in mouse PLN-R14del and human iPSC–derived cardiomyocytes, which showed already enhanced SERCA activity that could not be further stimulated by selective SERCA2a activators.23,24
The effect of PLN-R14del on SERCA2a function was also investigated in studies with human embryonic kidney 293 (HEK-293) cells and hiPSC-CMs. Coexpression of either WT-PLN or PLN-R14del with SERCA1 in HEK-293 cells resulted in moderate SERCA inhibition, but a combination of both WT-PLN and mutant-PLN caused a superinhibitory effect on SERCA activity.2 However, different cotransfection ratios and combinations may cause different PLN and SERCA1 protein levels, hence data are difficult to interpret without proper quantification. Force generation was reduced in engineered heart tissue generated from PLN-R14del patient–derived hiPSC-CMs compared to the mutation-corrected isogenic control hiPSC-CMs.27 Nevertheless, calcium handling parameters were similar between PLN-R14del patient–derived hiPSC-CMs and control hiPSC-CMs. Therefore, it was suggested that differences in force could not be attributed to major alterations in calcium handling, but impairment of the S/ER-mitochondria compartment was suggested. Further investigations with the selective SERCA2a stimulator PST3093, showed that calcium dynamics of control hiPSC-CMs treated with this activator resembled the PLN-R14del CMs, whereas the latter were less responsive toward this activator.24 For this reason, it was argued that instead of superinhibition, PLN-R14del was less competent in SERCA2a inhibition.
Structural topology and conformational dynamics of WT-PLN and PLN-R14del with SERCA were also evaluated in isolated systems. To this end, the S/ER was reconstituted using proteoliposomes with lipid-to-protein and PLN-to-SERCA ratios similar to those of cardiac S/ER membranes.28 These experiments showed that SERCA activity was reduced both by WT-PLN and R14del-PLN, but the reduction in SERCA activity was much larger with WT-PLN. These results indicate that R14del-PLN lost part of its inhibitory activity and was therefore considered to be a mild loss-of-function mutation. Moreover, in the presence of equal amounts of WT-PLN and R14del-PLN, SERCA activity was similar as in the presence of R14del-PLN only, indicating that it has a dominant negative effect.28 Similar experiments were conducted in phospholipid membranes, in which PLN-R14del protein was shown to have a slightly reduced inhibitory effect in comparison to WT-PLN. Structural analysis using solid-state nuclear magnetic resonance (NMR) and circular dichroism spectroscopy indicated that the functional differences are related to rather subtle alterations in the structure and topology of the cytoplasmic domain of the PLN-R14del protein.29 In addition, this study confirmed that mutant PLN is resistant to phosphorylation by PKA. Solution and solid-state NMR methods were also used to analyze the structural dynamics and topology of the R14del mutation in correlation with SERCA activity assays. This study demonstrated that in an isolated system, removal of the Arg14 residue renders PLN a less potent inhibitor of SERCA and that phosphorylation of PLN-R14del at Ser16 does not relieve inhibition.30 So also in this setting, it was concluded that the R14 deletion is a loss-of-function mutant compared to WT-PLN. These isolated system experiments used skeletal SERCA1 instead of cardiac SERCA2; however, the skeletal muscle isoform is a well-established surrogate for structural and functional studies of the complex.28
Table 1 provides an overview of the publications on SERCA function in different models for PLN-R14del cardiomyopathy. Although evidence is inconclusive about SERCA activity being inhibited or rather enhanced in presence of PLN-R14del compared to WT-PLN, both forms of SERCA dysregulation can cause calcium-handling dysfunction. A dynamic range of calcium regulation is required to accomplish a cardiac reserve for conditions of exercise and rest. Likewise, both strong SERCA inhibition and robust relief are needed for a dynamic range in cardiac contraction and relaxation. When one of these aspects is lost, the dynamic responsiveness of calcium handling decreases, and cardiac complications can occur. This holds true for both absence of PLN phosphorylation31 (SERCA inhibition) and PLN pentamer stabilization32 (enhanced SERCA activity compared to WT-PLN conditions) (see also Figure 1). Therefore, independent of the direction of SERCA regulation by PLN-R14del, these data indicate the occurrence of calcium handling dysfunction in PLN-R14del cardiomyopathy.
Table 1.
Publications on SERCA Function in Different Models for PLN-R14del Cardiomyopathy
| Year | Group | Model | Speciesa | PLN-R14del Effect on SERCA Function | Ref. # |
|---|---|---|---|---|---|
| 2006 | Kranias | Transgenic PLN-R14del mouse | M | SERCA superinhibition when WT-PLN is present. | 2 |
| HEK-293 cells | H | SERCA superinhibition when WT-PLN is present. | |||
| 2012 | Kranias | R14del mouse model in PLN-KO background | M | No SERCA superinhibition in absence of WT-PLN. | 18 |
| 2012 | Young | Proteoliposomes | H | Partial loss-of-function in presence and absence of WT-PLN. | 28 |
| 2014 | Middleton | Phospholipid membranes | H | Reduced inhibitory effect on SERCA. | 29 |
| 2015 | Veglia | DPC-d38 micelles and lipid membranes | H | Loss-of-function mutant (compared to WT-PLN). | 30 |
| 2021 | Hansen | hiPSC-CMs | H | SERCA function largely unaffected. | 27 |
| 2021 | Zaza | hiPSC-CMs | H | Loss-of-function. | 24 |
| 2021 | Bakkers | PLN-R14del zebrafish | Z | Longer Ca2+ transients and slower decay (indicative of enhanced SERCA inhibition). | 25 |
| 2021 | Kranias | hPLN-R14del mouse | H | Prolonged τ in RV (not LV) cardiomyocytes, indicative of enhanced SERCA inhibition. The propensity to arrhythmia may be increased. | 19 |
| 2022 | Kranias and Sanoudou | hPLN-R14del mouse | H | Increased binding to SERCA and HAX-1, which may enhance SERCA inhibition (not directly tested). | 42 |
| 2023 | Kranias and Sadayappan | hPLN-R14del mouse | H | Prolonged τ in RV cardiomyocytes (indicative of enhanced SERCA inhibition). Normal calcium handling in LV cardiomyocytes. | 20 |
| 2023 | Zaza | PLN-R14Δ/+mouse | M | Loss-of-function. | 23 |
| 2023 | Silljé | PLN-R14Δ/Δand R14Δ/+mouse | M | Enhanced calcium reuptake. | 5 |
| 2023 | Robia | HEK-293 cells | H | Loss-of-function via PLN pentamer stabilization and decreased binding affinity for SERCA. | 37 |
Bold represents data indicative of PLN loss-of-function of SERCA inhibition (either partial or complete); italic represents data indicative of SERCA superinhibition.
Ca2+ = calcium; DPC-d38 = dodecylphosphocholine-d38; H = human; HAX-1 = HS-1-associated protein X-1; HEK-293 cells = human embryonic kidney 293 cells; hiPSC-CMs = human induced pluripotent stem cell–derived cardiomyocytes; hPLN-R14del mouse = humanized PLN-R14del mouse model; KO = knockout; LV left ventricle; M = mouse, PLN = phospholamban; PLN-R14Δ/+ mouse = heterozygous PLN-R14del knock-in mouse model; PLN-R14Δ/Δ mouse = homozygous PLN-R14del knock-in mouse model; RV = right ventricle; SERCA = sarco/endoplasmic reticulum calcium adenosine triphosphatase; WT = wild type; Z = zebrafish.
Species column indicates from which species the used PLN-R14del sequence originates.
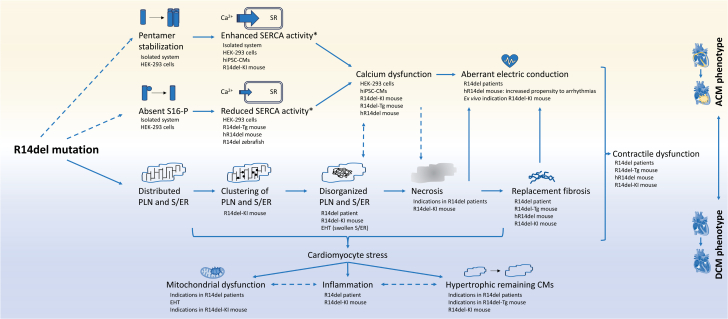
Figure 1.
Schematic Summary of the Potential Mechanisms Involved in the Disease Development of PLN-R14del Cardiomyopathy
The models supporting the findings (and optionally patient validation) are listed below each finding. Proven consequences or interactions are indicated by a solid arrow, whereas likely but not yet proven consequences or interactions are indicated by a dashed arrow. ∗The activity of sarco/endoplasmic reticulum calcium adenosine triphosphatase (SERCA) in presence of PLN-R14del compared to SERCA activity in the presence of wild-type PLN. ACM = arrhythmogenic cardiomyopathy; Ca2+ = calcium; DCM = dilated cardiomyopathy; EHT = engineered heart tissue derived from hiPSC-CMs; HEK-293 cells = human embryonic kidney 293 cells; hiPSC-CM = human induced pluripotent stem cell–derived cardiomyocytes; hR14del mouse = humanized PLN-R14del mouse model; PLN = phospholamban; R14del-KI = PLN-R14del knock-in mouse model; R14del-Tg = transgenic/overexpressing PLN-R14del mouse model; S16-P = phosphorylation of the protein kinase A phosphorylation site of PLN; S/ER = sarco/endoplasmic reticulum; SR = sarcoplasmic reticulum. Several icons from BioRender.com were used to design this figure.
The finding that the R14-deletion might be a partial or complete loss-of-function mutation would be in line with other known PLN mutations that cause DCM. The PLN-Leu-39-stop mutation produces a truncated PLN protein that, when introduced in HEK-293 cells, resulted in an unstable protein and no inhibition of SERCA.33 Consistent with these findings, PLN could not be detected in the heart of a patient homozygous for the mutation. Whereas this mutation may have pleiotropic effects in heterozygous carriers (unaffected, HCM, or DCM), in the homozygous state it resulted in DCM and premature death.33,34 In addition, a missense mutation in PLN cytoplasmic domain (R9C) was shown to cause DCM.35 PLN can exist both as a monomeric and a pentameric form in the S/ER, and the pentameric form is believed to have little to no effect on SERCA activity. The R9C mutation in PLN was shown to shift this equilibrium to the pentameric form, and this mutation caused in monomeric state a loss-of-function mutant with a partial inhibitory effect on SERCA and a total loss-of-function in the pentameric state.36 The mutation seemed to increase the stability of the PLN pentameric assembly, thereby preventing binding to SERCA. Moreover, PLN pentamers have been shown to provide a preferred PKA substrate and compete with monomers for the kinase, thereby reducing monomer phosphorylation and maximizing SERCA inhibition.32 Therefore, SERCA activity is higher in the presence of PLN-R9C compared to the presence of WT-PLN.36 In patients, the mutation triggered DCM and led to premature death.35 These mutations demonstrate that PLN loss-of-function mutations can result in disease in patients.
Even though PLN knockout mice do not develop cardiomyopathy, humans will develop disease in the absence of functional PLN protein. PLN-R14del was recently shown to have increased affinity for homo-oligomerization and decreased binding affinity for SERCA compared to WT.37 These data suggest that, like PLN-R9C, the R14del mutation shifts the equilibrium towards a pentameric form, decreasing its ability to regulate SERCA. In addition, the R14del mutation reduced the rate of PLN unbinding from the pentamer after a transient calcium elevation, limiting the rate of re-binding to SERCA. PLN-R14del might therefore be a partial loss-of-function mutation. Whilst WT-PLN protein is affected by the R9C mutation, the R14del mutation does not seem to affect WT-PLN protein, since calcium reuptake in heterozygous mouse PLN-R14del cardiomyocytes was intermediate between WT and homozygous PLN-R14del cardiomyocytes.5 These data suggest that the PLN-R14del mutation is probably not a full loss-of-function mutation and that other mechanisms are likely to contribute to disease development in PLN-R14del cardiomyopathy.
In summary, the latest functional and structural data on the p.Arg14del variant of PLN point toward (partial) loss-of-function and enhanced calcium uptake, rather than SERCA superinhibition (Figure 1, Central Illustration). Whereas reduced calcium uptake, owing to SERCA reduction, is generally believed to be arrhythmogenic and enhanced calcium uptake to be antiarrhythmogenic, recent data show that this relationship is more nuanced and dynamic. In particular, a moderate SERCA activity increase can also induce calcium sparks and promote arrhythmogenic calcium waves.38 Moreover, to adapt to changing conditions, a dynamic range of SERCA activity modulation is important to provide sufficient cardiac reserve,32 and this will be diminished if PLN-mediated SERCA regulation is hampered. Thus, dysfunctional calcium regulation may underly the pathogenic PLN-R14del phenotype, but as we will discuss later, we believe that abnormal S/ER clustering is the more important pathogenic factor in this disease.
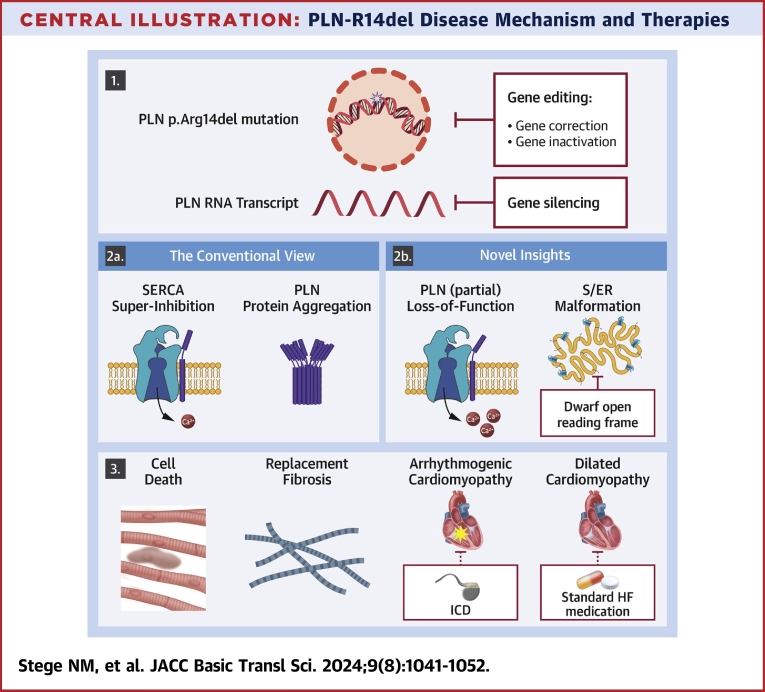
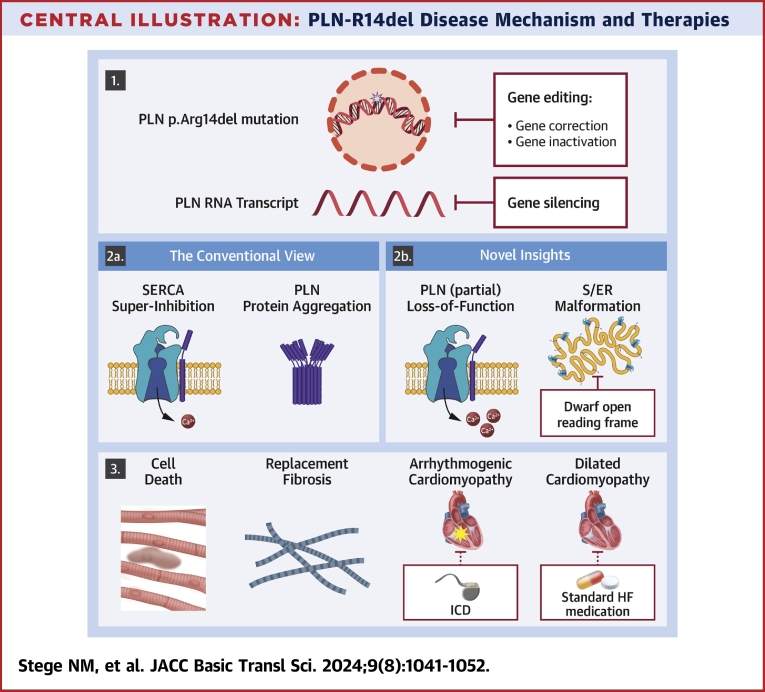
Central Illustration.
PLN-R14del Disease Mechanism and Therapies
The Central Illustration presents both original and novel insights into PLN-R14del disease mechanisms and potential therapeutic targets. Panel 1 illustrates the genetic mutation and proposes potential treatment approaches such as gene therapy or gene silencing of PLN mRNA. In panel 2a, the conventional view of the PLN-R14del disease mechanism is depicted, while in panel 2b, our novel insights and potential targeting of abnormal S/ER clusters by DWORF are highlighted. Panel 3 portrays the resulting cell death and replacement fibrosis contributing to ACM and DCM, with potential symptom management through interventions like ICD or standard HF medication, although these do not address the underlying disease itself. Ca2+ = calcium; HF = heart failure; ICD = implantable cardioverter-defibrillator; PLN = phospholamban; SERCA2a = sarco/endoplasmic reticulum calcium adenosine triphosphatase 2a; S/ER = sarco/endoplasmic reticulum.
The Effect of PLN-R14del on PLN Protein Distribution
In myocardial sections obtained from explanted hearts of patients carrying the PLN p.Arg14del mutation, abnormal cytoplasmic distribution and perinuclear accumulation of PLN protein was observed.39 This aberrant PLN protein distribution was not observed in myocardial sections obtained from explanted hearts of patients with ischemic cardiomyopathy. Immunohistochemistry for PLN performed in cardiac specimens of Dutch PLN p.Arg14del mutation carriers also showed abnormal PLN protein staining (large elongated perinuclear, sometimes circumnuclear) in cardiomyocytes in both ventricles in all examined hearts (presence in 20 of 20 examined PLN-R14del hearts).40 Because this abnormal PLN localization was not present in PLN-negative cases of idiopathic and genetic DCM or in cases of desmosomal ACM, Te Rijdt et al40 concluded that PLN p.Arg14del cardiomyopathy is characterized by PLN aggregates. As we will explain later, this is now believed to constitute abnormal S/ER clusters, rather than PLN aggregates. Thus, cardiac PLN immunostaining was proven to be a highly sensitive and specific method for diagnosing PLN-R14del cardiomyopathy.41
In vitro, normal ER-staining patterns were described for PLN-R14del protein that was coexpressed with WT-PLN in HEK-293 cells.2 When exclusively PLN-R14del protein was expressed in HEK-293 cells, mutant PLN localized with Na+/K+-adenosine triphosphatase at the plasma membrane where it stimulated Na+/K+-adenosine triphosphatase activity.18 Therefore, it was initially suggested that the PLN-R14del mutation may lead to mislocalization of PLN from the S/ER to the sarcolemma. However, a more recent study did not confirm this finding and normal subcellular S/ER distribution of PLN-R14del was reported in HEK-293 cells in the absence of WT-PLN.42 Moreover, H9C2 myoblasts that overexpressed PLN-R14del protein showed a similar colocalization with SERCA as compared with WT-PLN. Therefore, it was concluded that no changes were observed in the subcellular distribution of PLN-R14del or its colocalization to SERCA. A hiPSC-CM study also showed similar perinuclear PLN and SERCA protein localization in both WT-PLN and PLN-R14del cardiomyocytes, albeit staining was weaker in PLN-R14del cardiomyocytes.27 More elaborate quantification of cellular distribution of the hiPSC-CMs showed that the PLN p.Arg14del mutation only slightly decreased PLN/SERCA colocalization, but remarkably decreased the perinuclear density of both proteins.24 Probably, the absence of a well-developed S/ER structure in hiPSC-CMs, which have an immature phenotype, and in non-cardiomyocytes may hinder the formation of abnormal perinuclear PLN clusters that are so evidently present in adult cardiomyocytes in patient heart tissue.
Clustering of PLN has not been reported for the humanized PLN-R14del mouse model, and when PLN-R14del was overexpressed in a PLN-knockout background, the protein failed to colocalize with SERCA.18 Consistent with patient observations, the PLN-R14del knock-in mouse model showed the formation of perinuclear PLN clusters. In homozygous mice, extensive clustering of PLN was observed at 8 weeks of age, whereas the heterozygous mouse model showed a limited number of PLN clusters at 20 months of age.43 Interestingly, homozygous mice presented PLN cluster formation before onset of functional impairment or other structural abnormalities, indicating that this process might be involved in disease onset.44 Using this mouse model, it was recently shown that mutant PLN protein starts to form small clusters (speckles) that continue to progress into large PLN clusters, which consist of severe disorganized S/ER.5 In line with these observations, pathologically dilated S/ER structures and indications for disturbed ER/mitochondria contact sites were observed in engineered heart tissues generated from human patient hiPSC-CMs.27 Moreover, single-cell RNA sequencing of hiPSC-CMs that overexpressed PLN-R14del revealed the induction of the unfolded protein response pathway, and this study also showed improvement of hiPSC-CM function upon inhibition of the unfolded protein response.45 Activation of the unfolded protein response stress pathway of the S/ER could be related to the malformation of this organelle, as observed in the engineered heart tissues and a mouse model for this disease. Together these studies emphasize a more prominent role of pathological S/ER alterations as underlying disease mechanism of PLN-R14del cardiomyopathy. Importantly, the finding that large PLN clusters consist of severe disorganized S/ER was validated in PLN-R14del patient–derived cardiac tissue, whereas these structures were absent in cardiac tissue from patients with non-PLN-R14del DCM.5 This study also demonstrated that subsequent cardiomyocyte death occurs via necrosis. In particular, necrosis occurred after p62 accumulation in cardiomyocytes with large PLN S/ER clusters. Importantly, accumulation of p62 in cardiomyocytes with S/ER PLN clusters has also been observed in cardiac tissue of patients.40 The occurrence of cardiomyocyte necrosis was confirmed by release of cardiac troponin in the circulation and specific staining of necrotic cardiomyocytes in PLN-R14del mice.5 This also explains the excessive replacement fibrosis observed in these mice and also in cardiac tissue of patients with PLN-R14del cardiomyopathy. The observed cardiomyocyte loss, excessive replacement fibrosis, and hypertrophy of remaining cardiomyocytes will culminate in severe cardiac dysfunction. These data indicate essential involvement of PLN-R14del–induced S/ER cluster formation in disease development. Table 2 provides an overview of the publications on PLN protein localization in different models for PLN-R14del cardiomyopathy and in cardiac tissue of patients.
Table 2.
Publications on PLN Protein Localization in Different Models for PLN-R14del Cardiomyopathy
| Year | Group | Model | Speciesa | PLN-R14del Protein Localization | Ref. # |
|---|---|---|---|---|---|
| 2006 | Kranias | PLN-R14del patient tissue | H | PLN localization is not shown. | 2 |
| hPLN-R14del mouse | H | PLN localization is not shown. | |||
| HEK-293 overexpressing PLN-R14del | H | Normal ER-staining patterns. | |||
| 2012 | Kranias | R14del mouse model in PLN-KO background | M | PLN did not colocalize with SERCA2a. | 18 |
| HEK-293 overexpressing PLN-R14del | H | PLN localized with NKA at the plasma membrane. | |||
| 2015 | Kranias and Hajjar | PLN-R14del patient tissue | H | Abnormal cytoplasmic PLN distribution and perinuclear accumulation. | 39 |
| hiPSC-CMs | H | PLN predominantly polarized at one side of the cytoplasm. | |||
| 2016 | Suurmeijer | PLN-R14del patient tissue | H | Perinuclear PLN aggregation, colocalizing with autophagy markers. Aggresome-like ultrastructure. | 40 |
| 2017 | Suurmeijer | PLN-R14del patient tissue | H | Perinuclear PLN aggregation, diagnostic application. | 41 |
| 2020 | Silljé and De Boer | PLN-R14Δ/Δmouse | M | Extensive perinuclear PLN aggregation at 8 wks. | 43 |
| PLN-R14Δ/+mouse | M | Limited perinuclear PLN aggregation at 20 mo. | |||
| 2021 | Silljé and De Boer | PLN-R14Δ/Δmouse | M | PLN aggregates present before onset of functional impairment or other structural abnormalities. | 44 |
| 2021 | Hansen | hiPSC-CMs | H | Normal distribution and SERCA2a colocalization. | 27 |
| PLN-R14del patient tissue | H | Perinuclear PLN protein aggregates. | |||
| 2021 | Zaza | hiPSC-CMs | H | Decreased perinuclear density of PLN and SERCA. | 24 |
| 2022 | Kranias and Sanoudou | HEK-293 cells | H | Normal distribution and SERCA1 colocalization. | 42 |
| H9c2 cells | H | Normal distribution and SERCA2a colocalization. | |||
| 2023 | De Boer and Silljé | PLN-R14Δ/Δmice | M | Small PLN clusters progress into large clusters, which cause severe disorganization of the S/ER. | 5 |
| PLN-R14del patient tissue | H | Large PLN clusters cause S/ER disorganization. |
Bold represents data indicative of aberrant PLN localization; and italic represents data indicative of normal PLN protein distribution.
ER = endoplasmic reticulum; NKA = Na+/K+-adenosine triphosphatase; SERCA2a = sarco/endoplasmic reticulum calcium adenosine triphosphatase 2a; S/ER = sarco/endoplasmic reticulum; other abbreviations as in Table 1.
Species column indicates from which species the used PLN-R14del sequence originates.
Together, these novel insights show that the pathognomonic structure in PLN-R14del cardiomyopathy is a malformed organelle (the S/ER) instead of common insoluble cytoplasmic aggregates or fibrils composed of incorrectly folded proteins (Central Illustration). Therefore, the originally coined term “PLN protein aggregates” appears to be a misnomer. Instead, the pathological process should be referred to as PLN-R14del–induced S/ER disorganization. Further investigations are required to answer several remaining questions, including the exact nature of the PLN stained speckles that are observed throughout the entire cardiomyocyte before severe S/ER clustering appears. Live-cell imaging of isolated cardiomyocytes will be required to track these temporal and spatial changes and can provide better insights in the exact process. Moreover, it can define whether these clusters are still continuous with the S/ER network, and whether other organelles (golgi, endosomes, or the nuclear envelope) are also affected. In addition, the development of specific antibodies that can distinguish WT-PLN and PLN-R14del from each other, will be needed to evaluate whether predominantly mutant PLN is localized to these S/ER clusters or WT-PLN as well.
Mechanistic Interplay Between the ACM and DCM Phenotypes
Although speculative, it is intriguing to suggest that distinct disease phenotypes, namely ACM and DCM, might be triggered by diverse underlying mechanisms, as illustrated in Figure 1. The PLN-R14del–mediated structural alterations in the S/ER, leading to cardiomyocyte cell death and subsequent replacement fibrosis, could account for the observed DCM phenotype in patients with PLN-R14del cardiomyopathy. In PLN-R14del mutation carriers, fibrosis is also an early feature, which can already be observed in carriers that still have a preserved LV ejection fraction.46 Despite uncertainties regarding the direction of SERCA regulation by PLN-R14del, calcium dysregulation may cause arrhythmias and contribute to the ACM phenotype. PLN-R14del was shown to cause a transformation from an aligned S/ER network throughout the entire cardiomyocyte, to a perinuclear cluster of S/ER. Although not proven by data yet, this pathological change in S/ER distribution may cause local intracellular differences in calcium handling, which could contribute to the ACM phenotype. Moreover, fibrosis serves as a potent arrhythmogenic substrate and could be another important underlying factor in ACM development. Therefore, it is conceivable that a combination of these diverse underlying mechanisms lead to the development of the ACM phenotype. Although a strict demarcation between ACM and DCM phenotypes does not align with clinical presentation, where many patients exhibit both phenotypes, different disease mechanisms could potentially coexist and contribute to this complexity.
Treatment Strategies Reconsidered
Potential treatment strategies to target PLN cardiomyopathy have been extensively reviewed.47,48 Precision medicine approaches targeting the PLN-R14del gene, with the aim of repairing or disrupting the pathogenic allele, have demonstrated efficacy in model systems.39,49 Such approaches directly target the gene defect itself and will likely be the ultimate treatment strategy for PLN-R14del cardiomyopathy (Central Illustration). This may either involve correction of the pathogenic allele using CRISPR/Cas9 technology, including prime editing, or destroying the pathogenic allele by introducing frame shifts or stop codons. In the latter case, 1 WT allele will still be present, and this should be sufficient (50% of WT-PLN expressed). However, translation of this therapy to the human clinical situation will require significant time investment and optimization to minimize detrimental off-target effects and to obtain selective viral targeting. Moreover, it is unlikely that all cardiomyocytes will be targeted using this approach and unedited cardiomyocytes will still be prone to disease pathology. Silencing strategies have also great potential, and antisense oligonucleotides have been shown to be very effective in a PLN-R14del mouse model.50,51 This strategy has the advantage that viral vectors are not required and that there is no risk of potential irreversible DNA damage, because only PLN RNA is targeted. Even non–mutation-specific PLN targeting is effective based on mouse studies, but in humans the silencing effect may need to be restricted as full removal of PLN is most likely detrimental in humans. The advantage of this approach is that it could also be used for other types of heart failure in which SERCA activity might be hampered. Whether mutation-specific silencing of PLN-R14del is feasible has so far not been investigated.
The above-mentioned strategies directly target PLN or the RNA transcript and can be fruitful without proper knowledge of the underlying disease process. For strategies aiming to target more downstream processes, the situation becomes more complex. Based on the PLN-R14del SERCA superinhibitory theory, targeting was focused on restoring S/ER calcium reuptake. However, given the current uncertainty surrounding the direction in which PLN-R14del affects SERCA2a activity and the growing evidence supporting a partial or complete loss of SERCA inhibition by PLN-R14del, a reassessment of this intervention strategy is warranted. Therefore, small molecules that stimulate SERCA2a-mediated calcium S/ER uptake, such as PST3093, may not be effective in PLN-R14del cardiomyopathy, but they might be useful in other types of heart failure in which SERCA activity is reduced. Of note, a recent report showed there was no significant decrease in SERCA or PLN protein in heart failure;52 however, this does not exclude altered SERCA activity in heart failure pathogenesis. Whereas DWORF is known as a SERCA stimulator, the cardioprotective effects of DWORF overexpression were not exerted via enhanced SERCA activity in a mouse model for PLN-R14del cardiomyopathy. Instead, it was revealed that DWORF overexpression delayed PLN-R14del disease development by diminishing S/ER malformation (Central Illustration). This novel, PLN-R14del–specific, cardioprotective effect of DWORF explains why DWORF overexpression could effectively delay PLN-R14del disease development, whereas small molecules that selectively enhance calcium handling seemed unsuccessful. Although we cannot rule out the possibility that SERCA stimulating compounds might have positive effects in preventing S/ER malformation in a manner similar to DWORF, there is currently no supporting evidence for such effects. Similarly, interventions such as overexpressing SERCA2a may have limited effects in patients with PLN-R14del cardiomyopathy, and it cannot be excluded that such interventions may even stimulate S/ER malformation in PLN-R14del cardiomyopathy. Strategies to prevent PLN dephosphorylation, such as protein phosphatase 1 phosphatase inhibition may be hampered by the same issues. Again, such interventions may be effective in other heart failure etiologies, but in PLN-R14del cardiomyopathy with its specific disease mechanism, efficacy is questionable. Therefore, considering the uncertainties around PLN-R14del calcium-handling defects, caution with such strategies is needed and even more so because the mechanism they target may not be causative of PLN-R14del pathophysiology.
Currently, PLN-R14del–induced S/ER malformation has gained importance as a primary underlying disease mechanism, thus other treatments targeting this defect must be considered. Because the primary defect that causes the clustering is unknown and still very little is understood about how S/ER clusters form, this process should be actively studied further to enable the development of novel intervention strategies that directly target the initial stages of cluster formation. Several preclinical studies showed the cardioprotective potential of inhibiting or preventing S/ER clustering in PLN-R14del cardiomyopathy, indicating that dissecting the exact underlying disease mechanisms can be a rewarding task. DWORF overexpression, for example, which did not further enhance calcium reuptake in the PLN-R14del cardiomyopathy mouse model, did exert protective actions and extended life span by diminishing pathogenic PLN-R14del S/ER cluster formation.5 In addition, PLN silencing approaches showed great promise in restoring proper S/ER morphology and were shown to successfully halt disease development in a PLN-R14del mouse model.50,51 Because S/ER clustering is strongly accelerated in homozygous PLN-R14del mice as compared to heterozygous mice, an allele dose-dependent effect is evident. Therefore, overexpression of WT-PLN may also emerge as a potential therapy, capable of shifting the allelic balance.
To summarize, redirecting attention from calcium dysregulation to interventions aimed at restoring or maintaining proper S/ER structure in PLN-R14del cardiomyopathy holds promise for novel therapeutic strategies and may improve our understanding of the relatively unexplored field of S/ER morphology regulation.
Conclusions
In the last decade, PLN-R14del cardiomyopathy has gained substantial scientific interest. Initially, the primary disease pathology was attributed to calcium dysregulation, a fundamental process governing cardiac contraction and relaxation, but recent advancements have unveiled a more intricate and complex understanding. Whereas the R14del variant of PLN causes alterations in calcium handling, the precise defect is uncertain and instead of PLN-R14del causing superinhibition of SERCA, several studies now suggest the opposite, Showing that the mutant protein has partial or complete loss-of-function as a SERCA inhibitor and may even cause enhanced calcium S/ER reuptake (Central Illustration). Furthermore, even though dysfunctional calcium handling will likely contribute to disease development, it may not be the main disease driver of PLN-R14del cardiomyopathy. Instead, recent evidence shows a more prominent role for abnormal PLN protein distribution as a primary underlying disease mechanism. PLN-R14del has been shown to disrupt normal S/ER structure and morphology, leading to cardiomyocyte death. The paradigm shift from SERCA superinhibition as the predominant disease driver to S/ER disorganization urges re-evaluation of therapeutic approaches that target downstream disease mechanisms to address this intriguing multifaceted cardiomyopathy. Interventions that target the gene defect or the PLN messenger RNA will prove their efficacy when clinically applicable. However, therapies focused solely on calcium handling may prove to be less effective than initially anticipated for PLN-R14del cardiomyopathy. Recent insights suggest that targeting the PLN-R14del–mediated S/ER cluster formation could be more effective in delaying disease development. Therefore, dissecting the exact underlying disease mechanisms is a crucial and potentially rewarding task, that may yield novel targets for innovative therapeutic solutions to battle this disease and maybe other heart failure conditions.
Funding Support and Author Disclosures
This work was supported by grants from the Netherlands Heart Foundation (CVON PREDICT2, grant 2018-30; CARMA, grant 01-003-2022-0358); the Leducq Foundation (Cure-PLaN), EU Horizon grant Geremy, the Netherlands Heart Institute, the PLN Foundation. It was also supported by National Institutes of Health grant HL171221 to Dr Makarewich. Dr de Boer has received grant support from the Netherlands Heart Foundation (grants 2017-21; 2017-11; 2018-30; 2020B005; 01-003-2022-0358) and the European Research Council (ERC CoG 818715); has received research grants and/or fees from AstraZeneca, Abbott, Boehringer Ingelheim, Cardior Pharmaceuticals GmbH, Novo Nordisk, and Roche; has had speaker engagements with and/or received fees from and/or served on an advisory board for Abbott, AstraZeneca, Cardior Pharmaceuticals GmbH, Novo Nordisk, and Roche; and received travel support from Abbott, Cardior Pharmaceuticals GmbH, and Novo Nordisk. Dr van der Meer has received grant support from the European Research Council (ERC CoG 101045236, DISSECT-HF); has received consulting fees and/or grants from Novartis, Pharmacosmos, Vifor Pharma, AstraZeneca, Pfizer, Pharma Nord, BridgeBio, Novo Nordisk, Daiichi Sankyo, Boehringer Ingelheim, and Ionis. Dr Silljé has received research grants from AstraZeneca, Novo Nordisk and Ionis Pharmaceuticals. Dr Stege has reported that she has no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
References
- 1.van der Zwaag P.A., van Rijsingen I.A.W., de Ruiter R., et al. Recurrent and founder mutations in the Netherlands–phospholamban p.Arg14del mutation causes arrhythmogenic cardiomyopathy. Neth Heart J. 2013;21(6):286–293. doi: 10.1007/s12471-013-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haghighi K., Kolokathis F., Gramolini A.O., et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci U S A. 2006;103(5):1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Zwaag P.A., van Rijsingen I.A.W., Asimaki A., et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14(11):1199–1207. doi: 10.1093/eurjhf/hfs119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groeneweg J.A., van der Zwaag P.A., Olde Nordkamp L.R.A., et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy according to revised 2010 task force criteria with inclusion of non-desmosomal phospholamban mutation carriers. Am J Cardiol. 2013;112(8):1197–1206. doi: 10.1016/j.amjcard.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Stege N.M., Eijgenraam T.R., Oliveira Nunes Teixeira V., et al. DWORF extends life span in a PLN-R14del cardiomyopathy mouse model by reducing abnormal sarcoplasmic reticulum clusters. Circ Res. 2023;133(12):1006–1021. doi: 10.1161/CIRCRESAHA.123.323304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hof I.E., van der Heijden J.F., Kranias E.G., et al. Prevalence and cardiac phenotype of patients with a phospholamban mutation. Neth Heart J. 2019;27(2):64–69. doi: 10.1007/s12471-018-1211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeWitt M.M., MacLeod H.M., Soliven B., McNally E.M. Phospholamban R14 deletion results in late-onset, mild, hereditary dilated cardiomyopathy. J Am Coll Cardiol. 2006;48(7):1396–1398. doi: 10.1016/j.jacc.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Cheung C.C., Healey J.S., Hamilton R., et al. Phospholamban cardiomyopathy: a Canadian perspective on a unique population. Neth Heart J. 2019;27(4):208–213. doi: 10.1007/s12471-019-1247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X., Xu Y., Sun J., Wang L., Guo X., Chen Y. The phenotypic characteristic observed by cardiac magnetic resonance in a PLN-R14del family. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-73359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabata T., Kuramoto Y., Ohtani T., et al. Phospholamban p.Arg14del cardiomyopathy: a Japanese case series. Intern Med. 2022;61(13):1987–1993. doi: 10.2169/internalmedicine.8594-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.te Rijdt W.P., Hoorntje E.T., de Brouwer R., et al. Rationale and design of the PHOspholamban RElated CArdiomyopathy intervention STudy (i-PHORECAST) Neth Heart J. 2022;30(2):84–95. doi: 10.1007/s12471-021-01584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López-Ayala J.M., Boven L., van den Wijngaard A., Peñafiel-Verdú P., van Tintelen J.P., Gimeno J.R. Phospholamban p.arg14del mutation in a Spanish family with arrhythmogenic cardiomyopathy: evidence for a European founder mutation. Rev Esp Cardiol (Engl Ed) 2015;68(4):346–349. doi: 10.1016/j.rec.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Posch M.G., Perrot A., Geier C., et al. Genetic deletion of arginine 14 in phospholamban causes dilated cardiomyopathy with attenuated electrocardiographic R amplitudes. Heart Rhythm. 2009;6(4):480–486. doi: 10.1016/j.hrthm.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Eisner D.A., Caldwell J.L., Kistamás K., Trafford A.W. Calcium and excitation-contraction coupling in the heart. Circ Res. 2017;121(2):181–195. doi: 10.1161/CIRCRESAHA.117.310230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bers D.M. Cardiac excitation-contraction coupling. Nature. 2002;415(6868):198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 16.Simmerman H.K.B., Collins J.H., Theibert J.L., Wegener A.D., Jones L.R. Sequence analysis of phospholamban: identification of phosphorylation sites and two major structural domains. J Biol Chem. 1986;261(28):13333–13341. [PubMed] [Google Scholar]
- 17.Fujii J., Maruyama K., Tada M., MacLennan D.H. Expression and site-specific mutagenesis of phospholamban: studies of residues involved in phosphorylation and pentamer formation. J Biol Chem. 1989;264(22):12950–12955. [PubMed] [Google Scholar]
- 18.Haghighi K., Pritchard T., Bossuyt J., et al. The human phospholamban Arg14-deletion mutant localizes to plasma membrane and interacts with the Na/K-ATPase. J Mol Cell Cardiol. 2012;52(3):773–782. doi: 10.1016/j.yjmcc.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haghighi K., Gardner G., Vafiadaki E., et al. Impaired right ventricular calcium cycling is an early risk factor in R14del-phospholamban arrhythmias. J Pers Med. 2021;11(6):502. doi: 10.3390/jpm11060502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar M., Haghighi K., Koch S., et al. Myofilament alterations associated with human R14del-phospholamban cardiomyopathy. Int J Mol Sci. 2023;24(3):2675. doi: 10.3390/ijms24032675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogalska M.E., Vafiadaki E., Erpapazoglou Z., et al. Isoform changes of action potential regulators in the ventricles of arrhythmogenic phospholamban-R14del humanized mouse hearts. Metabolism. 2023;138 doi: 10.1016/j.metabol.2022.155344. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W., Yuan Q., Qian J., et al. The presence of Lys27 instead of Asn27 in human phospholamban promotes sarcoplasmic reticulum Ca2+-ATPase superinhibition and cardiac remodeling. Circulation. 2006;113(7):995–1004. doi: 10.1161/CIRCULATIONAHA.105.583351. [DOI] [PubMed] [Google Scholar]
- 23.Maniezzi C., Eskandr M., Florindi C., et al. Early consequences of the phospholamban mutation PLN-R14del+/- in a transgenic mouse model. Acta Physiol (Oxf) 2024;240(3) doi: 10.1111/apha.14082. [DOI] [PubMed] [Google Scholar]
- 24.Badone B., Ronchi C., Lodola F., et al. Characterization of the PLN p.Arg14del mutation in human induced pluripotent stem cell-derived cardiomyocytes. Int J Mol Sci. 2021;22(24) doi: 10.3390/ijms222413500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamel S.M., van Opbergen C.J.M., Koopman C.D., et al. Istaroxime treatment ameliorates calcium dysregulation in a zebrafish model of phospholamban R14del cardiomyopathy. Nat Commun. 2021;12(1):7151. doi: 10.1038/s41467-021-27461-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorski P.A., Trieber C.A., Ashrafi G., Young H.S. Regulation of the sarcoplasmic reticulum calcium pump by divergent phospholamban isoforms in zebrafish. J Biol Chem. 2015;290(11):6777–6788. doi: 10.1074/jbc.M114.585604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuello F., Knaust A.E., Saleem U., et al. Impairment of the ER/mitochondria compartment in human cardiomyocytes with PLN p.Arg14del mutation. EMBO Mol Med. 2021;13(6) doi: 10.15252/emmm.202013074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceholski D.K., Trieber C.A., Young H.S. Hydrophobic imbalance in the cytoplasmic domain of phospholamban is a determinant for lethal dilated cardiomyopathy. J Biol Chem. 2012;287(20):16521–16529. doi: 10.1074/jbc.M112.360859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes E., Middleton D.A. Comparison of the structure and function of phospholamban and the arginine-14 deficient mutant associated with dilated cardiomyopathy. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0106746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vostrikov V.V., Soller K.J., Ha K.N., Gopinath T., Veglia G. Effects of naturally occurring arginine 14 deletion on phospholamban conformational dynamics and membrane interactions. Biochim Biophys Acta. 2015;1848(1 Pt 1):315–322. doi: 10.1016/j.bbamem.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu G., Lester J.W., Young K.B., Luo W., Zhai J., Kranias E.G. A single site (Ser16) phosphorylation in phospholamban is sufficient in mediating its maximal cardiac responses to beta-agonists. J Biol Chem. 2000;275(49):38938–38943. doi: 10.1074/jbc.M004079200. [DOI] [PubMed] [Google Scholar]
- 32.Funk F., Kronenbitter A., Hackert K., et al. Phospholamban pentamerization increases sensitivity and dynamic range of cardiac relaxation. Cardiovasc Res. 2023;119(7):1568–1582. doi: 10.1093/cvr/cvad037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haghighi K., Kolokathis F., Pater L., et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111(6):869–876. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afana A.S., Vasiliu L., Sascău R., et al. Phospholamban p.Leu39∗ cardiomyopathy compared with other sarcomeric cardiomyopathies: age-matched patient cohorts and literature review. J Cardiovasc Dev Dis. 2024;11(2):41. doi: 10.3390/jcdd11020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt J.P., Kamisago M., Asahi M., et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299(5611):1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 36.Ha K.N., Mastersona L.R., Hou Z., et al. Lethal Arg9Cys phospholamban mutation hinders Ca2+-ATPase regulation and phosphorylation by protein kinase A. Proc Natl Acad Sci U S A. 2011;108(7):2735–2740. doi: 10.1073/pnas.1013987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cleary SR, Teng ACT, Kongmeneck AD, et al. Dilated cardiomyopathy variant R14del increases phospholamban pentamer stability, blunting dynamic regulation of cardiac calcium handling. Preprint. Posted online May 28, 2023. bioRxiv 2023:2023.05.26.542463. https://doi.org/10.1101/2023.05.26.542463
- 38.Sato D., Uchinoumi H., Bers D.M. Increasing SERCA function promotes initiation of calcium sparks and breakup of calcium waves. J Physiol. 2021;599(13):3267–3278. doi: 10.1113/JP281579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karakikes I., Stillitano F., Nonnenmacher M., et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.te Rijdt W.P., van Tintelen J.P., Vink A., et al. Phospholamban p.Arg14del cardiomyopathy is characterized by phospholamban aggregates, aggresomes, and autophagic degradation. Histopathology. 2016;69(4):542–550. doi: 10.1111/his.12963. [DOI] [PubMed] [Google Scholar]
- 41.te Rijdt W.P., van der Klooster Z.J., Hoorntje E.T., et al. Phospholamban immunostaining is a highly sensitive and specific method for diagnosing phospholamban p.Arg14del cardiomyopathy. Cardiovasc Pathol. 2017;30:23–26. doi: 10.1016/j.carpath.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Vafiadaki E., Haghighi K., Arvanitis D.A., Kranias E.G., Sanoudou D. Aberrant PLN-R14del Protein Interactions Intensify SERCA2a Inhibition, Driving Impaired Ca2+ Handling and Arrhythmogenesis. Int J Mol Sci. 2022;23(13):6947. doi: 10.3390/ijms23136947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eijgenraam T.R., Boukens B.J., Boogerd C.J., et al. The phospholamban p.(Arg14del) pathogenic variant leads to cardiomyopathy with heart failure and is unresponsive to standard heart failure therapy. Sci Rep. 2020;10(1):9819. doi: 10.1038/s41598-020-66656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eijgenraam T.R., Boogerd C.J., Stege N.M., et al. Protein aggregation is an early manifestation of phospholamban p.(Arg14del)-related cardiomyopathy: development of PLN-R14del-related cardiomyopathy. Circ Heart Fail. 2021;14(11) doi: 10.1161/CIRCHEARTFAILURE.121.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feyen D.A.M., Perea-Gil I., Maas R.G.C., et al. Unfolded protein response as a compensatory mechanism and potential therapeutic target in PLN R14del cardiomyopathy. Circulation. 2021;144(5):382–392. doi: 10.1161/CIRCULATIONAHA.120.049844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Te Rijdt W.P., Ten Sande J.N., Gorter T.M., et al. Myocardial fibrosis as an early feature in phospholamban p.Arg14del mutation carriers: phenotypic insights from cardiovascular magnetic resonance imaging. Eur Heart J Cardiovasc Imaging. 2019;20(1):92–100. doi: 10.1093/ehjci/jey047. [DOI] [PubMed] [Google Scholar]
- 47.Deiman F.E., Bomer N., van der Meer P., Grote Beverborg N. Review: precision medicine approaches for genetic cardiomyopathy: targeting phospholamban R14del. Curr Heart Fail Rep. 2022;19(4):170–179. doi: 10.1007/s11897-022-00558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vafiadaki E., Glijnis P.C., Doevendans P.A., Kranias E.G., Sanoudou D. Phospholamban R14del disease: the past, the present and the future. Front Cardiovasc Med. 2023;10 doi: 10.3389/fcvm.2023.1162205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dave J., Raad N., Mittal N., et al. Gene editing reverses arrhythmia susceptibility in humanized PLN-R14del mice: modelling a European cardiomyopathy with global impact. Cardiovasc Res. 2022;118(15):3140–3150. doi: 10.1093/cvr/cvac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grote Beverborg N., Später D., Knöll R., et al. Phospholamban antisense oligonucleotides improve cardiac function in murine cardiomyopathy. Nat Commun. 2021;12(1):5180. doi: 10.1038/s41467-021-25439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eijgenraam T.R., Stege N.M., Oliveira Nunes Teixeira V., et al. Antisense therapy attenuates phospholamban p.(Arg14del) cardiomyopathy in mice and reverses protein aggregation. Int J Mol Sci. 2022;23(5):2427. doi: 10.3390/ijms23052427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ragone I., Barallobre-Barreiro J., Takov K., et al. SERCA2a protein levels are unaltered in human heart failure. Circulation. 2023;148(7):613–616. doi: 10.1161/CIRCULATIONAHA.123.064513. [DOI] [PMC free article] [PubMed] [Google Scholar]