Abstract
The proto-oncogene pp60c-Src (c-Src) is activated in many types of cancer and contributes to the transformed phenotype of the tumor, although its role is not yet fully understood. Here we report that active Src elevates the levels of β-catenin by enhancing cap-dependent translation. Src induces phosphorylation of the eukaryotic initiation factor 4E via the Ras/Raf/ERK pathway and the phosphorylation of its inhibitor 4E-BP1 via the PI3K/mTOR pathway. Activated Src enhances the accumulation of nuclear β-catenin and enhances its transcriptional activity, elevating target genes such as cyclin D1. This novel activation of the Wnt pathway by Src most probably contributes to the oncogenic phenotype of cancer cells.
Activation of the proto-oncoprotein pp60c-Src (c-Src) is the hallmark of many types of cancer, especially highly metastatic ones (12, 13, 22). Although many correlative studies suggest a connection between c-Src activation and tumor development and progression, there are only a few reports on the role of c-Src activation in tumor development in vivo. Studies published recently show that c-Src activation plays central roles in both metastasis of breast cancer (27) and the development of skin tumors (24) in vivo. Thus, the prevalence of activated c-Src in aggressive tumors is well documented, but the molecular aspects of its oncogenic activity are not yet fully understood. Recently, we reported that activated c-Src elevates the levels of the transcription factor hypoxia-inducible factor 1α (HIF-1α) by the enhancement of HIF-1α translation (15). We wanted to examine whether activated Src specifically affects the translation of HIF-1α or whether this effect is actually more general and affects other signaling proteins. We were stimulated by the report that inhibition of eukaryotic translation initiation factor 4E (eIF4E) by overexpression of its inhibitor, eIF4E-BP1, inhibits transformation by v-Src (37) and by the finding that overexpression of eIF4E can lead to cell transformation (17).
Cap-dependent initiation of translation is a highly regulated process which requires the coordinated action of several initiation factors, initiation complexes, and their inhibitors. The rate-limiting step is executed by the initiation factor eIF4E, which is involved in the binding of capped mRNA (10, 34, 44). This initiation factor is regulated by phosphorylation on serine 209, which enhances its affinity to the capped mRNA and thus enhances translation initiation (25, 44). It is also regulated by the inhibitory proteins eIF4E-BP1 and eIF4E-BP2, which bind to eIF4E, inhibiting its binding to eIF4G (9, 23). Phosphorylation of eIF4E-BP1 and eIF4E-BP2 induces their dissociation from eIF4E, allowing eIF4E to bind eIF4G (9, 23). eIF4E can then recruit capped mRNA to the initiation complex (23). It is well established that activated Src enhances both the Ras/Raf/extracellular signal-regulated kinase (ERK) and the phosphatidylinositol 3-kinase (PI3K)/mTOR pathways (13, 31). Also, v-Src transformation results in high levels of eIF4E phosphorylation (8). eIF4E can be activated by the Ras/Raf/ERK pathway (32, 49) through Mnk1 and Mnk2 and by PI3K through mTOR (11). Here we show that active Src enhances protein synthesis probably by the phosphorylation of the translation initiation factors eIF4E and eIF4E-BP through the PI3K-mTOR and ErkMAPK pathways. This enhancement results in the accumulation of certain proteins, such as β-catenin transcription factor. Consequently, the expression of downstream target genes, such as the β-catenin target cyclin D1, is also enhanced. We propose that the enhancement of cap-dependent translation by oncoproteins, especially protein tyrosine kinases (PTKs) like activated c-Src, up-regulate the synthesis of signaling proteins like HIF-1α and β-catenin and therefore enhance the transformed phenotype. The possible mechanism for the selective up-regulation is discussed.
MATERIALS AND METHODS
Cell culture.
NIH 3T3, SrcNIH, CSH12, and HEK-293 cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). HT29 (human colon carcinoma) and Saos-2 (human osteosarcoma) cells were grown in McCoy-5A medium supplemented with 10% FCS. All media were supplemented with penicillin and streptomycin.
Src kinase inhibitor.
4-Amino-5-(4-methylphenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP1), 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP2), and 4-amino-7-phenylpyrazol[3,4-d]pyrimidine (PP3) were used as described previously (38).
Transient transfections and reporter assays.
Transient transfections were performed either with Fugene 6 (according to the manufacturer's instructions [Roche]) or with CaPO4 (as described in reference 4).
To examine the effects of Src on β-catenin expression, CSH12 cells were transfected in six-well plates with 0.2 μg of Myc-β-catenin expression plasmid, 0.4 μg of pEGFP plasmid (for normalization) and 1 μg of kinase-dead (KD) or active Src plasmids, as indicated. In addition, 0.4 μg of HA-GSK3 plasmid and 0.2 μg of flag-Axin plasmids were added in order to prevent saturation of the β-catenin degradation machinery. 48 h after transfection, cells were lysed with sample buffer.
To examine if active Src, active MEK1 (Δ3 MEK1), or active PI3K (myristoylated PI3K) can counteract the inhibition of β-catenin expression by KD Src, CSH12 or HEK-293 cells were transfected as described above, with 0.2 μg of Myc-β-catenin expression plasmid, 0.4 μg of pEGFP plasmid (for normalization), 0.4 μg of HA-GSK3 plasmid, 0.2 μg (CSH12) or 0.3 μg (HEK-293) of flag-Axin plasmid, 0.5 μg of KD Src plasmid and 0.6 μg of either active Src, active MEK1 (Δ3 MEK1), or active PI3K (pCMV5-Myristoylated-HA-PI3K) plasmids as indicated. In all the reporter gene experiments, cells were lysed 48 h after transfection with reporter lysis buffer (Promega) according to the manufacturer's instructions, and luciferase activity was measured and normalized to β-galactosidase (β-Gal) activity. Transcription from the reporter containing repeats of the TCF/LEF element was measured using the Topflash/Fopflash reporter system (16). NIH 3T3, SrcNIH, and CSH12 cells (8 × 104 cells/well) were transfected with 1 μg of Topflash luciferase reporter or Fopflash luciferase reporter in duplicates. PP1 was added 24 h after transfection. Forty-eight hours after transfection, Topflash luciferase activity was normalized to Fopflash luciferase activity. Transcription from the cyclin D1 promoter was measured using the −1745-luc construct containing the promoter of cyclin D1 (19, 41). CSH12 cells were transfected with 0.5 μg of −1745-luc construct together with 0.5 μg of cytomegalovirus (CMV)-β-Gal (for normalization) and 1 μg of Src constructs as indicated in the figure legend. In order to examine if the effects of Src are mediated by the β-catenin pathway, CSH12 cells were transfected with 0.2 μg of −1745-luc construct and 0.2 μg of CMV-β-Gal, as well as one of the following: 1.8 μg pUSE(−), or 1 μg pUSE(−) plus 0.8 μg active Src, or 1 μg flag-Axin plus 0.8 μg active Src, or 1.2 μg pUSE(−) plus 0.6 μg KD Src, or 1.2 μg β-catenin plus 0.6 μg KD Src, as indicated in the figure. Luciferase activities were normalized to β-Gal activities. Every experiment was repeated three times, with duplicate samples each time. The figures show the averages from duplicate samples, from a representative experiment. To examine the effect of 4EBP1 on transcription from the Topflash/Fopflash system, HEK-293 cells were cotransfected with 1 μg of either Topflash or Fopflash, plus 1 μg of either HA-4EBP1 or pUSE(−) (empty plasmid). Cells were lysed after 48 h. Results were normalized as described above.
Immunoblotting.
Cells were washed 3 times with PBS, then lysed with sample buffer (10% glycerol, 0.2 M Tris HCl [pH 6.8], 5% β-mercaptoethanol, 3% sodium dodecyl sulfate [SDS]) and boiled for 5 min.
Lysates were loaded on Whatman 3MM paper squares. Paper squares were stained with Coomassie and washed five times for 6 min with destain solution (20% methanol 7% acetic acid). Staining was extracted from papers with 3% SDS, and protein amounts were determined using a bovine serum albumin calibration curve, reading the absorbance at 590 nm. The whole-cell lysate (WCL) was then subjected to SDS-PAGE and transferred to nitrocellulose. The membranes were blocked in low-fat (1%) milk diluted 1:20 into TBST (10 mM Tris HCl [pH 7.5], 50 mM NaCl, 0.1% Triton X-100) for 30 min, followed by incubation for 1.5 h with the primary antibody (indicated in figure legends). Membranes were then washed extensively with TBST and immunoreactive proteins were detected by incubation with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG; 1:10,000; Jackson Immuno Research) for detection of monoclonal antibodies or horseradish peroxidase-conjugated anti-rabbit or anti-goat IgG (1:10,000; Jackson Immuno Research) for detection of polyclonal antibodies. Proteins were visualized using enhanced chemiluminescence (ECL).
Antibodies.
Anti-Src (monoclonal antibody [MAb] 327), anti-actin (1:2,000; Santa Cruz), anti-phospho-serine 209 of eIF4E (p-eIF4E) (1:1,000; cell signaling), anti-phospho-Thr-70 of eIF4E BP1 (p-4E BP1) (1:1,000; cell signaling), anti-S6 (1:2,500; cell signaling), anti-eIF4E (1:1,000; cell signaling), anti-HIF-1α (1:250; Transduction Laboratories), anti-myc antibody (9E10; 1:1,000; Santa Cruz), anti-green fluorescent protein (GFP; 1:5,000; Santa Cruz), anti-β-catenin (1:6,000 [Transduction Laboratories] or 1:2,000 [Sigma]), anti-cyclin D1 (1:1,000; M20; Santa Cruz), anti-p27 (1:500; Transduction Laboratories), anti-Shc (1:2,000; Transduction Laboratories), and anti-α-tubulin (1:40,000; Sigma) were used.
Northern blotting.
Cells were grown on 10-cm dishes and treated with PP1 for 2 h or 24 h as indicated in the Results. RNA was prepared using Tri reagent (Sigma), and 10 μg total RNA were denatured and loaded on a 1% agarose gel containing 8% formaldehyde and ethidium bromide. After electrophoresis, the gel was filmed in order to verify loading of equal amounts of RNA.
Following capillary blotting onto a nylon membrane, and cross-linking, the blot was hybridized overnight at 42°C with 32P-labeled DNA probe, prepared with the Rediprime kit (Amersham). After three washes at 50°C with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS, the blot was exposed to film (Fuji).
Accumulation experiments.
Saos-2 cells (2 × 105) were seeded on six-well plates and 24 h later were treated with PP1 (20 μM), LY294002 (20 μM), PD98059 (50 μM), or rapamycin (50 nM; Calbiochem) for 30 min. Medium was then replaced with medium containing N-acetyl-Leu-Leu-Nle-CHO (ALLN; (200 μM; Calbiochem) as well as the inhibitors mentioned above for an additional 5 h. Cells were then lysed with sample buffer.
35S labeling.
To measure protein synthesis in the presence of inhibitors, CSH12 cells (2 × 105 cells/well) were seeded on six-well plates and 24 h later were starved for methionine and cysteine for 1 h in the presence of kinase inhibitors. Cells were then labeled with 35S-methionine and 35S-cysteine Promix (Amersham), 50 μCi/ml. Cells were labeled for 10 min at 37°C in the presence of the indicated inhibitors. Cells were then washed with phosphate-buffered saline (PBS) and lysed with sample buffer. Lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes by Western blotting. The membranes were exposed to film. After exposure, membranes were probed with anti-actin and anti-Src antibodies to verify loading of equal amounts of protein. To measure protein synthesis of long-lived proteins, CSH12 cells (2 × 105 cells/well) were seeded on six-well plates and 24 h later were starved for methionine and cysteine for 1 h in the presence of kinase inhibitors. Cells were then labeled with 35S-methionine and 35S-cysteine Promix (Amersham), 50 μCi/ml, at 37°C for 1 h in the presence of the indicated inhibitors. Cells were then washed with PBS and incubated for 2 or 4 h (as indicated in the figure) in medium containing excess (5 mM) nonradioactive l-methionine (Sigma). Cells were then washed with PBS and lysed with sample buffer. The lysates were loaded onto paper squares and the amount of radioactive label incorporated was determined, as described above. To detect the synthesis of β-catenin, NIH 3T3, SrcNIH, or CSH12 cells (2 × 106 cells/dish) were seeded on 10-cm dishes (Nunc). Twenty-four hours later, cells were washed with PBS and starved for methionine and cysteine as described above and labeled with 100 μCi/ml Promix for 45 min in the presence of 200 μM ALLN. Cells were then washed with PBS and lysed with RIPA buffer (20 mM HEPES [pH 7.4], 125 mM NaCl, 1% Nonidet P-40, 1% deoxycholate, 0.1% SDS, 5 mM NaF, 100 μM NaVO3, 1 mM EDTA, 2 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 5 μg/ml leupeptin). After Bradford protein quantification, β-catenin was immunoprecipitated using (4 μg/500 μg lysate) α-β-catenin antibody (Transduction Laboratories). After additional washes the beads were mixed with 2× sample buffer and boiled for 5 min. After Western blotting and blocking, membranes were exposed as described above. For detection of β-catenin protein, the membrane was probed with anti β-catenin antibody (1:6,000; Transduction Laboratories). For measuring of total protein synthesis rates, NIH 3T3, SrcNIH, and CSH12 cells (140,000 cells/well) were seeded on six-well plates (Nunc). Twenty-four hours later, cells were washed with PBS and the medium was replaced with methionine- and cysteine-deficient medium for 1 h. Medium was then replaced with medium containing 100 μCi/ml 35S-Met/Cys (Promix; Amersham) in the presence of 200 μM ALLN. Cells were labeled for 45 min at 37°C. Cells were then washed with PBS and lysed with sample buffer. To measure synthesis rates, lysates were loaded on 3MM Whatmann paper squares. Paper squares were stained with Coomassie blue and washed five times for 6 min each with destain solution (20% methanol, 7% acetic acid). Then the paper squares were dried, and radioactivity was counted with scintillation solution in a beta counter. Another set of identical samples was run in parallel; stain was extracted from these papers with 3% SDS, and protein amounts were determined from the absorbance at 590 nm, against a bovine serum albumin calibration curve.
Nuclear/cytoplasmic fractionation.
For nuclear/cytoplasmic fractionation we either used the NE-PER nuclear and cytoplasmic extraction kit (Pierce) according to the manufacturer instructions or followed a fractionation protocol for NIH 3T3 cells previously described in (47). Briefly, 106 NIH 3T3, SrcNIH and CSH12 cells were grown on 10-cm plates. A day later, cells were starved with medium containing 0.2% serum for 24 h and were then trypsinyzed and collected by centrifugation at 650 × g for 10 min in 4°C. Collected cells were washed three times with 1× PBS, suspended in buffer A (10 mM HEPES, pH 7.4, 5 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol, protease inhibitors), and centrifuged as described above. Cells were suspended in a five-cell pack volume in buffer A and homogenized with a Dounce homogenizer. These homogenates were used as WCL. WCL were then centrifuged at 1,000 × g for 10 min, and the supernatant constituted the cytoplasmic fraction. The pellets were washed three times with buffer B (10 mM HEPES, 10 mM NaCl, 1.5 mM MgCl2, 0.5% deoxycholate, 1% Tween 40, and protease inhibitors). In between washes, pellets were homogenized 5 times. Pellets were then centrifuged at 1,000 × g for 10 min and resuspended in sample buffer. Protein concentrations were determined as described above, and the fractions were subjected to Western analysis as described above.
Affinity precipitation.
For affinity precipitation, clarified cell extracts were incubated with Sepharose beads (Pharmacia) to which either control glutathione S-transferase (GST) protein or a GST-E-cadherin fusion protein was bound. After incubation at 4°C for 1 h the Sepharose beads were collected by centrifugation in a microcentrifuge, washed three times with immunoprecipitation buffer, and solubilized in Laemmli sample buffer for SDS-PAGE and Western immunoblot analysis. The GST fusion protein for E-cadherin consists of bacterial glutathione S-transferase fused in frame to the entire cytoplasmic domain of E-cadherin (30). The GST-E-cadherin fusion protein and the control GST protein were purified from bacterial lysates. For normalization, samples from cell lysates prior to pull down (WCL) were also run on SDS-PAGE and the membrane was probed with anti-tubulin and anti-β-catenin antibodies.
RESULTS
Active Src enhances protein synthesis.
To examine the role of activated Src in the regulation of protein synthesis, we measured overall protein synthesis in metabolic labeled CSH12 cells, under inhibition of Src. CSH12 are NIH 3T3 cells that overexpress the constitutively active HER1/HER2 chimeric receptor and therefore possess constitutively active c-Src (18, 28).
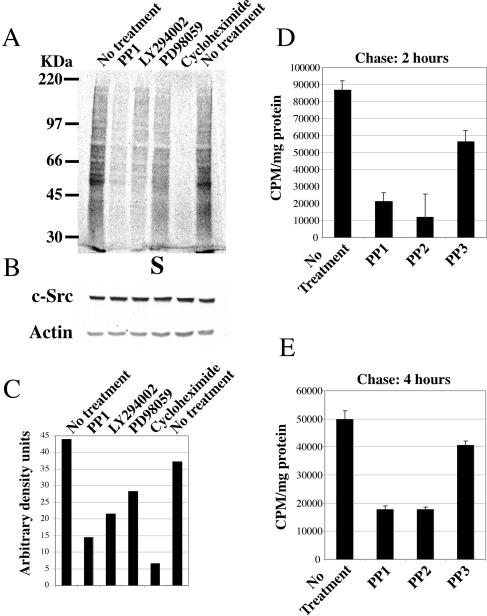
The Src kinase inhibitors PP1 and PP2 (Fig. 1A) inhibited overall protein synthesis threefold. PI3-K inhibitor LY294002, and cycloheximide had a similar inhibitory effect to that exerted by PP1. No change in the levels of c-Src or actin was detected (Fig. 1B). Figure 1C shows the quantitaion of the 35S labeling of CSH12 cells from Fig. 1A. Other cancer cell lines that are sensitive to Src inhibition yielded similar results, as described in (15). These cell lines include HT29 and Saos-2. The former is a colon cancer line, a cancer in which the role of activated Src is documented (6, 20, 22). The latter is an osteosarcoma line. The only phenotype of Src knock out mice is osteopetrosis, caused by defects in osteoclast function (45). This data, together with data accumulated in our lab (unpublished) indicate a role for Src in the survival of Saos-2 cells.
FIG. 1.
Src kinase activity regulates protein synthesis. (A) 35S labeling of CSH12 cells. Cells were seeded on six-well plates (Nunc) (2 × 105 cells/well) and after 24 h the cells were metabolically labeled with the 35S-labeled methionine-cysteine Promix as described in Materials and Methods. Equal amounts of protein from each sample were separated on SDS-10% PAGE, transferred to a nitrocellulose membrane, and the membrane was exposed to film. (B) After radioactive exposure, the membrane was probed with anti-Src (MAb 327) and anti-actin, to verify that equal amounts of protein were loaded. (C) Quantification of 35S labeling of CSH12 cells. Quantification of whole-lane mean density, using the MacBass 2.5 phosphorimager program (D) CSH12 cells (2 × 105 cells/well) were seeded on six-well plates. Twenty-four hours later, the cells were starved for methionine and cysteine for 1 h in the presence of PP1, PP2, or PP3 as indicated. The cells were then labeled with 35S-labeled methionine-cysteine Promix, 50 μCi/ml, at 37°C for 1 h, in the presence of the indicated inhibitors. Cells were then washed with PBS, and incubated for 2 or 4 h (E) in medium containing excess (5 mM) nonradioactive l-methionine. The cells were then washed with PBS and lysed with sample buffer. Radioactivity was measured as described in Materials and Methods. Error bars represent standard error of the mean of three independent experiments.
Since actin and Src itself are proteins with long half-lives, we wished to examine whether the effect of Src on protein synthesis depends on the length of the labeling, maybe reflecting the differences in protein turnover times. We therefore employed a pulse-chase procedure and treated the cells with a longer pulse (1 h) of 35S-labeled methionine-cysteine (Promix), followed by a chase for 2 h or 4 h in the presence of nonradioactive methionine (Fig. 1D and E). Short-lived proteins are degraded during the chase, so that only proteins that last longer than the chase time retain label. We observed that even after a chase of 4 h, there was much less residual 35S-labeled methionine-cysteine label in cells treated with the Src inhibitors PP1 and PP2 (Fig. 1E). Thus, the effect of Src on translation seems to be general, and influences the synthesis of both short and long-lived proteins.
Src activity regulates the phosphorylation of eIF4E, eIF4E-BP1 and S6 ribosomal protein.
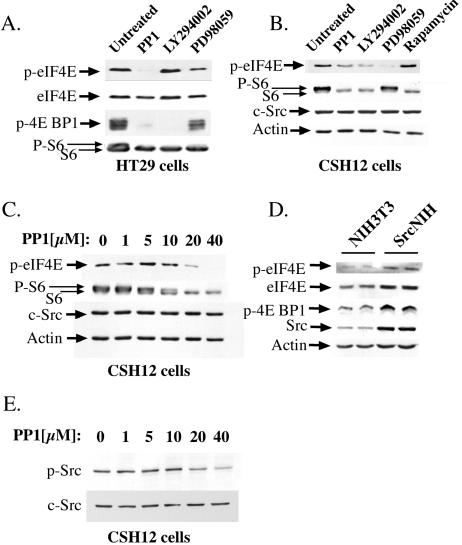
Since most mRNAs are translated in the cell by cap-dependent translation (10), we examined in more detail the relationship between cap-dependent translation and Src activation. Towards this end, we examined the effects of the Src kinase inhibitor PP1, on the phosphorylation status of the translation initiation factor eIF4E, its binding protein eIF4E-BP1, and S6 ribosomal protein. PP1 dramatically reduced the phosphorylation of eIF4E on serine 209, in HT29, Saos-2, and CSH12 cells (Fig. 2A through C and data not shown). The decrease in eIF4E phosphorylation is in correlation to the decrease in Src phosphorylation (Y416) in CSH12 cells (Fig. 2E). Moreover, PP1 strongly inhibited phosphorylation of eIF4E-BP1 on threonine 70, in HT29 and Saos-2 cells (Fig. 2A and data not shown). In addition, PP1 inhibited the phosphorylation of S6 protein in HT29, Saos-2, and CSH12 cells (Fig. 2A through C and data not shown). In SrcNIH cells, which are NIH 3T3 cells transformed with active Src (Y529F) (21), the levels of phosphorylated eIF4E-BP1 and eIF4E were higher than in the parental NIH 3T3 cell line (Fig. 2D). The higher levels of phosphorylated eIF4E in SrcNIH cells may be at least in part also accounted for by the elevated level eIF4E expression (Fig. 2D). These results strongly suggest that active Src up-regulates the activities of the components involved in the initiation of translation, and therefore has a strong effect on the rate of cap-dependent translation.
FIG. 2.
The effects of Src, MEK1, mTOR, and PI3-kinase inhibitors on the phosphorylation of eIF4E, eIF4E BP1 and S6 proteins. (A) HT29 cells were seeded on six-well plates (Nunc) (4 × 105 cells/well). Twenty-four hours later, the medium was replaced with medium containing the indicated inhibitors for 3 h (20 μM PP1, 20 μM LY294002 or 50 μM PD98059). Cells were then lysed with sample buffer and the lysates subjected to SDS-PAGE. After Western blotting, the membrane was probed with the following antibodies: anti-phospho-Ser-209 of eIF4E (p-eIF4E), anti-phospho-Thr-70 of eIF4E BP1 (p-4E BP1), anti-S6, and after stripping, anti-eIF4E. The higher band detected with anti-S6 represents its phosphorylated form. (B) CSH12 cells were seeded on six-well plates (2 × 105 cells/well; Nunc) and after 24 h cells were treated for 3 h with 20 μM PP1, 20 μM LY294002, 50 μM PD98059, or 50 nM rapamycin. Cells were then lysed with sample buffer. After Western blotting, the membrane was probed with anti- phospho-serine 209 of eIF4E, anti-S6, anti-Src and anti-actin. (C) CSH12 cells were seeded on six-well plates (2 × 105 cells/well; Nunc) and after 24 h cells were treated for 3 h with PP1 at the indicated concentrations. After Western blotting, the membrane was probed as in B. (D) NIH 3T3 and SrcNIH cells were seeded on six-well plates (2 × 105 cells/well; Nunc) and after 24 h were lysed. After Western blotting, the blot was probed as for panel A. (E) CSH12 cells were seeded on six-well plates and after 24 h were treated for 3 h with PP1 at the indicated concentrations. After Western blotting, the membrane was probed with anti-phospho-Src (Y416) and anti-Src.
The PI3K/mTOR and the MEK1/ERK pathways mediate the Src-induced enhancement of cap-dependent protein synthesis.
The PI3K/mTOR and the Ras/Raf/MEK1/ERK pathways, two central pathways downstream of active Src, mediate the contribution of Src to the transformed phenotype of cancer cells (31). The activation of both the PI3K/mTOR and the Ras/Raf/ERK pathways has been shown to enhance protein synthesis (11, 32, 34). Inhibition of PI3K by LY294002, or of MEK1 by PD98059, inhibited protein synthesis, as seen in Fig. 1A through C. Similar effects were seen in Saos-2 and HT29 cells treated with Wortmannin, another inhibitor of PI3K, or with rapamycin, an mTOR inhibitor (15). These pathways probably affect protein synthesis by inhibiting the phosphorylation of the translation initiation factor, eIF4E, and of its inhibitor, eIF4E-BP1 (Fig. 2). The PI3K inhibitor, LY294002, affected the phosphorylation of eIF4E on serine 209 in CSH12 cells but not in HT29 cells, whereas the MEK1 inhibitor, PD98059, inhibited phosphorylation at this site in both cell lines (Fig. 2A and B). LY294002 strongly inhibited the phosphorylation of eIF4E-BP1 on threonine 70, in both HT29 and Saos-2 cells (Fig. 2A and data not shown). The mTOR inhibitor, rapamycin, also inhibited phosphorylation at this site (data not shown). LY294002 and rapamycin inhibited the phosphorylation of the S6 protein—as did PD98059, albeit to a lesser extent—in HT29, Saos-2 and CSH12 cells (Fig. 2B and data not shown).
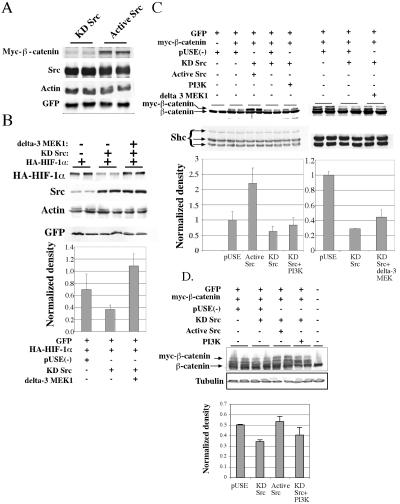
We have recently shown that Src enhances the translation of HIF-1α by enhancement of cap-dependent translation (15). We examined whether the protein levels of the transcription factor β-catenin is also affected by Src. Cotransfection of inactive Src (KD Src) together with β-catenin decreased the expression of this protein (Fig. 3A), while GFP (transfection control), Src or actin (loading controls) was unaffected. To examine if the MEK1/ERK and the PI3K pathways mediate the effects of Src on translation, we inhibited the synthesis of HA-HIF-1α and myc-β-catenin, by cotransfection with kinase-dead Src (KD-Src), and examined whether active MEK1 or PI3K could counteract the reduction in the levels of myc-β-catenin and HA-HIF-1α. We found that co-expression of active MEK1 (ΔMEK1) eliminated the inhibition of HA-HIF-1α caused by KD-Src (Fig. 3B). Active MEK1 partially counteracted the inhibition of myc-β-catenin expression by KD Src in CSH12 cells (Fig. 3C). Active PI3K had a very small, but reproducible, effect in counteracting the inhibition of myc-β-catenin expression by KD Src, both in CSH12 (Fig. 3C) and HEK-293 cells (Fig. 3D). These results suggest that MEK1 is downstream of Src in the enhancement of HIF-1α and of β-catenin synthesis. These results also indicate that the translation of β-catenin is activated by Src, through the Ras/Raf/MEK1/Erk pathway, which seems to be more prominent than the PI3K/mTOR pathway in this respect, and that neither of these pathways solely mediate the regulation of β-catenin translation by Src.
FIG. 3.
Active MEK1 overrides the translational inhibition caused by expression of kinase-dead Src. (A) CSH12 cells were transfected in six-well plates with 0.2 μg of Myc-β-catenin expression plasmid, 0.4 μg of HA-GSK3 plasmid, 0.2 μg of flag-Axin plasmid, 0.4 μg of pEGFP plasmid, and 1 μg of KD or active Src plasmids as indicated. 48 h after transfection, cells were lysed with sample buffer, the lysates were subjected to SDS-PAGE and, after Western blotting, were probed with anti-Myc antibody (9E10; 1:1,000; Santa Cruz), anti-GFP (1:5,000; Santa Cruz), anti-Src, and anti-actin as in Fig. 2. (B) CSH12 cells were grown in six-well plates and cotransfected with 50 ng of HA-HIF-1α expression plasmid and GFP expression plasmid, as well as empty plasmid, or plasmids coding for KD Src, or KD Src plus active MEK1 (ΔMEK1) as indicated. Forty-eight hours later, cells were lysed with sample buffer and after Western blotting the membrane was probed with anti-HIF-1α, anti-Src anti-GFP, and anti-actin. (C) CSH12 cells were grown on six-well plates and cotransfected with 200 ng of myc-β-catenin, 400 ng HA-GSK3β, 200 ng Flag-Axin, and GFP expression plasmids, as well as empty plasmid, KD Src plasmid, or KD Src plasmid (500 ng) plus active Src plasmid, active MEK1 (ΔMEK1) plasmid, or a myr-PI3K plasmid (600 ng) as indicated. Forty-eight hours later, cells were lysed with sample buffer and after Western blotting the membrane was probed with anti Shc (1:1,000; Santa Cruz) as a loading control and anti β-catenin (1:2,000; Santa Cruz). Results were normalized to Shc. Standard deviation between duplicates is shown. (D) HEK-293 cells were grown on six-well plates and cotransfected with 200 ng of myc-β-catenin, 400 ng HA-GSK3β, 300 ng Flag-Axin and 0.5 μg GFP expression plasmids, as well as 1.1 μg of empty plasmid (pUSE), or 500 ng KD Src plasmid, plus 600 ng of either empty plasmid, active Src plasmid, or a myr-PI3K plasmid, as indicated. Forty-eight hours later, cells were lysed with sample buffer and run on SDS-8% PAGE. After blotting, the membrane was probed with anti-β-catenin (1:6,000; Transduction Laboratories), and anti-α-tubulin (1:40,000; Sigma). Results were normalized to tubulin. Standard deviation between duplicates is shown.
Active Src enhances the synthesis and activity of β-catenin.
We examined the effect of active Src on the synthesis of β-catenin. We show that Src affects the rate of synthesis as well as the steady-state levels of β-catenin.
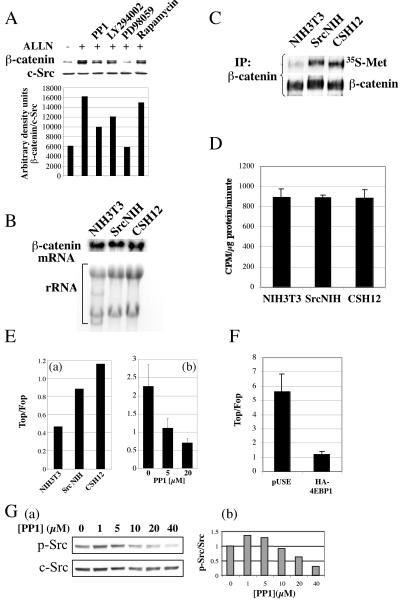
The transcription factor β-catenin, which is degraded by the ubiquitin-proteasome system, has a half-life of ∼20 min (26). The level of β-catenin protein is regulated by both its rates of synthesis and rate of its degradation. In order to distinguish between these two effects on the steady state level of β-catenin, we blocked the degradation of β-catenin by means of a proteasome inhibitor, ALLN. In the presence of ALLN, pharmacological inhibitors of Src, PI3K, and MEK1 prevented the accumulation of β-catenin (Fig. 4A). Thus, activated Src and its downstream pathways enhance the synthesis of β-catenin, rather than its stability. These results are supported by the finding that the incorporation of 35S-labeled methionine-cysteine into endogenous β-catenin was higher in SrcNIH and CSH12 cells than in the parental NIH 3T3 cell line (Fig. 4C). Total translation rates in these cell lines were examined in the same labeling period and were the same (Fig. 4D). We examined the mRNA levels of β-catenin in these cell lines and found no differences (Fig. 4B). Taken together, these results demonstrate that activation of Src elevates the rate of β-catenin protein synthesis.
FIG. 4.
Active Src enhances the synthesis of β-catenin protein and increases β-catenin dependent transcriptional activation. (A) Saos-2 cells (4 × 105 cells/well) were seeded on six-well plates (Nunc). After 24 h, the medium was replaced with medium lacking or containing an inhibitor (20 μM PP1, 20 μM LY294002, 50 μM PD98059, or 50 nM rapamycin) for 30 min. These media were then replaced with media containing the respective inhibitor and a proteasome inhibitor (200 μM ALLN; Calbiochem) for an additional 5 h. Cells were then lysed with sample buffer and after Western blotting were probed with anti-β-catenin and anti-c-Src as in Fig. 2. (B) NIH 3T3, SrcNIH, and CSH12 cells (106) were grown in 10-cm dishes. After 24 h RNA was extracted and 20 μg of total RNA were subjected to Northern blotting. The blot was hybridized with a probe for β-catenin (C) NIH 3T3, SrcNIH, and CSH12 cells were grown in 10-cm dishes. After 24 h the medium was replaced with methionine and cysteine deficient medium for 1 h, and later 100 μCi/ml of 35S-labeled Met-Cys Promix was added with 200 μM ALLN for 45 min as described in Materials and Methods. Proteins were extracted, size fractionated in SDS-PAGE, and blotted. The membrane was exposed to film and afterwards probed for β-catenin protein using anti-β-catenin antibody. (D) NIH 3T3, SrcNIH, and CSH12 cells were seeded in six-well plates (140,000 cells/well). Twenty-four hours later, cells were labeled with 35S-labeled methionine-cysteine Promix for 45 min and total protein synthesis rates were measured as described in Materials and Methods. Error bars represent duplicates from a representative experiment out of two independent experiments. (E) (a) NIH 3T3, SrcNIH, and CSH12 cells were transfected with Topflash or Fopflash encoding plasmids (see Materials and Methods) and Topflash activity was normalized to Fopflash activity. (b) CSH12 cells were transfected with Topflash- or Fopflash-encoding plasmids and 24 h later were treated with PP1 at the indicated concentrations for an additional 24 h. (F) HEK-293 cells were seeded on six-well plates (200,000 cell/well). Twenty-four hours later, cells werecotransfected with 1 μg/well of Topflash or Fopflash and 1 μg/well of either empty plasmid (pUSE) or HA-4EBP1. Forty-eight after transfection, Topflash activity was normalized to Fopflash activity. Error bars represent duplicates from a representative experiment out of two independent experiments. (G) (a) CSH12 cells were treated for 24 h with PP1 at the indicated concentration. After Western blotting, the membrane was probed with anti-phospho-Src (Y416) and anti-Src. (b) Phospho-Src band densities were quantitated using the NIH image 1.6 program, and the results were normalized to c-Src.
We next examined the effect of Src activity on transcriptional activation by β-catenin, using the Topflash/Fopflash reporter system. The Topflash reporter contains three Tcf/Lef/β-catenin DNA binding sites and a minimal promoter, while in the Fopflash reporter the three binding sites are mutated (16). Normalization of transcription (represented by luciferase activity) from the Topflash reporter to that from the Fopflash reporter yields the net effect of the Wnt/β-catenin pathway on transcription. In CSH12 and SrcNIH cells, β-catenin dependent transcriptional activation was higher than in the parental NIH 3T3 cells (Fig. 4E, panel a). Moreover, PP1, the Src kinase inhibitor, inhibited β-catenin dependent transcriptional activation in a dose-dependent manner (Fig. 4E, panel b). We also examined the dose response decrease in Src activity in CSH12 treated with PP1 in these times (24 h), and as Fig. 4G (panels a and b) shows, Src phosphorylation (Y416) decreased at 10 μM PP1.
We examined whether the eIF4E machinery is involved in the activation of β-catenin translation. Therefore, we cotransfected HEK-293 cells with Top/Fopflash reporters and either empty plasmid or a plasmid coding for eIF4E binding protein (HA-4EBP1). As shown in Fig. 4F, eIF4E binding protein inhibits β-catenin dependent transcription.
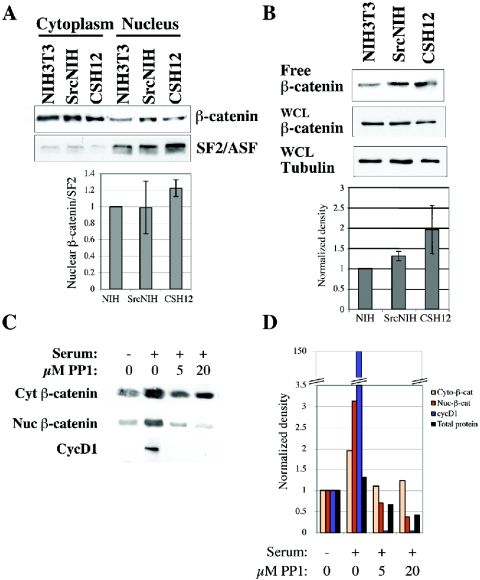
Most of the β-catenin protein is cytoplasmic, bound to cadherins in the membrane and is highly stable (5, 29, 30). Nuclear β-catenin represents a small portion of the β-catenin protein, but it is the fraction that participates in transcriptional activation (5, 29, 48). We therefore examined the levels of nuclear and free β-catenin in NIH 3T3, SrcNIH, and CSH12 cells. The levels of nuclear β-catenin were higher in SrcNIH and CSH12 cells than in NIH 3T3 cells, as determined by nuclear/cytoplasmic fractionation (Fig. 5A). Nuclear β-catenin levels were normalized to the levels of the nuclear splicing factor SF2/ASF (SF2/ASF is a splicing factor found only in the nucleus). However, this effect was modest and almost nonapparent in SrcNIH cells. Therefore we measured the fraction of β-catenin not bound to cadherins (“free β-catenin”), by pull-down with a GST-E-cadherin cytoplasmic tail (30). The levels of free β-catenin were normalized to tubulin levels in the whole-cell lysate. As can be seen in Fig. 5B, we observed a modest increase in free β-catenin levels in SrcNIH and CSH12 cells as compared to NIH 3T3 cells, similar to the results obtained in the cell fractionation experiments. Taken together, these results strongly suggest that Src activity regulates the level of free transcriptionally active β-catenin.
FIG. 5.
Src activity affects nuclear or free β-catenin levels. (A) NIH 3T3, SrcNIH, and CSH12 cells were seeded (106 cells/10-cm plate) and 24 h later were starved for 48 h in medium containing 0.2% serum. Cells were then lysed and fractionated into cytoplasmic and nuclear fractions as described in Materials and Methods. After Western blotting, the membrane was probed with the indicated antibodies (see Materials and Methods). Results were normalized to SF2/ASF nuclear marker levels. Error bars represent standard error of the mean of three independent experiments. (B) NIH 3T3, SrcNIH, and CSH12 cells were seeded and starved as described for panel A. Cells were lysed with RIPA buffer and cleared lysates were affinity-purified with GST-E-cadherin cytoplasmic tail (see Materials and Methods). The beads were boiled in sample buffer, and after Western blotting, the membrane was probed with the anti β-catenin antibody. Samples from whole-cell lysates (prior to pull-down) were also subjected to Western analysis, and the membrane was probed with anti-β-catenin and anti-α-tubulin antibodies. Results were normalized either to tubulin or to Shc. Error bars represent standard error of the mean of three independent experiments. (C) SrcNIH cells were seeded (6 × 105 cells) on 10-cm plates and starved (0.2% serum) for 24 h. Cells were activated with medium containing 10% serum for 24 h in the absence or presence of the Src kinase inhibitor PP1 at the indicated concentrations. Cells were lysed and fractionated into cytoplasmic and nuclear fractions using the NE-PER nuclear and cytoplasmic extraction kit (Pierce) according to the manufacturer's instructions. Equal volumes of each sample were loaded and subjected to SDS-PAGE. Protein quantities in each sample were determined and are shown in the graph. After Western blotting, the membrane was probed with antibodies to cyclin D1 or β-catenin. (D) The density of the bands was measured using NIH IMAGE 1.6 program and the graphs show the fold change in the density compared to the levels before activation (time zero). Shown is a representative experiment out of two independent experiments.
To further examine the effect of Src on β-catenin translation, we examined if Src activity affects β-catenin protein levels in cells activated with serum. Interestingly, Src inhibition blocked the accumulation of nuclear β-catenin but had a very small effect on the stable cytoplasmic pool of β-catenin (Fig. 5C and D). The β-catenin target gene cyclin D1 was induced after serum addition, and was inhibited almost completely upon PP1 addition (Fig. 5C).
We also examined the effect of Src activity on another protein, IκB, and showed that active Src enhanced its translation, while KD Src or PP1 inhibited its translation (see Fig. S1A through C in the supplemental material). As for HIF-1α (15) and β-catenin (see below), active Src had no effect on IκB half-life (see Fig. S1D in the supplemental material) or mRNA levels (see Fig. S1E in the supplemental material).
Active Src up-regulates target genes of β-catenin.
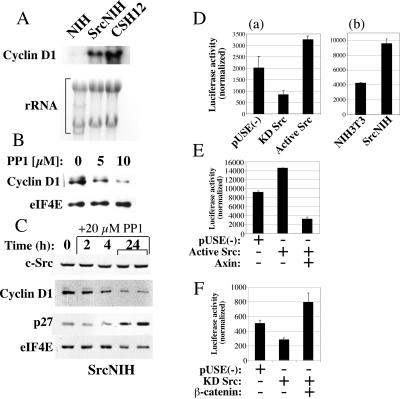
One of the best-known target genes of β-catenin is cyclin D1 (41, 46). Cyclin D1 controls the entry of cells into the cell cycle and is overexpressed in many types of cancer (35, 40, 46). We find that levels of cyclin D1 mRNA were higher in CSH12 and SrcNIH cells than in the parental NIH 3T3 cells (Fig. 6A). Treatment of SrcNIH cells with the Src kinase inhibitor PP1 reduced the levels of cyclin D1 protein in a dose-dependent manner (Fig. 6B). PP1 treatment had a rapid effect on cyclin D1 protein levels, which were reduced by 50% within 2 to 4 h, subsequent to the addition of the inhibitor, demonstrating the fast effect of Src inhibition on cyclin D1 levels (Fig. 6C). The level of p27 protein, an inhibitor of the cell cycle, was elevated after PP1 treatment (Fig. 6C). This may be accounted for by p27 being translated independently of the eIF4E machinery, by an internal ribosome entry site sequence at the 5′ untranslated region (UTR) of p27 mRNA (23). However, it was previously shown that v-Src down-regulates p27 by inhibition of transcription and by MEK-dependent proteolysis (36). The rise in cyclin D1 levels is not due to an increase in protein stability, since the half-life of cyclin D1 in NIH 3T3 is identical to the half-life in CSH12 cells (data not shown).
FIG. 6.
Src elevates the transcription of the β-catenin target gene, cyclin D1. (A) NIH 3T3, SrcNIH, and CSH12 cells (106) were grown in 10 cm dishes. After 24 h, total RNA was extracted and 20 μg of RNA were subjected to Northern blotting. The blot was hybridized with a probe for cyclin D1. (B) CSH12 cells (2 × 105/well) were seeded and treated with PP1 at the indicated concentrations for 24 h. After Western blotting the membrane was probed with anti-cyclin D1 and anti-eIF4E as in Fig. 2. (C) SrcNIH cells (2 × 105/well) were seeded and treated with PP1 for the indicated times (duplicates are shown at 24 h). After Western blotting, the membrane was probed with anti-cyclin D1, anti-eIF4E, anti c-Src, and anti p27. (D) (a) CSH12 cells were cotransfected with encoding luciferase reporter tethered to the cyclin D1 promoter (−1,745-bp) CMV-β-Gal control plasmid and an empty vector [pUSE(−)] or plasmids encoding the Src mutants, as indicated. Luciferase activity was normalized to β-Gal activity. Experiments were repeated three times, with duplicates in each experiment. (b) The cyclin D1 promoter (−1,745-bp)-luc reporter plasmid was transfected into NIH 3T3 and SrcNIH cells and the luciferase activity was normalized to that of β-Gal. (E) CSH12 cells were cotransfected with the cyclin D1 promoter (−1745 bp)-luc reporter, CMV-β-Gal control plasmid, and one or a pair of the following: luciferase and β-Gal empty plasmid [pUSE(−)], active Src, or active Src and axin. (F) In another experiment, cells were cotransfected with the reporters and one or a pair of the following as indicated: empty plasmid [pUSE(−)], KD Src, and β-catenin. Forty-eight hours after transfection luciferase activity was measured and normalized to β-Gal activity. All samples were prepared in duplicates. Shown are results derived from three independent experiments.
To look directly at the effects of activated Src on transcription from the cyclin D1 promoter, we employed a construct in which a luciferase reporter was fused to the cyclin D1 promoter. Active Src enhanced transcription from the cyclin D1 promoter, whereas KD Src inhibited transcription (Fig. 6D, panel a). In SrcNIH cells, the transcription from the cyclin D1 promoter was higher than in the parental NIH 3T3 cells (Fig. 6D, panel b). Moreover in CSH12 cells, cotransfection with Axin, which induces the degradation of β-catenin, counteracted the activation of the cyclin D1 promoter by active Src (Fig. 6E). Furthermore, coexpression of β-catenin bypassed the inhibition by KD Src of transcription from the cyclin D1 promoter (Fig. 6F). These results suggest that activated Src elevates the transcription and expression of a β-catenin target gene, the cyclin D1 gene, an important contributor to proliferation and transformation. Other mechanisms contribute also to Src mediated induction of cyclin D1, as it was shown by others that v-Src induces cyclin D1 expression in a Stat3-dependent manner (42).
DISCUSSION
In this study we show for the first time that persistent Src activation results in enhanced rates of protein synthesis, leading to elevated levels of expression of β-catenin. The ErkMAPK pathway has been implicated in the regulation of translation in various systems (39). Many growth factors, cytokines, and oncogenes can activate translation through this pathway, as well as through the PI3K/mTOR pathway (for a review, see reference 11). Here we show that the effect of activated Src is mediated by the PI3K/mTOR and the MEK1/ERKMAPK pathways (Fig. 1A, 2, and 3). The recruitment of capped mRNA by eIF4E is considered to be a rate-limiting step in cap-dependent initiation of translation. Our results show that eIF4E status is controlled by Src activity both through phosphorylation of eIF4E itself, as reported for v-Src transformed cells (8), and through phosphorylation of eIF4E-BP (Fig. 2). We found that inhibition of Src activity resulted in a three- to fivefold inhibition of protein translation (Fig. 1).
We found that Src activates the translation of β-catenin (Fig. 4). The levels of both nuclear β-catenin and free β-catenin (not bound to cadherin) were modestly higher in cells that possess high activity of Src (Fig. 5A and B). Accumulation of nuclear β-catenin (which is the form of β-catenin that is active in transcription) after serum stimulation was inhibited by PP1 (Fig. 5C). This inhibition probably accounts for the inhibitory effect of PP1 on the transcriptional activity of β-catenin (Fig. 4E). Both the inhibitions of eIF4E phosphorylation (Fig. 2) and beta catenin dependent transcripion (Fig. 4E) as a result of PP1 were in correlation with the decrease of Src phosphorylation at these times of treatment.
The elevation in β-catenin levels in the presence of active Src, in cell lines which possess high activity of Src, as well as the decrease in β-catenin levels, as a result of pharmacological inhibitors of Src and its downstream pathways, stems from regulation of its rate of synthesis (Fig. 4C) and is not a result of changes in transcription or stability (Fig. 4A and B).
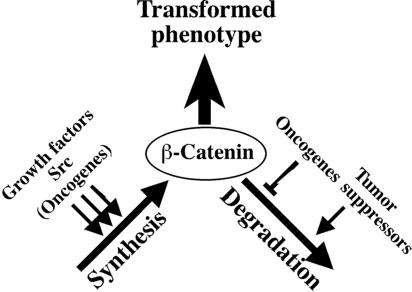
Our results are best accounted for by a new mechanism for the regulation of β-catenin and its target genes. Until now, the only known mechanism for regulating the amount of β-catenin protein was by the control of its degradation (14). Here we have shown that Src activation leads to enhanced synthesis of β-catenin, which in turn would be expected to lead to enhanced transcription of β-catenin target genes. Indeed, we found that the increased synthesis of β-catenin in cells possessing activated Src led to activation of transcription from a reporter containing the TCF/LEF/β-catenin binding elements (Fig. 4E), as well as activation of transcription of cyclin D1, a β-catenin target (Fig. 6). Another target gene of the Wnt signaling pathway is c-myc. Similarly to cyclin D1, c-Myc levels are higher in cells that possess high Src activity (data not shown). Src was also shown to regulate myc expression by mechanisms that include Stat3 (2), but also through Abl, Shc, and Vav2 (reviewed in reference 3). Clearly, the regulation of cyclin D1 and c-myc by Src depends on more than one mechanism.
We propose that Src increases β-catenin translation through the activation of the eIF4E machinery by both the PI3K/mTOR and Ras/Raf/MEK1/ERK pathways. In cell lines that possess high activity of Src, the levels of phosphorylation of eIF4E and its inhibitor, 4EBP1, were higher. Furthermore, we show that both PI3K and active MEK are necessary for the restoration of β-catenin levels in the presence of KD Src, though the Ras/Raf/MEK1/ERK pathway seems to be more prominent. Finally, 4EBP1, the inhibitor of eIF4E, reduces activity from the Top/Fopflash system (Fig. 4F), which indicate that indeed β-catenin translation is under the control of eIF4E machinery. The involvement of the Wnt signaling pathway in cancer has been amply demonstrated (1, 7). Thus, the activation of β-catenin/Wnt signaling by Src could play an important role in transformation (Fig. 7).
FIG. 7.
Src contributes to the regulation of β-catenin by enhancing its synthesis. The steady state levels of β-catenin protein are determined by the balance between the rates of degradation and synthesis. Any protein or oncogene that enhances translation, or stimulus that inhibits degradation, will elevate the levels of β-catenin, which contributes to the transformed phenotype.
Our results imply that Src up-regulates the signaling proteins Hif1α (15), β-catenin, and IκB (see the supplemental material). The common mechanism may be the preferential recruitment of existing mRNA molecules coding for these proteins to polysomes by the activated translational machinery. Increased cap-dependent translation by the activation of Src and presumably of other protein tyrosine kinases can therefore lead to transformation. In support of this hypothesis, it has been reported that enhancement of cap-dependent translation by overexpression of the eIF4E protein can transform cells (17, 43) and that inhibition of cap-dependent translation, by the expression of eIF4E-BP, can reverse transformation by various oncogenes (37). Furthermore, a recent study published while this work was under revision shows that Ras and Akt activation contribute to the transformed phenotype of glioblastoma by the differential recruitment of existing mRNAs to polysomes. The mRNAs most affected by this differential recruitment regulate growth, transcription, cell-cell interaction, and morphology (33). Our findings show that Src enhances cap-dependent translation of signaling proteins like β-catenin, IκB (this study), and HIF-1α (12). This activity of Src may play a central role in transformation.
Supplementary Material
Acknowledgments
We thank Eli Keshet for fruitful discussions and reagents; Yinon Ben-Neria (Haddasah Medical School, The Hebrew University of Jerusalem) for the Myc-β-catenin, HA-IκBα, Flag-Axin, and HA-GSK3 plasmids; and Nahum Sonenberg (McGill University, Montreal, Quebec, Canada) for eIF4E and 4EBP1 expression plasmids.
This study was partially supported by a grant from the Israel Science Foundation (ISF), Jerusalem, Israel.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bienz, M., and H. Clevers. 2000. Linking colorectal cancer to Wnt signaling. Cell 103:311-320. [DOI] [PubMed] [Google Scholar]
- 2.Bowman, T., M. A. Broome, D. Sinibaldi, W. Wharton, W. J. Pledger, J. M. Sedivy, R. Irby, T. Yeatman, S. A. Courtneidge, and R. Jove. 2001. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA 98:7319-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bromann, P. A., H. Korkaya, and S. A. Courtneidge. 2004. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 23:7957-7968. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conacci-Sorrell, M., J. Zhurinsky, and A. Ben-Ze'ev. 2002. The cadherin-catenin adhesion system in signaling and cancer. J. Clin. Investig. 109:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis, L. M., C. A. Staley, W. Liu, R. Y. Fleming, N. U. Parikh, C. D. Bucana, and G. E. Gallick. 1998. Down-regulation of vascular endothelial growth factor in a human colon carcinoma cell line transfected with an antisense expression vector specific for c-src. J. Biol. Chem. 273:1052-1057. [DOI] [PubMed] [Google Scholar]
- 7.Fodde, R., R. Smits, and H. Clevers. 2001. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer 1:55-67. [DOI] [PubMed] [Google Scholar]
- 8.Frederickson, R. M., K. S. Montine, and N. Sonenberg. 1991. Phosphorylation of eukaryotic translation initiation factor 4E is increased in Src-transformed cell lines. Mol. Cell. Biol. 11:2896-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 11.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 12.Irby, R. B., W. Mao, D. Coppola, J. Kang, J. M. Loubeau, W. Trudeau, R. Karl, D. J. Fujita, R. Jove, and T. J. Yeatman. 1999. Activating SRC mutation in a subset of advanced human colon cancers. Nat. Genet. 21:187-190. [DOI] [PubMed] [Google Scholar]
- 13.Irby, R. B., and T. J. Yeatman. 2000. Role of Src expression and activation in human cancer. Oncogene 19:5636-5642. [DOI] [PubMed] [Google Scholar]
- 14.Kang, D. E., S. Soriano, M. P. Frosch, T. Collins, S. Naruse, S. S. Sisodia, G. Leibowitz, F. Levine, and E. H. Koo. 1999. Presenilin 1 facilitates the constitutive turnover of beta-catenin: differential activity of Alzheimer's disease-linked PS1 mutants in the beta-catenin-signaling pathway. J. Neurosci. 19:4229-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karni, R., Y. Dor, E. Keshet, O. Meyuhas, and A. Levitzki. 2002. Activated pp60c-Src leads to elevated hypoxia-inducible factor (HIF)-1alpha expression under normoxia. J. Biol. Chem. 277:42919-42925. [DOI] [PubMed] [Google Scholar]
- 16.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 17.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J., T. J. Dull, I. Lax, J. Schlessinger, and A. Ullrich. 1989. HER2 cytoplasmic domain generates normal mitogenic and transforming signals in a chimeric receptor. EMBO J. 8:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, R. J., C. Albanese, R. J. Stenger, G. Watanabe, G. Inghirami, G. K. Haines III, M. Webster, W. J. Muller, J. S. Brugge, R. J. Davis, and R. G. Pestell. 1999. pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J. Biol. Chem. 274:7341-7350. [DOI] [PubMed] [Google Scholar]
- 20.Li, S., S. Ke, and R. J. Budde. 1996. The C-terminal Src kinase (Csk) is widely expressed, active in HT-29 cells that contain activated Src, and its expression is downregulated in butyrate-treated SW620 cells. Cell Biol. Int. 20:723-729. [PubMed] [Google Scholar]
- 21.Lin, P. H., S. Shenoy, T. Galitski, and D. Shalloway. 1995. Transformation of mouse cells by wild-type mouse c-Src. Oncogene 10:401-405. [PubMed] [Google Scholar]
- 22.Mao, W., R. Irby, D. Coppola, L. Fu, M. Wloch, J. Turner, H. Yu, R. Garcia, R. Jove, and T. J. Yeatman. 1997. Activation of c-Src by receptor tyrosine kinases in human colon cancer cells with high metastatic potential. Oncogene 15:3083-3090. [DOI] [PubMed] [Google Scholar]
- 23.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3:707-716. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto, T., J. Jiang, K. Kiguchi, L. Ruffino, S. Carbajal, L. Beltran, D. K. Bol, M. P. Rosenberg, and J. DiGiovanni. 2003. Targeted expression of c-Src in epidermal basal cells leads to enhanced skin tumor promotion, malignant progression, and metastasis. Cancer Res. 63:4819-4828. [PubMed] [Google Scholar]
- 25.Minich, W. B., M. L. Balasta, D. J. Goss, and R. E. Rhoads. 1994. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc. Natl. Acad. Sci. USA 91:7668-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munemitsu, S., I. Albert, B. Rubinfeld, and P. Polakis. 1996. Deletion of an amino-terminal sequence beta-catenin in vivo and promotes hyperphosporylation of the adenomatous polyposis coli tumor suppressor protein. Mol. Cell. Biol. 16:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myoui, A., R. Nishimura, P. J. Williams, T. Hiraga, D. Tamura, T. Michigami, G. R. Mundy, and T. Yoneda. 2003. C-SRC tyrosine kinase activity is associated with tumor colonization in bone and lung in an animal model of human breast cancer metastasis. Cancer Res. 63:5028-5033. [PubMed] [Google Scholar]
- 28.Osherov, N., and A. Levitzki. 1994. Epidermal-growth-factor-dependent activation of the src-family kinases. Eur. J. Biochem. 225:1047-1053. [DOI] [PubMed] [Google Scholar]
- 29.Papkoff, J. 1997. Regulation of complexed and free catenin pools by distinct mechanisms. Differential effects of Wnt-1 and v-Src. J. Biol. Chem. 272:4536-4543. [PubMed] [Google Scholar]
- 30.Papkoff, J., B. Rubinfeld, B. Schryver, and P. Polakis. 1996. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol. Cell. Biol. 16:2128-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penuel, E., and G. S. Martin. 1999. Transformation by v-Src: Ras-MAPK and PI3K-mTOR mediate parallel pathways. Mol. Biol. Cell 10:1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyronnet, S. 2000. Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1. Biochem. Pharmacol. 60:1237-1243. [DOI] [PubMed] [Google Scholar]
- 33.Rajasekhar, V. K., A. Viale, N. D. Socci, M. Wiedmann, X. Hu, and E. C. Holland. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12:889-901. [DOI] [PubMed] [Google Scholar]
- 34.Raught, B., and A. C. Gingras. 1999. eIF4E activity is regulated at multiple levels. Int. J. Biochem. Cell Biol. 31:43-57. [DOI] [PubMed] [Google Scholar]
- 35.Ridley, A. J. 2001. Cyclin' round the cell with Rac. Dev. Cell 1:160-161. [DOI] [PubMed] [Google Scholar]
- 36.Riley, D., N. O. Carragher, M. C. Frame, and J. A. Wyke. 2001. The mechanism of cell cycle regulation by v-Src. Oncogene 20:5941-5950. [DOI] [PubMed] [Google Scholar]
- 37.Rousseau, D., A. C. Gingras, A. Pause, and N. Sonenberg. 1996. The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene 13:2415-2420. [PubMed] [Google Scholar]
- 38.Schindler, T., F. Sicheri, A. Pico, A. Gazit, A. Levitzki, and J. Kuriyan. 1999. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell 3:639-648. [DOI] [PubMed] [Google Scholar]
- 39.Servant, M. J., E. Giasson, and S. Meloche. 1996. Inhibition of growth factor-induced protein synthesis by a selective MEK inhibitor in aortic smooth muscle cells. J. Biol. Chem. 271:16047-16052. [DOI] [PubMed] [Google Scholar]
- 40.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 41.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinibaldi, D., W. Wharton, J. Turkson, T. Bowman, W. J. Pledger, and R. Jove. 2000. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene 19:5419-5427. [DOI] [PubMed] [Google Scholar]
- 43.Smith, M. R., M. Jaramillo, Y. L. Liu, T. E. Dever, W. C. Merrick, H. F. Kung, and N. Sonenberg. 1990. Translation initiation factors induce DNA synthesis and transform NIH 3T3 cells. New Biol. 2:648-654. [PubMed] [Google Scholar]
- 44.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell. Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 45.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 46.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 47.Tsuchiya, N., H. Fukuda, T. Sugimura, M. Nagao, and H. Nakagama. 2002. LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur. J. Biochem. 269:2927-2933. [DOI] [PubMed] [Google Scholar]
- 48.van Es, J. H., N. Barker, and H. Clevers. 2003. You Wnt some, you lose some: oncogenes in the Wnt signaling pathway. Curr. Opin. Genet. Dev. 13:28-33. [DOI] [PubMed] [Google Scholar]
- 49.Waskiewicz, A. J., J. C. Johnson, B. Penn, M. Mahalingam, S. R. Kimball, and J. A. Cooper. 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19:1871-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.