Summary
Background
Cholelithiasis-induced acute cholangitis (CIAC) is an acute inflammatory disease with poor prognosis. This study aimed to create machine-learning (ML) models to predict the outcomes of patients with CIAC.
Methods
In this retrospective cohort and ML study, patients who met the both diagnosis of ‘cholangitis’ and ‘calculus of gallbladder or bile duct’ according to the International Classification of Disease (ICD) 9th revision, or met the diagnosis of ‘calculus of bile duct with acute cholangitis with or without obstruction’ according to the ICD 10th revision during a single hospitalization were included from the Medical Information Mart for Intensive Care database, which records patient admissions to Beth Israel Deaconess Medical Center, MA, USA, spanning June 1, 2001 to November 16, 2022. Patients who were neither admitted in an emergency department nor underwent biliary drainage within 24 h after admission, had an age of less than 18, or lost over 20% of the information were excluded. Nine ML methods, including the Logistic Regression, eXtreme Gradient Boosting (XGBoost), Light Gradient Boosting Machine, Adaptive Boosting, Decision Tree, Gradient Boosting Decision Tree, Gaussian Naive Bayes, Multi–Layer Perceptron, and Support Vector Machine were applied for prediction of in-hospital mortality, re-admission within 30 days after discharge, and mortality within 180 days after discharge. Patients from Zhongda Hospital affiliated to Southeast University in China between January 1, 2019 and July 30, 2023 were enrolled as an external validation set. The area under the receiver operating characteristic curve (AUROC) was the main index for model performance assessment.
Findings
A total of 1156 patients were included to construct models. We performed stratified analyses on all patients, patients admitted to the intensive care unit (ICU) and those who underwent biliary drainage during ICU treatment. 13–16 features were selected from 186 variables for model training. The XGBoost method demonstrated the most optimal predictive efficacy, as evidenced by training set AUROC of 0.996 (95% CI NaN–NaN) for in-hospital mortality, 0.886 (0.862–0.910) for re-admission within 30 days after discharge, and 0.988 (0.982–0.995) for mortality within 180 days after discharge in all patients, 0.998 (NaN–NaN), 0.933 (0.909–0.957), and 0.988 (0.983–0.993) in patients admitted to the ICU, 0.987 (0.970–0.999), 0.908 (0.873–0.942), and 0.982 (0.971–0.993) in patients underwent biliary drainage during ICU treatment, respectively. Meanwhile, in the internal validation set, the AUROC reached 0.967 (0.933–0.998) for in-hospital mortality, 0.589 (0.502–0.677) for re-admission within 30 days after discharge, and 0.857 (0.782–0.933) for mortality within 180 days after discharge in all patients, 0.963 (NaN–NaN), 0.668 (0.486–0.851), and 0.864 (0.757–0.970) in patients admitted to the ICU, 0.961 (0.922–0.997), 0.669 (0.540–0.799), and 0.828 (0.730–0.925) in patients underwent biliary drainage during ICU treatment, respectively. The AUROC values of external validation set consisting of 61 patients were 0.741 (0.725–0.763), 0.812 (0.798–0.824), and 0.848 (0.841–0.859), respectively.
Interpretation
The XGBoost models could be promising tools to predict outcomes in patients with CIAC, and had good clinical applicability. Multi-center validation with a larger sample size is warranted.
Funding
The Technological Development Program of Nanjing Healthy Commission, and Zhongda Hospital Affiliated to Southeast University, Jiangsu Province High-Level Hospital Construction Funds.
Keywords: Acute cholangitis, Cholelithiasis, MIMIC, XGBoost, Prognosis, Prediction model
Research in context.
Evidence before this study
Cholelithiasis-induced acute cholangitis (CIAC) is an acute inflammatory disease with poor prognosis. Although researches on the prognostic risk factors of CIAC have become increasingly profound, there is currently no systematic model to integrate these risk factors and accurately predict various outcomes. We searched PubMed and the Cochrane Library for peer reviewed articles published up to July 15, 2024 using the search terms “(CIAC OR cholelithiasis-induced acute cholangitis OR acute cholangitis) and (prediction model)”, with no language restriction. We could not find any effective models and clinical application tools for predicting the outcomes of patients with CIAC.
Added value of this study
To the best of our knowledge, it is the first time that applicable models based on the eXtreme Gradient Boosting (XGBoost) were developed for predicting in-hospital mortality, re-admission within 30 days after discharge, and mortality within 180 days after discharge in patients with CIAC, and their performance was validated in internal and external validation cohorts. These models have been translated into convenient webpages to facilitate the utility of clinicians. In addition, several laboratory indicators were proposed for the first time to correlate with prognosis of CIAC.
Implications of all the available evidence
Our XGBoost models firstly built a series of user–friendly online prediction platforms that can track multiple short- or long-term clinical outcomes in patients with CIAC, which warrant further validation with multi-center cohorts and a larger sample size.
Introduction
Acute cholangitis (AC) is an infectious disease arising from either partial or complete obstruction within the biliary system. According to recent international clinical guidelines,1 the diagnostic parameters of AC was considered to be associated with several key elements: fever, jaundice, indicators of systemic inflammation, abnormal hepatic function, radiological anomalies, and/or biliary obstruction. Globally, cholelithiasis afflicts approximately 10–15% of the adult populace,2 with one in five of them encountering gallstone-related complications, while the remainder remain largely asymptomatic.3 AC represents a consequential complication of cholelithiasis, with the latter constituting the main etiological factor for AC onset.4 Some scholars found that over half of the patients admitted to the Intensive Care Unit (ICU) due to severe acute cholangitis (SAC) were caused by cholelithiasis (53%), followed by tumors (22%).5
AC occurs rapidly and may progress to SAC within a brief timeframe. Sepsis occurs in approximately 10–29% of patients with AC,6 and 5% of them will develop septic shock.7 Therefore, adequate antibiotic therapy to mitigate the occurrence of sepsis is a key step in the management of patients with acute cholangitis.8 Moreover, timely intervention for restoring biliary drainage, particularly when administered within 48 h of obstruction onset, significantly helps patient prognosis.5,9 Nevertheless, despite favorable responses to antibiotic treatment and effective biliary drainage in most cases, contemporary researches underscore a 7-day mortality rate of up to 10% in SAC cases, with a 30-day in-hospital mortality rate reaching 36.25%.10 Also, re-admission within 30 days after discharge and mortality within 180 days after discharge are other two main variables for assessing prognosis of SAC.11,12
Taking the disease severities of AC into consideration, there have been increasing number of researches for the aims of predicting prognostic outcomes of AC. Lina Pan et al. devised a nomogram for predicting in-hospital mortality within 30 days among patients with AC admitted to the ICU.13 This nomogram incorporated factors such as oxygen saturation (SpO2), Glasgow Coma Scale (GCS), the ratio of aspartate aminotransferase (AST) to alanine aminotransferase (ALT), potassium concentration, serum albumin, blood glucose, partial thromboplastin time (PTT), and the presence of peripheral vascular disease. Similarly, Qingqing Liu et al. developed a risk-prediction nomogram comprised five variables for sepsis in patients with AC.14 In addition, O Inan et al. conducted a study in line with the Tokyo severity grading system and identified several factors correlated with clinical outcomes in geriatric patients.15 These studies highlighted the identification of various factors that contribute to the prognosis of AC, but the model algorithms used in the researches may not be able to meet the precision requirements of modern diagnosis and treatment. In 2016, Schneider J et al. constructed a Random Forest model using 22 clinical variables to stratify patients into different treatment pathways based on their risk of mortality.16 However, their research only studied the single outcome of in-hospital mortality, and the features ultimately included in the model were not selected after extensive screening, thus lacking sufficient persuasiveness. Currently, the field still lacks comprehensive models that can integrate clinical factors systematically and quantify their predictive power for outcomes in a robust manner. Further research is required to enhance our understanding of these prognostic factors and improve the accuracy of outcome prediction in SAC.
The utilization of machine-learning (ML) models within clinical contexts has garnered increasing attention and acknowledgment. As an emerging technological paradigm, ML is extensively employed across various medical domains, owing to its capacity for formulating robust risk models and augmenting predictive efficacy.17 Compared to traditional scoring systems like sepsis-related organ failure (SOFA) and acute physiology score (APS) II, ML models exhibit a markedly expanded capability to accommodate a larger array of predictors.18 The progression in computational prowess ensures that they largely mitigate the limitations inherent in conventional scoring systems during the calculation phase, such as massive input variables requirement, poor sensitivity and specificity, large fluctuation of prediction results, reliance on practitioner experience, and cumbersome process.19 Moreover, contemporary ML models, exemplified by the eXtreme Gradient Boosting (XGBoost),20 are increasingly regarded as viable alternatives to traditional linear models like logistic or Cox regression for predicting various clinical outcomes.21 Traditional linear models necessitate manual selection of variables and lack the ability to address nonlinearities within clinical applications, while, XGBoost can engender superior models by combining weaker models with more adaptable data processing mechanisms.22
By comparing the performance indicators and clinical utility of ML models, with multi-dimensional clinical characteristics inputted, this study aimed to develop a series of ML models to predict the outcomes of patients with cholelithiasis-induced acute cholangitis (CIAC) and provide clinicians with efficacious prospective decision support tools for practice.
Methods
Data source
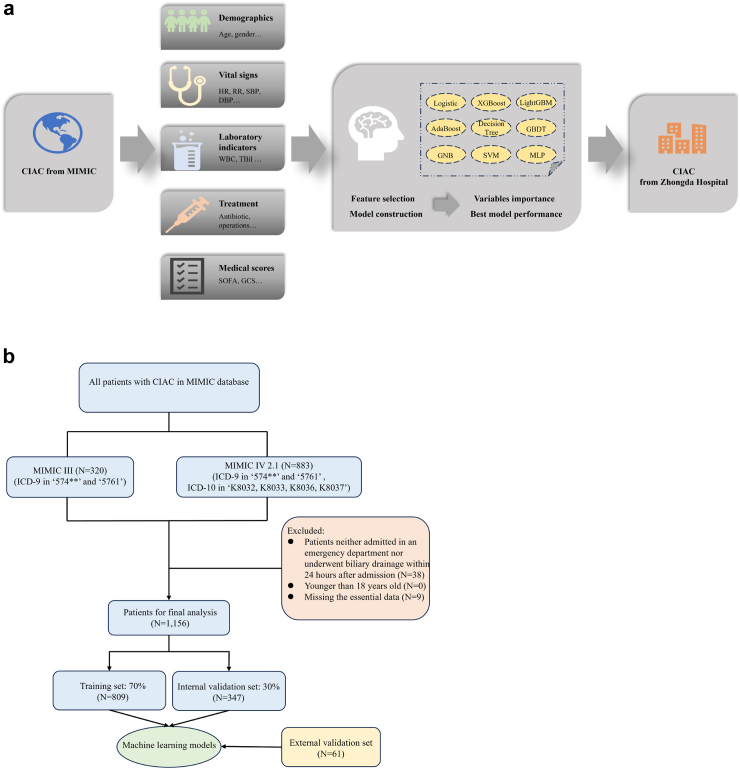
The training and internal validation set data used in ML model construction were retrieved from the Medical Information Mart For Intensive Care (MIMIC) database, which records patient admissions to Beth Israel Deaconess Medical Center (BIDMC) spanning from June 1, 2001 to November 16, 2022, including MIMIC III and IV version2.1.23,24 Detailed information of all patients with CIAC during their hospitalization, such as time of discharge and admission, basic demographic characteristics, vital signs, laboratory and microbiological results, fluid balance records, course of medication, survival data and more are available in the MIMIC database (Certificate number: 52598832). Additionally, data from Zhongda Hospital affiliated to Southeast University, Nanjing, China, was collected as an external validation set, spanning from January 1, 2019 to July 30, 2023. The research methodology adheres to the principles declared in the Declaration of Helsinki and its subsequent revisions, as confirmed by the approval obtained from the Ethics Committee (2019ZDSYLL093-P01, 2022ZDSYLL456-P01). Informed consent was waived for the retrospective study, and this study adheres to STROBE guidelines. An overview of the study workflow is depicted in Fig. 1.
Fig. 1.
The overall flowchart of the study (a). The algorithm chart of the study (b). CIAC, cholelithiasis-induced acute cholangitis; MIMIC, Medical Information Mart for Intensive Care; HR, heart rate; RR, respiratory rate; SBP, systolic blood pressure; DBP, diastolic blood pressure; WBC, white blood cell; TBil, total bilirubin; SOFA, sepsis-related organ failure; GCS, glasgow coma scale; ICD, International Classification of Disease.
Diagnostic criteria of SAC from external validation patients
The diagnostic criteria for patients from Zhongda hospital is consistent with the currently internationally recognized TG18/TG13 diagnostic criteria.1
Case inclusion criteria from MIMIC database
Patients who met the both diagnosis of ‘cholangitis’ and ‘calculus of gallbladder or bile duct’, the diagnostic code of which are ‘5761’ and ‘574∗∗’ (∗ can represent any number between 0 and 9) according to the International Classification of Disease 9th revision (ICD-9), or met the diagnosis of ‘calculus of bile duct with acute cholangitis with or without obstruction’, the diagnostic code of which are ‘K8032, K8033, K8036, K8037’ according to the International Classification of Disease 10th revision (ICD-10) during a single hospitalization were considered for inclusion. The excluded criteria were: (1) patients neither admitted in an emergency department nor underwent biliary drainage within 24 h after admission; (2) patients under the age of 18 years; (3) patients with essential information missed or over 20% of the information lost.
Data collection
The following parameters were systematically gathered across all patients diagnosed with CIAC: (1) demographic characteristics, encompassing gender and age; (2) treatment and clinical management, including operations, renal replacement therapy, arterial catheterization, intravenous catheterization, mechanical ventilation, use of intravenous antibiotics and vasopressors; (3) comorbidities and complications, including hypertension, diabetes, acute kidney injury, sepsis and acute pancreatitis; (4) laboratory test results, comprising blood routine, arterial blood gas analysis, kidney function, electrolytes, liver function, coagulation function, and microbial cultures from bile and blood specimens; (5) others, including length of hospitalization, length of ICU stay and severity grade of cholangitis. For variables measured repeatedly during hospitalization, the maximum, average, and minimum values of them were involved, as outlined in Table 1. For example, ALT_max, ALT_avg, and ALT_min represent, respectively, the maximum, average, and minimum value of ALT measured throughout the hospitalization.
Table 1.
Baseline characteristics of all the patients included.
| Characteristics | n (%), mean ± SD (range), or med [IQR] |
|---|---|
| Age (years) | 76 [63–85] |
| Male, n (%) | 604 (52.2%) |
| Female, n (%) | 552 (47.8%) |
| Length of stay in hospital (days) | 6 [4–8] |
| Length of stay in ICU (hours) | 21 [0–52] |
| Re-admission within 30 days after discharge, n (%) | 262 (22.7%) |
| Death in hospital, n (%) | 52 (4.5%) |
| Follow-up after discharge until death, n (%) | 343 (29.7%) |
| Death within 180 days after discharge, n (%) | 166 (14.4%) |
| Severity of cholangitis, n (%) | |
| Mild | 252 (21.8%) |
| Moderate | 197 (17.0%) |
| Severe | 707 (61.2%) |
| Surgical operations, n (%) | 339 (29.3%) |
| Endoscopy, n (%) | 995 (86.1%) |
| Interventional operations, n (%) | 108 (9.3%) |
| Surgical or interventional or endoscopic operations, n (%) | 1072 (92.7%) |
| Endoscopy combined with surgical operations, n (%) | 275 (23.8%) |
| Endoscopy combined with interventional operations, n (%) | 79 (6.8%) |
| Endoscopy combined with another operations, n (%) | 326 (28.2%) |
| Surgical or interventional operations other than endoscopy, n (%) | 77 (6.7%) |
| Endoscopic, surgical, and interventional procedures were performed during a single hospitalization, n (%) | 28 (2.4%) |
| Duration of intravenous antibiotic use (hours) | 119 [68–233.5] |
| Blood culture positive, n (%) | 219 (18.9%) |
| Bile culture positive, n (%) | 37 (3.2%) |
| Hypertension, n (%) | 598 (51.7%) |
| Diabetes, n (%) | 342 (29.6%) |
| Acute pancreatitis, n (%) | 247 (21.4%) |
| Anion gap _ max (mEq/L) | 16 [15–18] |
| Anion gap _ avg (mEq/L) | 13.5 [12.2–15] |
| Anion gap _ min (mEq/L) | 11 [9–13] |
| Bicarbonate _ max (mEq/L) | 27 [25–30] |
| Bicarbonate _ avg (mEq/L) | 24.6 [22.4–26.5] |
| Bicarbonate _ min (mEq/L) | 22 [19–24] |
| Chloride _ max (mEq/L) | 108 [105–111] |
| Chloride _ avg (mEq/L) | 104.7 [102.5–107] |
| Chloride _ min (mEq/L) | 101 [98–104] |
| Creatinine _ max (mg/dL) | 1.1 [0.8–1.6] |
| Creatinine _ avg (mg/dL) | 0.9 [0.7–1.3] |
| Creatinine _ min (mg/dL) | 0.8 [0.6–1] |
| Glucose _ max (mg/dL) | 140 [115–183] |
| Glucose _ avg (mg/dL) | 109.3 [95.4–127] |
| Glucose _ min (mg/dL) | 83 [72–95] |
| Potassium _ max (mEq/L) | 4.3 [4–4.7] |
| Potassium _ avg (mEq/L) | 3.8 [3.6–4.1] |
| Potassium _ min (mEq/L) | 3.4 [3.1–3.6] |
| Sodium _ max (mEq/L) | 142 [140–144] |
| Sodium _ avg (mEq/L) | 139.3 [137.4–141] |
| Sodium _ min (mEq/L) | 137 [134–139] |
| Urea nitrogen _ max (mg/dL) | 21 [15–33] |
| Urea nitrogen _ avg (mg/dL) | 15.9 [11.2–25.2] |
| Urea nitrogen _ min (mg/dL) | 11 [7–17] |
| Hematocrit _ max (%) | 36.4 ± 4.8 (25.1–56.6) |
| Hematocrit _ avg (%) | 32.6 [29.8–36.1] |
| Hematocrit _ min (%) | 30.3 ± 5.5 (10–45.3) |
| Hemoglobin _ max (g/dL) | 12.1 ± 1.7 (7.5–18.3) |
| Hemoglobin _ avg (g/dL) | 10.9 [9.9–12.1] |
| Hemoglobin _ min (g/dL) | 10.1 ± 1.9 (3.8–15.4) |
| MCH _ max (pg) | 31.1 [29.6–32.4] |
| MCH _ avg (pg) | 30.4 [29.1–31.7] |
| MCH _ min (pg) | 29.9 [28.5–31.2] |
| MCHC _ max (%) | 34 [33.1–34.9] |
| MCHC _ avg (%) | 33.2 [32.5–34] |
| MCHC _ min (%) | 32.4 [31.5–33.2] |
| MCV _ max (fL) | 93 [90–97] |
| MCV _ avg (fL) | 91.3 [88–95.4] |
| MCV _ min (fL) | 90 [86–93] |
| PLT _ max (K/μL) | 236 [170–322] |
| PLT _ avg (K/μL) | 189.8 [142–251.9] |
| PLT _ min (K/μL) | 152 [109–207] |
| RDW _ max (%) | 14.7 [13.9–16.2] |
| RDW _ avg (%) | 14.5 [13.6–15.6] |
| RDW _ min (%) | 14.1 [13.3–15.1] |
| RBC _ max (m/μL) | 4 ± 0.6 (3.6–4.4) |
| RBC _ avg (m/μL) | 3.6 ± 0.6 (3.2–4) |
| RBC _ min (m/μL) | 3.3 ± 0.6 (2.9–3.8) |
| WBC _ max (K/μL) | 13.6 [9.4–19.5] |
| WBC _ avg (K/μL) | 9.7 [7.2–12.9] |
| WBC _ min (K/μL) | 6.7 [5–8.9] |
| TBil _ max (mg/dL) | 4 [2–6] |
| TBil _ avg (mg/dL) | 2.3 [1.3–4] |
| TBil _ min (mg/dL) | 1.2 [0.7–2.2] |
| ALT _max (IU/L) | 167 [83.3–320.8] |
| ALT _avg (IU/L) | 105.8 [55.9–185] |
| ALT _ min (IU/L) | 54.5 [28–107] |
| ALP _ max (IU/L) | 244 [155–387.5] |
| ALP _ avg (IU/L) | 195.2 [132.1–296.4] |
| ALP _ min (IU/L) | 151.5 [104–235] |
| AST _max (IU/L) | 145 [74–282.5] |
| AST _avg (IU/L) | 80 [48.4–134] |
| AST _min (IU/L) | 34.5 [23–56] |
| Magnesium _ max (mg/dL) | 2.2 [2–2.4] |
| Magnesium _ avg (mg/dL) | 1.9 [1.8–2.1] |
| Magnesium _ min (mg/dL) | 1.7 [1.5–1.9] |
| Calcium _ max (mg/dL) | 8.7 [8.4–9.2] |
| Calcium _ avg (mg/dL) | 8.3 [8–8.7] |
| Calcium _ min (mg/dL) | 7.9 [7.4–8.4] |
| Phosphate _ max (mg/dL) | 3.5 [3–4.2] |
| Phosphate _ avg (mg/dL) | 2.9 [2.5–3.3] |
| Phosphate _ min (mg/dL) | 2.2 [1.8–2.6] |
| INR (PT) _ max | 1.4 [1.2–1.8] |
| INR (PT) _ avg | 1.3 [1.2–1.5] |
| INR (PT) _ min | 1.2 [1.1–1.3] |
| PT _ max (seconds) | 15.3 [13.3–19.3] |
| PT _ avg (seconds) | 14.3 [13–16.2] |
| PT _ min (seconds) | 13.1 [12.2–14.3] |
| PTT _ max (seconds) | 33.1 [28.7–42.8] |
| PTT _ avg (seconds) | 31.2 [27.4–36.8] |
| PTT _ min (seconds) | 28.7 [25.4–31.9] |
| Lipase _ max (IU/L) | 54 [20–564] |
| Lipase _ avg (IU/L) | 46 [18.6–264.4] |
| Lipase _ min (IU/L) | 26.5 [15–100] |
| Basophils _ max (%) | 0.3 [0.1–0.6] |
| Basophils _ avg (%) | 0.2 [0.1–0.4] |
| Basophils _ min (%) | 0.1 [0–0.3] |
| Eosinophils _ max (%) | 1.2 [0.2–2.8] |
| Eosinophils _ avg (%) | 0.9 [0.2–2] |
| Eosinophils _ min (%) | 0.4 [0–1.5] |
| Lymphocytes _ max (%) | 10.8 [6.6–16] |
| Lymphocytes _ avg (%) | 9.2 [5.9–12.8] |
| Lymphocytes _ min (%) | 7 [4–10.9] |
| Monocytes _ max (%) | 5.1 [3.7–7] |
| Monocytes _ avg (%) | 4.4 [3.2–5.9] |
| Monocytes _ min (%) | 3.8 [2.2–5.1] |
| Neutrophils _ max (%) | 85.3 [80.8–90.1] |
| Neutrophils _ avg (%) | 82.3 [78–86.9] |
| Neutrophils _ min (%) | 79.7 [72.6–86] |
| Albumin _ max (g/dL) | 3.2 [2.9–3.6] |
| Albumin _ avg (g/dL) | 3.1 ± 0.5 (1.3–4.9) |
| Albumin _ min (g/dL) | 3 [2.6–3.3] |
| LD _ max (IU/L) | 257 [180–437.6] |
| LD _ avg (IU/L) | 223.2 [172–311.4] |
| LD _ min (IU/L) | 188.8 [151–259.9] |
| PH _ max (units) | 6 [5.4–6.6] |
| PH _ avg (units) | 5.9 [5.4–6.5] |
| PH _ min (units) | 5.8 [5.1–6.3] |
| Specific gravity _ max | 1.018 [1.009–1.034] |
| Specific gravity _ avg | 1.017 [1.012–1.03] |
| Specific gravity _ min | 1.018 [1.01–1.031] |
| Lactate _ max (mmol/L) | 1.8 [1–2.9] |
| Lactate _ avg (mmol/L) | 1.6 [1.1–2.2] |
| Lactate _ min (mmol/L) | 1.3 [0.9–1.9] |
| Amylase _ max (IU/L) | 75.9 [25–295] |
| Amylase _ avg (IU/L) | 72.4 [25.6–212] |
| Amylase _ min (IU/L) | 55 [20.9–195.7] |
| NLR _ avg | 9 [6.1–14.7] |
| PLR _ avg | 21.8 [13.6–35.5] |
| MLR _ avg | 0.5 [0.3–0.7] |
| SII _ avg (K/μL) | 1796.7 [1065.8–3113.9] |
| SIRI _ avg (%) | 39.6 [26.4–61.3] |
Total (n = 1156).
SD, standard deviation; IQR, inter-quartile range; ICU, intensive care unit; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MCV, mean corpuscular volume; PLT, platelet count; RDW, red blood cell distribution width; RBC, red blood cells; WBC, white blood cells; TBil, total bilirubin; ALT, alanine aminotransferase; ALP, alkaline phosphatase; AST, aspartate aminotransferase; INR (PT), international normalized ratio of prothrombin time; PT, prothrombin time; PTT, partial thromboplastin time; LD, lactate dehydrogenase; NLR, neutrophil-lymphocyte ratio; PLR, platelet-lymphocyte ratio; MLR, monocyte-lymphocyte ratio; SII, platelet∗neutrophil/lymphocyte; SIRI, neutrocyte∗monocyte/lymphocyte.
For patients admitted to ICU, additional indicators were collected on their first day in the ICU, including anthropometric measurements (height and weight), physiological parameters (blood glucose level, blood oxygen saturation, body temperature, heart rate, respiratory rate, systolic and diastolic blood pressure, and mean arterial pressure). Additionally, various scores reflecting disease severity were incorporated into the assessment, including but not limited to the Systemic Inflammatory Response Syndrome (SIRS), GCS, Logistic Organ Dysfunction System (LODS), APS III, Model for End-Stage Liver Disease (MELD), Oxford Acute Severity of Illness Score (OASIS), and SOFA. All the variables are shown in Supplementary Table S1.
For patients who underwent biliary drainage during ICU treatment, the interval between admission and biliary drainage was calculated independently. Description of variables can be found in Supplementary Table S2. The data comprising the external validation set originated from the electronic medical record (EMR) system of Zhongda Hospital. Measures were undertaken to harmonize the data units with those existing in the MIMIC database. For example, when measuring total bilirubin in blood, the unit used by Zhongda Hospital is μmol/L, while mg/dL is used in MIMIC database, and the conversion formulas is that 1 mg/dL is equal to 17.1 μmol/L. The unit conversion details were presented in Supplementary Table S3.
Clinical and model outcomes
This study concentrated on three primary endpoints, including in-hospital mortality, re-admission within 30 days after discharge and mortality within 180 days after discharge.
Statistical analysis
Continuous variables were reported as either mean ± standard deviation (with range) or median (with interquartile range) and were compared utilizing either the Student's t-test or the Wilcoxon rank-sum test. Categorical variables were expressed as frequency and percentage, and comparisons were conducted using either the chi-square test or Fisher's exact test, as appropriate.
Data processing
To mitigate bias stemming from missing data, factors exhibiting over 20% missing values were excluded during the data collection phase. The multiple imputation (MI) technique was implemented to address missing values within the remaining variables for analysis. The MI method involves comprehensively considering inter-variable relationships, assigning multiple plausible values to missing entries, and generating multiple interchangeable datasets.25 Ten-fold cross-validation was performed to reduce overfitting and enhance model stability. In this approach, the complete dataset was randomly divided into ten folds, with each fold serving as an internal validation set during model development, while the remaining folds were employed as training sets. This procedure was replicated ten times. Within the training subset of patients with CIAC, variables independently associated with outcomes were identified by employing univariate logistic regression. A P-value cut-off of 0.1 was then chosen for feature selection, which balances the need to avoid missing potentially important features (Type II errors) with the goal of creating an inclusive initial model, constituting a crucial stage in model construction. This approach is particularly useful in high-dimensional data contexts and exploratory phases of model building. The SelectFromModel algorithm utilized herein is a model-driven feature selection method, adept at identifying the most salient features from a given model. Its basic principle is to rank importance of features based on the model, subsequently select those exceeding the given threshold and delete those less than the threshold value, so as to foster model accuracy and diminish feature dimensionality.26
Algorithms
A total of nine ML algorithms were included for model construction, namely Logistic Regression (LR), eXtreme Gradient Boosting (XGBoost), Light Gradient Boosting Machine (LightGBM), Adaptive Boosting (AdaBoost), Decision Tree, Gradient Boosting Decision Tree (GBDT), Gaussian Naive Bayes (GNB), Multi–Layer Perceptron (MLP), and Support Vector Machine (SVM). A total of nine imputed datasets were generated corresponding to various clinical outcomes and patient treatment groups respectively, and the nine ML algorithms were applied to each dataset, with the results of each dataset summarized individually.
Model evaluation indexes
The area under the receiver operating characteristic curve (AUROC) was the main index for model performance assessment. Additionally, various performance metrics including accuracy, sensitivity, specificity, Positive Predictive Value (PPV), Negative Predictive Value (NPV), F1 score, and Kappa value were scrutinized. Furthermore, the area under the precision–recall curve (AUPRC) was assessed, complemented by an evaluation of clinical utility through decision curve analysis (DCA). The statistical analyses and modeling procedures were implemented utilizing Python (version 3.8), with a significance threshold set at a two-sided P value of less than 0.05.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. There was no commercial support. All authors had full access to the data in the study and YF had final responsibility for the decision to submit for publication.
Results
Patient characteristics
According to the inclusion criteria, a total of 1203 patients with CIAC were included. Subsequent to the application of exclusion criteria, 47 patients were omitted, resulting in the final inclusion of 1156 patients, as delineated in Fig. 1, with details presented in Table 1. Among these patients, there were 52 deaths during hospitalization (4.5%), 262 cases (22.7%) requiring re-admission within 30 days after discharge, and 166 deaths (14.4%) within 180 days after discharge. Notably, the differences in characteristics between patients with distinct prognostic outcomes were detailed in Supplementary Table S4. Our analysis revealed that the advanced age (P = 0.021, P < 0.001) and the absence of interventional procedures (P = 0.040, P = 0.031) were associated with heightened risks of re-admission within 30 days after discharge and mortality within 180 days after discharge. Meanwhile, factors including the length of hospitalization (P = 0.009, P < 0.001), severity grading of cholangitis (P < 0.001, P < 0.001), duration of intravenous antibiotic administration (P < 0.001, P < 0.001), and blood culture results (P = 0.026, P = 0.001) independently influenced the re-admission rate within 30 days after discharge, as well as mortality within 180 days after discharge. Furthermore, it should be emphasized that the occurrence of acute pancreatitis (P = 0.006) during hospitalization was linked to an increased probability of in-hospital mortality. In fact, among patients re-admitted within 30 days after discharge, there may be concurrent deaths within 180 days (Supplementary Table S5, n = 262). By analyzing cases that fit both outcome categories (Supplementary Table S6, Supplementary Table S7), it was found that these patients had higher age (P < 0.001, OR = 0.16), cholangitis severity grade (P = 0.008, OR = 15.38), and several higher laboratory tests during hospitalization (P < 0.050) than re-admitted patients without mortality within 180 days after discharge. In addition, patients re-admitted within 30 days had a higher mortality within 180 days after discharge (Supplementary Table S8, P < 0.001). According to the consistency analysis (Kappa = 0.049, P = 0.065), re-admission within 30 days after discharge and mortality within 180 days after discharge are independent outcomes in this patient cohort.
The duration of ICU stay exhibited a significant correlation with the occurrence of all three primary outcomes. Patients admitted to the ICU during hospitalization (Supplementary Table S1, n = 652) had a higher risk of in-hospital mortality and mortality within 180 days after discharge (Table 2, P < 0.001, P < 0.001), in comparison to those placed in standard ward settings. However, they displayed reduced probabilities of re-admission within 30 days after discharge (P < 0.001). Within the ICU-admitted patient subgroup (Supplementary Table S9), the duration of intravenous antibiotic administration emerged as an independent prognostic determinant for all three aforementioned outcomes (P = 0.012, P = 0.012, P < 0.001). Furthermore, the advanced age (P = 0.002), prolonged hospitalization (P = 0.008), extended ICU stay (P < 0.001), higher severity of cholangitis (P < 0.001), and absence of endoscopic therapy (P = 0.014) were identified as positive factors predisposing patients to mortality within 180 days after discharge. The onset of acute pancreatitis (P = 0.018) emerged as an independent determinant of death during ICU stay.
Table 2.
Different outcomes corresponding to various patient categories.
| Outcomes | All (n = 1156) | Patients admitted to the ICU |
Patients underwent operations during ICU treatment |
||||
|---|---|---|---|---|---|---|---|
| Yes (n = 652) | No (n = 504) | P-value | Yes (n = 614) | No (n = 38) | P-value | ||
| In-hospital mortality | 52 (4.5%) | 50 (7.7%) | 2 (0.4%) | <0.001 | 43 (7.0%) | 7 (18.4%) | 0.020 |
| Re-admission within 30 days after discharge | 262 (22.7%) | 116 (17.8%) | 146 (29.0%) | <0.001 | 112 (18.2%) | 5 (13.1%) | 0.293 |
| Mortality within 180 days after discharge | 166 (14.4%) | 136 (20.9%) | 30 (6.0%) | <0.001 | 122 (19.9%) | 14 (36.8%) | 0.014 |
ICU, intensive care unit.
Biliary drainage is a critical treatment for patients with CIAC. A total of 614 patients underwent various operative procedures during their ICU treatment, including surgical, interventional and endoscopic operations. As described in Table 2, patients underwent biliary drainage during ICU treatment had an in-hospital mortality rate of 7.0%, with corresponding re-admission rates within 30 days after discharge and mortality within 180 days after discharge up to 18.2% and 19.9%, respectively. There was no statistically significant difference observed in the re-admission rates within 30 days after discharge (P = 0.29) between patients who underwent biliary drainage and those who did not. However, biliary drainage demonstrated efficacy in mitigating the mortality rates among ICU patients, both during the hospitalization period (P = 0.020) and within 180 days after discharge (P = 0.014). The statistical correlations between clinical variables and outcomes were shown in Supplementary Table S10.
Feature selection
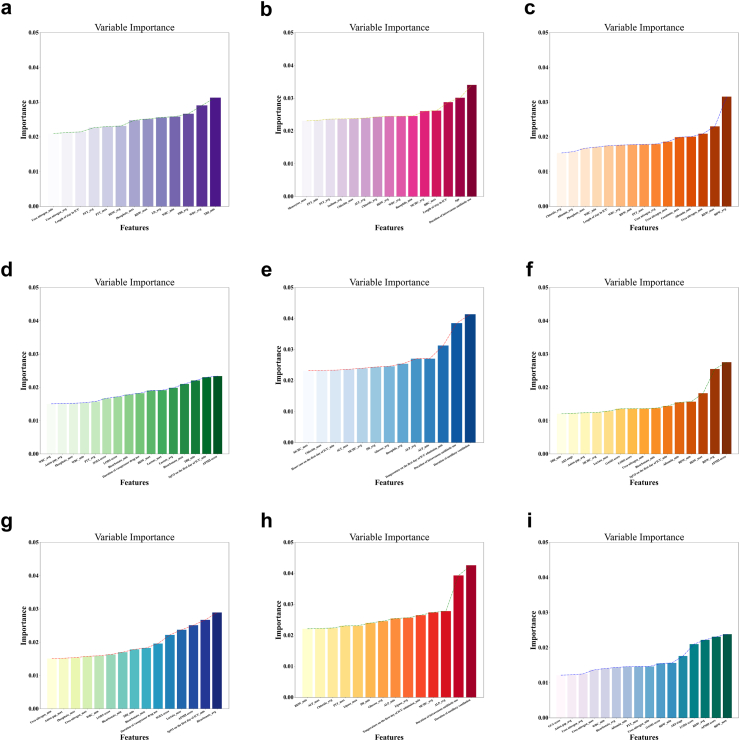
A total of 186 clinical variables were counted and classified according to the type of data distribution, as shown in Supplementary Table S11. Univariate logistic regression was employed to scrutinize all the variables alongside associated outcomes. Variables with a P value less than 0.1 were documented in Supplementary Table S12, and were analyzed for feature importance ranking and filtration. The outcomes of feature selection, conducted utilizing the SelectFromModel algorithm, were illustrated in Fig. 2.
Fig. 2.
Feature selection based on the SelectFromModel algorithm. The horizontal axis represents the name of each variable, while the vertical axis represents the significance attributed to each variable. Outcomes of all the patients (a–c): in-hospital mortality, re-admission within 30 days after discharge and mortality within 180 days after discharge. Outcomes of patients admitted to the ICU (d–f). Outcomes of patients underwent biliary drainage during ICU treatment (g–i). ICU, intensive care unit; PTT, partial thromboplastin time; RDW, red blood cell distribution width; LD, lactate dehydrogenase; WBC, white blood cells; TBil, total bilirubin; ALT, alanine aminotransferase; RBC, red blood cells; SOFA, sepsis-related organ failure; LODS, logistic organ dysfunction system; APSIII, acute physiology score iii; ALP, alkaline phosphatase; MCHC, mean corpuscular hemoglobin concentration; AKI, acute kidney injury; OASIS, oxford acute severity of illness score.
After a comprehensive analysis on all the patients, 13 key variables were found have most influence on the occurrence of in-hospital mortality. These variables, in order of significance, comprise total bilirubin (TBil)_min, white blood cells (WBC)_avg, TBil_avg, WBC_min, lactate dehydrogenase (LD)_avg, red blood cell distribution width (RDW)_max, Phosphate_max, RDW_avg, PTT_max, PTT_avg, duration of ICU stay, urea nitrogen_avg, and urea nitrogen_min. Similarly, a hierarchical assessment revealed the 15 variables most closely linked to re-admission within 30 days after discharge, namely, duration of intravenous antibiotic administration, age, duration of ICU stay, red blood cells (RBC)_max, mean corpuscular hemoglobin concentration (MCHC)_avg, basophils_min, WBC_avg, RDW_avg, chloride_avg, ALT_avg, chloride_max, sodium_avg, PTT_avg, PTT_min, and monocytes_max. Likewise, a set of 15 factors, including albumin_avg, albumin_min, chloride_avg, creatinine_max, duration of ICU stay, phosphate_max, PTT_max, RDW_avg, RDW_max, RDW_min, urea nitrogen_avg, urea nitrogen_max, urea nitrogen_min, WBC_avg, and WBC_min, emerged as most significantly associated with mortality within 180 days after discharge. In addition, 16, 13, 15 and 15, 14, 16 variables were selected for all patients admitted to the ICU and patients underwent biliary drainage during ICU treatment, when constructing prediction models of in-hospital mortality, re-admission within 30 days after discharge, and mortality within 180 days after discharge, respectively. Details of selected variables were listed in Supplementary Table S13.
Simultaneously, feature selection was performed on patients admitted to the ICU and those underwent biliary drainage during their ICU treatment. In addition to the variables mentioned above, some other features were also screened out and entered the model construction process, such as acute kidney injury (AKI) stage, LODS score, APS III score, GCS score, OASIS score, SOFA score, duration of auxiliary ventilation, duration of vasopressor drug administration, SpO2 on the first day of ICU admission, temperature on the first day of ICU admission, PH, anion gap, bicarbonate, glucose, lactate, alkaline phosphatase, and lipase.
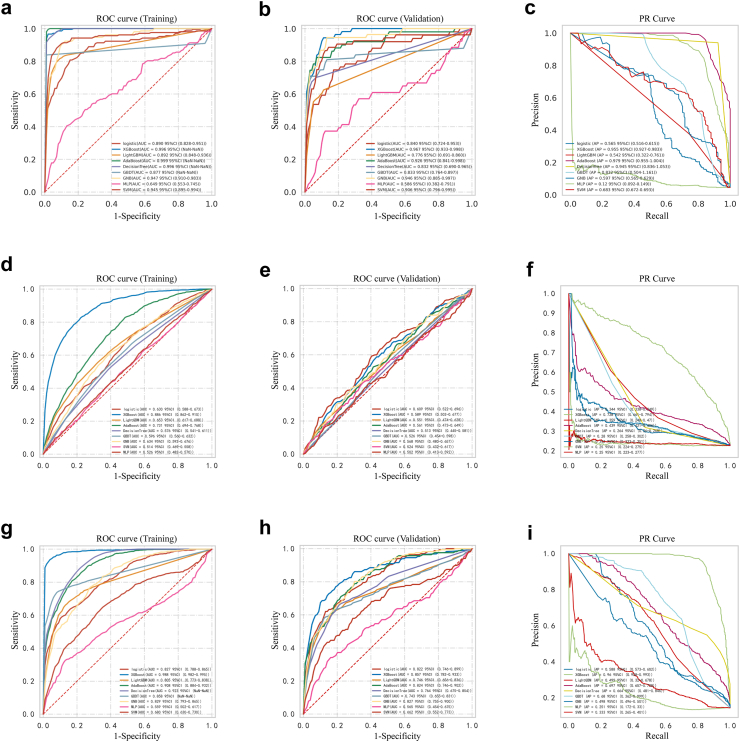
Comparisons of model performance
A total of nine ML models were employed to predict outcomes for all the patents with CIAC. The predictive efficacy of models was presented in Fig. 3, as depicted by receiver operating characteristic (ROC) and precision-recall (PR) curves. For patients who were admitted to the ICU and those underwent biliary drainage during ICU treatment, outcomes were predicted as shown in Supplementary Figure S1. Comprehensive performance metrics detailing the predictive capabilities of the ML models across various prognostic outcomes, stratified by patient classifications, were delineated in Supplementary Tables S14–S22.
Fig. 3.
Receiver operating characteristic curve and Precision-Recall curve of nine models. Outcomes of all the patients: in-hospital mortality (a–c), re-admission within 30 days after discharge (d–f), and mortality within 180 days after discharge (g–i). XGBoost, eXtreme Gradient Boosting; LightBGM, Light Gradient Boosting Machine; AdaBoost, Adaptive Boosting; GBDT, Gradient Boosting Decision Tree; GNB, Gaussian Naive Bayes; MLP, Multi-Layer Perceptron; SVM, Support Vector Machine.
Taking each metric of the models into account, XGBoost was found to be the most proficient in predicting all three outcomes. When predicting in-hospital mortality for all the patients, XGBoost model has the best discriminative performance as evidenced by training set AUROC of 0.996 (95% CI NaN–NaN), with the highest AUROC value (0.967, 95% CI 0.933–0.998), accuracy (0.968, 95% CI 0.961–0.975), sensitivity (0.964, 95% CI 0.892–1.035), PPV (0.665, 95% CI 0.553–0.777), F1 Score (0.783, 95% CI 0.693–0.872), and Kappa value (0.623, 95% CI 0.553–0.694) in the internal validation set. Meanwhile, the specificity of XGBoost stood at 0.899 (95% CI 0.860–0.939) and the NPV was 0.983 (95% CI 0.980–0.986). Likewise, when predicting re-admission within 30 days after discharge, the XGBoost model presented the highest AUROC value of 0.886 (95% CI 0.862–0.910) in the training set, along with leading performance across various metrics compared to the other ML models, encompassing accuracy (0.813, 95% CI 0.773–0.853), sensitivity (0.808, 95% CI 0.777–0.840), specificity (0.816, 95% CI 0.768–0.864), PPV (0.569, 95% CI 0.493–0.646), NPV (0.934, 95% CI 0.923–0.945), F1 score (0.666, 95% CI 0.609–0.723), and Kappa value (0.541, 95% CI 0.458–0.624), followed by the internal validation set AUROC of 0.589 (0.502–0.677). Similarly, when predicting mortality within 180 days after discharge, XGBoost consistently outperformed its counterparts across all evaluated metrics. As described in Supplementary Table S16, the training set AUROC value, accuracy, sensitivity, specificity, PPV, NPV, F1 score, and Kappa value were 0.988 (95% CI 0.982–0.995), 0.962 (95% CI 0.935–0.990), 0.953 (95% CI 0.915–0.992), 0.965 (95% CI 0.939–0.992), 0.834 (95% CI 0.718–0.950), 0.990 (95% CI 0.983–0.997), 0.887 (95% CI 0.805–0.968), and 0.861 (95% CI 0.763–0.959), respectively, with the AUROC of 0.857 (0.782–0.933) in the internal validation set. In addition, as shown in Supplementary Figure S1, corresponding to three different clinical outcomes, the training set AUROC of the XGboost models reached 0.998 (95% CI NaN–NaN), 0.933 (95% CI 0.909–0.957), and 0.988 (95% CI 0.983–0.993) in patients admitted to the ICU, 0.987 (95% CI 0.970–0.999), 0.908 (95% CI 0.873–0.942), and 0.982 (95% CI 0.971–0.993) in patients underwent biliary drainage during ICU treatment, respectively. Meanwhile, the internal validation set AUROC reached 0.963 (NaN–NaN), 0.668 (0.486–0.851), and 0.864 (0.757–0.970) in patients admitted to the ICU, 0.961 (0.922–0.997), 0.669 (0.540–0.799), and 0.828 (0.730–0.925) in patients underwent biliary drainage during ICU treatment, respectively. Regarding the PR curve analysis results (Fig. 3), the AUPRC values of the XGBoost models were 0.955 (95% CI 0.927–0.983), 0.728 (95% CI 0.661–0.794), and 0.960 (95% CI 0.928–0.993) in all the patients, respectively. The optimal ML models for predicting various clinical outcomes based on patient classifications were summarized in Table 3.
Table 3.
Optimal machine-learning algorithms for predicting various outcomes corresponding to patient categories.
| Outcomes | ROC training (AUROC) | ROC internal validation (AUROC) | PR curve (AUPRC) | DCA |
|---|---|---|---|---|
| All-hospdeath | Adaboost (0.999) | XGBoost (0.967) | Adaboost (0.979) | XGBoost |
| All-1month | XGBoost (0.886) | Logistic (0.609) | XGBoost (0.728) | XGBoost |
| All-180death | XGBoost (0.988) | XGBoost (0.857) | XGBoost (0.960) | XGBoost |
| ICU-hospdeath | Adaboost (0.999) | XGBoost (0.963) | XGBoost (0.992) | XGBoost |
| ICU-1month | XGBoost (0.933) | Adaboost (0.687) | XGBoost (0.811) | XGBoost |
| ICU-180death | XGBoost (0.988) | XGBoost (0.864) | XGBoost (0.964) | XGBoost |
| ICU-operation-hospdeath | XGBoost (0.987) | XGBoost (0.961) | XGBoost (0.925) | XGBoost |
| ICU-operation-1month | XGBoost (0.908) | Adaboost (0.683) | XGBoost (0.758) | XGBoost |
| ICU-operation-180death | XGBoost (0.982) | XGBoost (0.828) | XGBoost (0.924) | XGBoost |
ICU, intensive care unit; ROC, Receiver Operating Characteristic curve; AUROC, area under the receiver operating characteristics curve; PR, Precision-Recall; AUPRC, area under the precision-recall curve; DCA, Decision Curve Analysis; XGBoost, eXtreme Gradient Boosting; AdaBoost, Adaptive Boosting; Logistic, Logistic regression.
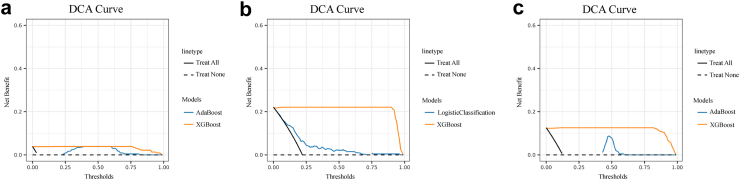
Moreover, to further demonstrate the clinical utility of XGBoost, we selected the ML model having best predictive power other than XGboost in the process of predicting various clinical outcomes based on patient classifications. The selected model may not be consistent across each predicted category, as shown in Table 3. For example, when predicting the in-hospital death for all patients, Adaboost was considered to be second only to XGboost after considering various metrics of model. It was therefore used to further compare the clinical efficacy with that of XGBoost. As shown in the DCA curve (Fig. 4), XGBoost demonstrated a superior net benefit than that of Adaboost. Supplementary Figure S2 presented additional DCA curves for patients admitted to the ICU and those underwent biliary drainage during ICU treatment. These findings underscored the XGBoost as the optimal model with favorable clinical applicability.
Fig. 4.
The decision curve analyses of XGBoost and the model with the most disputable predictive efficacy apart from XGBoost. Outcomes of all the patients: in-hospital mortality (a), re-admission within 30 days after discharge (b), and mortality within 180 days after discharge (c). XGBoost, eXtreme Gradient Boosting; AdaBoost, Adaptive Boosting.
Validation of the XGBoost models
A total of 61 patients (five deaths in hospital, 15 re-admissions within 30 days after discharge, 12 deaths within 180 days after discharge) from January 2019 to July 2023 in Zhongda Hospital were enrolled as an external validation cohort, with detailed information shown in Supplementary Table S23. The XGBoost models were validated based on the 14 to 16 top-ranking risk factors and got the AUROC values of 0.741 (95% CI 0.725–0.763), 0.812 (95% CI 0.798–0.824), 0.848 (95% CI 0.841–0.859), proving the favorable generalizability of our models (Supplementary Figure S3).
Application of the model
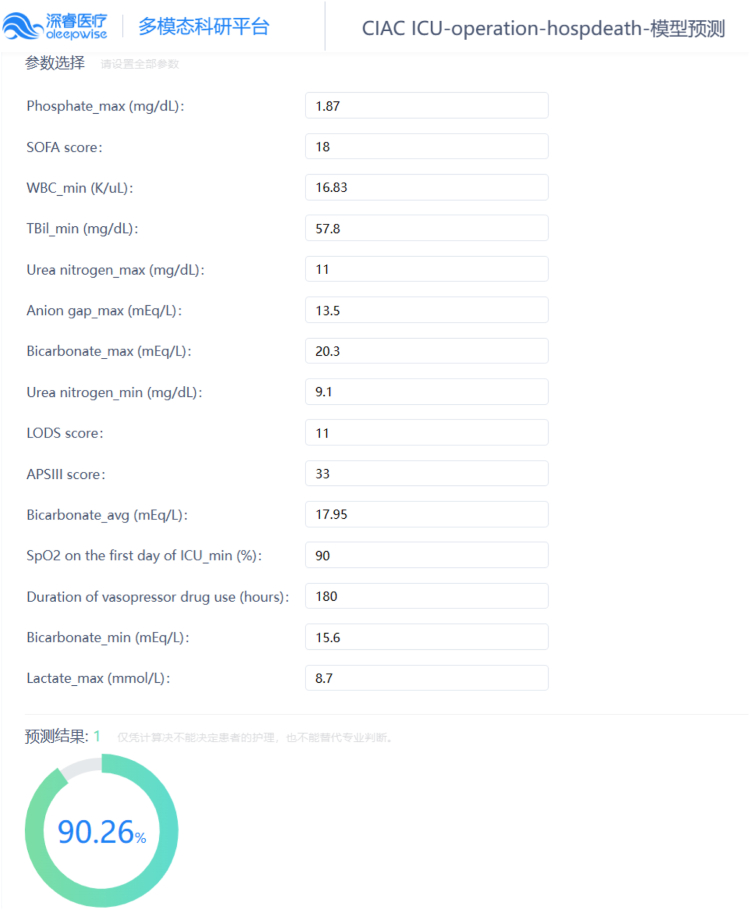
We have developed a series of online platforms (Table 4), providing access to web tools tailored to the classification of patients with CIAC and the targeted outcome events. By inputting clinical feature data directly into designated text fields on the webpage, users can obtain the desired prediction outcomes conveniently (Fig. 5).
Table 4.
Links to the web tools for predicting different outcomes of patients with CIAC.
CIAC, cholelithiasis-induced acute cholangitis; ICU, intensive care unit.
Fig. 5.
An example of web tool usage. The prediction of in-hospital mortality was conducted by inputting 15 clinical parameters pertaining to a single patient underwent biliary drainage during ICU treatment, thereby indicating an unfavorable prognosis.
Discussion
Although researches on the prognostic risk factors of CIAC have become increasingly profound, there is currently no systematic model to integrate these risk factors and accurately predict various outcomes. In this study, we utilized 13–16 clinical parameters to establish models based on the XGBoost, demonstrating that our models can better predict multiple clinical outcomes of CIAC.
Our study has several characteristics: In terms of prognostic research on CIAC, (1) we first combined and applied three elements in the same study: online database, offline collected real clinical data, and ML methods. (2) The clinical variables collected included not only individual data that can only be collected at a fixed time point but also indicators dynamically measured during hospitalization (with maximum, minimum, and average values involved). This allowed the models to function throughout the whole treatment process, providing continuous tracking predictions that change with the disease during hospitalization. (3) The included clinical elements were multidimensional. Various objective laboratory examination indicators, process of clinical treatment and care, as well as several disease scores that need to be calculated manually were involved. By adding these clinically convenient data, the accuracy of the model prediction was further improved. (4) Our team believed that traditional disease scoring systems have undergone many years of clinical practice verification and have their unique advantages. Therefore, instead of complete abandonment, we incorporated them as ordinary variables into the ML models, which differs from other studies. (5) The models we created can predict both short-term and long-term prognosis of patients. (6) The patients included in our study come from different countries. Although the models were built based on a single MIMIC database, satisfactory results were still achieved when using data from patients in China for external validation. (7) For the first time, we created a series of user-friendly online prediction platforms for endoscopists and patients worldwide that can also be internalized into the EMR. Despite that the information of the patients involved in model construction and validation came from only two regions, there are no significant regional differences in the diagnosis and treatment process of AC between countries, therefore, our models have universal applicability. Clinical workers only need to click a few times to extract specific data of each patient and then obtain the final prediction results. Even without the EMR, we enable real-time prognosis prediction based on part of the patient's baseline data in an outpatient clinic by inputting these parameters into the program through the network. This will facilitate the rapid triage of emergency patients, the efficient screening of high-risk patients and reasonably assignment of specialist physicians. The visual interface display allows patients to have a clearer understanding of the severity of disease, helping to achieve more convenient doctor-patient communication and nursing cooperation.
With the development of ML, the XGBoost provides better method for establishing medical prediction models. In previous studies, scholars have found that the XGBoost model has outstanding performance in predicting acute kidney injury in patients with diabetic ketoacidosis.27 To our surprise, after comprehensive comparison of various model performance indicators, the XGBoost models were found to have the best predictive performance and clinical utility in all nine combinations of CIAC populations corresponding to outcomes. The previously prognosis model was the Random Forest curve proposed by Schneider J et al.,16 with AUROC, sensitivity, and specificity of 0.91, 0.829, and 0.851, (which lower than those of the XGBoost model in our study, 0.967, 0.964, and 0.899 respectively) for predicting in-hospital mortality of CIAC. The performance of the model proposed by Li-Na Pan et al., in 2023,13 a nomogram for predicting mortality of AC in ICU (with the AUROC of training and validation set were 0.896 and 0.847 respectively), is not as good as the model constructed based on the XGBoost in our study (with the AUROC of 0.998 and 0.963). Therefore, the prognostic prediction models for CIAC we constructed can be considered more reliable than those proposed in previous studies.
We found that, for all the patients, on one hand, the length of ICU stay is positively correlated with in-hospital mortality and mortality within 180 days after discharge, on the other hand, the longer the ICU stay of patients, the less likely they are to be re-admitted within 30 days after discharge. This finding is consistent with the experience of clinicians. Patients who need to stay in the ICU for long-term monitoring are often those with severe conditions and a higher risk of death. Adequate length of ICU treatment increases the likelihood of being cured, hence reduces the risk of re-admission. 13–16 features were included in each model for the various populations corresponding to different outcomes of CIAC in our study. Most features are consistent with the factors related to the severity grading of AC proposed in the TG18 guidelines.1 At the same time, our study found a tight relationship between MCHC and re-admission within 180 days after discharge in patients with CIAC for the first time. The prognostic significance of MCHC has not been directly linked to AC before, but it has been confirmed to be associated with short-term death in serum biomarker studies of acutely admitted patients.28 RDW, as a frequently occurring indicator in complete blood cell count examinations, is a strong indicator of poor prognosis, while is often ignored by clinical workers. Interestingly, our study found that RDW is also a key feature in multiple models. For all the patients and those underwent biliary drainage during ICU treatment, RDW is the strongest predictive factor of re-admission within 180 days after discharge. High risk of malnutrition is associated with high RDW values, and patients with elevated RDW values demonstrate a robust responsiveness to nutritional interventions, which is much stronger than those with low RDW values.29 The discovery of these two new indicators may be used as supplements to the TG18 diagnostic guidelines.
In prior researches, single-center data were utilized to identify factors relevant to a single prognostic outcome of AC, with limited clinical dimensions explored and predominantly traditional linear model algorithms employed. Furthermore, due to the lack of data validation from different institutions, most previous prediction models tended to suffer from overfitting issues, making it challenging for a model developed by one institution to be applicable to another. In this context, our work integrating and enriching significant clinical features identified in previous studies, coupled with validation of prediction models using both online and offline data, is poised to become a significant milestone in developing universal, highly practical models applicable across multiple hospitals.
The corresponding diagnostic criteria of MIMIC III and IV version2.1 databases used in our study is the ICD-9 and ICD-10, respectively. Therefore, to avoid diagnostic bias, the updated ICD-11 was not used to replace ICD-9 and ICD-10 as inclusion criteria. In this study, some cases were placed in both positive outcome groups, that is, some patients were re-admitted within 30 days after a discharge and then died within 180 days after that discharge. This is in line with the actual clinical situation. The exclusion of data from this particular subset of patients would lead to bias in patient inclusion criteria and inaccurate estimates of outcome incidence in the process of exploring the occurrence of mortality within 180 days after discharge. Our study also had some limitations. Firstly, despite of the high quality of MIMIC database, and satisfactory performance of external validation set, our models were developed retrospectively based on the data from a single database, which introduced inherent biases in data collection. Secondly, the sample size of the external validation set is relatively small, which may affect the assessment of model performance on external data. As real-world clinical data varies across hospitals, establishing a stable prediction model well applied in multiple institutions proves challenging. We hope that future studies will include larger-scale, multi-center external validation cohorts to further verify the generalizability of the models. Thirdly, some key data were missed, such as the time interval between symptom onset and hospital admission, the timing of bile duct drainage, and the absence of laboratory markers like N-terminal pro-brain natriuretic peptide and C-reactive protein. Failure to further explore the database may also result in the omission of critical variables. Fourthly, the ML models we proposed lacks prospective validation, which is what our team will strive to achieve in future.
In conclusion, this study demonstrates that the XGBoost models could be promising tools to predict the occurrence of outcomes in patients with CIAC. Multicenter validation and large-scale prospective studies should be conducted to help verify our findings.
Contributors
SH was responsible for developing the models and drafting the manuscript. YZ undertook data collection and curation. YF contributed expertise in clinical study design and provided primary oversight of the analyses. YL participated in the literature review and validation of statistical results. The initial draft was reviewed by all the authors, who subsequently approved the final manuscript. SH, YZ, YL, SY, AZ, JL, XB, CY and YF had access to and verify the underlying study data. YF had final responsibility for the decision to submit for publication.
Data sharing statement
The dataset, code, algorithm files, and de-identified results used in this study are not publicly available. However, the data for this study can be shared upon reasonable request to the corresponding author.
Declaration of interests
All authors declare no competing interests.
Acknowledgements
This work was funded by Technological Development Program of Nanjing Healthy Commission (YKK22276) and Zhongda Hospital Affiliated to Southeast University, Jiangsu Province High-Level Hospital Construction Funds (2024GSPKY06).
Footnotes
Translation For the Chinese translation of the abstract see the Supplementary Materials section.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2024.102820.
Appendix A. Supplementary data
References
- 1.Kiriyama S., Kozaka K., Takada T., et al. Tokyo guidelines 2018: diagnostic criteria and severity grading of acute cholangitis (with videos) J Hepatobiliary Pancreat Sci. 2018;25(1):17–30. doi: 10.1002/jhbp.512. [DOI] [PubMed] [Google Scholar]
- 2.Cafasso D.E., Smith R.R. Symptomatic cholelithiasis and functional disorders of the biliary tract. Surg Clin North Am. 2014;94(2):233–256. doi: 10.1016/j.suc.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Gallaher J.R., Charles A. Acute cholecystitis: a review. JAMA. 2022;327(10):965–975. doi: 10.1001/jama.2022.2350. [DOI] [PubMed] [Google Scholar]
- 4.Gomi H., Takada T., Hwang T.L., et al. Updated comprehensive epidemiology, microbiology, and outcomes among patients with acute cholangitis. J Hepatobiliary Pancreat Sci. 2017;24(6):310–318. doi: 10.1002/jhbp.452. [DOI] [PubMed] [Google Scholar]
- 5.Lavillegrand J.R., Mercier-Des-Rochettes E., Baron E., et al. Acute cholangitis in intensive care units: clinical, biological, microbiological spectrum and risk factors for mortality: a multicenter study. Crit Care. 2021;25(1):49. doi: 10.1186/s13054-021-03480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annane D., Aegerter P., Jars-Guincestre M.C., Guidet B. Current epidemiology of septic shock: the CUB–Rea Network. Am J Respir Crit Care Med. 2003;168(2):165–172. doi: 10.1164/rccm.2201087. [DOI] [PubMed] [Google Scholar]
- 7.Fang F., Zhang Y., Tang J., et al. Association between source of infection and hospital mortality in patients who have septic shock. Am J Respir Crit Care Med. 2014;189(10):1204–1213. doi: 10.1164/rccm.201310-1875OC. [DOI] [PubMed] [Google Scholar]
- 8.Mayumi T., Okamoto K., Takada T., et al. Tokyo guidelines 2018: management bundles for acute cholangitis and cholecystitis. J Hepatobiliary Pancreat Sci. 2018;25(1):96–100. doi: 10.1002/jhbp.519. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y., Zhang Y.Q., Huang S.J., et al. Urgent one-stage endoscopic treatment for choledocholithiasis related moderate to severe acute cholangitis: a propensity score-matched analysis. World J Gastroenterol. 2024;30(15):2118–2127. doi: 10.3748/wjg.v30.i15.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.G. Diagnosis and management of acute cholangitis. Nat Rev Gastroenterol Hepatol. 2009;6(9):533–541. doi: 10.1038/nrgastro.2009.126. [DOI] [PubMed] [Google Scholar]
- 11.Oseran A.S., Wadhera R.K., Orav E.J., Figueroa J.F. Effect of medicare advantage on hospital Re-admission and mortality rankings. Ann Intern Med. 2023;176(4):480–488. doi: 10.7326/M22-3165. [DOI] [PubMed] [Google Scholar]
- 12.Novy E., Carrara L., Remen T., et al. Prognostic factors associated with six month mortality of critically ill elderly patients admitted to the intensive care unit with severe acute cholangitis. HPB (Oxford) 2021;23(3):459–467. doi: 10.1016/j.hpb.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Pan L.N., Pan S.A., Hong G.L., Chen K.W. A new nomogram for predicting 30-day in-hospital mortality rate of acute cholangitis patients in the intensive care unit. Emerg Med Int. 2023;2023 doi: 10.1155/2023/9961438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q., Zhou Q., Song M., et al. A nomogram for predicting the risk of sepsis in patients with acute cholangitis. J Int Med Res. 2020;48(1) doi: 10.1177/0300060519866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inan O., Sahiner E.S., Ates I. Factors associated with clinical outcome in geriatric acute cholangitis patients. Eur Rev Med Pharmacol Sci. 2023;27(8):3313–3321. doi: 10.26355/eurrev_202304_32102. [DOI] [PubMed] [Google Scholar]
- 16.Schneider J., Hapfelmeier A., Thöres S., et al. Mortality Risk for Acute Cholangitis (MAC): a risk prediction model for in-hospital mortality in patients with acute cholangitis. BMC Gastroenterol. 2016;16:15. doi: 10.1186/s12876-016-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haug C.J., Drazen J.M. Artificial intelligence and machine learning in clinical medicine, 2023. N Engl J Med. 2023;388(13):1201–1208. doi: 10.1056/NEJMra2302038. [DOI] [PubMed] [Google Scholar]
- 18.Churpek M.M., Yuen T.C., Winslow C., Meltzer D.O., Kattan M.W., Edelson D.P. Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med. 2016;44(2):368–374. doi: 10.1097/CCM.0000000000001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz B.A., Cui Z., Zhang M., et al. Deep-learning model for predicting 30-day postoperative mortality. Br J Anaesth. 2019;123(5):688–695. doi: 10.1016/j.bja.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ester M., Kriegel H.P., Xu X. Proceedings of the 22Nd ACM SIGKDD international conference on knowledge discovery and data mining. Vol. 785. Geographical Anal; 2022. XGBoost: a scalable tree boosting system. [Google Scholar]
- 21.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 22.Fleuren L.M., Klausch T.L.T., Zwager C.L., et al. Machine learning for the prediction of sepsis: a systematic review and meta–analysis of diagnostic test accuracy. Intensive Care Med. 2020;46(3):383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson A.E., Pollard T.J., Shen L., et al. MIMIC–III, a freely accessible critical care database. Sci Data. 2016;3 doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson A.E.W., Bulgarelli L., Shen L., et al. MIMIC–IV, a freely accessible electronic health record dataset. Sci Data. 2023;10(1):1. doi: 10.1038/s41597-022-01899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K.J., Simpson J.A. Introduction to multiple imputation for dealing with missing data. Respirology. 2014;19(2):162–167. doi: 10.1111/resp.12226. [DOI] [PubMed] [Google Scholar]
- 26.Uddin S., Ong S., Lu H. Machine learning in project analytics: a data-driven framework and case study. Sci Rep. 2022;12(1) doi: 10.1038/s41598-022-19728-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan T., Wang J., Li L., Kang J., Wang W., Zhang C. Predicting the risk factors of diabetic ketoacidosis-associated acute kidney injury: a machine learning approach using XGBoost. Front Public Health. 2023;11 doi: 10.3389/fpubh.2023.1087297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jawad B.N., Shaker S.M., Altintas I., et al. Development and validation of prognostic machine learning models for short- and long-term mortality among acutely admitted patients based on blood tests. Sci Rep. 2024;14(1):5942. doi: 10.1038/s41598-024-56638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haenggi E., Kaegi-Braun N., Wunderle C., et al. Red blood cell distribution width (RDW) - a new nutritional biomarker to assess nutritional risk and response to nutritional therapy? Clin Nutr. 2024;43(2):575–585. doi: 10.1016/j.clnu.2024.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.